Abstract
To study the function of ABCE1/Rli1 in vivo, we used ribosome profiling and biochemistry to characterize its contribution to ribosome recycling. When Rli1 levels were diminished, 80S ribosomes accumulated both at stop codons and in the adjoining 3′UTRs of most messenger RNAs. Frequently these ribosomes reinitiated translation without the need for a canonical start codon, as small peptide products predicted by 3′UTR ribosome occupancy in all 3 reading frames were confirmed by Western analysis and mass spectrometry. Eliminating the ribosome-rescue factor Dom34 dramatically increased 3′UTR ribosome occupancy in Rli1 depleted cells, indicating that Dom34 clears the bulk of unrecycled ribosomes. Thus, Rli1 is crucial for ribosome recycling in vivo and controls ribosome homeostasis. 3′UTR translation occurs in wild-type cells as well, and observations of elevated 3′UTR ribosomes during stress suggest that modulating recycling and reinitiation is involved in responding to environmental changes.
Keywords: ABCE1, Rli1, Dom34, ribosome recycling, reinitiation, termination, 3′UTR
INTRODUCTION
Ribosome recycling is a vital cellular process that dissociates post-termination ribosomes into small and large subunits for participation in new rounds of translation initiation. Recycling of ribosomes is essential for maintaining homeostasis of the pool of free ribosomes, and it has been well documented that mutations in ribosome rescue/recycling factors such as Dom34 and Hbs1 exhibit synthetic phenotypes with other deficiencies in ribosome production (Bhattacharya et al., 2010; Carr-Schmid et al., 2002).
In eukaryotes, recycling effectively begins with termination on recognition of a stop codon in the A site of the 80S ribosome by release factors eRF1 and GTP-bound eRF3. Following hydrolysis of GTP, eRF3 dissociates, leaving eRF1 in the A site poised to hydrolyze the peptidyl-tRNA in the P site. This peptide release step is temporally coupled through the action of Rli1 (yeast)/ABCE1 (mammals) to the first stage of recycling, where the 80S ribosome is separated into a free 60S subunit and a 40S subunit bound to deacylated tRNA and mRNA. In the second stage of recycling, the deacylated tRNA is removed from the ribosome with attendant dissociation of the remaining 40S·mRNA complex (Jackson et al., 2012) (Dever and Green, 2012).
In vitro studies in reconstituted mammalian and yeast translation systems defined this common pathway for ribosome recycling. While ribosome dissociation is promoted simply by eRF1 (and by the ribosome rescue factor Dom34) (Shoemaker et al., 2010), the rate of the reaction is greatly stimulated by ABCE1/Rli1 resulting in efficient dissociation of 60S subunits over a wide range of Mg+2 concentrations (Pisarev et al., 2010; Shoemaker and Green, 2011). The ribosome-splitting activity of ABCE1/Rli1 leaves mRNA and deacylated tRNA bound to the 40S subunit, and release of the tRNA in the second stage of recycling appears to be stabilized by eIF1, ligatin/eIF2D, or the interacting proteins MCT and DENR (Pisarev et al., 2007; Skabkin et al., 2010).
Yeast Rli1 also stimulates translation termination (Khoshnevis et al., 2010)(Shoemaker and Green, 2011), where its contribution is ATP-hydrolysis independent. The dual function of Rli1 in termination and recycling, gated by ATP hydrolysis, is consistent with its location in a cryo-EM structure of an 80S complex containing peptidyl-tRNA in the P site and eRF1 in the A site. In this structure, Rli1 interacts directly with eRF1 and with components of both the large and small ribosomal subunits in the intersubunit space (Preis et al., 2014).
The impact of depleting ABCE1/Rli1 on ribosome recycling in vivo has not been addressed previously, and earlier publications suggest roles for the yeast factor in ribosome biogenesis (Yarunin et al., 2005) (Strunk et al., 2012) and translation initiation (Dong et al., 2004). It is even plausible that in certain situations in the cell, destabilization of the subunit interface by eRF1 (Shoemaker et al., 2010) may be sufficient, with initiation factors acting to stabilize dissociated subunits (Pisarev et al., 2007), to provide recycling independently of Rli1.
Other studies have probed biochemically the possible consequences of deficiencies in ribosome recycling. Early studies suggested that post-termination ribosomes, generated by puromycin treatment, remain associated with mRNA transcripts and could resume translation (Freedman et al., 1968). Using a mammalian reconstituted translation system, it was found that un-recycled 80S ribosomes, where peptide had been released, can migrate upstream or downstream from the stop codon and form stable complexes at nearby triplets that are complementary to the deacylated tRNA remaining in the P site (cognate to the penultimate codon of the open reading frame or ORF) (Skabkin et al., 2013). Other studies with yeast in vitro translation extracts argued that ribosomes terminating at a “premature stop codon” are inefficiently recycled and can migrate to nearby AUG codons (Amrani et al., 2004). Even in bacteria, impairment of ribosome recycling factor (RRF) evokes scanning and reinitiation by post-termination ribosomes (Janosi et al., 1998). These studies provide fodder for thinking about the fate of ribosomes in the absence of sufficient recycling activity in vivo.
Rli1/ABCE1 can also function in ribosome splitting to rescue stalled elongating 80S ribosomes, acting in conjunction with the eRF1 and eRF3 paralogs Dom34(yeast)/PELO(mammals) and Hbs1(yeast)/HBS1L(mammals), respectively, to release stalled ribosomes in the no-go and non-stop mRNA decay pathways (Pisareva et al., 2011b; Shoemaker et al., 2010) (Tsuboi et al., 2012). Dom34/PELO·Hbs1 and Rli1/ABCE1 have also been implicated in subunit splitting of vacant 80S subunits (Pisareva et al., 2011a). Such 80S couples cannot function in translation initiation and, under starvation, accumulate in yeast cells stabilized by the Stm1 protein (Ben-Shem et al., 2011). Recently, evidence was provided that Dom34 promotes translational recovery from starvation (van den Elzen et al., 2014). Thus, Rli1 appears to operate in the recycling of terminating, stalled elongating, or long-term-storage 80S ribosomes.
Using ribosome profiling, we recently defined a role for Dom34·Hbs1 in recovering un-recycled 80S ribosomes in the 3′UTRs of ~10% of all yeast genes in a dom34Δ mutant (Guydosh and Green, 2014). While the origin of these 3′UTR ribosomes was unclear, a defect in ribosome recycling seemed plausible because the phenomenon was enhanced by treating cells with diamide, an oxidizing agent known to inactivate Fe-S cluster proteins (Philpott et al., 1993) such as Rli1 (Yarunin et al., 2005). It appeared that some 3′UTR ribosomes present in dom34Δ cells scanned, rather than translated, the 3′UTR and eventually accumulated at the beginning of the poly(A) tail. However, translation by a fraction of the 3′UTR ribosomes, either by read-through of the main ORF stop codon or reinitiation, was not excluded (Guydosh and Green, 2014).
In this study, we use ribosome profiling (Ingolia et al., 2012) and biochemistry to define the role of Rli1/ABCE1 in living cells. In an Rli1-depleted yeast strain (dubbed rli1-d), we find that 80S ribosomes accumulate at the stop codons and in the 3′UTRs of virtually all yeast genes, to a much greater extent than seen previously in dom34Δ cells. The distribution of 80S footprints strongly suggests that ribosomes in the 3′UTR of the rli1-d strain are frequently engaged in translation, displaying occupancy peaks that coincide with 3′UTR stop codons in all three reading frames. We detected the aberrant 3′UTR translation products for several genes and obtained strong evidence that they can originate from reinitiation in any frame rather than by read-through of the main ORF stop codon. We further find that reinitiation tends to occur in proximity to the main ORF stop codon, but that neither AUG codons nor triplets complementary to the penultimate P-site tRNA are preferred start sites. We show that overexpressing Rli1 diminishes the 3′UTR ribosomes detected previously in dom34Δ cells, demonstrating that they arise from incomplete recycling of terminating ribosomes at main ORF stop codons. Finally, the absence of Dom34 in rli1-d cells evokes an increase in ribosomes occupying main ORF stop codons and 3′UTRs, as well as a dramatic rise in the number arrested at the 3′UTR/poly(A) boundary; these observations indicate that Dom34 is critically required for rescuing ribosomes that escape normal termination/recycling. Our findings demonstrate that Rli1 is crucial for ribosome recycling in vivo and thus plays an essential role in controlling non-canonical 3′UTR translation.
RESULTS
Depletion of Rli1 evokes increased 80S occupancies in 3′UTRs transcriptome-wide
To determine the translational consequences of reduced Rli1 function in vivo, we conducted ribosome profiling of an rli1 degron strain that allows for conditional depletion of an unstable version of Rli1 tagged at the N-terminus with ubiquitin followed by an arginine residue (Park et al., 1992). Transcription of this engineered allele PGAL-UBI-R-FH-RLI1 is under the GAL promoter and therefore is repressed with glucose as carbon source (Dong et al., 2004). Western analysis confirmed that the degron protein is expressed at lower levels than Rli1 expressed from the RLI1 promoter on galactose medium, and is undetectable only 2h following a shift to glucose medium (Supplementary Fig. S1A–B). By ~8h on glucose medium, the degron mutant (rli1-d) ceases growth completely (Fig. S1C) and displays a ~40% reduction in polysomes compared to isogenic WT cells (Fig. S1D). Accordingly, we chose these conditions to culture rli1-d and WT cells for ribosome profiling experiments. Cells were harvested by rapid filtration, lysed while frozen in liquid N2, and thawed in the presence of cycloheximide to arrest elongation (Guydosh and Green, 2014). Following digestion by RNase I, 80S ribosomes were isolated by sedimentation through a sucrose density gradient. Ribosome footprint levels (on ORFs) were highly reproducible between biological replicates (Spearman’s R2 = 0.98 for rli1-d cells) (Fig. S2A). We also performed mRNA-Seq on the WT and rli1-d cells and found that the density of reads was broadly redistributed among coding sequences in the depletion strain and showed signs typical of stress, such as down-regulation of ribosomal proteins (Fig S2B–C). As expected, these changes in gene expression at the transcript level were correlated with (and often amplified by) the observed redistribution in footprint density.
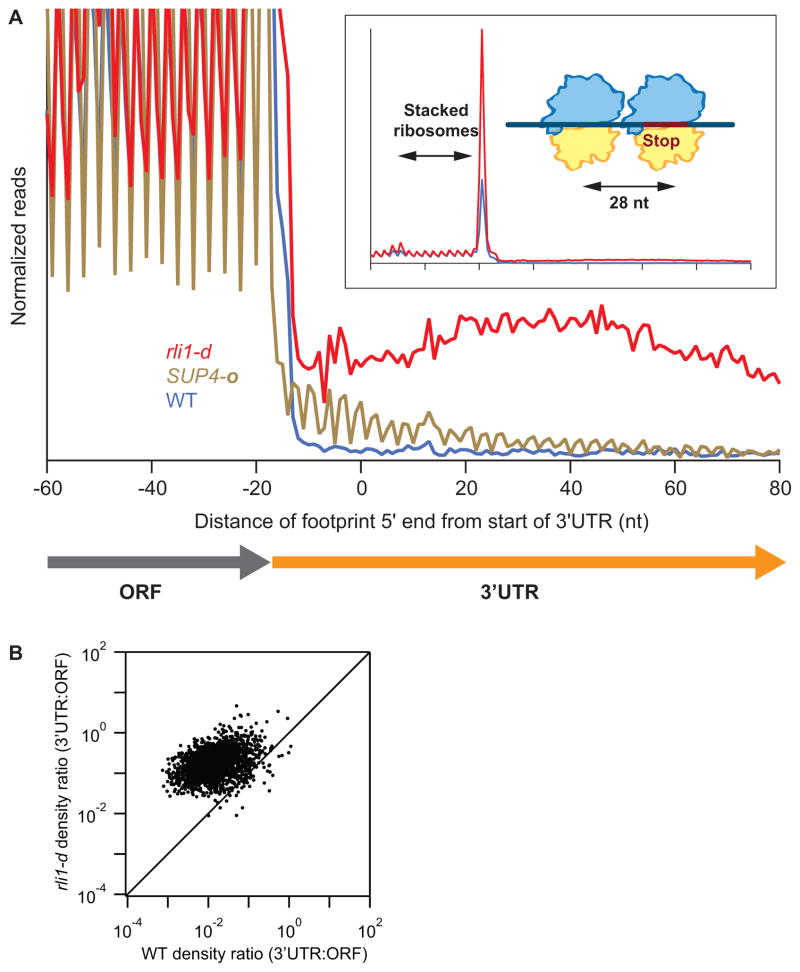
Examination of the average level of ribosome footprint reads for all genes aligned at stop codons in WT cells revealed a 3-nt periodicity in the ORF, indicating the translated reading frame, with a peak occurring at the stop codon (Fig. 1A), in accordance with previous results (Ingolia et al., 2009; Ingolia et al., 2011). The corresponding profile for rli1-d cells revealed two striking differences: the peak at stop codons was greatly magnified, and a second, smaller, peak appeared ~30 nt upstream. Both phenomena are consistent with a delay in termination or recycling at the stop codon accompanied by queuing of the trailing elongating ribosomes behind those stalled at the stop codon. Second, ribosome occupancies were elevated in 3′UTR regions, slightly rising for the first ~50 nt of the 3′UTR and gently falling off thereafter (Fig. 1A). Moreover, these 3′UTR ribosomes do not occupy a single reading frame like those in the coding sequence (no more than 35% of 28-nt reads mapped without mismatches to a single 3′UTR reading frame vs 94% in ORFs). A comparison of the ratio between ribosome density in 3′UTR vs. the ORF for each transcript revealed a broad increase in 3′UTR density, by roughly an order-of-magnitude on average, when Rli1 was depleted (Fig. 1B). We found that the level of ribosome density in 3′UTRs was correlated with that found in ORFs (Fig. S2D) and, between biological replicates, averaged between 21-34% of that found in ORFs (Fig. S2E). The general increase in 3′UTR occupancies seen in rli1-d cells is consistent with a genome-wide failure in termination or recycling at stop codons that leads to either a continuation of translation without termination (“read-through translation”), reinitiation of new translation in the 3′UTR, or termination followed by 80S “scanning.” We define scanning as the movement of unrecycled ribosomes (for brevity dubbed 80S post-termination complexes or post-TCs) along mRNA in the absence of peptide synthesis. In general, the averaged data from the rli1-d strain differed significantly from data derived from a strain expressing an ochre suppressor tRNA (SUP4-o) (Guydosh and Green, 2014). In that case, ribosome occupancy in 3′UTRs following UAA stop codons maintains the same reading frame as the main coding sequence and gradually decreases downstream as ribosomes reading past the main stop codon terminate translation and are recycled at downstream stop codons (Fig 1A). The absence of these trends in the rli1-d data point to a phenomenon distinct from simple read-through.
Figure 1. Ribosomes accumulate at stop codons and in 3′UTRs of most genes when Rli1 is depleted.
(A) Normalized average ribosome footprint occupancy (each gene equally weighted) from all genes aligned at their stop codons for WT, SUP4-o (genes with UAA stop codons only) and rli1-d cells. (Inset) Demagnified view of (A) with schematic depicting ribosome stalling and queuing at the stop codon. (B) Ratio of footprint densities in 3′UTRs to the respective ORFs is plotted for rli1-d cells versus WT cells, for genes with >5 rpkm in ORFs and >0.5 rpkm in 3′UTRs. Each point represents the data for 1 gene.
Accumulation of 80S ribosomes at 3′UTR-encoded stop codons is consistent with 3′UTR translation
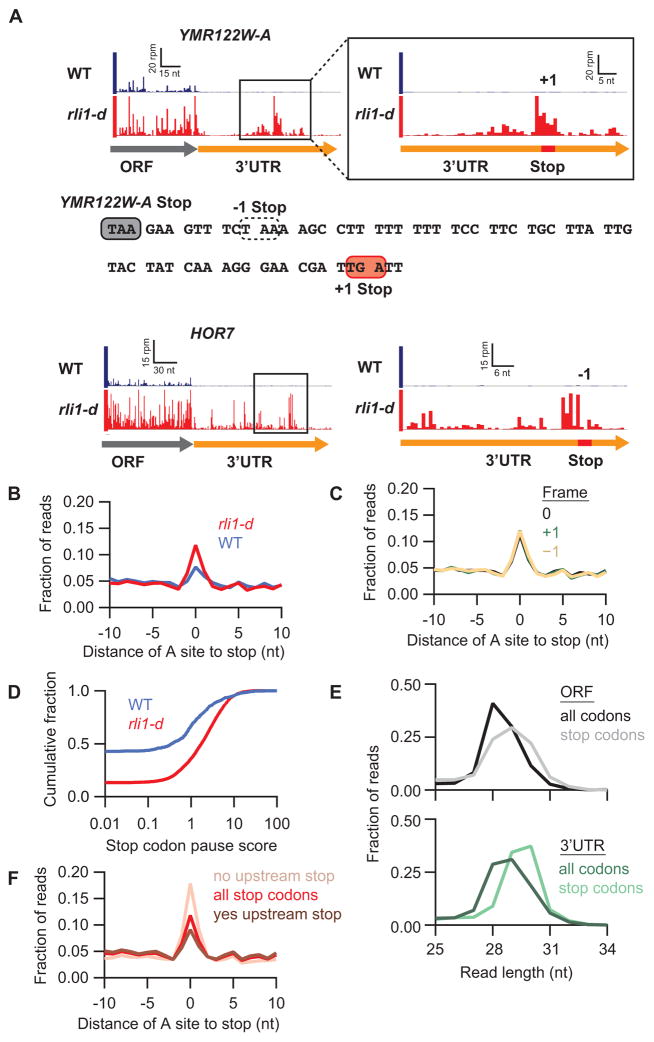
To evaluate whether ribosomes in 3′UTRs were translating, we examined the relative ribosome density across numerous 3′UTRs and found a notable enrichment in density on stop codons (Figs. 2A, S3). To get a sense for the typical shape of the peak at stop codons in 3′UTRs, we averaged ribosome density around all stop codons in 3′UTRs and found the average peak was 2–3 fold above background level in the rli1-d strain and lower in the WT strain (Fig. 2B). Interestingly, this average peak for the rli1-d strain appeared the same when we limited our averaging to each of the three reading frames relative to the main ORF (Fig. 2C). To quantify this phenomenon on the level of individual stop codons, we computed a pause score by taking the ratio of ribosome occupancy at each stop codon relative to the background density in its respective 3′UTR (Fig. 2D). We found that the median pause score increased ~5-fold in rli1-d cells relative to WT cells, emphasizing the global nature of this effect.
Figure 2. Ribosomes accumulate at stop codons within 3′UTRs in rli1-d cells.
(A) Ribosome footprints on genes YMR122W-A and HOR7. Boxed regions on the left are magnified on the right, showing schematically the positions of 3′UTR stop codons (red) in the indicated reading frames (+1 or −1). A portion of the YMR122W-A 3′UTR sequence is shown, highlighting the main ORF stop codon (filled grey box), −1 stop codon (dotted box), and the +1 stop codon showing a strong 80S peak (filled red box). (B) Average fraction of ribosome occupancy in a window surrounding stop codons in the 3′UTR (all frames included). (C) Same as rli1-d data in (B) but sorting data for each reading frame relative to the main ORF (0) frame. (D) Cumulative frequency histogram of pause scores on 3′UTR stop codons shows enhanced pausing in the rli1-d strain vs WT, for genes with >10 rpkm in 3′UTRs. (E) Size distributions of rli1-d footprints that mapped without mismatches to either spliced coding sequences (ORFs) or 3′UTRs. Mapping performed for full sequences or 34-nt windows starting 17-nt upstream of stop codons. (F) Same as rli1-d data in (B) but data are sorted by the presence or absence of an upstream, in-frame 3′UTR stop codon.
The apparent accumulation of 80S ribosomes at 3′UTR stop codons seems incompatible with scanning 80S ribosomes and more consistent with translating 80S ribosomes. A second feature consistent with 3′UTR translation is that the footprint peaks at 3′UTR stop codons are enriched in atypically long nuclease-protected fragments, as previously reported for canonical stop codons in the main ORFs (Ingolia et al., 2011) (Fig. 2E). Finally, when we evaluated the average ribosome occupancy for stop codons across the 3′UTR, we found that the presence of an upstream 3′UTR stop codon in the same reading frame tends to diminish the size of the 80S peak at a downstream stop codon whereas the absence of an upstream stop codon increases it (Fig. 2F). This feature is expected from 3′UTR translation because the presence of an in-frame upstream stop codon would tend to trigger termination and recycling of most ribosomes at the expense of stop codons located further downstream in the 3′UTR.
Histidine starvation evokes ribosome stalling at 3′UTR His codons in the manner expected for 3′UTR translation
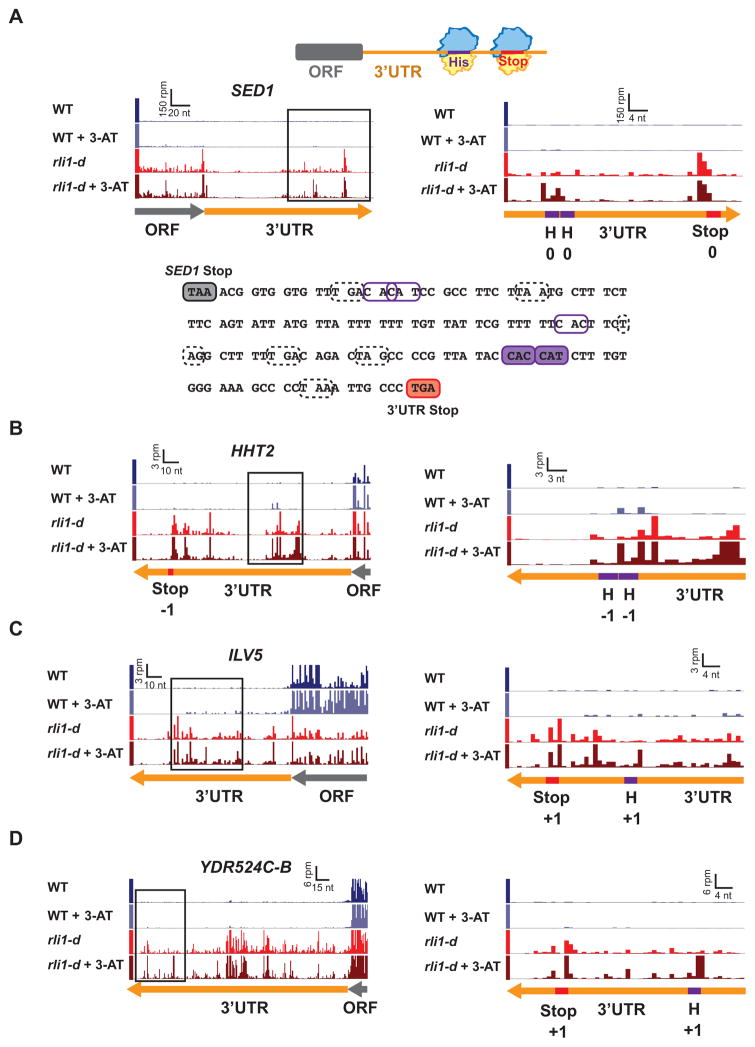
We showed previously that starvation of yeast cells for histidine with 3-aminotriazole (3-AT) evokes pausing of elongating 80S ribosomes, detected as 3-AT-enhanced ribosome occupancy at histidine codons in the ribosome profiles of ORFs genome-wide (Guydosh and Green, 2014). We exploited this phenomenon here to support the hypothesis that the 3′UTR ribosomes are translating. We established conditions for histidine starvation in Rli1-depleted cells by monitoring induction of eIF2α phosphorylation by protein kinase Gcn2—a well-established signature of amino acid starvation (Dever et al., 1992)—and determined that adding 3-AT after only 4h of Rli1 depletion evoked increased eIF2α-P that peaked after an additional 3h incubation in 3-AT medium (Fig. S4). As a control, we noted that ribosome profiling under these modified growth conditions did not prevent the appearance of peaks at stop codons in the 3′UTRs of rli1-d cells (Fig. 3A, 3-AT, WT vs. rli1-d). Importantly, 3-AT treatment of rli1-d cells evoked peaks at His codons upstream from (and in the same frame as) 3′UTR stop codons that exhibit strong peaks in untreated rli1-d cells (Fig. 3A–D). While the extent of histidine starvation was insufficient to systematically investigate pausing at the read depth of our dataset, we did find evidence of this phenomenon on well over 100 genes, further strengthening our hypothesis that 3′UTRs are translated when Rli1 is depleted (Table S1).
Figure 3. Histidine starvation evokes ribosome stalling at 3′UTR His codons in rli1-d cells.
(A) The schematic at the top depicts terminating ribosomes paused at stop codons and upstream elongating ribosomes paused at His codons in histidine-starved Rli1-depleted cells. Ribosome footprint reads in SED1 are presented as in Fig. 2A, showing the two His codons (H, purple) and predicted 3′UTR termination site (red), all in the 0 frame, in the enlargement on the right. These His codons (filled purple boxes) and in-frame stop codon (filled red box) are also highlighted in the SED1 3′UTR sequence, along with His codons (unfilled purple boxes) and stop codons (dotted boxes) in the other two reading frames that lack strong peaks. (B–D) Similar to (A) for the genes HHT2, ILV5, and YDR524C-B.
Direct detection of epitope-tagged 3′UTR translation products
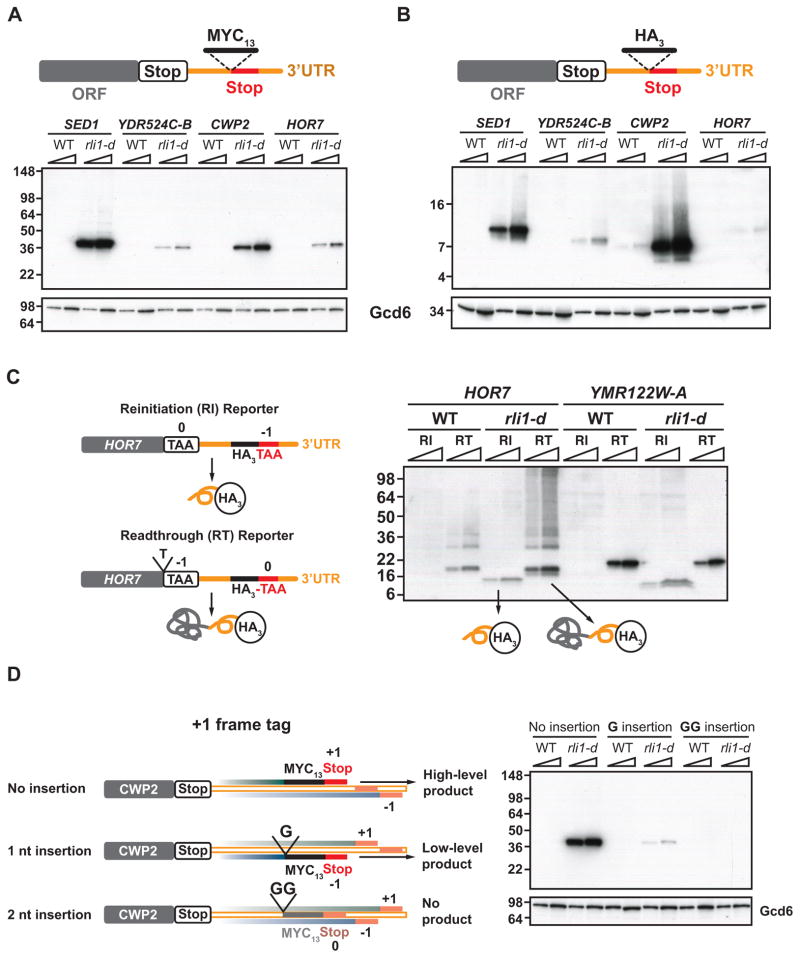
To differently evaluate our model for 3′UTR translation, we set out to detect polypeptides predicted to arise from 3′UTR translation in particular genes. To this end, we inserted the coding sequences for 13 tandem Myc epitopes immediately preceding, and in-frame with, 3′UTR stop codons in the endogenous loci of various genes exhibiting prominent peaks in ribosome density at 3′UTR stop codons in rli1-d cells. Western analysis revealed Myc13-tagged polypeptides of ~40kDa for 11 of 16 tagged genes in rli1-d cells, none of which were observed in identically tagged WT cells (Figs. 4A and S5A). These results provide direct evidence for translation of 3′UTR sequences in rli1-d cells in reading frames where ribosomes are stalled at stop codons. For the 5 genes where no Myc13-tagged product was detected, the predicted polypeptides might be subject to rapid proteolysis owing to unusual amino acid compositions encoded by normally untranslated 3′UTR sequences. In the course of our analysis, we realized that the 20.5kDa Myc13 epitope migrates anomalously as a polypeptide of ~40 kDa (Fig. S5B). Thus, our finding that the Myc13-tagged polypeptides from all 11 genes that produced stable 3′UTR translation products also have apparent MWs of ~40 kDa implies that only small portions of the tagged translation products are encoded by endogenous sequences at these genes. This in turn suggests that the tagged polypeptides originate from reinitiation after peptide release at the main stop codon, rather than read-through from the main ORF.
Figure 4. Detection of epitope-tagged 3′UTR translation products that result from reinitiation versus proteolysis of readthrough products in rli1-d cells.
(A–B) MYC13 (A) or HA3 (B) epitope tags were inserted just upstream of predicted 3′UTR termination sites in the chromosomal alleles of 4 candidate genes (depicted schematically) in WT and rli1-d strains. Tagged strains were cultured as in Fig. S1A and WCEs were subjected to Western analysis using antibodies against c-Myc or HA (upper blots) or Gcd6 (lower blots). Two amounts of extracts were loaded in each lane pair. Migration of MW standards is indicated on the left. ORF, main coding sequences. (C) HA3-tagged reinitiation (RI) and readthrough (RT) reporters for HOR7, and their predicted tagged products, are depicted schematically on the left. The RI reporters for HOR7 and YMR122W-A were those analyzed in (B), and the RT reporters were constructed from them by inserting a single nucleotide (T for HOR7) before the main ORF stop codon, putting that terminator out of frame, and placing the 3′UTR termination site in frame with the main ORF. Western analysis of the resulting strains was conducted as in (B). (D) To detect CWP2 3′UTR translation in different reading frames, single or tandem G nucleotides were inserted immediately preceding the MYC13 tag located adjacent to the predicted 3′UTR +1-frame termination site in the CWP2 reinitiation reporter analyzed in (A) and depicted here as “No insertion”, thus shifting the MYC13 tag and adjoining in-frame stop codon from the +1 frame into the −1 or 0 frames for the 1 nt and 2 nt insertions, respectively, as depicted schematically. The first stop codons encountered downstream of the tag coding sequences in the +1 (top), 0 (middle), and −1 (bottom) frames are indicated with filled red or orange boxes. For the 2 nt insertion/GG construct, the tag is shown partially transparent and the adjoining stop codon in orange versus red to indicate the absence of detectable reinitiation in the 0 frame. Tagged WT and rli1-d cells were analyzed as in (A).
To support this last conclusion, we pursued two different strategies. First, we inserted coding sequences for the much smaller HA3 epitope (that would not mask informative mobility differences) at the same 3′UTR locations used above for Myc13-tagging of 6 candidate genes. Importantly, their apparent MWs are now in the ~7–10kDa range (Fig. 4B & Fig. S5C). Given a mass of 4.8kDa for HA3, the observed MWs are considerably smaller than those predicted from read-through translation from the main ORF. Rather, their apparent MWs imply that only ~2–5kDa of the HA3-tagged products are encoded by endogenous sequences, which is within the range of masses predicted by reinitiation at sites close to the main ORF stop codons and subsequent termination occurring within the 3′UTR (see Table S2).
In the second strategy, we modified a subset of the aforementioned tagged alleles by mutating the main ORF stop codon resulting in a shift of the reading frame into that proposed to be utilized for 3′UTR translation in rli1-d cells. These mutations should produce large read-through products extended at the C-terminus by the 3′UTR-encoded peptides (plus epitope tags). In every instance, the engineered read-through product displayed the predicted MW, which was demonstrably larger than the corresponding 3′UTR product expressed from the parental tagged allele (Figs. 4C and S6A). This finding is inconsistent with read-through translation from the main ORF as the mechanism of 3′UTR translation. Moreover, the detection of only long read-through products from the engineered alleles in rli1-d cells excludes the possibility that the shorter products expressed from the parental tagged alleles arise from proteolytic cleavage of tagged C-terminal extensions from (hypothetical) read-through products. Hence, all of our results point to reinitiation following termination at the main ORF stop codon as the mechanism of 3′UTR translation.
Interestingly, ribosome profiling data from the CWP2 3′UTR in rli1-d cells reveals a peak at a second stop codon in the −1 frame, downstream of the “major” 3′UTR stop codon that terminates translation of a short peptide in the +1 frame (Fig. S6B–C). This second peak suggests that reinitiation occurs in more than one reading frame. To ask whether 3′UTR translation also occurs in the −1 frame, we modified the CWP2-3′UTR-MYC13 allele analyzed above by inserting a single G nucleotide immediately before the Myc13 coding sequences, thus shifting the epitope into the −1 frame (Fig. 4D, schematics, 1 nt insertion). The high-level +1 frame product generated by the parental construct was no longer detected, because the Myc13 coding sequences are now found in the −1 frame, and instead we observed a much less abundant Myc13-tagged product that is consistent with low-level 3′UTR translation in the −1 frame (Fig. 4D, 1G insertion vs. No insertion). By contrast, inserting two Gs before the Myc13 coding sequences of CWP2-3′UTR-MYC13 resulted in no detectable Myc13-tagged polypeptides (Fig. 4D, 2 nt/GG insertion), indicating that little or no translation occurs in the 0 frame of the CWP2 3′UTR. These interpretations were confirmed by examining an independent construct in which Myc13 coding sequences were inserted just upstream from the second “minor” stop codon in the −1 frame, which produces the low-abundance product attributed above to 3′UTR translation in the −1 frame (Fig. S6D, No insertion). In addition to providing clear evidence for 3′UTR translation in two different reading frames of the CWP2 3′UTR, the results of these and other experiments in Fig. S6D establish that 3′UTR translation in rli1-d cells adheres to the strict rules of frame maintenance that characterizes conventional translation elongation.
Reinitiation likely occurs at non-AUGs near the main ORF stop codon
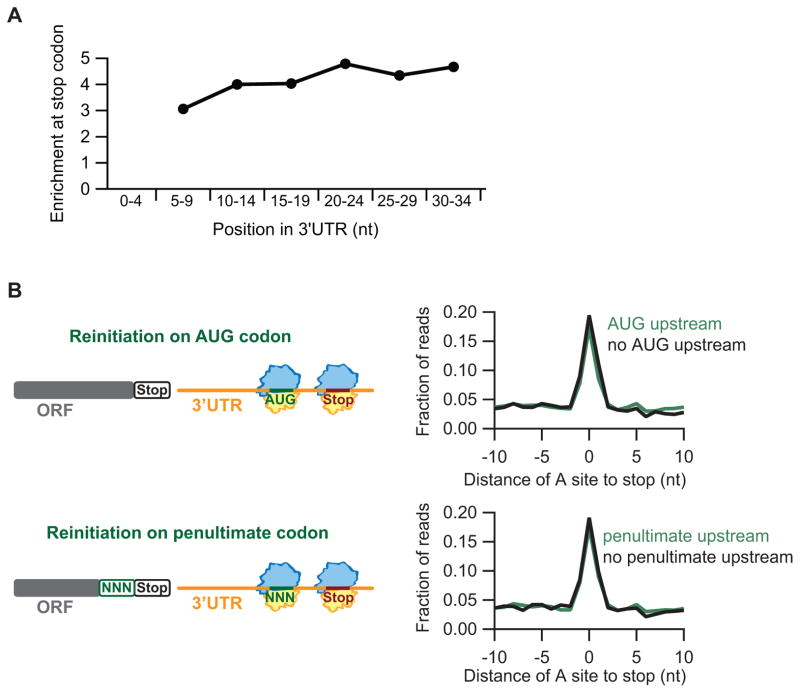
We next turned to the question of how and where reinitiation occurs. We again took advantage of the observation that stalls on 3′UTR stop codons in the ribosome profiling data can serve as signals of active translation. To determine whether translation tends to reinitiate immediately after the main ORF stop codon or whether the ribosome typically first scans some distance downstream, we analyzed the extent of ribosome stalling on stop codons within narrow (5-nt) windows downstream of the main stop. We only included the first stop codon in any frame in our analysis since we previously showed that stalling on subsequent stop codons is reduced (Fig. 2F). The amplitude of stalling in these windows achieved a relatively constant value shortly after the main stop (Fig. 5A). From this we conclude that most reinitiation events occur very close to the main stop (likely <10 nt in the downstream direction).
Figure 5. Reinitiation most likely occurs at non-AUGs near the main ORF stop codon.
(A) Average fraction of ribosome occupancy was computed for stop codons in 5-nt windows downstream of the main ORF stop codon, and peak enrichment was plotted versus the center of each window. Data for the 0–4 nt window could not be computed due to interfering reads from the pause on the main stop codon of the ORF. (B) Schematics (left) illustrate potential reinitiation mechanisms. Average fraction of ribosome occupancy (right) in a window surrounding 3′UTR stop codons, sorting data by the presence or absence of an upstream, in-frame AUG (top) or main-ORF penultimate codon (bottom) in the 3′UTR.
We next evaluated whether the presence or absence of an AUG in the 3′UTR would modulate the amplitude of ribosome stalling on downstream in-frame 3′UTR stop codons. To maximize any potential effect, we again limited the analysis to the first stop codon in any frame. Surprisingly, the presence or absence of an in-frame AUG had no influence on pause amplitude (Fig. 5B, upper); these results imply that reinitiation generally does not occur at AUG codons in the rli1-d strain. We performed a similar analysis to evaluate the presence of upstream in-frame triplets that could be decoded by the penultimate deacylated tRNA remaining in the P site of the 80S post-TC (Skabkin et al., 2013). We similarly found that the presence or absence of such in-frame triplets had no effect on the extent of stop codon pauses (Fig. 5B, lower). Thus, we have no evidence that reinitiation involves scanning by the 80S post-TC to a 3′UTR codon that can base pair with the recently deacylated P-site tRNA.
Mass spectrometry analysis of epitope-tagged 3′UTR translation products
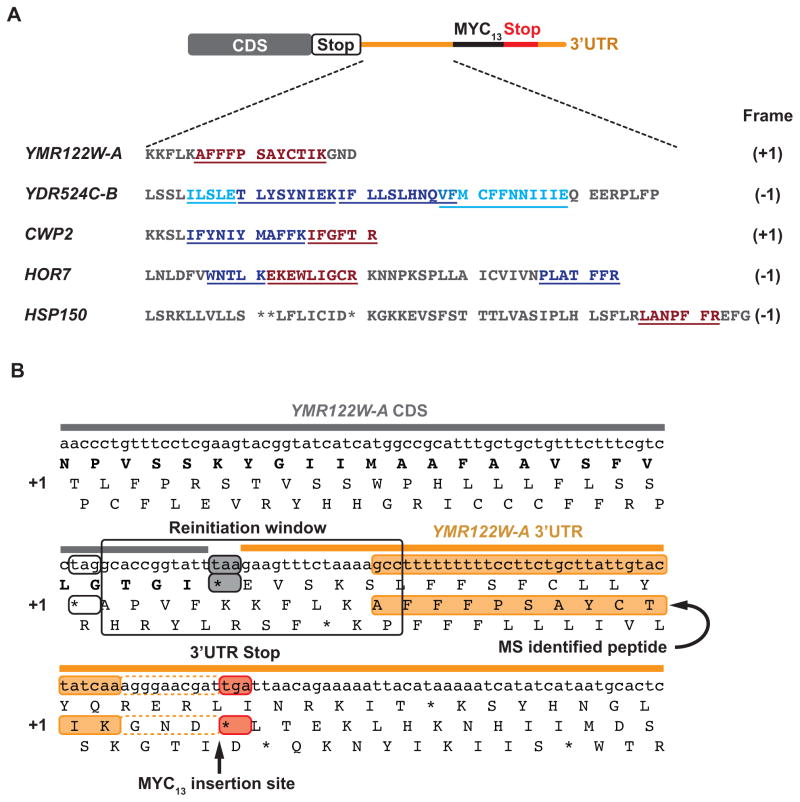
We next asked whether mass spectrometry (MS) could help us to better define reinitiation sites for 3′UTR peptide products. We gel-purifed Myc13-tagged reinitiation products from anti-Myc immune complexes isolated from rli1-d cells, digested them in-gel with trypsin or GluC, and analyzed the proteolytic fragments by MS. We identified tryptic peptides or “semi-tryptic” fragments (that lack a Lys/Arg residue at one end of the peptide) of the Myc13-tagged products for YMR122W-A, YDR524C-B, CWP2, HOR7, and HSP150, and additional “semi-GluC” fragments (that lack a Glu residue at one end) for YDR524C-B (Fig. 6A, underlined; Table S3); and confirmed their sequences by collisionally induced dissociation of the peptides. The identified peptide sequences are consistent with 3′UTR translation in the expected reading frames and are not predicted from the canonical yeast proteome. For YDR524C-B, HOR7, and YMR122W-A, we found no peptides in control gel slices prepared from the corresponding RLI1+ MYC13-tagged strains (data not shown), consistent with our inability to detect Myc13-tagged reinitiation products by Western analysis of these strains (Figs. 4A & S5A). These results lend further confidence to our conclusion that the 3′UTRs of these genes are translated in the predicted reading frames.
Figure 6. Mass spectrometry of immunoprecipitated Myc13-tagged 3′UTR translation products.
(A) Tagged 3′UTR translation products were immunopurified and resolved by SDS-PAGE, and peptide products of trypsin (SED1, YDR524C-B, CWP2, HOR7, HSP150 and YMR122W-A) or GluC (YDR524C-B) digestion were identified by LC-MS/MS analysis. Peptide sequences (underlined) were determined by peptide fragmentation-MS, highlighted in brick red for canonical tryptic peptides ending in Lys/Arg, respectively, and preceded by the corresponding codons (Lys/Arg). Shown in blue and cyan are “semi-tryptic” peptides lacking either a C-terminal Lys/Arg or the preceding Lys/Arg codons, and “semi-GluC” peptides lacking either a C-terminal Glu or a preceding Glu codon, respectively. (B) A portion of the main ORF and 3′UTR sequence of YMR122W-A translated in all 3 frames, showing the MS-identified tryptic peptide (in gold) translated in the +1 frame of the 3′UTR. The first upstream stop codon in the +1 frame of the ORF (unfilled black box), main ORF stop codon (grey), 3′UTR termination codon in the +1 frame (red), and window encompassing the deduced reinitiation site are indicated.
The results for the YMR122W-A tryptic peptide are particularly instructive because its N-terminal sequence begins only six codons downstream from the main ORF stop codon in the +1 frame (Fig. 6B, gold). The first amino acid of the observed peptide, Ala, defines the position furthest downstream where reinitiation could occur. Reading further upstream, the first stop codon in frame with the peptide is found 10 codons upstream of its N-terminus, defining the furthest possible upstream reinitiation site (Fig. 6B). The MS data are compatible with reinitiation at any of the +1 frame codons in this narrow 30-nt window surrounding the main ORF stop codon, which is notably devoid of both AUG codons and isoleucine codons cognate to the penultimate tRNA.
The tryptic peptides and “semi-tryptic” or “semi-GluC” peptides identified for CWP2, HOR7, and YDR524C-B allowed us to place the reinitiation sites close to the main ORF stop codons (Figs. S7A–B), compatible with reinitiation beginning within 4 or 5 codons downstream of the main ORF stop codons. As for YMR122W-A, there are no AUG codons or triplets cognate to the penultimate tRNA in the reinitiation windows defined for CWP2 and HOR7 (Fig. S7A–B). These observations are consistent with the findings from ribosome profiling data (Fig. 5A–B) that reinitiation frequently occurs relatively close to the main ORF stop codon at triplets that do not correspond to AUG nor the penultimate codon of the main coding sequence.
The tryptic peptide we identified in the HSP150 Myc13-tagged product is encoded considerably farther downstream of the main ORF stop codon (Fig. 6A). Reinitiation could occur either following an extended period of scanning from the main ORF stop codon, or following a prior reinitiation event that begins within 10 codons of the main ORF stop codon and terminates at one of the 3 distinct 3′UTR stop codons. Nevertheless, the peptides identified by MS for at least 4 of these 5 genes are consistent with the conclusion that reinitiation is not preceded by an extended period of scanning by the post-TC.
Dom34 rescues a large fraction of unrecycled 80S subunits to suppress aberrant 3′UTR translation in rli1-d cells
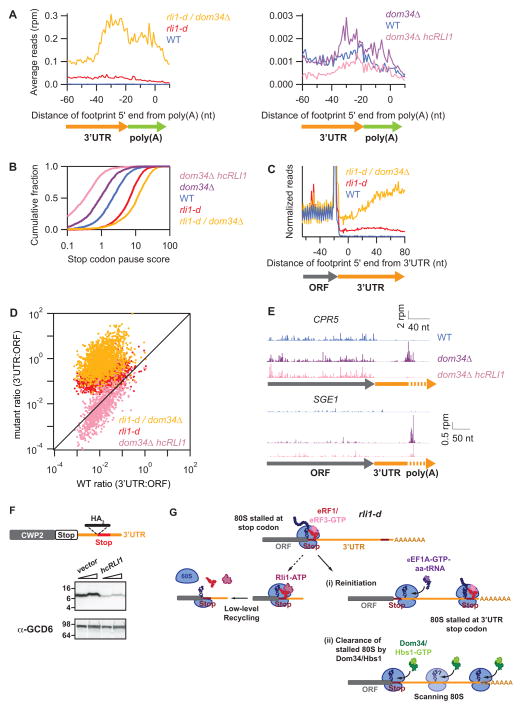
Our previous study suggested that Dom34 rescues ribosomes that escape normal recycling and ultimately accumulate at the junction between the 3′UTR and the poly(A) tail in dom34Δ cells (Guydosh and Green, 2014). To test whether Dom34 rescues ribosomes that evade recycling due to depletion of Rli1, we performed ribosome profiling of an rli1-d dom34Δ double mutant (Fig. S4C–E). Unlike the rli1-d single mutant, the double mutant displays substantial accumulation of 80S ribosomes at the 3′UTR/poly(A) boundary (Fig. 7A left), much as we observed in dom34Δ cells (Fig. 7A right) (Guydosh and Green, 2014). We observed a ~2.5-fold increase in ribosome pausing at main ORF stop codons compared to that seen in the rli1-d single mutant (Fig. 7B), and elevated average ribosome occupancy just downstream of the stop codon (ca. 30 nts) by a factor of ~4 (Fig. 7C). Strikingly, in the rli1-d dom34Δ double mutant, the average ribosome occupancies in 3′UTRs reach the level of those in coding regions (Figs. 7C–D), implying that a large fraction of ribosomes are sequestered in aberrant 3′UTR events.
Figure 7. Dom34 is critically required to rescue unrecycled ribosomes in vivo.
(A) Average ribosome occupancy from 3′UTRs aligned at the annotated site of polyadenylation for WT, rli1-d, and rli1-d dom34Δ cells (left panel) and WT, dom34Δ, and dom34Δ/hcRLI1 cells (right panel). (B) Cumulative histogram of pause scores on ORF stop codons computed by taking the ratio of local density at stop codons compared to the overall ORF, for genes with >100 rpkm in ORFs. (C) Normalized average ribosome footprint occupancy from all genes aligned at their stop codons for WT, dom34Δ, rli1-d, and rli1-d dom34Δ strains, analysis as in Fig. 1A. (D) Ratio of footprint densities between 3′UTRs and respective ORFs plotted for the indicated strains, for genes with >5 rpkm in ORFs and >0.5 rpkm in 3′UTRs, with each point representing 1 gene. (E) Ribosome footprints on CPR5 and SGE1. Approximate start site of 3′UTR is indicated. (F) The WT CWP2-3′UTR-HA3 strain was transformed with either empty vector (YEplac195) or hcRLI1 (YEplac195-RLI1). Transformed strains were grown as in Fig. S1A except SCGAL-U and SC-U media was used instead of YPGAL and YPD media to maintain selection for plasmids. WCEs were subjected to Western analysis using antibodies against HA (upper blots) or Gcd6 (lower blots). Two amounts of extracts were loaded in each lane pair.. (G) Schematic model depicting the fate of post-TCs on depletion of Rli1 in rli1-d cells. Recognition of the main ORF stop codon by eRF1/eRF3-GTP (top row) is followed by release of the completed polypeptide and dissociation of eRF3-GDP (not depicted). Any residual Rli1 could bind post-TCs and catalyze dissociation of the 60S subunit (middle row, left). However, many post-TCs are not recycled, migrate a short distance from the stop codon, reinitiate translation, and frequently terminate at a 3′UTR stop codon to produce a 3′UTR-encoded polypeptide (middle row, right). Such reinitiation events appear to be diminished by Dom34, potentially because post-TC ribosomes are rescued at the main ORF stop codons or as they begin scanning. Any ribosomes that reach the 3′UTR/poly(A) boundary by reinitiation or scanning are also rescued by Dom34.
The fact that depleting Rli1 in dom34Δ cells greatly elevates 80S species at 3′UTR/poly(A) boundaries supports the notion that aberrant complexes arise in dom34Δ cells because native levels of Rli1 are insufficient to recycle all ribosomes. Supporting this idea, the accumulation of 80S ribosomes at 3′UTR/poly(A) boundaries seen in dom34Δ cells is diminished by Rli1 overexpression (hcRLI1, Figs. 7A right and 7E), as would be expected if these aberrant 80S species result from failed recycling due to insufficient levels of Rli1. We also found that overexpression of Rli1 in dom34Δ cells (Fig. S4F) diminished ribosome pausing at main ORF stop codons well below that observed in WT cells (Fig. 7B), and similarly reduced 80S occupancies throughout the 3′UTR to levels below those observed in WT (Figs. 7D, points appearing below diagonal line). We also observed that low-level reinitiation products from CWP2 in WT cells (Fig. 4B) could be diminished by overexpressing Rli1 (Fig. 7F), adding additional evidence for Rli1 insufficiency in WT cells. These data imply that Dom34 normally compensates for this inherent recycling deficiency, resulting in the normally low levels of 80S ribosome 3′UTR occupancy observed in WT cells.
DISCUSSION
In this study, we set out to test the hypothesis that Rli1/ABCE1 is crucial for recycling 80S post-TCs in vivo and to determine the consequences of an acute loss of Rli1 function on the fate of ribosomes. First, ribosome profiling of cells depleted of Rli1 revealed dramatic phenotypes that support a critical role for Rli1 in ribosome recycling (Figure 7G, left pathway). These phenotypes include accumulation of 80S ribosomes both at stop codons of most annotated ORFs and throughout the 3′UTRs of yeast mRNAs. Second, and more surprisingly, the data reveal that following the stop-codon associated delay in the rli1-d strain, ribosomes can reinitiate in a region just upstream or downstream of the main stop codon and translate 3′UTR sequences. Such a 3′UTR translation model is supported by multiple lines of evidence. First, we identify numerous ribosome occupancy peaks that coincide with stop codons in the 3′UTR, and these stop codons can be found in any of the three reading frames relative to the main ORF (depending on the gene in question) (Figs. 2, S3). Importantly, the peak occupancy is diminished by the presence of another 3′UTR stop codon located upstream in the same reading frame (Fig. 2F), as expected for ribosomes translating the 3′UTR and preferentially terminating at the first in-frame stop codon encountered downstream from the main ORF. These ribosome density peaks at 3′UTR stop codons represent stalled 80S post-TCs that, for a second time, are inefficiently recycled by low levels of Rli1. The occurrence of 3′UTR translation was also supported by the fact that histidine starvation (with 3-AT) increased ribosome occupancy at 3′UTR histidine codons located upstream from, and in the same reading frame as, prominent stop codon-stall sites that we observed (Fig. 3). These translating ribosomes together with those scanning for reinitiation sites likely account for the overall increase in ribosome occupancy of 3′UTRs for nearly all genes when Rli1 is depleted (Fig. 1B). We found that nearly one-fifth of stop codons (3279 out of 18514 for genes with >3 rpkm in 3′UTR ribosome density) had a pause score >3, giving an indication of how many 3′ UTR ORFs may be translated (and that number is substantial).
Putative 3′UTR translation products were directly detected by western analysis and mass spectrometry after inserting coding sequences for epitope tags immediately prior to prominent stop codon-stall sites (Figs. 4 & 6). All of the epitope-tagged 3′UTR products we detected in rli1-d cells had electrophoretic mobilities consistent with reinitiation taking place near the main ORF stop codon. Mass spectrometry of 3′UTR translation products is also consistent with this conclusion. Finally, these biochemical results are in accordance with our computational analysis (Fig. 5A) indicating that most reinitiation events occur near the main ORF stop codon (likely within ~10 nt). Together these data provide compelling support for a model invoking the reinitiation of translation in the 3′UTR by unrecycled ribosomes at sites proximal to the main ORF stop codon (Figure 7G, right).
How does the reinitiation observed in rli1-d cells take place? Our results are consistent with a mechanism wherein termination and polypeptide release occur at the main ORF stop codon, but splitting of the 60S subunit from the 80S post-TC fails. The resulting 80S post-TC releases eRF1 from the A site, allowing the remaining P-site tRNA to adopt the P/E conformation required for scanning (Skabkin et al., 2013). The 80S post-TC undergoes a limited period of scanning, ultimately replacing the stop codon in the A site with a sense codon, which then recruits cognate eEF1A-GTP-aa-tRNA ternary complex. A pseudo-translocation event could then position this ternary complex in the P site to allow translation to resume by the canonical elongation pathway, akin to the translocation that occurs without peptide bond formation in translation initiation directed by the dicistrovirus IGR IRES (Thompson, 2012) and the reinitiation of translation that occurs in the “StopGo” mechanism of viral 2A protease (Atkins et al., 2007). Regardless of the exact mechanism of reinitiation, once a new round of translation begins, it terminates at the first in-frame stop codon in the 3′UTR to produce a short peptide product (Fig. 7G, right).
In principle, reinitiation could occur either immediately upstream or downstream of the main stop codon following a short bidirectional scanning process. We tested this possibility by examining out-of-frame reads just upstream of the main stop codon at the end of ORFs (Fig. S2F). Interestingly, 80S ribosome density was found to be enriched in alternate reading frames in the interval beginning <10 codons upstream of the main stop codon (Fig. S2F–G). This finding is consistent with a fraction of post-TCs scanning short distances upstream of the stop codon to find a favored site for reinitiation.
Our finding that reinitiation in rli1-d cells does not appear to involve scanning of the post-TC to a codon complementary to the penultimate P-site tRNA stands in contrast to observations made in a mammalian reconstituted termination system lacking ABCE1 and supplemented with added Mg2+ (Skabkin et al., 2013). While it seems possible that the P-site tRNA will remain and stabilize the 80S post-TC, perhaps what dictates the reinitiation event is driven in part by pairing interactions between the P-site tRNA and the available codon (near-cognate or even non-cognate) and in part by availability of the appropriate eEF1A-GTP-aa-tRNA ternary complex. Interestingly, non-cognate triplets are selected as landing sites for peptidyl-tRNA during some translational hopping events (Herr et al., 2004). The apparent absence of reinitiation at AUGs in rli1-d cells also differs from the AUG-dependent toeprints observed following recognition of a premature termination codon (Amrani et al., 2004). Unless the P-site tRNA is also lost from the post-TC, and the subunits are split, it is difficult to envision how canonical initiation with Met-tRNA loading by eIF2 would be the preferred mode of reinitiation in our system. Clearly, more work will be required to define what lingers on the post-TC 80S and how it might determine the mode of ribosome movement following a recycling failure.
Our ribosome profiling of the dom34Δrli1-d mutant revealed a striking increase in 3′UTR 80S occupancies compared to that seen in the rli1-d single mutant. These data suggest that Dom34 may control access of unrecycled ribosomes to the 3′UTR. The fact that ribosome occupancy is elevated at canonical stop codons raises the possibility that Dom34 controls 3′UTR access by dissociating ribosomes at stop codons, presumably after the dissociation of eRF1 (Becker et al., 2012; Shoemaker et al., 2010), and prior to the initiation of scanning. The increased ribosome occupancy throughout the 3′UTRs might be explained by Dom34 acting continuously as post-TC ribosomes scan to look for a favorable reinitiation site (Fig. 7G, right). In this scenario, the lower overall 3′UTR occupancy in the rli1-d strain (compared to the dom34Δrli1-d strain) is the result of fewer ribosomes escaping Dom34 rescue.
Previously, we identified 3′UTR-bound 80S ribosomes in dom34Δ cells stalled primarily at the poly(A) tail boundary. This species of 3′UTR ribosome is more abundant in the dom34Δ rli1-d mutant relative to the dom34Δ single mutant (Fig. 7A). These observations suggest that this class of ribosomes originates from 80S post-TCs that move down the mRNA after a failure in Rli1 recycling, as we had previously predicted (Guydosh and Green, 2014); these ribosomes may fail to reinitiate, and thus land at this terminal point. In some cases, these ribosomes could also be translating to this point if no stop codons occur downstream of the reinitiation point. These ideas are further strengthened by our finding that overexpressing Rli1 eliminates a majority of these 3′UTR ribosomes in the dom34Δ mutant (Fig. 7D, 7E).
Ribosome homeostasis is critical to cellular function; ribosome availability is directly linked to the rate of cell division (Maaloe, 1966) and altered levels of available ribosomes are known to be critical to promoting proliferation of cancer cells (Ruggero, 2012). It is intriguing that the level of Rli1 in WT cells is insufficient to ensure recycling of all post-TC complexes, as unmasked in dom34Δ cells and rescued by overexpression of Rli1. Perhaps complete recycling is simply unnecessary because Dom34 is normally present to remove the relatively small number of unrecycled post-TC complexes that escape Rli1 function. However, it seems possible that the WT level of Rli1 activity may be just below the threshold of sufficiency for complete recycling to allow low-level production of 3′UTR-encoded peptides at certain genes under specific conditions. We provided evidence for reinitiation in the 3′UTR of CWP2 that was attributable to Rli1 insufficiency even under optimal growth conditions in WT cells. Moreover, we interrogated previously published ribosome profiling data and found increased 3′UTR ribosome occupancy in WT cells undergoing various types of nutrient starvation (Fig. S2H). In each of these cases, ribosome footprint levels on RLI1 are considerably reduced relative to other genes (and thus likely Rli1 expression levels). These data suggest that increased 3′UTR translation could introduce new functions during times of stress, as with stop codon read-through in yeast infected with the PSI+ prion (True et al., 2004). As suggested previously (Skabkin et al., 2013), this phenomenon could also underlie the production of novel peptides for antigen presentation in the immune system (Schwab et al., 2003).
EXPERIMENTAL PROCEDURES
Plasmid construction and yeast strains
Plasmids and yeast strains used in this study are listed in Tables S4 and S5 and their constructions are described in Supplemental Information (SI).
Biochemical techniques
Polysome analysis was conducted as described previously (Valášek et al., 2001), as were immunoprecipitations of epitope-tagged proteins (Zhang et al., 2004) using aliquots of WCE with 5mg of total protein and 160 uL of myc- or HA-agarose (Santa Cruz). The buffer used for cell lysis and washes contained 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 15 mM MgCl2, 0.01% NP-40, 20% Glycerol, and protease inhibitors Complete Protease Inhibitor tablet – EDTA (Roche), AEBSF, and pepstatin A. Western analysis was conducted as described (Nanda et al., 2009) using 4–20% Tris-HCl, and 16.5% Tris-Tricine gels from Bio-Rad, and antibodies described in SI. Mass spectrometry was conducted by the Proteomics Center of Excellence at Northwestern University as described in the SI.
Ribosome footprint profiling
Ribosome footprints were prepared as described (Guydosh and Green, 2014) by using a protocol very similar that used by Ingolia and coworkers (Ingolia et al., 2012). Biological replicate datasets were created for rli1-d and rli1-d / dom34Δ strains. A technical replicate dataset was created for WT cells. All ribosome footprints that appear in this study were extracted from gel slices that included the range 25–34 nt. mRNA-Seq footprints were extracted from the range 40–60 nt from total cell lysate and not subject to poly(A) selection. Ribosome footprints were analyzed essentially as described (Guydosh and Green, 2014) with modifications as described in the Supplemental Information. Unless noted otherwise, data shown for a given strain represent a composite from all biological and technical replicates. Sequencing was performed on an Illumina HiSeq2000 or HiSeq2500 at UC Riverside or the Johns Hopkins Institute of Genetic Medicine.
The NCBI GEO accession number for the sequencing data (debarcoded fastq files and wig files) reported in this paper is GSE69414.
Supplementary Material
Acknowledgments
Proteomics data and analysis were performed by Susan Fishbain, Paige Gottlieb, Ioanna Ntai and Paul Thomas of the Proteomics Center of Excellence at Northwestern University. We thank Tom Dever, Jon Lorsch and members of our laboratories for many helpful suggestions during the course of this work. This study was funded in part by the Intramural Research Program of the NIH (AH) and by HHMI (RG).
Footnotes
AUTHOR CONTRIBUTIONS
DJY and NRG collected and analyzed the data and helped write the manuscript; and FZ purified tagged proteins for MS analysis. AGH and RG supervised the work and helped to write the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Atkins JF, Wills NM, Loughran G, Wu CY, Parsawar K, Ryan MD, Wang CH, Nelson CC. A case for “StopGo”: reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go) RNA. 2007;13:803–810. doi: 10.1261/rna.487907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The Structure of the Eukaryotic Ribosome at 3.0 A Resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr-Schmid A, Pfund C, Craig EA, Kinzy TG. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol Cell Biol. 2002;22:2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TD, Hinnebusch AG. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Fisher JM, Rabinovitz M. Puromycin interference of reticulocyte polyribosome disaggregation caused by tryptophan deficiency. J Mol Biol. 1968;33:315–318. doi: 10.1016/0022-2836(68)90297-0. [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Wills NM, Nelson CC, Gesteland RF, Atkins JF. Factors that influence selection of coding resumption sites in translational bypassing: minimal conventional peptidyl-tRNA:mRNA pairing can suffice. J Biol Chem. 2004;279:11081–11087. doi: 10.1074/jbc.M311491200. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of Mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- Janosi L, Mottagui-Tabar S, Isaksson LA, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaloe OaK, NO . Control of macromolecular synthesis. 1966. [Google Scholar]
- Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EC, Finley D, Szostak JW. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci USA. 1992;89:1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Haile D, Rouault TA, Klausner RD. Modification of a free Fe-S cluster cysteine residue in the active iron-responsive element-binding protein prevents RNA binding. J Biol Chem. 1993;268:17655–17658. [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. Embo J. 2011a doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. Embo J. 2011b;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A, Heuer A, Barrio-Garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, Beckmann R. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014;8:59–65. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D. Revisiting the nucleolus: from marker to dynamic integrator of cancer signaling. Sci Signal. 2012;5:pe38. doi: 10.1126/scisignal.2003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SR, Li KC, Kang C, Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science. 2003;301:1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011;108:E1392–1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell. 2013;51:249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR. Tricks an IRES uses to enslave ribosomes. Trends in microbiology. 2012;20:558–566. doi: 10.1016/j.tim.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Valášek L, Phan L, Schoenfeld LW, Valásková V, Hinnebusch AG. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recoginition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elzen AM, Schuller A, Green R, Seraphin B. Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress. EMBO J. 2014;33:265–276. doi: 10.1002/embj.201386123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. Embo J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.