Abstract
Malignant mesothelioma (MM) shows inactivation of the BRCA1-associated protein 1 (BAP1) gene. In this study, we found BAP1 mutations in 5 (26%) of the 19 cell lines that we established from Japanese MM patients, and examined functional differences between the WT and mutant BAP1. First, we studied the subcellular localization of BAP1, demonstrating that the WT primarily resides in the nucleus and that the mutant BAP1 is found in the cytoplasm of the cells. Transduction of the WT BAP1 vector into MM cells with homozygous deletion at the BAP1 3′ side resulted in both inhibition of cell proliferation and anchorage-independent cell growth, whereas BAP1 mutants of a missense or C-terminal truncated form showed impaired growth inhibitory effects. Next, we studied how BAP1 is involved in MM cell survival after irradiation (IR), which causes DNA damage. After IR, we found that both WT and mutant BAP1 were similarly phosphorylated and phospho-BAP1 localized mainly in the nucleus. Interestingly, BRCA1 proteins were decreased in the MM cells with BAP1 deletion, and transduction of the mutants as well as WT BAP1 increased BRCA1 proteins, suggesting that BAP1 may promote DNA repair partly through stabilizing BRCA1. Furthermore, using the MM cells with BAP1 deletion, we found that WT BAP1, and even a missense mutant, conferred a higher survival rate after IR compared to the control vector. Our results suggested that, whereas WT BAP1 suppresses MM cell proliferation and restores cell survival after IR damage, some mutant BAP1 may also moderately retain these functions.
Keywords: BAP1, malignant mesothelioma, mutation, subcellular localization, tumor suppressor gene
Malignant mesothelioma (MM) is an aggressive neoplasm which was primarily associated with widespread use of asbestos.1–5 It is usually resistant to conventional multimodal therapies including chemotherapy, radiotherapy, and surgical therapy. The prognosis of patients with MM remains very poor; the median survival of MM patients after diagnosis is only 7–12 months.2,4–7
Previous studies have identified frequent genetic alterations of the two tumor suppressor genes, CDKN2A and NF2, in MM.8–10 NF2 encodes merlin, an upstream regulator of the Hippo signaling pathway, and we recently reported that LATS2, SAV1, and/or AJUBA can also be inactivated in a subset of MMs, all of which encode the components of the Hippo signaling pathway.11–13
The BRCA1-associated protein 1 (BAP1) gene is located on chromosome 3p21.1, which encodes a protein of 729 amino acids. BAP1 has also been shown to be mutated in approximately 25% of MMs from Caucasian patients.14,15 Meanwhile, Yoshikawa et al.16 reported BAP1 mutations in 61% of MM from Japanese patients.
BAP1 is a nuclear-localized deubiquitinating (DUB) enzyme with an NH2-terminal ubiquitin COOH-terminal hydrolase (UCH) domain and a COOH-terminal domain which contains two nuclear localization signals (NLS).17 BAP1 has been suggested to act as a tumor suppressor with possibly three functions.18 First, BAP1 acts as a transcriptional factor, associating with host cell factor 1, YY1, and E2F1. The complex of these factors is recruited to various promoters to upregulate gene expression.19–22 Second, BAP1 acts as a component of the Polycomb repressive deubiquitinase (PR-DUB) complex, associating with ASXL1. The PR-DUB complex deubiquitinates the histone H2A (H2AK119ub1), leading to gene repression.23 Third, BAP1 contributes to the process of DNA repair. 24–26
The possible function BAP1 may have in the DNA repair process has not yet been made clear. The homologous recombination (HR) pathway, one of the major pathways of DNA repair, includes many proteins, some of which may be potential substrates for BAP1-mediated ubiquitin hydrolysis. Eletr et al.27 reported that BAP1 is phosphorylated at serine 592 following DNA damage, that phospho-BAP1 causes deubiquitination of H2AK119ub1, and that this signal promotes the HR pathway. Yu et al.26 also found six irradiation (IR)-induced phosphorylation sites in BAP1 and showed that mutation of these residues inhibits the recruitment of BAP1 to the double-strand break (DSB) sites. Furthermore, Ismail et al.25 suggested that BAP1 could function for HR repair in the BRCA1-related pathway, and they hypothesized that BAP1 enables BRCA1 to readily accumulate DSB sites. These three reports suggested that the DUB catalytic activity of BAP1 is critical for promoting DNA repair.
In this study, we investigated the BAP1 gene status using MM cell lines, most of which were established from Japanese MM patients. We further examined the functional differences between WT and mutant forms of BAP1 regarding subcellular localization, cell growth regulation, and the response to IR-induced cellular damage. We found that some mutant forms of BAP1 still retain partial activities of these functions.
Materials and Methods
Cell lines
Nineteen Japanese MM cell lines, namely ACC-MESO-1, -4, Y-MESO-8D, -9, -12, -14, -21, -22, -25, -26B, -27, -28, -29, -30, -45, -48, -61, -72, and -76, were established in our laboratory as reported previously and described elsewhere, and the cells at 10–15 passages were used for assays.28,29 Four MM cell lines including NCI-H28, NCI-H2052, NCI-H2373, and MSTO-211H, and one immortalized mesothelial cell line, MeT-5A, were purchased from ATCC (Rockville, MD, USA), and cells at 3–5 passages were used. NCI-H290 and NCI-H2452 were the kind gifts of Dr. Adi F. Gazdar (Hamon Center for Therapeutic Oncology, University of Texas Southwestern Medical Center, Dallas, TX, USA). All MM cell lines and MeT-5A were cultured as described in Data S1. Malignant mesothelioma tissue samples from patients for the establishment of cell culture were obtained according to the Institutional Review Board’s approved protocol and with written informed consent from each patient.
Antibodies
For Western blot and immunofluorescence analyses, mouse anti-BAP1 antibody (sc-28383) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and rabbit anti-phospho-BAP1 (Ser592; #9373), rabbit anti-BRCA1 (#9010), rabbit anti-phospho-BRCA1 (Ser1524; #9009), rabbit anti-Lamin-B1 (#12586), and rabbit anti-α-tubulin (#2125) antibodies were from Cell Signaling Technology (Danvers, MA, USA), and mouse anti-β-actin (clone AC74) was from Sigma (St. Louis, MO, USA).
Construction of expression vectors
cDNA fragments of WT or mutant BAP1 were amplified by PCR using PrimeSTAR Max DNA polymerase (Takara Bio, Otsu, Japan), and introduced into the pcDNA3.1 V5-His expression vector (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), thereby fusing these cDNAs with the V5-His sequence. The sequences of all constructs were confirmed. To generate BAP1 expressing lentiviral vector, cDNA coding for the human BAP1 tagged with V5-His was amplified by PCR and cloned into the pLL3.7 lentiviral vector with an infusion cloning system (Clontech, Mountain View, CA, USA).
Results
BAP1 mutations in MM
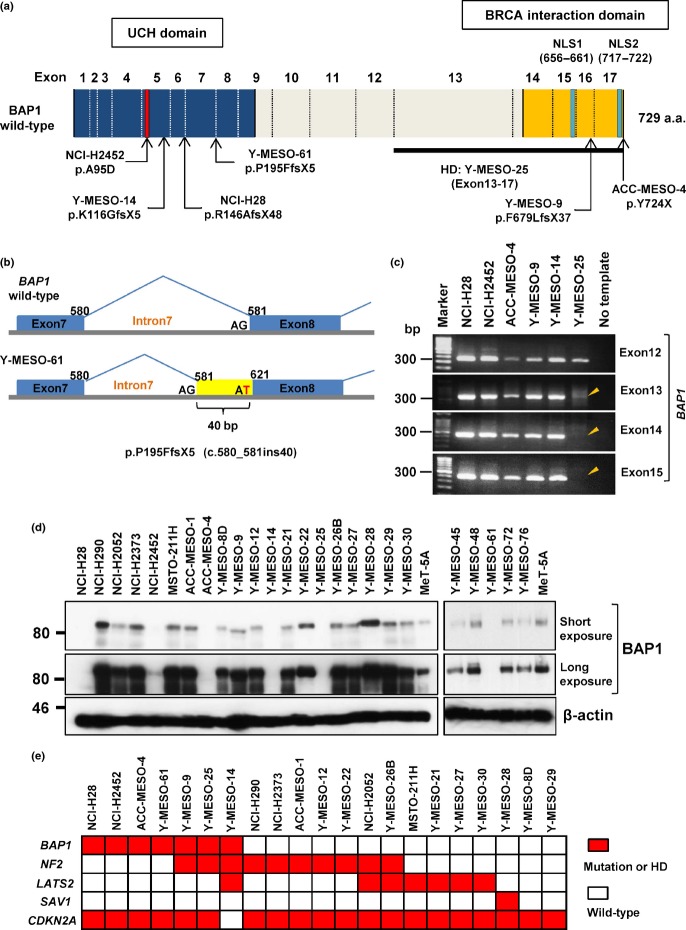
We carried out mutational analyses of BAP1 using 25 MM cell lines. Among 19 cell lines established from Japanese patients, four non-synonymous or insertion/deletion mutations were found; a nonsense mutation in ACC-MESO-4, two truncating mutations in Y-MESO-9 and Y-MESO-14, and a 40-bp insertion mutation at the intron 7–exon 8 junction in Y-MESO-61 (Fig.1a,b). We also carried out sequence analysis of cDNA synthesized from these cell line RNAs and confirmed the expression of the mutant RNAs (data not shown). As our previous study11 using array comparative genomic hybridization analysis detected a possible homozygous deletion (HD) in Y-MESO-25 (Fig. S1a), we carried out a genomic PCR analysis and confirmed that this cell line has HD of BAP1 at exons 13–17 (Fig.1c). In total, we found BAP1 mutations in 26% (5/19) of the Japanese MM cell lines (Table1). In addition, of six cell lines, the mutation status of which has been previously reported by another group,14 we confirmed the same mutations in two cell lines (NCI-H28 and NCI-H2452) and in the WT among four cell lines (Table1).
Figure 1.
BRCA1-associated protein 1 (BAP1) gene mutations in malignant mesothelioma (MM). (a) Schematic diagram of BAP1 mutations in MM cell lines. (b) Schematic diagram of an insertion mutation due to G to T change at the acceptor site of exon 8 in Y-MESO-61. (c) Genomic PCR analysis of exons of BAP1 detected homozygous deletion (HD) in Y-MESO-25 cells (arrowhead). (d) Western blot analysis of BAP1. Expression of β-actin was used as the control. (e) Summary of five gene mutation statuses in MM cell lines. Red boxes indicate inactivating mutation or HD. The mutation statuses of neurofibromatosis type 2 (NF2), large tumor suppressor homolog 2 (LATS2), Salvador homolog 1 (SAV1), and cyclin-dependent kinase inhibitor 2A (CDKN2A) were previously reported.11 NLS, nuclear localization signal; UCH, ubiquitin COOH-terminal hydrolase.
Table 1.
Summary of BAP1 mutations in 25 malignant mesothelioma cell lines
| Cell line | Race | Histological subtypes | Mutation | Amino acid change |
|---|---|---|---|---|
| NCI-H28 | Caucasian | Epithelial | C.438-24_438-2del23† | P.r146afsx48† |
| NCI-H290 | Unknown | ND | WT† | – |
| NCI-H2052 | Caucasian | Epithelial | WT† | – |
| NCI-H2373 | Unknown | ND | WT† | – |
| NCI-H2452 | Caucasian | Epithelial | C.284c>a† | P.a95d† |
| MSTO-211H | Caucasian | Biphasic | WT† | – |
| ACC-MESO-1 | Japanese | Epithelial | WT | – |
| ACC-MESO-4 | Japanese | Epithelial | C.2172delc | P.y724x |
| Y-MESO-8D | Japanese | Biphasic | WT | – |
| Y-MESO-9 | Japanese | Epithelial | C.2035_2036del2 | P.f679lfsx37 |
| Y-MESO-12 | Japanese | Epithelial | WT | – |
| Y-MESO-14 | Japanese | Biphasic | C.349_359del11 | P.k116gfsx5 |
| Y-MESO-21 | Japanese | Epithelial | WT | – |
| Y-MESO-22 | Japanese | Biphasic | WT | – |
| Y-MESO-25 | Japanese | Epithelial | Hd (exon13-17) | Del 417-729 |
| Y-MESO-26B | Japanese | Lymphohistiocytoid | WT | – |
| Y-MESO-27 | Japanese | Epithelial | WT | – |
| Y-MESO-28 | Japanese | Epithelial | WT | – |
| Y-MESO-29 | Japanese | Epithelial | WT | – |
| Y-MESO-30 | Japanese | Epithelial | WT | – |
| Y-MESO-45 | Japanese | ND | WT | – |
| Y-MESO-48 | Japanese | Epithelial | WT | – |
| Y-MESO-61 | Japanese | Epithelial | C.580_581ins40 | P.p195ffsx5 |
| Y-MESO-72 | Japanese | Epithelial | WT | – |
| Y-MESO-76 | Japanese | Epithelial | WT | – |
Reported by Bott et al. (2011). –, No change; HD, homozygous deletion; ND, not determined.
Next, we used Western blot analysis to confirm the inactivation status of BAP1 with the antibody that detects the COOH-terminus of BAP1. Among seven cell lines with a BAP1 mutation, four (NCI-H28, Y-MESO-14, Y-MESO-25, and Y-MESO-61) expressed no band, two (NCI-H2452 and ACC-MESO-4) expressed a very weak band, and one (Y-MESO-9) expressed a short-sized band of BAP1 protein (Fig.1d). Thus, these aberrant protein expression patterns were consistent with the mutation status.
We compared the mutation status of BAP1 with other genes we previously reported (Fig.1e).11 However, we found no significant associations between BAP1, CDKN2A, NF2, LATS2, and SAV1 mutations, which seem to be in agreement with other published reports.14
Effects of BAP1 mutation on its own nuclear localization
BAP1 contains two NLS at the COOH-terminus, and is primarily localized in the nucleus.17 We used immunofluorescence analysis to determine whether or not the BAP1 mutations have an effect on its subcellular localization, because four mutations were predicted to result in the loss of the both NLS and one in the NLS2 at the COOH-terminus among the seven mutations. As expected, NCI-H290 cells, which harbor WT BAP1, showed endogenous BAP1 expression primarily in the nucleus (Fig.2a). In contrast, Y-MESO-9 cells, which lack the NLS2, showed endogenous BAP1 mainly in the cytoplasm. These results suggested that nuclear localization of endogenous BAP1 is impaired primarily by the loss of NLS, which was consistent with a previous study.30
Figure 2.
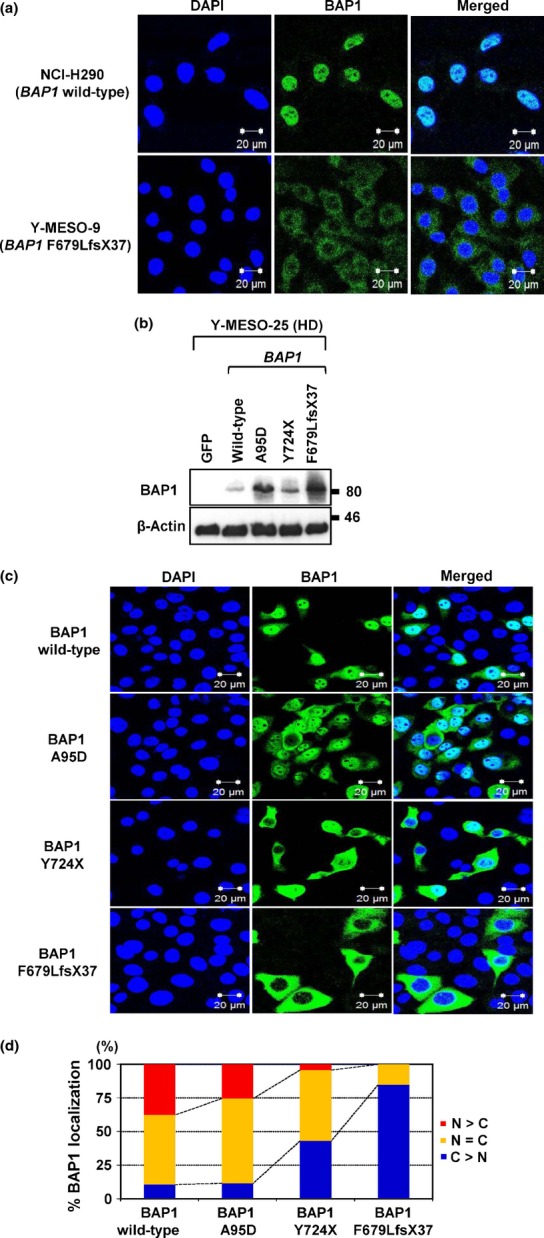
Effects of BRCA1-associated protein 1 (BAP1) mutation on its own nuclear localization. (a) Endogenous WT BAP1 in NCI-H290 showed nuclear localization, whereas mutant BAP1 (p.F679LfsX37) in Y-MESO-9 showed cytoplasmic translocation. (b) Protein expression of exogenously transduced WT or mutant BAP1 (A95D, Y724X, and F679LfsX37) vectors into the malignant mesothelioma cell line with BAP1 deletion (Y-MESO-25). (c) Immunofluorescence analysis of subcellular BAP1 localization with exogenously transduced BAP1 vectors into the Y-MESO-25 cells. (d) Percentages of subcellular localization (c) were calculated. Wild-type BAP1 was mainly localized in the nucleus (N), whereas BAP1 mutants showed translocation in the cytoplasm (C). HD, homozygous deletion.
To confirm whether these mutations were the main cause of the aberrant cellular localization of BAP1, we transduced WT or mutant BAP1 constructs into the MM cell with BAP1 deletion (Y-MESO-25 cell line) and examined their cellular localization (Fig.2b). As expected, WT BAP1 was preferentially found in the nucleus, whereas mutant BAP1 proteins, which were nonsense (Y724X) or truncating (F679LfsX37) forms, showed impaired nuclear localization with increased cytoplasmic localization. Interestingly, a modest increase in the cytoplasmic localization of BAP1 with A95D, a point mutation in the UCH domain, was also observed (Fig.2c,d).
BAP1 suppresses MM cell proliferation
To determine whether BAP1 has growth-suppressive activity against MM cells, we carried out cell proliferation assays. Transduction of the WT BAP1 vector inhibited cell proliferation of the Y-MESO-25 MM cell line with BAP1 deletion by approximately 50% compared to the cells with control vector (GFP) (Fig.3a). In contrast, the mutant BAP1 constructs showed reduced or no inhibitory effects according to the possible deterioration effects of the mutants (Fig.3a). With the colony formation assay, we also found that WT BAP1 suppressed the anchorage-independent cell growth of the MM cells with BAP1 deletion, whereas mutant BAP1 showed little or no reduced effect (Fig.3b,c).
Figure 3.
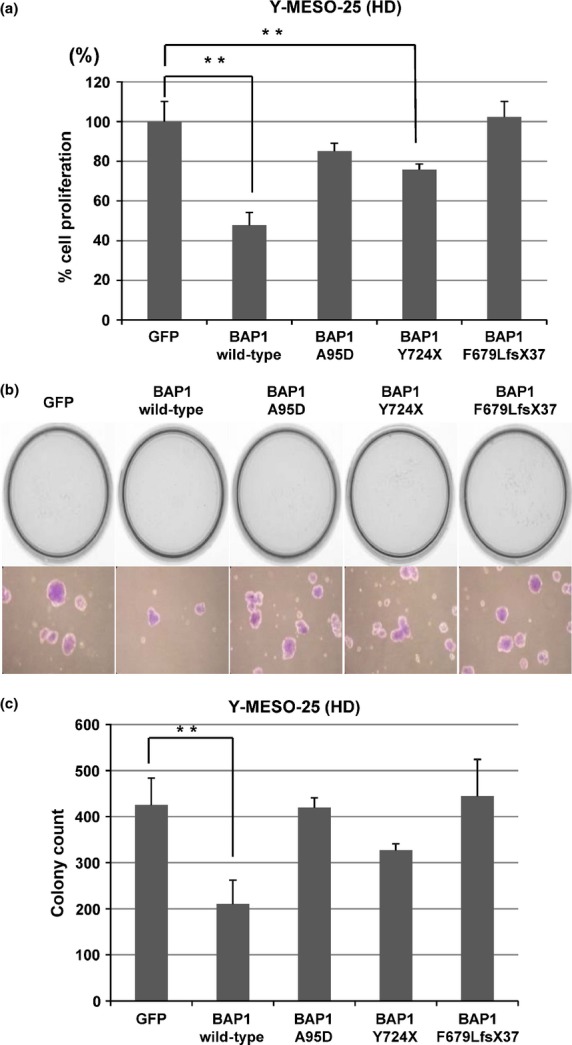
BRCA1-associated protein 1 (BAP1) suppresses cell proliferation of malignant mesothelioma cells. (a) Transduction of WT BAP1 vector inhibited cell proliferation in malignant mesothelioma cells with BAP1 deletion (Y-MESO-25). Bars, SD; columns, average. **P < 0.01 versus empty lentivirus control (GFP). (b) Transduction with BAP1 suppressed anchorage-independent colony formation of Y-MESO-25 cells. Representative results are shown (top) with a higher magnification of their representative colonies (bottom). (c) Graphic presentation of (b). Average and SD of triplicated experiments are shown in (c). **P < 0.01 versus empty lentivirus control (GFP). HD, homozygous deletion.
The Y-MESO-25 cell line also has the NF2 mutation, which was suspected to influence the growth-suppressive effects of BAP1. We introduced BAP1 vectors into the ACC-MESO-4 cell line, which harbors mutant BAP1 but WT NF2. We confirmed the significant growth-suppressive effects of WT BAP1 (approximately 40%) (Fig. S2), which was quite similar to the results observed in Y-MESO-25 cells (Fig.3a). Because both WT and mutant BAP1 showed consistently similar growth-suppressive effects regardless of NF2 mutation status, we think a possible growth-suppressive mechanism of BAP1 may be independent of the NF2 pathway.
BAP1-induced gene expression profile
BAP1 has been suggested to have multiple functions, including gene transcription. To determine whether WT BAP1 induces specific gene expression that is involved in the suppression of MM cell proliferation, we carried out a microarray analysis. With the transduction of WT BAP1 into two BAP1-mutated MM cell lines, we found that four pathways were significantly upregulated in both cell lines (Table S1). They were related to the extracellular region, integral to plasma membrane, cytokine activity, and inflammatory response (Tables S2,S3). As eight genes with altered expression, which were involved in these four pathways, were found by using the GenMAPP/MAPPFinder software, we used quantitative RT-PCR analysis and confirmed the up- or downregulation of these genes (Fig. S3). These results suggested that some of these pathways might be involved in the growth-suppressive effects of MM cells and BAP1 expression might influence the expressions of these genes. However, well-known strong growth-suppressive pathways or genes that have been defined in other common malignancies were not picked up in these analyses. Thus, these data suggest that the BAP1-regulated pathways may not have a strong growth-suppressive effect individually, which may require cumulative inactivation of the multiple pathways for effective growth-suppression of MM cells.
Irradiation of MM cells induces phosphorylation of BAP1
As one of the major cellular functions of BAP1 is thought to be DNA repair, we induced DNA damage in MM cells with X-ray irradiation to reveal how BAP1 is involved in DNA repair in MM cells. First, when MeT-5A cells, which harbor WT BAP1, were irradiated, we observed the strongest BAP1 phosphorylation level 1 h after IR, which then gradually decreased (Fig.4a). The phosphorylation level of WT BAP1 was also confirmed to be increased in proportion to IR doses (Fig.4b). In addition, with immunofluorescence analysis, the strong phosphorylation of the exogenously transduced WT BAP1 into the MM cell with BAP1 deletion was also observed in the nucleus after IR exposure, in the same manner as with endogenous BAP1 (Fig.4c). Using the MM cell with BAP1 deletion to confirm these results, we then carried out cellular fractionation and found that phospho-mutant-type BAP1 proteins tended to localize in the nucleus after IR like WT BAP1, whereas total mutant-type BAP1 proteins dominantly localized in the cytoplasm (Fig.4d). These results suggested that several types of mutant BAP1 proteins as well as the WT form might be recruited to IR-induced DSB sites, although the precise mechanism is yet to be determined.
Figure 4.
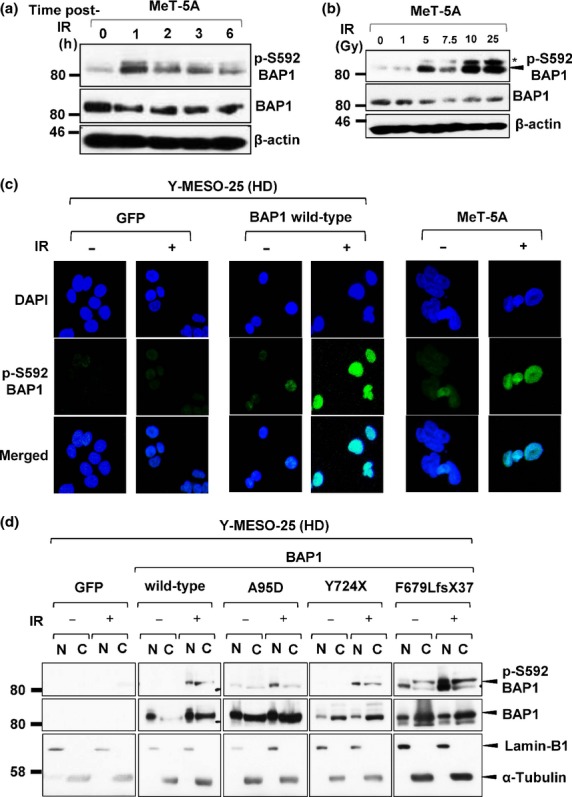
Irradiation (IR) induces phosphorylation of BRCA1-associated protein 1 (BAP1). (a) Western blot analysis with anti-pS592-BAP1 antibody revealed that endogenous WT BAP1 was phosphorylated after IR at 7.5 Gy. (b) BAP1 phosphorylation levels increased in proportion to IR doses. *Non-specific bands. (c) Immunofluorescence analysis showed dominant nuclear localization of phosphorylated-BAP1 protein at 1 h post-IR (7.5 Gy). (d) Western blot analysis of nuclear/cytoplasmic fractionation tested the localization of exogenously transduced BAP1 and its phosphorylated form in malignant mesothelioma cells with BAP1 deletion. HD, homozygous deletion.
BAP1 stabilizes BRCA1 protein and restores cell survival rates of MM cells with BAP1 deletion after IR
To determine the roles of BAP1 on DNA repair in MM cells, we next studied the expression levels of BAP1, and BRCA1, a DNA repair protein that has been suggested to associate with BAP1. We found that the expression levels of BRCA1 protein decreased in more BAP1-mutant cell lines (4 [57%] of 7) than BAP1-WT cell lines (6 [35%] of 17), although it was not statistically significant (Figs5a and S4). As real-time RT-PCR analysis indicated that BRCA1 mRNA expression levels in the three BAP1 mutant cell lines was the same as with MeT-5A cells and the levels in the other four were at least 50–80% (Fig.5b), these results suggested a possibility that BAP1 might be involved in the regulation of BRCA1 protein stability. To confirm this hypothesis, we transduced WT BAP1 into the MM cells with BAP1 deletion, which express a very small amount of BRCA1. Noticeably, we found that exogenous BAP1 significantly increased the BRCA1 protein level (Fig.5c). Furthermore, we found that all three mutant BAP1 forms increased BRCA1 protein levels (Fig.5c). In addition, regarding subcellular localization of BRCA1, we carried out an immunofluorescence analysis, and found that the subcellular location of BRCA1 proteins was not changed in cells by mutant BAP1 transduction (Fig. S5). In other words, while mutant BAP1 protein preferentially localizes in the cytoplasm, BRCA1 protein keeps residing in the nucleus of BAP1-mutant cells. This suggests that BAP1, regardless of WT or mutant form, may not be a strong determinant for subcellular localization of BRCA1.
Figure 5.
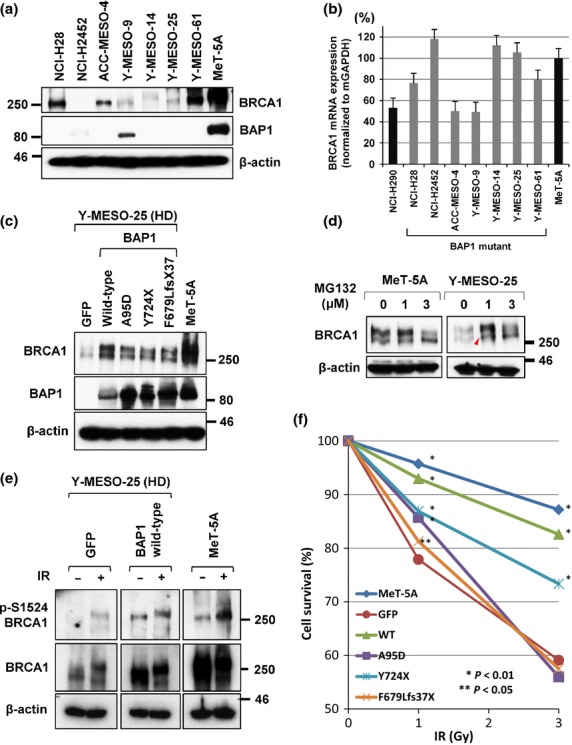
BRCA1-associated protein 1 (BAP1) stabilizes BRCA1 protein and restores cell survival rates of malignant mesothelioma (MM) cells with BAP1 deletion after irradiation (IR). (a) Western blot analysis showed low BRCA1 protein levels in BAP1-mutated MM cell lines. (b) Real-time RT-PCR analysis of BRCA1 mRNA expression in MM cell lines. Three cell lines, H2452, Y-MESO-14 and Y-MESO-25, which expressed very low BRCA1 protein, showed a similar BRCA1 mRNA level compared to MeT-5A cells. The other four cell lines expressed 50–80% levels of BRCA1 mRNA. (c) Western blot analysis showed that transduction of mutant as well as WT BAP1 vectors increased the levels of BRCA1 proteins in MM cells with BAP1 deletion. (d) BRCA1 protein level was increased after MG-132 treatment in Y-MESO-25 cells (arrowhead). (e) Exogenous BAP1 increased both BRCA1 protein and its phosphorylation levels. (f) Wild-type and mutant BAP1 restored the survival rates of MM cells with BAP1 deletion impaired with IR. *P < 0.01 and **P < 0.05 versus empty lentivirus control (GFP).
Jensen et al.17 previously suggested that BAP1 enhanced BRCA1-mediated inhibition of cancer cell growth, although they did not directly establish the possibility that BAP1 enhances the BRCA1 protein level. We hypothesized that BAP1 might deubiquitinate BRCA1 and protect it from proteasome-mediated degradation; this might explain the BRCA1 stabilization by the mutant BAP1 forms that retained the DUB activity. We used a proteasome inhibitor, MG-132, to confirm this idea, and found that the BRCA1 protein level was increased after MG-132 treatment (Fig.5d, arrowhead). This result supported the idea that BRCA1 protein expression might be negatively regulated by the proteasome system, which is suppressed by the DUB enzyme activity of BAP1. In addition, the level of phospho (activated)-BRCA1 decreased in Y-MESO-25 in comparison with BAP1 WT cell lines (Fig.5e), and this reduction of phospho-BRCA1 may impair the activity of DNA repair.
Finally, we examined whether or not BAP1 affects the survival rates of MM cells that were impaired by IR. We found a significantly low survival rate of BAP1-deleted cells (GFP) compared to the MeT-5A cells (Fig.5f). However, the WT BAP1 had a significantly strong activity to restore the survival rate of the MM cells with BAP1 deletion up to the same level as MeT-5A cells. Furthermore, we found that the mutant BAP1 of Y724X restored the survival rates significantly compared to GFP control at both 1 and 3 Gy. Meanwhile, BAP1 mutants of A95D and F679Lfs37X showed only marginal restoration (significant only at 1 Gy). These results indicate that WT BAP1 is one of the important components of the DNA repair pathway and even that some mutant BAP1 proteins retain the activity of this pathway in MM cells.
Discussion
In this study, we analyzed BAP1 mutations using a total of 25 MM cell lines, including 19 that we established, and found that 7 (28%) cell lines had an inactivating mutation. We examined the differences between WT and mutant form BAP1 in terms of their cellular functions, including subcellular localization, growth-suppressive activity, and cellular resistance against IR. We showed that, even though several mutant forms lack growth-suppressive activity, they preserve some functional activities of BAP1 in vitro.
It is worth noting that BAP1 can restore the protein expression of BRCA1. Although BRCA1 expression significantly decreased in an MM cell line with BAP1 deletion, Y-MESO-25, the transduction of the WT and even mutant forms of BAP1 induced upregulation of the BRCA1 protein level. This result suggests that BAP1 contributes to the stabilization of BRCA1 and that the COOH-terminal side of BAP1, even with a small mutation inside, may confer the stability of the BRCA1 protein, probably through the BAP1–BRCA1 interaction.
Mallery et al.31 suggested that the BRCA1/BARD1 heterodimer complex catalyzes the formation of polyubiquitin chains on itself, and thus the BRCA1 protein level may also be controlled by the ubiquitin–proteasome system. As a possible mechanism of BRCA1 stabilization by BAP1 transduction in this study, we speculated that the ubiquitination of BRCA1 protein might be directly deubiquitinated by BAP1. In this regard, however, Nishikawa et al.32 suggested that BAP1 inhibits the E3 ligase activity of BRCA1 in a manner independent of its deubiquitination activity. Hence, it will be necessary to determine the involvement of deubiquitination activity of BAP1 for BRCA1 stabilization in detail in a future study.
The MM cells with BAP1 deletion transduced with WT BAP1 showed a good survival rate that was comparable to MeT-5A at 1-Gy IR. This result strongly suggests that the recovery of the MM cells with BAP1 deletion from the IR damage is attributable not only to the exogenous expression of BAP1 itself but also to the upregulation of BRCA1. In addition, the MM cells with BAP1 deletion transduced with mutant BAP1 (Y724X) tended to have a better survival rate than the cells with control vector (GFP). This result supports the idea that some BAP1 mutants still partially retain important functions of BAP1 that affect cellular viability; in this experiment, mutant BAP1 contributed to the recovery of the DNA repair system together with the upregulation of BRCA1.
Previous studies indicated that several distinct domains are particularly important for the tumor suppressor activity of BAP1. For example, Ventii et al.30 showed that the NLS was necessary for BAP1 to act as a tumor suppressor. They used an in vivo implantation experiment with a BAP1-null human lung cancer cell line that was transduced with BAP1 vectors, and found that WT BAP1, but not mutant BAP1 lacking the NLS, showed significantly weaker tumor development. We also observed that NLS-lacking F679Lfs mutant BAP1, which was dominantly localized in the cytoplasm, had a significantly inferior growth-suppressive effect compared to the WT BAP1.
Mashtalir et al.33 also underscored the functional importance of the UCH-domain. While ubiquitination enzyme UBE2O monoubiquitinates the NLS of BAP1 and induces translocation (inactivation) of BAP1 from the nucleus to the cytoplasm, DUB activity of the UCH-domain of BAP1 counteracts the UBE2O activity mediated through the intramolecular interaction between the UCH-domain and COOH-terminal domain of BAP1. Thus, the autodeubiquitinating activity of the UCH-domain is crucial to its growth-suppressive function. Consistent with this, we found that the A95D mutation within the UCH-domain of BAP1 showed reduced growth-suppressive activity.
Additionally, it was intriguing to find that phosphorylation of mutant BAP1 seemed to induce its translocation from the cytoplasm to the nucleus, as Western blot analysis detected strong p-S592 BAP1 in the nucleus of the mutant F679LfsX37-transduced cells (Fig.4d, second lane from the right). We initially thought that this observation might have resulted from active transportation of phosphorylated BAP1 protein. As this BAP1 mutant form lacks NLS, we thought that this active transportation might have been induced in an NLS-independent manner. However, we currently think that this observation does not necessarily reflect nuclear transport of the phosphorylated mutant BAP1. As a fraction of the F679Lfs37X BAP1 mutant protein resides in the nucleus (Fig.4d, fourth lane from the right), even if much less abundant than in the cytoplasm, the remarkably increased p-S592 BAP1 signal of F679Lfs37X may just reflect the phosphorylation of the mutant BAP1 that has been localized in the nucleus from the beginning. In any event, the phosphorylation effect of BAP1 must be clarified in more detail in terms of changes in the activity and cellular localization of both WT and mutant BAP1.
In conclusion, the present study suggests that various types of BAP1 mutations as well as WT BAP1 may exert different levels of growth suppression and/or DNA repair activities with different levels of residual functions. This result indicates that various BAP1 mutations in MM cells may cause a different sensitivity to chemotherapy and/or radiation therapy. Therefore, our study suggests that BAP1 mutation status must be precisely determined in order to select an optimal therapeutic strategy for individual patients with MM.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This work was supported in part by the Japan Society for the Promotion of Science (Kakenhi 25290053), a Grant-in-Aid for Third-Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor and Welfare of Japan, the Takeda Science Foundation, and a Research Grant from the Princess Takamatsu Cancer Research Fund (14-24617), and P-DIRECT.
Supporting Information
Data S1. Supporting Materials and Methods.
Fig. S1. BAP1 gene mutation analyses.
Fig. S2. BAP1 suppresses cell proliferation of malignant mesothelioma cells that harbor BAP1-mutation but not NF2-mutation.
Fig. S3. Quantitative RT-PCR analysis for eight genes, detected as differentially expressed genes in microarray analysis.
Fig. S4. BRCA1 expression in malignant mesothelioma cell lines.
Fig. S5. Subcellular localization of endogenous BRCA1.
Table S1. Significantly increased pathways in ACC-MESO-4 and Y-MESO-25 BAP1 expression.
Table S2. Pathways or functional groups of genes differently expressed in ACC-MESO-4/WT BAP1 by GenMAPP/MappFinder.
Table S3. Pathways or functional groups of genes differently expressed in Y-MESO-25/WT BAP1 by GenMAPP/MappFinder.
References
- Carbone M, Bedrossian CW. The pathogenesis of mesothelioma. Semin Diagn Pathol. 2006;23:56–60. doi: 10.1053/j.semdp.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Pass HI, Vogelzang N, Hahn S, Carbone M. Malignant pleural mesothelioma. Curr Probl Cancer. 2004;28:93–174. doi: 10.1016/j.currproblcancer.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Ramos-Nino ME, Testa JR, Altomare DA, et al. Cellular and molecular parameters of mesothelioma. J Cell Biochem. 2006;98:723–34. doi: 10.1002/jcb.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med. 2005;353:1591–603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- Tsao AS, Wistuba I, Roth JA, Kindler HL. Malignant pleural mesothelioma. J Clin Oncol. 2009;27:2081–90. doi: 10.1200/JCO.2008.19.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BW, Musk AW, Lake RA. Malignant mesothelioma. Lancet. 2005;366:397–408. doi: 10.1016/S0140-6736(05)67025-0. [DOI] [PubMed] [Google Scholar]
- Vogelzang NJ, Rusthoven JJ, Symanowski J, et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J Clin Oncol. 2003;21:2636–44. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- Bianchi AB, Mitsunaga SI, Cheng JQ, et al. High frequency of inactivating mutations in the neurofibromatosis type 2 gene (NF2) in primary malignant mesotheliomas. Proc Natl Acad Sci USA. 1995;92:10854–8. doi: 10.1073/pnas.92.24.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido Y, Pass HI, Bader S, et al. Neurofibromatosis type 2 (NF2) gene is somatically mutated in mesothelioma but not in lung cancer. Cancer Res. 1995;55:1227–31. [PubMed] [Google Scholar]
- Xio S, Li D, Vijg J, Sugarbaker DJ, Corson JM, Fletcher JA. Codeletion of p15 and p16 in primary malignant mesothelioma. Oncogene. 1995;11:511–5. [PubMed] [Google Scholar]
- Murakami H, Mizuno T, Taniguchi T, et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011;71:873–83. doi: 10.1158/0008-5472.CAN-10-2164. [DOI] [PubMed] [Google Scholar]
- Tanaka I, Osada H, Fujii M, et al. LIM-domain protein AJUBA suppresses malignant mesothelioma cell proliferation via Hippo signaling cascade. Oncogene. 2015;34:73–83. doi: 10.1038/onc.2013.528. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Osada H, Murakami H, et al. YAP1 is involved in mesothelioma development and negatively regulated by Merlin through phosphorylation. Carcinogenesis. 2008;29:2139–46. doi: 10.1093/carcin/bgn200. [DOI] [PubMed] [Google Scholar]
- Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–72. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Sato A, Tsujimura T, et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci. 2012;103:868–74. doi: 10.1111/j.1349-7006.2012.02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Zauderer MG, Krug LM, et al. New strategies in pleural mesothelioma: BAP1 and NF2 as novel targets for therapeutic development and risk assessment. Clin Cancer Res. 2012;18:4485–90. doi: 10.1158/1078-0432.CCR-11-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletr ZM, Wilkinson KD. An emerging model for BAP1’s role in regulating cell cycle progression. Cell Biochem Biophys. 2011;60:3–11. doi: 10.1007/s12013-011-9184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–88. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi S, Ottosen S, Izrael-Tomasevic A, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–92. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mashtalir N, Daou S, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30:5071–85. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–7. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–9. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Davidson R, Gagne JP, Xu ZZ, Poirier GG, Hendzel MJ. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 2014;74:4282–94. doi: 10.1158/0008-5472.CAN-13-3109. [DOI] [PubMed] [Google Scholar]
- Yu H, Pak H, Hammond-Martel I, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014;111:285–90. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletr ZM, Yin L, Wilkinson KD. BAP1 is phosphorylated at serine 592 in S-phase following DNA damage. FEBS Lett. 2013;587:3906–11. doi: 10.1016/j.febslet.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Karnan S, Fukui T, et al. Genomic profiling of malignant pleural mesothelioma with array-based comparative genomic hybridization shows frequent non-random chromosomal alteration regions including JUN amplification on 1p32. Cancer Sci. 2007;98:438–46. doi: 10.1111/j.1349-7006.2006.00386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami N, Fukui T, Kondo M, et al. Establishment and characterization of four malignant pleural mesothelioma cell lines from Japanese patients. Cancer Sci. 2006;97:387–94. doi: 10.1111/j.1349-7006.2006.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventii KH, Devi NS, Friedrich KL, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–62. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery DL, Vandenberg CJ, Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002;21:6755–62. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Wu W, Koike A, et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2009;69:111–9. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- Mashtalir N, Daou S, Barbour H, et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell. 2014;54:392–406. doi: 10.1016/j.molcel.2014.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Materials and Methods.
Fig. S1. BAP1 gene mutation analyses.
Fig. S2. BAP1 suppresses cell proliferation of malignant mesothelioma cells that harbor BAP1-mutation but not NF2-mutation.
Fig. S3. Quantitative RT-PCR analysis for eight genes, detected as differentially expressed genes in microarray analysis.
Fig. S4. BRCA1 expression in malignant mesothelioma cell lines.
Fig. S5. Subcellular localization of endogenous BRCA1.
Table S1. Significantly increased pathways in ACC-MESO-4 and Y-MESO-25 BAP1 expression.
Table S2. Pathways or functional groups of genes differently expressed in ACC-MESO-4/WT BAP1 by GenMAPP/MappFinder.
Table S3. Pathways or functional groups of genes differently expressed in Y-MESO-25/WT BAP1 by GenMAPP/MappFinder.