Summary
It is widely accepted that the DNA, RNA and protein content of Enterobacteriaceae is regulated as a function of exponential growth rates; macromolecular content increases with faster growth regardless of specific composition of the growth medium. This phenomenon, called growth rate control, primarily involves regulation of ribosomal RNA and ribosomal protein synthesis. However, it was uncertain whether the global regulator ppGpp is the major determinant for growth rate control. Therefore, here we re-evaluate the effect of ppGpp on macromolecular content for different balanced growth rates in defined media. We find that when ppGpp is absent RNA/protein and RNA/DNA ratios are equivalent in fast and slow growing cells. Moreover, slow growing ppGpp deficient cells with increased RNA content, display a normal ribosomal subunit composition although polysome content is reduced when compared to fast growing wild type cells. From this we conclude that growth rate control does not occur in the absence of ppGpp. Also, artificial elevation of ppGpp or introduction of stringent RNA polymerase mutants in ppGpp deficient cells restores this control. We believe these findings strongly argue in favor of ppGpp and against redundant regulation of growth rate control by other factors in E. coli and other enteric bacteria.
Keywords: ppGpp, growth rate control, balanced growth, ribosomes, dksA
Introduction
Enteric bacteria were reported long ago to regulate their cell size, mass, and macromolecular content as a function of exponential growth rates regardless of the composition of the media needed to determine a given growth rate (Schaechter et al., 1958; Kjeldgaard et al., 1958) by a phenomenon called growth rate control, which is sometimes referred to as growth rate-determined control (Gaal and Gourse, 1990). For example, as the number of cell doublings per hour (μ) increases from 0.6 to 2, stable RNA increases 10-fold, cellular protein increases 4.5-fold, and DNA increases 2.2-fold (Bremer and Dennis, 1996). Therefore RNA/DNA as well as RNA/protein ratios are a measure of growth rate control. Major advances have been made in the molecular understanding of the individual cellular components with respect to structure, regulation and function. Neveretheless uncertainties still remain as to how much regulatory redundancy is needed for E. coli to achieve this regulation.
The rate of exponential growth under conditions unrestricted by substrate availability is commonly thought to be determined by adjustment of the number of ribosomes functioning at a high constant activity, rather than adjusting their protein synthetic activities (Bremer and Dennis, 1996; Neidhardt, 1999). This feature is reflected by an increase in stable RNA content (rRNA and tRNA) with growth rate. A consensus also exists that ribosomal protein content is regulated largely at the translational level by a feedback mechanism governed by nascent rRNA availability through shared ribosomal protein binding sites on ribosomal protein mRNA and rRNA (Keener and Nomura, 1996). Transcription of rRNA in E. coli arises from seven operons, each with two precisely spaced, highly conserved, strong promoters possessing unique properties. Over the years, many of these features have been thought to participate in rRNA transcription regulation along with additional factors (Condon et al., 1995; Cashel et al., 1996; Paul et al., 2004b; Potrykus and Cashel, 2008). One factor is a guanine nucleotide, ppGpp, long thought to be an effective inhibitor of in vivo rRNA and tRNA synthesis during amino acid starvation (Cashel and Gallant, 1968). Regulation by ppGpp is called stringent control. Subsequently, ppGpp has been shown to function as a global regulator responding to many sources of nutritional stress, not just amino acid deprivation (Potrykus and Cashel, 2008). These include starvation for sources of carbon, nitrogen, phosphate, iron, as well as limited fatty acid synthesis.
Transcriptional regulation by ppGpp is complex. Both positive and negative regulatory promoter-specific effects of ppGpp on transcription may involve DksA, a protein with structural similarities to the transcription elongation factors GreA, GreB, and TFIIS (Nickels and Hochschild, 2004; Perederina et al., 2004; Paul et al., 2004a; Paul et al., 2005). However, mechanistic details of the regulation of RNA polymerase by DksA and ppGpp are still emerging along with uncertainties due to reports of several RNA polymerase binding sites for ppGpp (Paul et al., 2004b, Haugen et al., 2008; Potrykus and Cashel, 2008).
Several features of the complex physiology of cells lacking ppGpp complicate studies of the effects of ppGpp on growth. First, ppGpp0 cells have multiple amino acid requirements that have not been precisely defined. Second, revertants that mimic ppGpp presence often take over slow growing ppGpp0 cultures. Here, we have bypassed these difficulties by determining the individual amino acid requirements so that the growth media can be defined. We have circumvented the problem with revertants by using special growth conditions.
We find that growth rate control of ppGpp0 cells during balanced growth is abolished. In addition, transitions from exponential growth to partial nutrient limitation and stationary phase are profoundly altered. These results substantially contribute to the understanding of bacterial growth physiology which is related to exploitation of different ecological niches and to life cycles of microbial pathogens.
Results and Discussion
Establishing conditions for unrestricted growth of ppGpp0 strains
Early studies of growth rate control in ppGpp0 strains were performed with media that contained all 20 amino acids [AA], sometimes varying growth rate by varying carbon sources (Hernandez and Bremer, 1993) or AA sources (pure amino acids, Casamino acids or yeast extract) (Gaal and Gourse, 1990). This was a technical necessity at the time because the specific multiple amino acid requirements of ppGpp0 strains were unknown. Here, we determined the minimal requirements for specific AA so that different combinations could be added in excess to minimal media containing different carbon sources that would support a range of growth rates for ppGpp0 cells (see Experimental Procedures and Table I). This was done so that exponential growth rates would be unrestricted, i.e. limited only by the cell's ability to use each nutrient, and not by exhaustion of the concentration of any one component to the point where it would impair growth, as defined classically (Schaechter et al., 1958). Unrestricted growth gives rise to balanced exponential growth, in which the rate of accumulation of each cellular component remains constant over time (Neidhardt, 1999).
Table I. Doublings/hour (μ) measured for wt and ppGpp0 strains in different media.
Wt strains are CF1648, CF7968; ppGpp0 are CF1693, CF12257. CF1648 and CF1693 carry rph- mutation.
| MEDIUMa | Doublings/hour (μ) | ||||
|---|---|---|---|---|---|
|
rph-1
CF1648 |
rph-1 relA251
spoT207 CF1693 |
CF7968 |
relA256
spoT212 CF12257 |
||
| 1 | LB | 2.14 | 1.94 | 2.61 | 2.31 |
| 2 | Medium A + Glucose 0.4%+ CAA 0.5% + W40 | 1.30 | 1.09-1.11 | 1.82 | |
| 3 | Medium A + Glucose 0.4% + 19 AA40 + S500 | 1.18 | 1.58 | ||
| 4 | M9 + glucose 0.2% + 19 AA40 + S400 | 1.09-1.33 | 1.43 | 1.82-1.88 | 1.54 |
| 4a | M9 + glucose 0.2% + 18 AA40 + S400 | 1.25 | 1.25 | ||
| 5 | M9 + glucose 0.2% + AA set + (GMNQY)40 | 1.25 | |||
| 6 | M9 + glucose 0.2% + AA set + (GNQY)40 | 1.05 | 0.78 | ||
| 7 | M9 + malate 0.2% + AA set | 0.71-0.78 | 0.69 | ||
| 8 | M9 + glucose 0.2% + AA set | 0.76 | 1.20-1.22 | 0.63 | |
| 9 | M9 + glucose 0.2% | 0.71-0.72 | 0.97-1.00 | ||
| 10 | M9 + glycerol 0.2% | 0.45-0.50 | 0.55-0.65 | ||
CAA = Casamino acids
AA set = (DEFHILTV)40+ S400
Medium 4a is all AA – M (medium used for S35-Met labeling)
Subscript numbers indicate concentration used (μg/ml)
We also took special precautions to minimize the extent of contamination by spontaneous mutants in RNA polymerase (stringent mutants) that very frequently overtake ppGpp0 cultures during slow growth (see Experimental Procedures). These precautions also entailed the use of small cultures in early exponential phase of growth to eliminate the need for prolonged growth and enrichment for stringent mutants. This was facilitated by use of fluorescent assays for RNA and DNA content with detection limits (5 ng for RNA and 12.5 ng DNA) that are about 1000 times more sensitive than older chemical or UV assays. Although measurement of individual components per cell is desirable, we elected to use ratios to characterize macromolecular content rather than presenting individual values for RNA, DNA or protein. In a given sample, ratios normalize for differences in cell number, because elongated ppGpp0 cells have variable recoveries after centrifugation (Xiao et al., 1991; Magnusson et al., 2007).
Measurement of RNA/DNA ratios in ppGpp0 strains
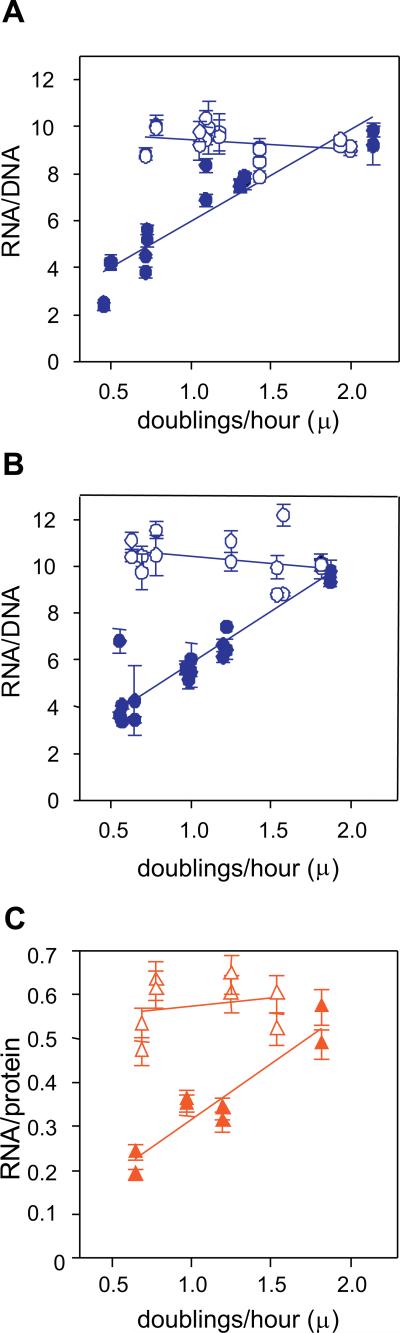
Figure 1A compares the effect of a ppGpp-deficiency on the RNA/DNA ratios found when unrestricted growth rates (μ) are varied for an isogenic strain pair. These ratios for the wild type MG1655 (strain CF1648) increase with faster growth rate and are in near agreement with published ratios for wild type E. coli B/r strains (Bremer and Dennis, 1996). In contrast, the ppGpp deficient strain (CF1693) has RNA/DNA ratios that do not seem to change appreciably over this range of growth rates but instead are maintained at a high value that is equivalent to the fast growing wild type [wt] strain.
Figure 1. A ppGpp deficiency abolishes regulation of RNA/DNA and RNA/protein ratios as a function of growth rate.
Isogenic strains differing in the presence of ppGpp were grown in different media (Table I) to achieve a range of balanced growth rates from about 120 min/doubling (μ = 0.5) to about 30 min/doubling (μ = 2). Cellular RNA and DNA content were measured by a fluorescent assay. Filled symbols correspond to wt strains, open symbols correspond to ppGpp0 strains. The solid lines represent linear regressions calculated with SigmaPlot software. (A) the wt (CF1648) and mutant (CF1693 = CF1648 ΔrelA251 ΔspoT207) strain are basically the same as used in earlier conflicting reports regarding the role of ppGpp in growth rate control (Gaal and Gourse, 1990; Hernandez and Bremer, 1993). (B) the wt (CF7968) and mutant (CF12257) strains used here are thought to be improved over the strains used in (A) with respect to the absence of polar effects due to insertions (ΔrelA256 ΔspoT212) and elimination of the leaky uracil requirement (see text for details) (C) Selected cell lysate samples from (B) were assayed to measure RNA/protein ratios.
The behavior in Fig. 1A differs from conflicting results published earlier (Gaal and Gourse, 1990; Hernandez and Bremer, 1993). The strains used here are identical to the strains used earlier except our strains do not carry P1rrnB- lacZ fusions on the chromosome. These strains have two drawbacks. One is that CF1693 has partial orf deletion-insertion alleles in the two genes responsible for ppGpp synthesis (relA and spoT), with the potential of polar effects on downstream genes. Another drawback is that these strains have a frame shift mutation in rph, which alters pyrE expression to give a leaky uracil requirement that in turn might affect RNA synthesis (Jensen, 1993). Yet, the media used in earlier experiments, or in Figure 1, were generally not supplemented with uracil.
Because of these considerations, similar experiments were performed with strains CF7968 (wt) and CF12257 (ppGpp0) that do not have these drawbacks (Figure 1B). CF12257 has complete in frame deletions of relA and spoT orfs without insertions. This strain pair is also constructed to be rph+. Nevertheless the RNA/DNA ratios behave similarly for both isogenic strain pairs. The absence of ppGpp results in a constant RNA/DNA ratio of about 10, regardless of growth rate. The wt strains reach the ratio of 10 only at the very fast growth rates. The published RNA/DNA ratio for the wt at the same fast growth rate is 9.2 (Bremer and Dennis, 1996). This validates our methodology.
Measurement of RNA/protein ratios in ppGpp0 strains
An increase in the cellular RNA content with increasing growth rate in wt strains is known to reflect a corresponding increase in the number of ribosomes operating at a relative constant efficiency (~80%) and with tRNA also increasing in parallel with ribosomes (Bremer and Dennis, 1996). The question arises whether the high RNA content of slow growing ppGpp0 strains reflects an increase in functional ribosomes. One approach to answer this question would be to measure cellular protein content because as the number of functional ribosomes increases, their catalytic products (synthesized proteins) also increase accordingly. Thus, in ppGpp0 strains it might be expected that the protein content should be the same during fast and slow growth. Proteomic studies have revealed that the protein content in ppGpp0 strains is independent of fast or slow growth (Magnusson et al., 2003).
In wt cells the RNA/protein ratios are known to increase with growth rate from 0.2 (μ = 0.6 doublings/hr) to 0.39 (μ = 2 doublings/hr) (Bremer and Dennis, 1996). Figure 1C shows that for the wt (CF7968) RNA/protein ratios parallel RNA/DNA ratios as both values increase with faster growth. RNA/protein ratios observed here at slow growth rates are identical to published values (0.2) and are somewhat higher than published values for fast growing wt cells (0.5 vs. 0.39).
Slow growing ppGpp0 cells seem to have the same amount of protein as slow growing wt cells, yet a higher RNA content. For the ppGpp0 strain, RNA/protein values remain high (about 0.55) and appear relatively constant whether during slow or fast growth, as if their protein synthesizing machinery is present at levels equivalent to very fast growing wt cells (Fig. 1C). It is expected that during growth rate control in wt cells the regulatory role of ppGpp is to limit the number of ribosomes in proportion to nutrient availability, which limits growth. In contrast, during balanced growth slow growing ppGpp0 cells unexpectedly have the RNA and protein ratio equivalent to fast growing cells. The difference in balanced growth rates under the two conditions suggests that the nature of the substrates limits growth rather than the ribosomal number. In this case, the ability to synthesize either the amino acids missing in the slow growth medium or charged tRNAs could be impaired. High content of RNA and low protein level in slow growing ppGpp0 cells implies that either only a fraction of all ribosomes is fully active or all ribosomes are partially active. We therefore attempted to characterize the functionality of ribosomes in ppGpp0 strains at slow and fast growth rates.
Ribosomal RNA subunit composition and polysome profiles
First, we compared the rates of protein accumulation at both growth rates by in vivo labeling with S35-methionine. Previous work has shown no defect in transport of S35-methionine leading to specific activity differences during ppGpp induction in the same strain (Svitil et. al., 1993). This allowed us to estimate protein accumulation rates without considering ppGpp effects on methionine uptake. We find that the rate of protein accumulation at fast growth rates is about 2.5 fold faster than at slow growth rate regardless of the presence of ppGpp (Fig. S1). Therefore protein accumulation is in agreement with growth rates for both ppGpp0 and wt strains. However, the protein accumulation result is in contrast to the RNA/protein ratios, which are ppGpp dependent and not growth rate dependent. We conclude that ribosomes synthesize protein with a less than normal efficiency in slow growing ppGpp0 cells.
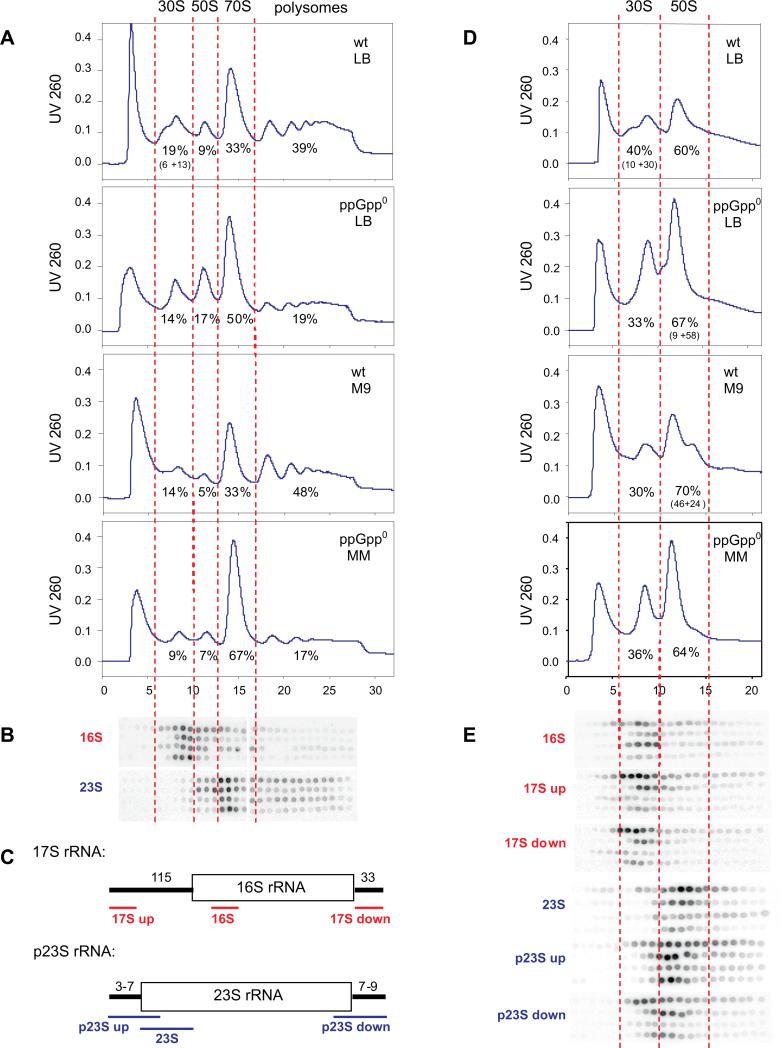
In order to assess what fraction of ribosomal RNA is engaged in protein synthesis, we assayed polysome profiles. This was done by sucrose gradient centrifugation under associating conditions (10 mM MgCl2) where the polysomes should be intact (Fig. 2A). Chloramphenicol was added just before samples were harvested to ensure ribosomes would not run off mRNA.
Figure 2. A ppGpp deficiency lowers polysomes abundance without ribosome maturation defects.
(A) Sucrose gradient profiles of wt (CF1648) and ppGpp0 (CF1693) strains grown in rich (LB) and poor media(M9= medium #9; MM= medium #8 (Table I)) under associating conditions monitored by A260. Percent abundance is indicated below peaks of 30S, 50S, 70S and polysomes. Values in parenthesis indicate fractions of unusual minor peaks. Percentages and error estimates for samples from three independent cultures for each panel are presented in Supplementary Table I. The peak at the very top of the gradient contains non-ribosomal UV adsorbing material; its abundance is not reproducible and thus not considered here. (B) Fractions collected from panel (A) were also subjected to dot blot analysis with 16S and 23S probes. The red dashed lines align the fractions analyzed. Groups of rows reflect the same strain and medium order as profiles in panel (A). (C) Schematic representation of the probes used, not drawn to scale. The positions of probes relative to mature 16S (red) and 23S (blue) sequences are shown. (D) The same samples as in (A) but subjected to dissociating gradient conditions, where polysomes and 70S particles are converted to ribosomal subunits. (E) As panel (B) but also including probes for premature subunits, as depicted in panel (C).
The content of active ribosomes in fast growing wt is the same as fast growing ppGpp0 and slow growing ppGpp0 cells. If we assume that all 70S and polysomes are active in the protein synthesis and 30S and 50S free subunits visualized in our scans are not active, then the ribosomal fraction that is active during fast growth of wt is 72% and in ppGpp0 is 69%. During slow growth, that fraction in both strains increases to 81% and 84% respectively. It is noteworthy that it takes twice as many cells of slow growing wt as for slow growing ppGpp0 cells to obtain similar amount of A260 units sedimenting as ribosomes. This comprises a direct assay for ribosomal content to supplement estimates of rRNA production from RNA/DNA and RNA/protein ratios.
Although the content of active ribosomes is the same, the fraction of polysomes is different. It is evident that the percentage of ribosomes in the form of polysomes is higher in wt cells (39% for fast growth (LB) and 48% for slow growth (M9 minimal)) than in ppGpp0 cells (19% and 17%, respectively), regardless of growth rate. The monosome fraction (70S) behaves in an opposite fashion, 33% for wt at both growth rates, and 50% and 67% for ppGpp0. To verify the presence of 16S and 23S in appropriate peaks, Northern blots with oligonucleotide probes were performed, as shown in Fig. 2B and C. Although it would appear from panel B that there is more 23S than 16S present in the 70S fraction, we attribute this disparity as due to different specific activity of each probe.
There are many examples where accumulation of premature ribosomes compromises protein synthesis. One is the accumulation of the 20, 30 and 43S RNA (‘relaxed particles’), lacking some ribosomal proteins, when RNA accumulates in relA mutants starved for amino acids (Sypherd, 1965a, b; Nakada and Unowsky, 1966; Nakada 1967). Another example is mutants in DEAD-box helicases, where 40S particles accumulate (Charollais et al., 2003; Peil et al., 2008).
In contrast, high ribosomal content in slow growing ppGpp0 cells is not associated with ribosomal maturation defects. In order to assess ribosomal subunit maturation, we performed ribosome sedimentation profiles of the same samples with gradients that contain 1 mM MgCl2, which dissociate polysomes and monosomes into 30S and 50S ribosomal subunits (Fig. 2D). Probes for both mature and premature 16S and 23S rRNA were used in Northern blots to study subunit maturation (Fig. 2E). The 30S subunit profiles of ppGpp0 strains appear normal. However, the wt cells show a slower sedimenting shoulder in the 30S fraction, which we assign from probe hybridizations to be 17S rRNA. This shoulder is especially evident under fast growth conditions, and was even detected under associating gradient conditions (Fig. 2A). We attribute this to chloramphenicol, which transiently lowers ppGpp levels, thus increasing rRNA abundance without the accompanying synthesis of ribosomal proteins. Ribosomal synthesis of ppGpp0 cells is not further stimulated by addition of chloramphenicol. Similar experiments with wt strains grown in LB but without chloramphenicol addition, reveal the premature 30S shoulder is not present (data not shown).
We also find that there is a small shoulder (about 1/7th of the 50S peak) for only the ppGpp0 strain grown in LB, which appears to contain both types of premature 23S rRNA. In other fractions, the amount of premature 23S rRNA does not seem to be significant, as judged by A260 tracings, although detectable by probe hybridization. In contrast, the wt strain has a significant shoulder which comprises about half of the 50S peak, but only for slow growing cells. This suggests that only the wt strain growing slowly has significant accumulation of premature 50S subunit.
Overall, these data suggest that the high accumulation of ribosomal RNA in slow growing ppGpp0 cells is not accompanied by significant maturation defects. Moreover, the fraction of ribosomes that is functional (polysomes and 70S) is the same in wt and ppGpp0 cells. The differences in polysome and 70S content could be due to increased mRNA levels in ppGpp0 cells, assuming that less mRNA and more ribosomes would give longer polysomes. We predict this population of mRNA to be enriched for σ70 dependent transcripts of both housekeeping and ribosomal protein genes, and depleted of stress related messages despite nutritional limitation. In summary, ppGpp0 cells have more ribosomes than needed at slow growth rates and that the overall rate of translation is reduced in these ribosomes when compared to wild type cells.
ppGpp is sufficient for growth rate control
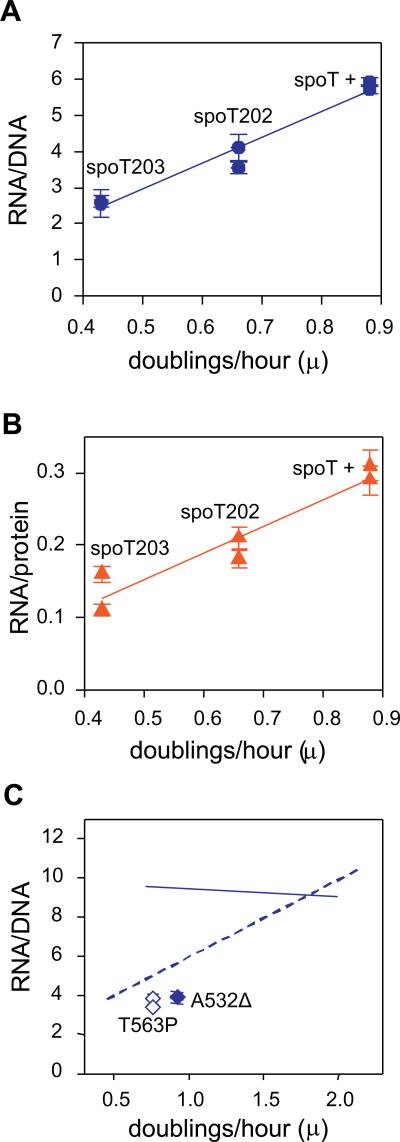
The high RNA/DNA and RNA/protein ratios of slow growing ppGpp0 cells can be taken to argue that ppGpp is necessary for growth rate control. The question whether ppGpp is also sufficient, can be addressed by studying the effects of artificially elevating ppGpp levels without nutrient starvation. Mutants in spoT have been described that allow such an elevation of basal levels of ppGpp: the T78I spoT202 = 5 fold, the R140CspoT203= 7 fold increase (Sarubbi et al., 1988). Indeed, Figure 3A and B, shows that RNA/DNA as well as RNA/protein ratios change appropriately in a manner that can be correlated with growth rates associated with increased ppGpp basal levels. This suggests that manipulating ppGpp basal levels provokes growth rate control, at least over a range of slow growth rates studied here (μ= 0.4 to 0.9).
Figure 3. Gratuitous induction of ppGpp or stringent RNA polymerase mutants restore growth rate control.
Gratuitous induction of ppGpp in the absence of starvation is achieved by point mutations in spoT, which in turn slows growth rate. (A) RNA/DNA ratios were measured as in Figure 1A. Solid line represents linear regression calculated with SigmaPlot software. The slope of the line approximates the value shown in Fig. 1 for wt cells. The strains were grown in medium #9 (Table I) (B) RNA/protein ratios were measured as in Figure 1C. The solid line represents linear regression calculated with SigmaPlot software. The slope of the line again approximates the value shown in Figure 1C for wt cells, given that the spoT203 mutant grows more slowly than the wt in the poorest medium. (C) Two stringent RNA polymerase mutants in a ppGpp0 host (rpoB A532Δ = B3449, and rpoB T563P = B3370) were grown in medium # 8 (Table I). Solid and dashed lines represent linear regressions obtained for ppGpp0 and wt strains in Figure 1A, respectively.
Mutants of RNA polymerase in ppGpp0 strains have been described that phenotypically behave as if ppGpp is present (Zhou and Jin, 1998; Murphy and Cashel, 2003). These have been called stringent mutants. We have tested two such mutants (Figure 3C), and find RNA/DNA ratios to be appropriate for a wt growth rate, even in the absence of ppGpp. Apparently the deregulation of RNA/DNA ratios in the absence of ppGpp can be reversed by these RNA polymerase mutants. Mechanistically it is usually assumed that both ppGpp and stringent mutants confer a similar conformational change in RNAP that results in common regulatory phenotypes. The fact that the stringent mutants restore RNA/DNA ratios to wild type levels, gives further credence to the role of ppGpp in growth rate control at the level of transcription.
The effects of transitions from balanced growth to stationary phase in the absence of ppGpp
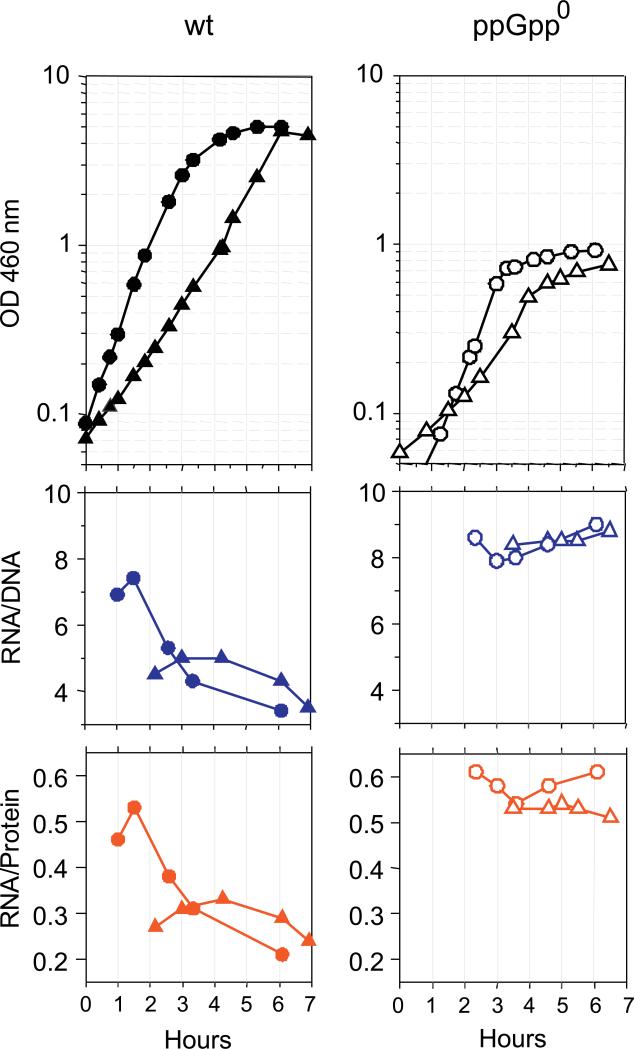
The growth conditions so far studied here involved unrestricted growth. Biologically the question arises as to what happens when cells enter stationary phase. Would high RNA/DNA and RNA/protein ratios persist in slow and fast growing ppGpp0 cells? We addressed this question by following the cell transition into stationary phase after growth in both rich and minimal media (Figure 4). The RNA/DNA ratios as well as the RNA/protein ratios initially measured during exponential growth shown in Figure 4 are consistent with the data for balanced growth shown in Figure 1. In fast growing wt cells both RNA/DNA and RNA/protein ratios are at high values until OD460 = 0.6, but markedly decrease at higher cell densities where growth begins to slow. Nutrient limitation leads to ppGpp accumulation, which inhibits ribosomal RNA and ribosomal protein synthesis. In slow growing wt cells, exponential growth persists to a much higher cell density (OD460=5) and RNA/DNA and RNA/protein ratios remain low, consistent with high ppGpp levels. In contrast, ppGpp0 cells maintain RNA/DNA and RNA/protein ratios at constant high levels regardless of initial growth rates or transitions into stationary phase. It seems that high RNA/DNA ratios persist for at least 3 hours after growth stops under these conditions. We take this to mean that in the absence of growth, RNA formed by ppGpp0 cells is metabolically stable, at least initially, as it would be if rRNA was packaged into ribosomes.
Figure 4. Changes in RNA/DNA and RNA/protein ratios during entry into stationary phase.
Filled symbols represent wt strains (CF7968), open symbols represent ppGpp0 strains (CF12257). Both strains were grown in rich (circles) or in poor (triangles) growth media (as defined in Table I). For wt: medium # 9 (poor) and #4 (rich); for ppGpp0: #6 (poor) and # 4 (rich). Upper panels depict changes in cell density, middle panels RNA/DNA ratios and lower panels RNA/protein ratios.
The effects of DksA on transitions from balanced growth to stationary phase
Negative regulation of stable RNA promoters by ppGpp is proposed to involve DksA acting in this instance as a synergistic co-factor to destabilize RNA polymerase-DNA open complexes enough to limit transcription initiation (Paul et al., 2004b). We examined RNA/DNA ratios in a dksA mutant during fast and slow unrestricted balanced growth (Figure S2). At all growth rates, cells that lacked DksA had RNA/DNA ratios as elevated as those observed for ppGpp0 strains (Figure 1). We conclude that at unrestricted growth rates, the absence of DksA results in deregulation of RNA/DNA ratios to approximately the same extent as the absence of ppGpp.
Does the same conclusion apply to restricted growth and growth phase transitions? In the previous experiments growth has been followed in different media for each strain, supporting similar growth rates. In the experiments shown in Figure 5, a single, undefined LB medium is used because it supports a transition from fast growth to stationary phase that is incremental, as one after another nutrient is exhausted or secondary metabolites are re-utilized (Sezonov et al., 2007). Growth in LB is predicted to begin with uniformly high RNA/DNA ratios because of initial fast growth, regardless of ppGpp or the state of the dksA allele.
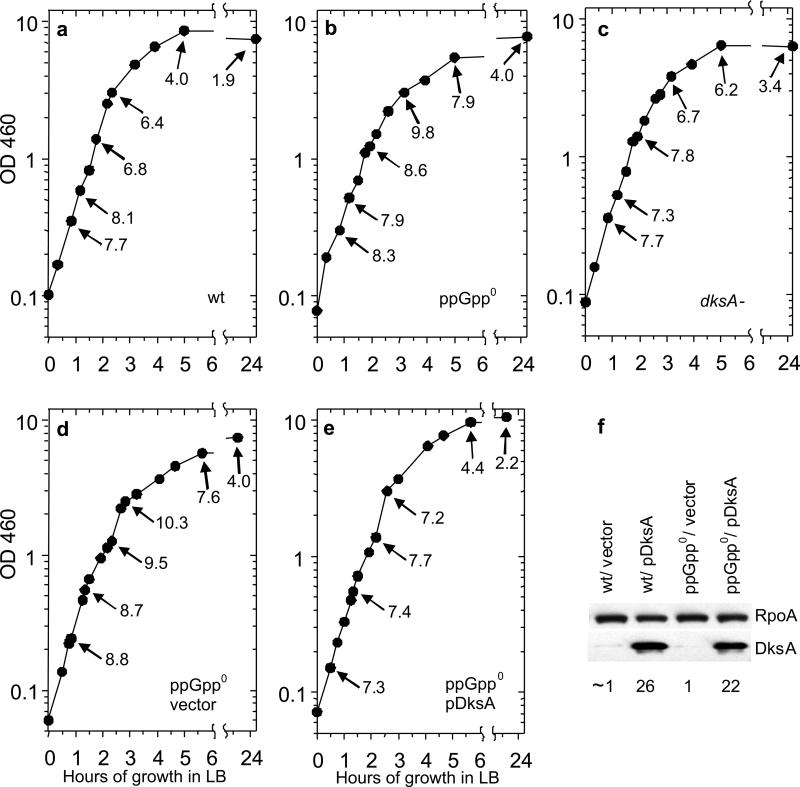
Figure 5. Changes in RNA/DNA ratios during entry into stationary phase in LB.
Numbers with arrows represent RNA/DNA ratios measured at the indicated points of growth. Panels (A) and (B) allow the comparison of the effects of ppGpp, contrasted with the deficiency of dksA (wt is CF1648; ppGpp0 is CF1693; dksA is CF9239) (C). Panels (D) and (E) compare the effects of DksA overexpression on a multicopy plasmid from its natural promoter (pDksA= pJK537) and pBR322 (vector control). The extent of overexpression of DksA at early stationary phase is shown by the Western blot with an RpoA loading control, in panel (F). Quantitation relative to wt vector control is shown beneath each lane.
Sampling LB cultures at OD460 values less than 0.6 indeed results in high RNA/DNA values that range between 7.3 and 8.3 for wild type, ppGpp0 (dksA+), and dksA− (ppGpp+) strains (Figure 5 A-C). As growth of the wt strain begins to slow, its RNA/DNA ratio drops to 4.0 while overnight nongrowing wt cultures have very low RNA/DNA ratios of 1.9. For the ppGpp0 strain at a similar point of slow growth, the RNA/DNA ratio remains relatively high (7.9); the RNA/DNA ratio drops overnight to 4.0, which is significantly higher than the wt (Figure 5B). The dksA mutant strain shows RNA/DNA ratios that are intermediate between those of the wild type and ppGpp0 strains both at the point where growth slows from exponential to stationary, and for overnight growth (Figure 5C). Evidently, the absence of DksA exerts a slightly less severe regulatory defect on RNA/DNA ratios during stationary phase entry in LB medium than a deficiency of ppGpp. The above results show that RNA/DNA ratios do reflect expected transitional changes in growth rate during restricted growth.
It has been reported that excess DksA in some instances can mimic the ability of ppGpp to regulate gene expression (Brown et al., 2002; Potrykus et al., 2006; Magnusson et al., 2007; Aberg et al., 2008; Blankschien et al., 2008). Therefore we overexpressed DksA from a multicopy plasmid bearing the dksA gene driven by its natural promoter in a ppGpp0 strain and monitored RNA/DNA ratios. Fig. 5E reveals that the ratios during growth phase transitions are restored to wt values, but restoration occurs with a large excess (22-fold) of DksA relative to wt (Fig. 5F). The DksA levels obtained here are similar to those achieved in the reports cited above. Apparently, DksA can substitute for ppGpp in the growth rate control, but only at high nonphysiological levels. Under physiological conditions, DksA levels remain low and constant throughout all phases of growth (Brown et al., 2002; Paul et al., 2004a).
Concluding remarks
In summary, we take these results to mean that ppGpp, acting through DksA, is the major determinant of growth rate control since slow growing ppGpp0 strains achieve RNA/DNA and RNA/protein ratios equivalent to fast growing wt cells. We verified that high RNA levels achieved in the absence of ppGpp are not accompanied by noticeable ribosome maturation defects. Why do our results differ from those of Gaal and Gourse (1990), who also measured RNA/protein ratios as a function of growth rate and found the absence of ppGpp had no effect? They also differ from those of Hernandez and Bremer (1993), who found the constitutive rRNA content of ppGpp0 cells was fixed at an unexpectedly low level normally seen only in slow growing wt cells rather than the high levels expected for the absence of a major negative regulator of rRNA synthesis. One possible explanation is that these differences are due to spontaneous suppressor mutants enriched in slow growing cultures of ppGpp0 strains, especially after overnight growth, although the authors indicate they checked for revertants. Our methodology takes specific precautions to avoid this problem (see Experimental Procedures). Neither the use of what should be the same strains or attempt to use their growth conditions has changed our conclusions.
We cannot exclude the possibility of some minor determinants of growth rate control in addition to ppGpp. There is evidence that the number of ribosomes formed during fast growth of wt cells is not the maximum that can be synthesized from the seven ribosomal operons in E. coli. Two ribosomal operons can be deleted with barely noticeable effects on growth rate (Condon et al., 1993). The interpretation is that an unexpressed potential (2/7ths) for rRNA synthesis exists in fast growing wt cells. Here, the maximum RNA/DNA or RNA/protein ratios we observe is the equivalent of what is found for fast growing cells. If ppGpp was responsible for regulating the usually unexpressed potential, then in the absence of ppGpp we should have seen a significantly higher RNA/DNA ratio at all growth rates. It is not known what mechanisms regulate the excess potential for ribosomal synthesis, which might involve ribosomal feed back inhibition, RNA polymerase packing on rRNA operons, promoter gating or be a consequence of rRNA promoters not being fully saturated with RNA polymerase (Baracchini and Bremer, 1991; Condon et al., 1995; Gummesson et. al., 2009; Keener and Nomura, 1996).
With this qualification, we view our results as an important step in understanding the role of ppGpp in growth rate control and cell physiology. We hope that this work resolves the controversy that is at least 20 years old. The significance of our findings is that they greatly simplify what was previously an intricate view of growth rate control due to many redundant elements. Instead of robust regulatory systems for ribosomal RNA, it appears that a single regulatory circuit (ppGpp and DksA) links nutrient abundance to growth rate control. The ability of bacterial cells to sense environmental changes through ppGpp is an emerging field. Roles for ppGpp have been reported in Gram-positive and Gram-negative bacteria (Potrykus and Cashel, 2008).
Recently, ppGpp has been found to be necessary for the survival of pathogens in the course of infection inside host cells through a variety of mechanisms (Dalebroux et. al., 2010). The fundamental reasons for this dependence is unknown and our findings contribute to better understanding of physiological changes occurring in the bacterial cell in the absence of ppGpp.
Experimental Procedures
Strains and media
Two pairs of isogenic E. coli K-12, MG1655 strains are used to compare effects of ppGpp0: CF1648-CF1693 (Xiao et al., 1991) and CF7968-CF12257 (Brown et al., 2002; Harinarayanan et al., 2008). An additional ppGpp+ isogenic strain pair with a dksA deletion is CF1648-CF9240 (Brown et al., 2002). The plasmid used for DksA overexpression was pJK537 (Kang and Craig, 1990). LB broth contains 5 g/l NaCl; glucose minimal medium A and M9 were prepared as described (Miller, 1972), supplemented with 1μg/ml thiamine, and M9 was modified to contain 2 μM FeSO4. Amino acid solutions were sterilized by filtration and added to heat sterilized media. The different media necessary to allow a range of unrestricted growth rates for various strains are presented in Table I.
Defining amino acid requirements that allow unrestricted growth of ppGpp0 strains at 37°C
Derivatives of MG1655 lacking ppGpp have complex AA requirements: growth occurs on M9 glucose supplemented with all 20 AA but in several instances cells grow poorly, if at all, when a full complement is missing one or another AA (Xiao et al., 1991). These instances suggested possible requirements for seven AA (F,H,I,L,S,T,V), yet growth did not occur in the presence of only these. Adding all 20 AA, 40 μg/ml each (AA40), allowed overnight growth at low yield (OD460~ 0.2) whereas increasing the AA levels to 400 μg/ml gave high yields (OD460~ 1.0) and a moderate growth rate. Screening such cultures with 20 AA40 by additionally supplementing with a single AA at 400 μg/ml identified serine as the only enhancer of growth yield, OD460≈1.0. This level of serine also enhanced growth yield in the absence of either glucose or NH4Cl, indicating serine can be used as a source of carbon or nitrogen (data not shown). We found some batches of serine contain an inhibitor that gives long lag period before growth resumes, due to an inhibitor because the length of the lag is proportional to the serine concentration (data not shown). When the six AA (FHILTV)40 were combined with serine at 400 μg/ml (S400), overnight growth still did not occur. Therefore, screening was repeated with (FHILTV)40 + S400 adding single AA. Four AA (DENQ)40 were found to slightly stimulate growth yield; supplementing (FHILTV)40 + S400 with pairwise combinations revealed DE, NE or QD supported overnight growth to OD460 of nearly 1.0. Thus, (DEFHILTV)40 + S400 was adopted as the standard set of minimal amino acid supplements. A similar screen of individual AA with (DEFHILTV)40 + S400 media to increase yield identified G, M or Y. Doubling times for wild type in all AA + S400 ranged from 33 min (CF7968) to 45 min (CF1648) while for ppGpp0 the range was 39 to 42 min. In LB doubling times for wild type ranged from 23-28 min and for ppGpp0 from 26 to 30 min.
Avoiding revertants
Liquid growth procedures were modified to minimize takeover of slow growing ppGpp0 cultures by RNA polymerase suppressor mutants, which spontaneously occur at a frequency of about 10−5 (Murphy and Cashel, 2003). Liquid cultures were inoculated (OD460 ≤0.05) by suspensions of a few colonies grown on the same test media but on plates, taking care to avoid suppressor contamination. Liquid growth at 37°C was achieved with 25 ml cultures in 250 ml flasks, shaken at 175 rpm. The frequency of revertants was measured as the ratio of titers on minimal and on LB plates after the experiment. Revertant colony titers that exceeded 15% were arbitrarily judged sufficient to invalidate an experiment. This occurred in about 1/3rd of experiments where cultures were inoculated from an overnight liquid culture and rarely by the modified growth protocol.
Sampling and lysis
Only culture media that could support exponential growth to at least OD460 =0.8 were used. We assume that sampling well before the detectable slowing of exponential growth in defined media reflects unrestricted growth that is not limited by the amount of any particular component. For undefined media with many components at varying concentrations, this assumption may not be valid for all but very low cell densities, before the least abundant but most efficiently utilized component is consumed. This can occur even though apparent growth rates are faster and growth appears exponential (Sezonov, et al. 2007).
Sample volumes were taken at cell densities OD460 = 0.25 and 0.5, so as to contain 1.4 optical density units. Cell samples were chilled on ice, pelleted for 5 min at 3716 × g, washed with 1 ml of cold 0.9% NaCl and resuspended in 1ml of the same solution. Then 0.8 ml of the suspension was added to a tube containing 0.2 ml 0.2% SDS and 50 mM EDTA (pH 8) solution, equilibrated in a 95-100°C water bath. This gives a clear lysate in 15 s, that was immediately chilled on ice and stored at −20°C. The hot SDS lysis method (Sarmientos et al., 1983) is modified to reduce SDS levels enough to avoid interference with the RNA-specific fluorescent dye assays yet high enough to completely lyse a fixed number of cells.
DNA and RNA Assays
Assays with dyes that fluoresce upon binding specifically to RNA or DNA are about 1,000 times more sensitive than classical chemical assays with acid hydrolysates for ribose (orcinol) or deoxyribose (diphenylamine). This sensitivity allows smaller culture volumes and shorter growth periods before assay. RNA determinations were made with the Quant-it RNA Assay (InVitrogen-Molecular Probes) using 620 nm/680 nm excitation/emission with a 5 ng detection limit: each RNA assay requires 20 μl of a 1/20 dilution of sample lysate and is run concurrently with an RNA standard curve. Similarly, the Quant-it DNA Assay (InVitrogen-Molecular Probes) was used at 485nm/528nm for excitation/emission with a 12.5 ng detection limit; 10 μl of undiluted lysate is used for each DNA assay. Protein assays with the BCA reagent (Pierce) required 50 μl of lysate. A Synergy HT 96 well microplate scanning spectrofluorimeter (Bio-Tek) was used to measure fluorescence with the Synergy KC4 software for statistical analyses. Each sample assayed at least in duplicate had the following standard deviations: RNA 3.0%; DNA 1.8%; protein 4%. Macromolecular composition per cell is expressed as either RNA/DNA or RNA/protein ratios with each component measured as μg/OD460.
Preparation of cell lysates and ribosome profiles
Cell lysates and polysomes fractionation was adapted from that reported by (Jiang et al., 2007) and (Shin and Dever, 2007). Briefly, cells were grown at 37°C in 100 ml of appropriate media (with the exception of wt strain in poor medium, which had to be grown in 200 ml in order to obtain similar A260 units of RNA). At OD460~0.4, chloramphenicol was added to 200 μg/ml for 1 min, the cells were quickly cooled on ice and then harvested by centrifugation. The pellets were then washed with 10 ml of ice-cold 0.9% NaCl, followed by wash with ice-cold 10 ml of the lysis buffer (10 mM Tris-Cl (pH 7.5), 10 mM MgCl2, 30 mM NH4Cl, 2 mM DTT, 100 μg/ml chloramphenicol). The cells were then resuspended in 400 μl of the same lysis buffer, 0.5 volume of acid-washed glass beads (Sigma) was added and the cells were disrupted by 5 cycles of 1 min vortexing and 1 min rest on ice. The samples were then spun at 12,500 × g at 4°C, for 10 minutes. The amount of ribosomes in the supernatant was assessed by A260. A sample of cells was checked for suppressor mutants, before chloramphenicol was added. Each experiment was repeated with three independent cultures.
Two types of sucrose gradients were used. For associating conditions: 10 mM Tris-Cl (pH 7.5), 10 mM MgCl2, 100 mM NH4Cl, 2 mM DTT. For dissociating conditions: 10 mM Tris-Cl (pH 7.5), 1 mM MgCl2, 200 mM NH4Cl, 2 mM DTT. Approximately 2 A260 units were loaded onto 12 ml 7-47% sucrose gradients, prepared with Gradient Master (BioComp). An aliquot of the same sample was applied to both types of gradients. The samples were spun in SW41 rotor (Beckman) at 18,300 rpm, 4°C, for 15 hrs 10 min.
Fractions (0.5 ml) were collected with the use of ISCO piercing apparatus connected to BioLogic chromatography system (BioRad), at the rate of 0.5 ml/min. Data was collected with LP DataView software (BioRad) and area under individual peaks was calculated with the use of OriginPro 8.1 software. The baseline for each plot was established in the following manner: for associating conditions, two lowest points in the graph were connected to form a straight line (at the beginning of 30S peak, and before the first polysome peak); for dissociating conditions, a line parallel to the x-axis but intersecting the lowest point on the graph (before 30S peak) was drawn.
Dot Blot Northerns
Dot blots were performed as described in (Bugl et al., 2000) with some modifications. Briefly, 8 μl from each fraction were incubated for 15 min at 55°C in a final concentration of 2.2 M formaldehyde, 50% (v/v) formamide, 10 mM MOPS, 4 mM NaCl, 0.5 mM EDTA (pH 7.0), final volume 30 μl, and cooled on ice. Next, 2 μl of each sample were spotted manually on dry GeneScreen Plus membranes (Perkin Elmer). The samples were then UV crosslinked, and the membranes were baked for 30 min at 80°C under vacuum. Pre-hybridization was performed for 2 hrs with UltrahybOligo buffer (Ambion) at 42°C. Hybridization was carried out over night at the same temperature with γP32-labeled oligonucleotide probes. The membranes were then washed twice for 20 min at room temperature with 2×SSC, 0.5% SDS, dried and exposed with phosphoimager screens.
The probes used were: 16S: 5’GCGTTCAATCTGAGCCATGATCAAACTCTTC3’, 17Sup: 5’AGAATCCCGTATCTTCGAGTGCCCACA3’, 17Sdown: 5’GTGTGAGCACTACAAAGTACGCTTCTTTAAGGTAAGG3’, 23S: 5’ATCCACCGTGTACGCTTAGTCGCTTAACC3’, p23up: 5’GTGTACGCTTAGTCGCTTAACCTCACAAC3’, p23down: 5’CGGCGTTGTAAGGTTAAGCCTCACGG3’.
Western Blot Assays
Western blots were performed as described (Brown et al., 2002).
Supplementary Material
Acknowledgements
We thank Dr. R. D'Ari for encouragement and many discussions during the course of this study. We especially acknowledge the constructive exchanges of views with Dr. V.J. Hernandez. Dr. R. Maraia, A. Crawford, Dr. T.E. Dever and Dr. B.S. Shin provided help with the polysomes methodology. We also thank Drs. D. Vinella, R. Harinarayanan and the Friday Seminar for helpful advice.
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development intramural program of the National Institutes of Health.
References
- Aberg A, Shingler V, Balsalobre C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol Microbiol. 2008;67:1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x. [DOI] [PubMed] [Google Scholar]
- Baracchini E, Bremer H. Control of rRNA synthesis in Escherichia coli at increased rrn gene dosage. Role of guanosine tetraphosphate and ribosome feedback. J Biol Chem. 1991;266:11753–11760. [PubMed] [Google Scholar]
- Blankschein MD, Potrykus K, Grace E, Choudhary A, Vinella D, Cashel M, Herman C. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 2008;5(1):e1000345. doi: 10.1371/journal.pgen.1000345. doi:10.1371/journal.pgen.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JCA, Jakob U. RNA methylation under heat shock control. Mol Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington DC: 1996. pp. 1553–1569. [Google Scholar]
- Brown L, Gentry D, Elliott T, Cashel M. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol. 2002;184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1968;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry D, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington DC: 1996. pp. 1458–1496. [Google Scholar]
- Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- Condon C, French S, Squires C, Squires CL. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 1993;12:4305–4315. doi: 10.1002/j.1460-2075.1993.tb06115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Gourse RL. Guanosine 3'-diphosphate 5'-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummesson B, Magnusson LU, Lovmar M, Kvint K, Persson O, Ballesteros M, Farewell A, Nystrom T. Increased RNA polymerase availability directs resources towards growth at the expense of maintenance. EMBO J. 2009;28:2209–2219. doi: 10.1038/emboj.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harinarayanan R, Murphy H, Cashel M. Synthetic growth phenotypes of Escherichia coli lacking ppGpp and transketolase A (tktA) are due to ppGpp-mediated transcriptional regulation of tktB. Mol Microbiol. 2008;69:882–94. doi: 10.1111/j.1365-2958.2008.06317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez VJ, Bremer H. Characterization of RNA and DNA synthesis in Escherichia coli strains devoid of ppGpp. J. Biol. Chem. 1993;268:10851–10862. [PubMed] [Google Scholar]
- Jensen KJ. The Escherichia coli K-12 “wild types” W3110 and MG1665 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J Bacteriol. 2007;189:3434–3444. doi: 10.1128/JB.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Craig E. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt FC,, editor. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington DC: 1996. pp. 1417–1431. [Google Scholar]
- Kjeldgaard NO, Maaloe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:607–616. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Nystrom T, Farewell A. Underproduction of sigma 70 mimics a stringent response. A proteome approach. J Biol Chem. 2003;278:968–973. doi: 10.1074/jbc.M209881200. [DOI] [PubMed] [Google Scholar]
- Miller JH. A short course in Bacterial Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1972. [Google Scholar]
- Murphy H, Cashel M. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 2003;371:596–601. doi: 10.1016/S0076-6879(03)71044-1. [DOI] [PubMed] [Google Scholar]
- Nakada D. Functional activity of ribosomes formed in vivo from “relaxed particles”. Biochim. Biophys. Acta. 1967;145:664–670. doi: 10.1016/0005-2787(67)90125-6. [DOI] [PubMed] [Google Scholar]
- Nakada D, Unowsky J. In vitro formation of functional ribosomes from the “relaxed particles”. Proc Natl Acad Sci U S A. 1966;56:659–663. doi: 10.1073/pnas.56.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC. Bacterial growth: constant obsession with dN/dt. J. Bacteriol. 1999;181:7405–7408. doi: 10.1128/jb.181.24.7405-7408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Hochschild A. Regulation of RNA polymerase through the secondary channel. Cell. 2004;118:281–284. doi: 10.1016/j.cell.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004a;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu rev Genet. 2004b;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl Acad Sci U.S.A. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Virumae K, Remme J. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 2008;275:3772–3782. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary -channel structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Vinella D, Murphy H, Szalewska-Palasz A, D'Ari R, Cashel M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J Biol Chem. 2006;281:15238–15248. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- Sarmientos P, Sylvester JE, Contente S, Cashel M. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell. 1983;32:1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- Sarubbi E, Rudd KE, Cashel M. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Sezonov G, Joseleau-Petit D, D'Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Dever TE. Molecular genetic structure-function analysis of translation initiation factor eIF5B. Methods Enzymol. 2007;429:185–201. doi: 10.1016/S0076-6879(07)29009-3. [DOI] [PubMed] [Google Scholar]
- Svitil AL, Cashel M, Zyskind JW. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J. Biol. Chem. 1993;268:2307–11. [PubMed] [Google Scholar]
- Sypherd PS. Accumulation of ribonucleoprotein particles in a relaxed mutant of Escherichia coli. J Bacteriol. 1965a;90:403–410. doi: 10.1128/jb.90.2.403-410.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sypherd PS. Formation of ribosomes from precursor ribonucleoprotein particles. J Bacteriol. 1965b;90:411–417. doi: 10.1128/jb.90.2.411-417.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl Acad Sci U.S.A. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.