Abstract
There are many layers of regulation governing DNA replication to ensure that genetic information is accurately transmitted from mother cell to daughter cell. While much of the control occurs at the level of origin selection and firing, less is known about how replication fork progression is controlled throughout the genome. In Drosophila polytene cells, specific regions of the genome become repressed for DNA replication, resulting in underreplication and decreased copy number. Importantly, underreplicated domains share properties with common fragile sites. The Suppressor of Underreplication protein SUUR is essential for this repression. Recent work established that SUUR functions by directly inhibiting replication fork progression, raising several interesting questions as to how replication fork progression and stability can be modulated within targeted regions of the genome. Here we discuss potential mechanisms by which replication fork inhibition can be achieved and the consequences this has on genome stability and copy number control.
Keywords: common fragile sites, DNA replication, Drosophila, endocycle, genome stability, polyploidy, underreplication
Introduction
Proper regulation of the DNA replication program is necessary not only to ensure faithful transmission of genetic information from mother cell to daughter cell, but also to prevent genomic aberrations that are associated with numerous diseases such as cancer. While we know a great deal about the regulatory mechanisms that control DNA replication at the level of initiation of DNA replication [1, 2], the degree to which regulation of replication fork progression and stability contribute to accurate DNA replication remains poorly understood. Because replication origins are not uniformly distributed, large genomic regions are dependent on the activity of a single replication fork emanating from a distant origin for their duplication [3, 4]. Large origin-less regions of the genome are associated with chromosome fragility at common fragile sites, which are regions of the genome prone to breakage upon replication stress [5–8]. It is thought that if a replication fork stalls or collapses within these large origin-less regions of the genome, no nearby replication forks are able to rescue it within the timing confines of S phase. Importantly, not all large origin-less regions of the genome are common fragile sites, presumably because they are more amenable to fork progression[5]. Therefore, identification of factors necessary to promote replication fork progression and stability within common fragile sites is likely to uncover fundamental processes necessary to maintain genome stability.
Common fragile sites have been identified in organisms as diverse as yeast and humans [6]. Drosophila polytene cells contain many common fragile sites that have been mapped precisely. These are regions of the genome repressed for DNA replication that become underreplicated [9, 10]. As in mammalian cells, these common fragile sites are large origin-less regions of the genome, prone to DNA damage and display cell-type specificity [11–14]. It is currently unknown if these same regions are fragile sites in non-polytene Drosophila cell types. A key feature of common fragile sites in Drosophila polytene cells is that a single protein, Suppressor of Underreplication or SUUR, is required for the vast majority of underreplication, thus providing a tractable system to study replication dynamics within common fragile sites [12, 15, 16].
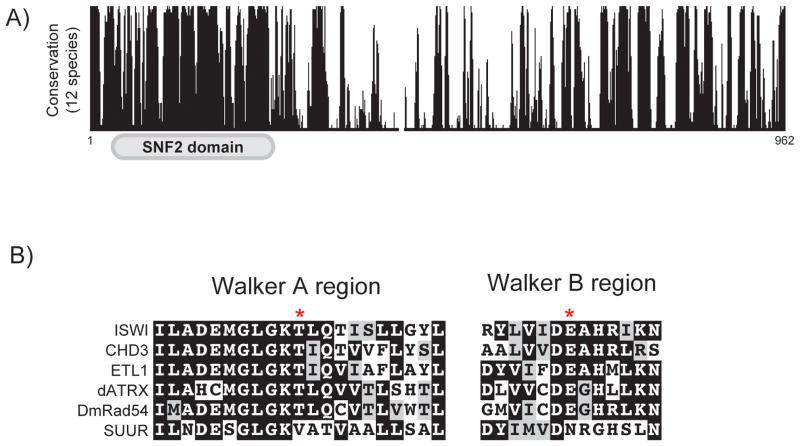
SUUR is conserved in Drosophila species and has a recognizable SNF2 domain at its N terminus (Fig. 1A)[15]. SNF2 represents a family of chromatin remodeling enzymes that couple the energy of ATP hydrolysis to move, eject, or position nucleosomes [17]. Interestingly, residues necessary for ATP binding and hydrolysis are not conserved in SUUR orthologs from distantly related Drosophila species, which strongly suggests that SUUR has lost its ability to bind and hydrolyze ATP (Fig. 1B)[15].
Figure 1.
Conservation of SUUR. A: Conservation of SUUR across twelve Drosophila species. Alignments and conservation scores were generated with UCSC genome browser. B: Conservation of the Walker A and B regions from multiple SNF2 domain-containing proteins in Drosophila. Asterisks mark the sites of key residues involved in ATP binding and hydrolysis that are not conserved in SUUR. Alignments were generated using ClustalW and BoxShade.
The association of SUUR with chromatin has been mapped in Drosophila cultured cells by DamID. DamID is a way to monitor protein association with chromatin by fusing a protein of interest to the DNA adenine methylase, Dam, of E. coli. Dam methylates adenosine residues within GATC consensus sequences. Therefore, the association of a protein of interest with chromatin can be indirectly assayed by monitoring the methylation status of GATC sequences upon the expression of a candidate protein-Dam fusion. In Drosophila cultured cells, SUUR was shown to have a similar binding profile to the linker histone H1 and several other chromatin proteins that are associated with repressive chromatin [18]. It is important to note, however, that DamID cannot distinguish between proteins that simultaneously bind the same site, proteins that bind the same site in a mutually exclusive manner, or proteins that are statically versus dynamically associated with chromatin, and it does not provide information about protein abundance.
Although it was clear that SUUR directly influenced replication dynamics within common fragile sites of the Drosophila genome, the mechanism by which it promotes underreplication, and thus chromosome fragility, was unknown. A recent study has now demonstrated that SUUR functions by targeting active replication forks and inhibiting their progression [14]. This work has revealed that the DNA replication program, and thus DNA copy number, can be modulated through direct inhibition of replication fork progression.
This work has challenged the prevailing view that SUUR is a component of a repressive chromatin subtype that constitutes nearly half of the Drosophila genome [18]. Because these studies were performed using DamID there is currently no direct evidence that SUUR is a direct component of a specialized chromatin subtype. If SUUR associates with replication forks within late-replicating regions of the genome that contain these repressive chromatin proteins, its DamID profile would be similar to those chromatin proteins, without it being a component of a repressive chromatin subtype. Thus, the SUUR DamID profile should be reevaluated in light of the dynamic association of SUUR with replication forks.
Many fascinating questions have been raised by studying the effect SUUR has on replication fork progression and chromosome fragility. First, how can SUUR, or any protein for that matter, target replication forks for inhibition within specific regions of the genome? Second, once SUUR has been recruited to replication forks, through what molecular mechanism does it inhibit fork progression? Finally what could be the potential benefit of reducing copy number within specific regions of the genome? Here we speculate on the molecular mechanism by which SUUR could directly influence replication fork progression and what studying SUUR can reveal about fundamental aspects of genome duplication that still remain largely unknown.
SUUR targets specific regions of the genome
To duplicate a metazoan genome thousands of replication forks must function in concert. Targeting a few replication forks for inhibition within specific regions of the genome presents a formidable challenge for any protein. Importantly, the DNA replication program is not random, but rather origin firing and replication fork progression occur in an orchestrated manner. It is well established that regions of the genome replicate at defined times during S phase, a phenomenon known as replication timing [19]. Although the function of replication timing remains enigmatic, the structure of chromatin has a significant role in establishing the replication-timing program. In general, open and active euchromatic regions of the genome replicate early in S phase, whereas condensed chromatin such as heterochromatin replicates late in S phase [20, 21].
Because genome duplication occurs in a temporally ordered manner, regions of the genome that activate replication origins early in S phase will have completed replication of early domains while replication forks emanating from origins that fire late in S phase are still actively progressing. One mechanism by which fork progression could be influenced within specific genomic regions is to modulate fork activity as a function of S phase progression. For example, replication fork progression could be inhibited specifically within heterochromatin by changing fork activity late in S phase. This is an attractive model to explain how SUUR influences replication fork progression within underreplicated domains. SUUR is not associated with all replication forks, but is recruited to elongating replication forks late in S phase, and underreplicated domains are generally late replicating regions of the genome [14, 22]. This association pattern would inhibit forks that are active in the late part of S phase, while excluding replication forks that have completed replicating genomic regions early in S phase.
The above model raises the question of how replication fork dynamics could be modulated at distinct times in S phase. One possibility is that changes in kinase activity throughout S phase influence the activity of proteins at the replication fork. Alternatively, localization of proteins to the replication fork could change during S phase. CDK2 (cyclin-dependent kinase 2) is a cell cycle regulated kinase that affects many aspects of the DNA replication program [2]. CDK2 is active when associated with a Cyclin, which during S phase is either Cyclin E or Cyclin A. CycE/CDK2 activity is high at the beginning of S phase and decreases as S phase progresses, whereas CycA/CDK2 activity has the inverse activity pattern [23–25]. Changes in CDK2 activity, or substrate specificity, throughout S phase could directly affect the activity or composition of replication forks.
SUUR has two modes of recruitment to chromatin. First is the constitutive localization to heterochromatin upon entry into the endocycle that is irrespective of the phase of the endo cycle [22]. Second, SUUR shows an S phase-dependent localization to sites of underreplication and replication forks [14, 22]. Interestingly, a hypomorphic allele of cyclin E was identified because it abolished underreplication of heterochromatin in Drosophila nurse cells. This is consistent with the possibility that SUUR recruitment to heterochromatin and/or replication forks is influenced by CDK2 [26]. Furthermore, CycA has been proposed to influence replication dynamics in a specific endocycling cell type in Drosophila [27]. Therefore, it is possible that SUUR activity, and thus underreplication, could be directly influenced by CDK activity.
The recruitment of SUUR to replication forks, however, may not be the sole requirement to promote underreplication. SUUR associates with more genomic regions than are actually underreplicated [11, 15]. Also, tethering SUUR to a specific site is not sufficient to promote underreplication [11]. Thus, SUUR is necessary but insufficient to trigger underreplication. One possibility is that SUUR-mediated inhibition of replication fork progression could be sensitive to genomic position or chromatin environment. This model would require the combination of SUUR association with replication forks in a specific chromatin context, such as histone H1-containing chromatin, to permit SUUR’s inhibitory activity. For example, SUUR could block an activity at the replication fork required to facilitate progression through specific regions of the genome or chromatin subtypes.
SUUR-mediated inhibition of replication fork progression is highly sensitive to the dosage of SuUR. By utilizing a model system to study replication fork progression in metazoans, it was shown that just two extra copies of SuUR resulted in a nearly 50% decrease in replication fork progression [14]. Furthermore, two extra copies of SuUR leads to excessive DNA damage and additional sites of underreplication in endocycling cells of the larval salivary gland [13]. These phenotypes are worsened as the gene dosage of SuUR increases [13]. Together, these results suggest that SUUR activity, and perhaps its regulated recruitment to replication forks, is highly sensitive to dosage.
Through what molecular mechanism does SUUR inhibit replication fork progression?
Once SUUR is recruited to a replication fork a key question is: through what molecular mechanism does SUUR inhibit fork progression? This has implications beyond understanding the molecular details of underreplication, as studying SUUR is likely to uncover fundamental processes contributing to replication fork progression, stability and chromosome fragility. Here we elaborate on two non-mutually exclusive mechanisms that SUUR could utilize to inhibit replication fork progression.
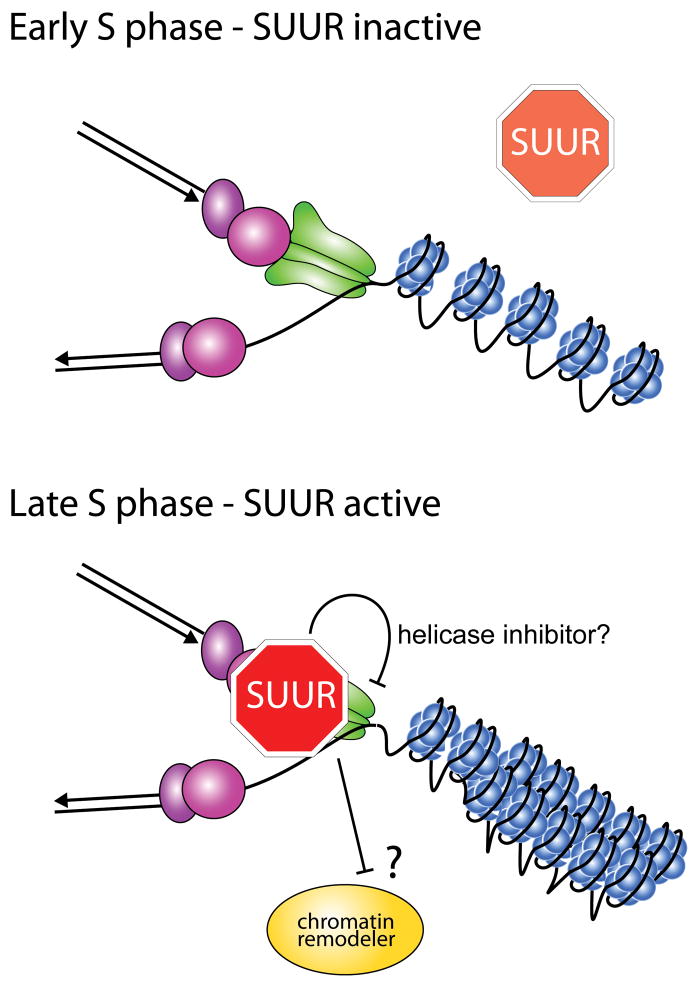
Chromatin-remodeling complexes have been shown to localize to replication forks and likely have multiple roles at replication forks [28]. Chromatin remodeling is necessary to reestablish chromatin state and epigenetic information in the wake of replication forks. Additionally, chromatin remodelers have been shown to affect replication fork progression. These remodelers may be necessary to promote fork progression and stability through regions of the genome, such as condensed chromatin, that are more difficult to replicate [29–32]. Through its defective chromatin-remodeling domain, SUUR could inhibit the normal chromatin remodeling activity at the fork by acting competitively with chromatin remodeling complexes. This competition would, in turn, result in fork inhibition and destabilization within regions of condensed chromatin (Fig. 2). Although appealing, it is not known if the SNF2 domain of SUUR is necessary to promote underreplication. Overexpression of the C-terminal portion of SUUR is sufficient to inhibit DNA replication, but it is unclear if overexpression bypasses an essential function of the SNF2 domain [33].
Figure 2.
Potential models by which SUUR could inhibit active replication forks. Depicted is a model in which SUUR becomes recruited to replication forks late in S phase. Once at a replication fork, SUUR could prevent the decondensation of highly condensed regions of chromatin (e.g. heterochromatin) through competition with a chromatin remodeling factor. Also, SUUR could directly inhibit a key activity associated with replication fork progression, such as helicase activity. It is important to note that these two models of SUUR function are not mutually exclusive.
One way to inhibit replication fork progression is to block the activity of a critical component of the replication fork. A component of the replicative helicase, CDC45, was shown to associate with SUUR [14]. Although it is unknown if this association is direct, or if it occurs in the context of a replication fork, it raises the possibility that SUUR directly interferes with helicase activity (Fig. 2). Further association studies will be necessary to address this possibility.
What is the biological function of underreplication?
Unlike many proteins that function to promote replication fork progression through various chromatin structures and replication impediments, SUUR has the opposite function in that it inhibits fork progression. This appears to be an extreme example of replication timing where replication is indefinitely delayed. One outstanding question that remains to be answered is the benefit of repressing DNA replication within specific regions of the genome. This is an important issue because SUUR-mediated repression of DNA replication is associated with DNA damage and genome instability within underreplicated domains.
In S. cerevisiae, replication fork progression is coordinated with the highly transcribed rDNA region to prevent conflicts between the replication and transcription machineries [34, 35]. This occurs through a replication fork barrier-like mechanism where the Fob1 protein physically prevents replication forks from entering the rDNA locus. The genes within Drosophila underreplicated domains, however, are generally transcriptionally silent so there does not seem to be the need to coordinate replication and transcription within these regions of the genome [11, 12].
Another possibility was that underreplication was an extreme mechanism to repress transcription of genes that reside within these domains. This could be important in the context of the endocycle when ploidy can be increased 1,000-fold, as is the case for the larval salivary gland. This was an intriguing possibility given that pro-apoptotic genes reside within underreplicated domains and apoptosis is silenced in endocycling cells [11, 36]. Genome-wide measurements of transcript levels showed no significant changes when comparing wild-type and SuUR mutant salivary glands in which replication is restored [11]. Thus, underreplication is not an essential strategy to repress transcription.
One interesting observation is that in the absence of SUUR, there is constitutive DNA damage within heterochromatin, whereas DNA damage in the euchromatic underreplicated domains is largely dependent on SUUR [13, 14]. This suggests that in the absence of SUUR, replication of heterochromatin during the endocycle is problematic. One reason for this could be due to the repetitive nature of heterochromatin. If a double-strand (DSB) break is produced within heterochromatin it appears to be moved outside of the heterochromatic domain for repair [37, 38]. During the endocyle, this problem is compounded by the presence of ~1,000 copies of highly repetitive heterochromatin. One possible way to avoid this is to simply prevent replication of this difficult-to-replicate region of the genome. Also, heterochromatin accounts for approximately 30 percent of the Drosophila genome [39]. Endocycling cells are generally terminally differentiated cells that are thought to be highly metabolically active. Underreplication could be an economic strategy that diverts resources into cellular metabolism rather than duplication of silent chromatin.
Final thoughts and conclusions
Outside of Drosophila species, no obvious SUUR homologs have been identified based on sequence homology, but it is likely that functional homologs exist. To identify potential functional homologs, it will be necessary to investigate processes that could require the need to control replication fork progression, such as replication timing, at a more mechanistic level. Although it is still largely unknown why replication timing exists, it is an evolutionarily conserved process subject to cell-type specific regulation [40, 41]. Interestingly, imprinted genes often display parent-of-origin specific difference in their replication-timing pattern [42, 43]. It seems possible that some degree of replication timing could be controlled through modulation of replication fork progression. Recently it has been shown that at least a subset of polyploid trophoblast giant cells of the mouse placenta undergo a modest degree of underreplication [44]. Perhaps this system can be exploited to reveal the long anticipated functional homolog of SUUR.
Acknowledgments
We would like to thank Tom DiCesare for graphical support. We thank Boryana Petrova for comments on the manuscript. J.N. is supported by an NIH K99 award 1K99GM104151 and a Margaret and Herman Sokol postdoctoral award. T.L.O.-W. is supported by NIH grant GM57960 and an American Cancer Society Research Professorship.
References
- 1.Bell SP, Kaguni JM. Helicase loading at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5:a010124. doi: 10.1101/cshperspect.a010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddiqui K, On KF, Diffley JFX. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol. 2013;5:a012930. doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman TJ, Mamun MA, Nieduszynski CA, Blow JJ. Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts. Nucleic Acids Res. 2013;41:9705–9718. doi: 10.1093/nar/gkt728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, et al. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Letessier A, Millot GA, Koundrioukoff S, Lachagès A-M, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–3. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 6.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 7.Norio P, Kosiyatrakul S, Yang Q, Guan Z, et al. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20:575–87. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 8.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–6. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 9.Rudkin GT. Non replicating DNA in Drosophila. Genetics. 1969;61:227–38. [PubMed] [Google Scholar]
- 10.Spradling A, Orr-Weaver T. Regulation of DNA replication during Drosophila development. Annu Rev Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- 11.Sher N, Bell GW, Li S, Nordman J, et al. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 2012;22:64–75. doi: 10.1101/gr.126003.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordman J, Li S, Eng T, MacAlpine D, et al. Developmental control of the DNA replication and transcription programs. Genome Res. 2011;21:175–81. doi: 10.1101/gr.114611.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreyeva EN, Kolesnikova TD, Belyaeva ES, Glaser RL, et al. Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res. 2008;16:851–62. doi: 10.1007/s10577-008-1244-4. [DOI] [PubMed] [Google Scholar]
- 14.Nordman JT, Kozhevnikova EN, Verrijzer CP, Pindyurin AV, et al. DNA copy-number control through inhibition of replication fork progression. Cell Rep. 2014;9:841–9. doi: 10.1016/j.celrep.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, et al. The Drosophila Suppressor of Underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics. 2002;160:1023–34. doi: 10.1093/genetics/160.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, et al. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci USA. 1998;95:7532–7. doi: 10.1073/pnas.95.13.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nature. 2006;7:437–47. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 18.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert DM, Takebayashi SI, Ryba T, Lu J, et al. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol. 2010;75:143–53. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol. 2002;14:377–83. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 21.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolesnikova TD, Posukh OV, Andreyeva EN, Bebyakina DS, et al. Drosophila SUUR protein associates with PCNA and binds chromatin in a cell cycle-dependent manner. Chromosoma. 2013;122:55–66. doi: 10.1007/s00412-012-0390-9. [DOI] [PubMed] [Google Scholar]
- 23.Zielke N, Kim KJ, Tran V, Shibutani ST, et al. Control of Drosophila endocycles by E2F and CRL4CDT2. Nature. 2011;480:123–7. doi: 10.1038/nature10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 25.Budirahardja Y, Gönczy P. Coupling the cell cycle to development. Development. 2009;136:2861–72. doi: 10.1242/dev.021931. [DOI] [PubMed] [Google Scholar]
- 26.Lilly MA, Spradling AC. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10:2514–26. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- 27.Sallé J, Campbell SD, Gho M, Audibert A. CycA is involved in the control of endoreplication dynamics in the Drosophila bristle lineage. Development. 2012;139:547–57. doi: 10.1242/dev.069823. [DOI] [PubMed] [Google Scholar]
- 28.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–67. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 29.Cohen SM, Chastain PD, Rosson GB, Groh BS, et al. BRG1 co-localizes with DNA replication factors and is required for efficient replication fork progression. Nucleic Acids Res. 2010;38:6906–19. doi: 10.1093/nar/gkq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirbu BM, McDonald WH, Dungrawala H, Badu-Nkansah A, et al. Identification of proteins at active, stalled, and collapsed replication forks using isolation of Proteins on Nascent DNA (iPOND) coupled with mass spectrometry. J Biol Chem. 2013;288:31458–67. doi: 10.1074/jbc.M113.511337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Contreras AJ, Ruppen I, Nieto-Soler M, Murga M, et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013;3:1105–16. doi: 10.1016/j.celrep.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alabert C, Bukowski-Wills J-C, Lee S-B, Kustatscher G, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–93. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolesnikova TD, Makunin IV, Volkova EI, Pirrotta V, et al. Functional dissection of the Suppressor of UnderReplication protein of Drosophila melanogaster: identification of domains influencing chromosome binding and DNA replication. Genetica. 2005;124:187–200. doi: 10.1007/s10709-005-1167-3. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–30. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–76. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 36.Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, et al. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008;22:3158–71. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiolo I, Minoda A, Colmenares SU, Polyzos A, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–44. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–31. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 39.Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–91. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiratani I, Ryba T, Itoh M, Rathjen J, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–69. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhar V, Mager D, Iqbal A, Schildkraut CL. The coordinate replication of the human beta-globin gene domain reflects its transcriptional activity and nuclease hypersensitivity. Mol Cell Biol. 1988;8:4958–65. doi: 10.1128/mcb.8.11.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon I, Tenzen T, Reubinoff BE, Hillman D, et al. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–32. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- 43.Göndör A, Ohlsson R. Replication timing and epigenetic reprogramming of gene expression: a two-way relationship? Nat Rev Genet. 2009;10:269–76. doi: 10.1038/nrg2555. [DOI] [PubMed] [Google Scholar]
- 44.Hannibal RL, Chuong EB, Rivera-Mulia JC, Gilbert DM, et al. Copy number variation is a fundamental aspect of the placental genome. PLoS Genet. 2014;10:e1004290. doi: 10.1371/journal.pgen.1004290. [DOI] [PMC free article] [PubMed] [Google Scholar]