Abstract
Rhabdoid histology in clear cell renal cell carcinoma is associated with a poor prognosis. The prognosis of patients with clear cell renal cell carcinoma may also be influenced by molecular alterations. The aim of this study was to evaluate the association between histologic features and salient molecular changes in rhabdoid clear cell renal cell carcinoma. We macrodissected the rhabdoid and clear cell epithelioid components from 12 cases of rhabdoid clear cell renal cell carcinoma. We assessed cancer related mutations from 8 cases using a clinical next generation exome sequencing platform. The transcriptome of rhabdoid clear cell renal cell carcinoma (n=8) and non-rhabdoid clear cell renal cell carcinoma (n=37) was assessed by RNA-seq and gene expression microarray. VHL (63%) showed identical mutations in all regions from the same tumor. BAP1 (38%) and PBRM1 (13%) mutations were identified in the rhabdoid but not the epithelioid component and were mutually exclusive in 3/3 cases and 1 case, respectively. SETD2 (63%) mutations were discordant between different histologic regions in 2/5 cases, with mutations called only in the epithelioid and rhabdoid components, respectively. The transcriptome of rhabdoid clear cell renal cell carcinoma was distinct from advanced stage and high grade clear cell renal cell carcinoma. The diverse histologic components of rhabdoid clear cell renal cell carcinoma, however, showed a similar transcriptomic program, including a similar prognostic gene expression signature. Rhabdoid clear cell renal cell carcinoma is transcriptomically distinct and shows a high rate of SETD2 and BAP1 mutations and a low rate of PBRM1 mutations. Driver mutations in clear cell renal cell carcinoma are often discordant across different morphologic regions whereas the gene expression program is relatively stable. Molecular profiling of clear cell renal cell carcinoma may improve by assessing for gene expression and sampling tumor foci from different histologic regions.
INTRODUCTION
Renal cell carcinoma is a lethal genitourinary malignancy with cancer-specific death arising primarily from the clear cell subtype of renal cell carcinoma (1). Histologic differentiation of clear cell renal cell carcinoma, expressed as the tumor grade, is an independent prognostic factor that has been incorporated into various prognostic nomograms (2–4). Clear cell renal cell carcinoma shows substantial morphologic grade variation within a given tumor: the pathologically assigned grade is based on the least differentiated or highest grade component (2). For example, clear cell renal cell carcinoma may manifest a biphasic pattern with a better differentiated, low grade clear cell epithelioid (E) component and a de-differentiated, high grade component that shows plump cells with eosinophilic cytoplasm that resemble rhabdomyoblasts, so-called rhabdoid histologic features. Rhabdoid features in clear cell renal cell carcinoma are associated with a dismal prognosis with most patients presenting with metastases and a median survival of less than 1 year (5–7).
Others using molecular methods have shown that patients with clear cell renal cell carcinoma can be stratified into good and poor prognosis groups based on the mutational profile (8) and gene expression signature of their neoplasms (9, 10). However, the intratumoral genetic heterogeneity of clear cell renal cell carcinoma, as evidenced by multiregion sequencing that established its subclonal architecture, poses a challenge since a single sample does not capture the genomic landscape of the entire tumor (11). Pathologists who select lesional tissues for molecular profiling are aware of the heterogeneity of morphologic grades within clear cell renal cell carcinoma. It is unknown, however, if different histologic components (regional grades) within clear cell renal cell carcinoma show different molecular attributes. The potential limitations of single biopsy sampling also have not been evaluated in a clinical setting using archival formalin-fixed, paraffin-embedded tissues.
The aim of this study was to evaluate cancer-related mutations and gene expression profiles within different regional grades of rhabdoid clear cell renal cell carcinoma using archival tissues in a clinical setting. We show that rhabdoid clear cell renal cell carcinoma is molecularly distinct, but driver mutations in renal cell carcinoma are not distributed uniformly across different morphologic regional tumor grades and cannot be captured reliably with a single biopsy of the tumor. The gene expression program between different regional histologies of rhabdoid clear cell renal cell carcinoma, by contrast, is more stable. These findings have implications for molecular profiling of clear cell renal cell carcinoma for prognostic or predictive purposes.
MATERIALS AND METHODS
Patient and tumor characteristics
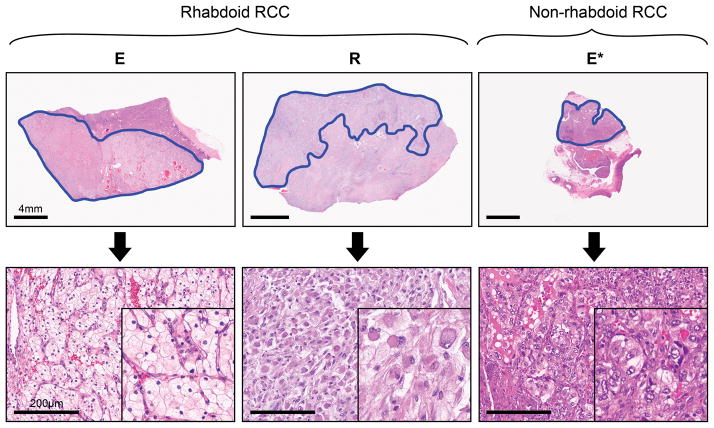
Formalin-fixed paraffin embedded samples from 12 patients with rhabdoid clear cell renal cell carcinoma resected between 1996–2013 at The University of Texas MD Anderson Cancer Center (Houston, Texas) were selected (Supplementary Table 1). Inclusion criteria included: clear cell histology of the parent clear cell epithelioid (E) tumor; > 5 × 5 mm area of pure rhabdoid, sarcomatoid or clear cell epithelioid component on a slide; and > 60% cancer cells within a lesional focus. Samples from the epithelial component of clear cell renal cell carcinoma without rhabdoid or sarcomatoid foci were used as a reference for gene expression profiling (n=37). Sections from normal adjacent renal parenchyma were used as controls for mutational analyses. Lesional foci were marked on H&E stained slides with all cases reviewed by at least two genitourinary pathologists (Figure 1). Clinicopathologic characteristics of the patient cohorts with respect to the different molecular profiling platforms are summarized in Table 1. The study was performed with approval from the institutional review board (IRB # LAB 08-670).
Figure 1. Biphasic components of rhabdoid renal cell carcinoma and non-rhabdoid renal cell carcinoma macrodissected.
The paired clear cell epithelioid (E) and rhabdoid (R) components of rhabdoid renal cell carcinoma and the epithelial (E*) component of non-rhabdoid Fuhrman grade 3 renal cell carcinoma, macrodissected as illustrated above (H&E stain, scale bars are 4mm and 200μm in upper and lower panels, respectively).
Table 1.
Clinicopathologic characteristics of rhabdoid and non-rhabdoid clear cell renal cell carcinoma (RCC) cases
| No. | Platform | Pretreatment | Age, years | Rhabdoid histology (%) | Stage | Fuhrman grade | Died of Disease | Follow up, wks | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| I | II | III | IV | 1 | 2 | 3 | 4 | ||||||||
| Gene expression | |||||||||||||||
|
| |||||||||||||||
| Rhabdoid clear cell RCC | n=4 | RNAseq | S, n=1; None, n=3 | 60 (35–72) | 35 (15–60) | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 3 (75%) | 66 (13–107) |
| Non-rhabdoid clear cell RCC | n=15 | RNAseq | None, n=15 | 64 (46–85) | 0 (0) | 0 | 0 | 7 | 8 | 0 | 0 | 9 | 6 | 8 (53%) | 224 (2–452) |
| Rhabdoid clear cell RCC | n=4 | cDNA microarray | None, n=4 | 52 (48–57) | 55 (10–90) | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 4 | 4 (100%) | 35 (8–108) |
| Non-rhabdoid clear cell RCC | n=22 | cDNA microarray | None, n=22 | 69 (56–88) | 0 (0) | 0 | 0 | 7 | 15 | 0 | 4 | 10 | 8 | 16 (73%) | 53 (1–653) |
|
| |||||||||||||||
| Cancer related mutations | |||||||||||||||
|
| |||||||||||||||
| Rhabdoid clear cell RCC | n=8 | NGS exome | S, n=1; A, n=1; None, n=6 | 52 (35–73) | 28 (7–90) | 0 | 0 | 2 | 6 | 0 | 0 | 0 | 8 | 6 (75%) | 26 (7–87) |
S: Sunitinib; A: Axitinib; Data are expressed as median (range) or n (%)
Genomic DNA extraction and Next Generation Sequencing and Analysis
Extraction of DNA from formalin-fixed, paraffin-embedded tumor sections was performed using the PicoPure DNA extraction kit (Arturus, Mountain View, CA, USA) and further purified using AMPureXP kit (Agentcourt Biosciences, Beverly, MA, USA). Purified DNA was quantified using Qubit DNA HS assay kit (Life Technologies, Carlsbad, CA, USA).
Sequencing of 409-cancer related genes involved target capture and sequencing of the exonic areas of 409 genes as previously described (12). Briefly, starting from 60ng DNA, a genomic library for the 409-gene panel was prepared using Ion AmpliSeq Comprehensive Cancer Panel (Life Technologies) following the manufacturer’s instructions. The library was clonally amplified on to Ionsphere beads using Ion PI Template OT2 kit V2 and the Ion One Touch 2 system (Life Technologies). Sequencing of the samples was performed using Ion Proton high capacity sequencer using Proton I chip and Ion PI sequencing 200Kit V2. Sequence alignment and variant calling were performed using Torrent Suite v3.6.2. To inspect the mutation calls and confirm their authenticity, sequencing reads were visualized using Integrative Genomics Viewer (13). An in-house designed software program (OncoSeek) was used to filter the germline variants and compare the mutations between the different tumor components (12).
RNA-seq based gene expression profiling and analysis
We assessed epithelioid/rhabdoid pairs (n=4) and non-rhabdoid renal carcinoma (n=15) cases with paired-end sequencing on an Illumina HiSeq2000 platform to generate gene expression data. The raw data are deposited in Gene Expression Omnibus (GSE59066). Lesional foci were macrodissected from formalin-fixed, paraffin-embedded blocks corresponding to marked hematoxylin-eosin stained slides using a disposable punch biopsy instrument (Miltex). RNA for all samples was extracted using the RNeasy formalin-fixed, paraffin-embedded kit. RNA from 15 non-rhabdoid renal carcinoma cases was pooled into 5 sets of 3 whereas RNA was extracted and kept separate for each epithelioid and rhabdoid component. Sequencing libraries for the HiSeq run were prepared using the Ovation RNA-Seq formalin-fixed, paraffin-embedded System (Nugen). Synthesis, barcoding, size selection, multiplexed sequencing, and analysis were performed by the Genome and RNA Profiling Core at the Baylor College of Medicine (Houston, Texas).
RNA-seq reads were aligned using the TopHat suite for splice-aware alignment. Htseq-count of the Python package HTSeq was used to count reads that mapped to each gene. The gene counts were used to detect differential gene expression by fitting generalized linear models using the edgeR Bioconductor package. Counts per million (CPM) values were log transformed as log2 (CPM+1) and plotted. Values for log transformed expression were then compared between E and R using a homoscedastic student’s t-test. Data were analyzed across sample subtypes and P values were adjusted for multiple tests using the Benjamini-Hochberg procedure. For supervised analyses, we interrogated probe sets that represented known good-prognosis (clear cell type A) and poor-prognosis (clear cell type B) gene expression signatures of clear cell renal cell carcinoma (9, 10).
Microarray based gene expression profiling and analysis
We assessed gene expression on independent epithelioid/rhabdoid pairs (n=4) and non-rhabdoid renal carcinoma (n=22) cases using a cDNA microarray platform (Illumina). The raw expression data are deposited in Gene Expression Omnibus (GSE59266). Total cellular RNA was isolated from core punch specimens according to the manufacturer’s protocol (Epicentre Biotechnologies) after de-paraffinization and proteinase K treatment. RNA samples were normalized using the Ribogreen RNA quantitation kit (Life Technologies) for the whole-genome cDNA-mediated annealing, selection, extension, and ligation HT assay. Normalized RNA was converted to cDNA and incubated on Illumina HumanHT-12v4 BeadChips. The slides were scanned using a BeadArray Reader and the signal intensities were quantified using GenomeStudio software.
We log 2-transformed the data and normalized them using quantile normalization. We performed hierarchical clustering and principal component analyses to identify batch effects. We performed 2-sample t-tests to compare epithelioid vs. rhabdoid, epithelioid/rhabdoid vs. non-rhabdoid renal carcinoma, epithelioid/rhabdoid vs. Fuhrman grade 3 non-rhabdoid renal carcinoma, epithelioid/rhabdoid vs. Fuhrman grade 4 non-rhabdoid renal carcinoma. We used beta-uniform mixture models to adjust for multiple tests. For supervised analyses, we extracted data for 86 clear cell type A and 24 clear cell type B genes from the normalized expression data (9, 10).
Statistical analysis
P-values were two-sided unless otherwise specified. A p value ≤0.05 was considered statistically significant except in multiple comparisons, in which the false discovery rate required q≤0.05.
RESULTS
Rhabdoid renal cell carcinoma shows frequent BAP1 and SETD2 mutations and infrequent PBRM1 mutations
Based on our analysis of 409 cancer related genes, we found few of these being mutated in our cohort of rhabdoid clear cell renal cell carcinoma (Supplementary Table 2). All of the significant mutations identified in this cohort have been described previously in clear cell renal cell carcinoma and belong mostly to the family of tumor suppressor genes involved in chromatin remodeling and histone modification located at 3p21-25 (14, 15) as well as 1 case with an mTOR mutation. No mutations in other known driver genes in clear cell renal cell carcinoma were detected in this cohort—eg. PIK3CA, KDM5C, TP53, PTEN or TCEB.
Our data represent the most extensive mutational examination of this aggressive subgroup of renal cell carcinoma cases (Tables 2A and 2B and Supplementary Table 2). VHL was mutated in 63% of patients with high mutation frequencies in SETD2 (63%) and BAP1 (38%) and a low PBRM1 (13%) mutation rate. The BAP1 and PBRM1mutations were mutually exclusive; no BAP1/PBRM1 double mutant tumors were identified.
Table 2A.
Driver mutations in RCC detected relative to regional histologic grade
| Gene | Regional histologic grade
|
Patients (n=8) | ||||
|---|---|---|---|---|---|---|
| Rhabdoid | Sarcomatoid | Epithelioid | ||||
|
| ||||||
| R (n=8) | Sr (n=3) | G2 (n=3) | G3 (n=4) | G4 (n=1) | ||
| VHL | 5 (63%) | 3 (100%) | 2 (67%) | 3 (75%) | 0 (0%) | 5 (63%) |
| PBRM1 | 1 (13%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (13%) |
| SETD2 | 4 (50%) | 2 (67%) | 1 (33%) | 2 (50%) | 1 (100%) | 5 (63%) |
| BAP1 | 3 (38%) | 2 (67%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (38%) |
| mTOR | 0 (0%) | 1 (33%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (13%) |
Table 2B.
Somatic mutation profiles in rhabdoid, epithelioid and sarcomatoid components of ccRCC
| Case | Region | Gene | Location | Type |
|---|---|---|---|---|
| E | Rhabdoid | |||
| THBS1 | Exon 4 | SNV | ||
| ROS1 | Splice Site | SNV | ||
| Epithelioid_G2 | ||||
| THBS1 | Exon 4 | SNV | ||
| ROS1 | Splice Site | SNV | ||
| C | Rhabdoid | |||
| VHL | Exon 3 | Indel | ||
| VHL | Exon 3 | SNV | ||
| BAP1 | Exon 14 | Indel | ||
| PTPRD | Exon 46 | SNV | ||
| Epithelioid_G2 | ||||
| VHL | Exon 3 | Indel | ||
| VHL | Exon 3 | SNV | ||
| DPYD | Exon 22 | SNV | ||
| D | Rhabdoid | |||
| SETD2 | Exon 3 | SNV | ||
| Epithelioid_G3 | ||||
| SETD2 | Exon 3 | SNV | ||
| ZNF384 | Exon 6 | Indel | ||
| G | Rhabdoid | |||
| VHL | Exon 3 | SNV | ||
| Epithelioid_G3 | ||||
| VHL | Exon 3 | SNV | ||
| F | Rhabdoid | |||
| PAX8 | Exon 9 | SNV | ||
| TCF12 | Exon 14 | SNV | ||
| CSF1R | Exon 3 | SNV | ||
| PBRM1 | Exon 17 | SNV | ||
| CBL | Exon 11 | SNV | ||
| Epithelioid_G4 | ||||
| TET1 | Exon 4 | SNV | ||
| SETD2 | Exon 3 | SNV | ||
| SETD2 | Exon 3 | SNV | ||
| ERCC5 | Exon 3 | SNV | ||
| H | Rhabdoid | |||
| VHL | Splice Site | SNV | ||
| SETD2 | Exon 16 | SNV | ||
| MLL3 | Exon 16 | SNV | ||
| TET1 | Exon 12 | SNV | ||
| BAP1 | Exon 16 | SNV | ||
| Epithelioid_G3 | ||||
| VHL | Splice Site | SNV | ||
| SMARCA4 | Exon 10 | Indel | ||
| HIF1A | Exon 13 | Indel | ||
| Sarcomatoid | ||||
| VHL | Splice Site | SNV | ||
| MLL3 | Exon 16 | SNV | ||
| BAP1 | Exon 16 | SNV | ||
| SMARCA4 | Exon 10 | Indel | ||
| I | Rhabdoid | |||
| ARID1A | Exon 20 | SNV | ||
| BAP1 | Exon 14 | Indel | ||
| SETD2 | Exon 6 | SNV | ||
| ERCC4 | Exon 11 | SNV | ||
| MYH11 | Exon 4 | SNV | ||
| RNF213 | Exon 29 | SNV | ||
| VHL | Exon 2 | Indel | ||
| PDE4DIP | Exon 15 | SNV | ||
| Epithelioid_G3 | ||||
| ARID1A | Exon 20 | SNV | ||
| VHL | Exon 2 | Indel | ||
| SETD2 | Exon 6 | SNV | ||
| ERCC4 | Exon 11 | SNV | ||
| MYH11 | Exon 4 | SNV | ||
| PDE4DIP | Exon 15 | SNV | ||
| Sarcomatoid | ||||
| ARID1A | Exon 20 | SNV | ||
| BAP1 | Exon 14 | Indel | ||
| SETD2 | Exon 6 | SNV | ||
| VHL | Exon 2 | Indel | ||
| ERCC4 | Exon 11 | SNV | ||
| MYH11 | Exon 4 | SNV | ||
| RNF213 | Exon 29 | SNV | ||
| MTOR | Exon 26 | SNV | ||
| PDE4DIP | Exon 15 | SNV | ||
| J | Rhabdoid | |||
| AFF3 | Exon 19 | SNV | ||
| VHL | Exon 2 | Indel | ||
| SETD2 | Exon 20 | SNV | ||
| PIK3CB | Exon 22 | SNV | ||
| Epithelioid_G2 | ||||
| VHL | Exon 2 | Indel | ||
| SETD2 | Exon 20 | SNV | ||
| PIK3CB | Exon 22 | SNV | ||
| WHSC1 | Exon 17 | SNV | ||
| FGFR4 | Exon 8 | SNV | ||
| DNMT3A | Exon 23 | SNV | ||
| CSMD3 | Exon 7 | SNV | ||
| Sarcomatoid | ||||
| VHL | Exon 2 | Indel | ||
| SETD2 | Exon 20 | SNV | ||
| PIK3CB | Exon 22 | SNV | ||
| ERBB4 | Exon 24 | SNV | ||
| TAF1L | Exon 1 | SNV | ||
Notably, mutation frequencies differ from those reported for clear cell renal cell carcinoma in The Cancer Genome Atlas (14), Memorial Sloan Kettering Cancer Center (16) and Japanese cohorts (15) which showed the following mutation spectra: BAP1 (10.1%, The Cancer Genome Atlas; 7.5%, Japan; 6.3%, Memorial Sloan Kettering); SETD2 (11.6%, The Cancer Genome Atlas; 11.3%, Japan; 7.4%, Memorial Sloan Kettering); PBRM1 (32.9%, The Cancer Genome Atlas; 26.4%, Japan; 30.3%, Memorial Sloan Kettering). However, given that rhabdoid clear cell renal cell carcinomas are aggressive, high-grade tumors and that SETD2 mutations have been correlated with survival in The Cancer Genome Atlas cohort and BAP1 mutations have been correlated to grade, stage and survival in The Cancer Genome Atlas and Memorial Sloan Kettering cohorts, we analyzed in silico the mutational profile of the highest grade tumors using The Cancer Genome Atlas data. We found the incidence of driver mutations in Fuhrman grade 4 clear cell renal cell carcinoma to be as follows: VHL (46.3%), BAP1 (20.9%), PBRM1 (32.8%), SETD2 (16.4%). Only BAP1 showed significantly increased mutation frequency between grade 3 and grade 4 clear cell renal cell carcinoma (P=0.02, chi-squared test); the other drivers showed non-significant changes in mutation frequency (Supplementary Table 3).
Driver mutations are not uniformly distributed across different morphologic regional grades
We sequenced morphologically distinct regions from the tumors of 8 patients, with two samples per patient derived from a lower grade clear cell epithelioid focus and a higher grade rhabdoid focus in each case. In 3 patients, we sequenced an additional region with sarcomatoid histology. We found in 6 (75%) patients that the histologically different tumor foci showed genotypic differences in terms of cancer-related mutations (Table 2B and Supplementary Table 2).
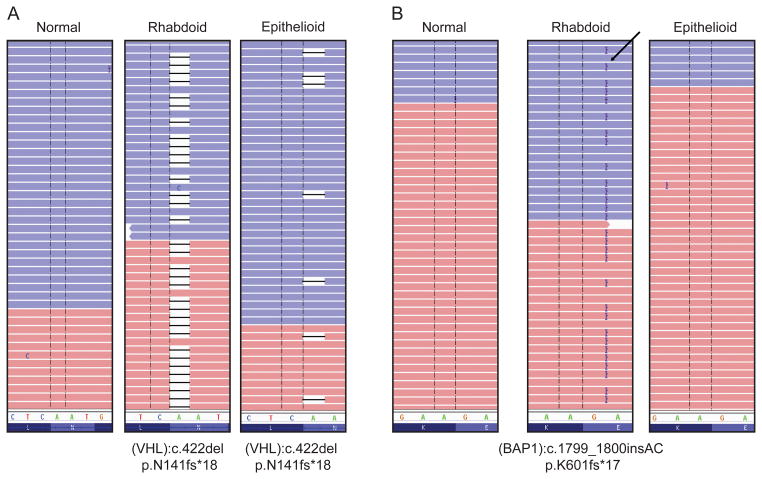
Only the VHL mutation was uniformly distributed among the different regions. Thus, an identical VHL mutation was present in the various morphologic tumor foci for a given patient (Figure 2A), though the allelic frequencies for VHL differed (Supplementary Table 2). The other clear cell renal cell carcinoma driver genes showed a variant pattern of mutations across different histologic regions. BAP1 and PBRM 1 mutations were discordant between the epithelioid and rhabdoid components in 3/3 cases and 1 case, respectively, with mutations identified only in the rhabdoid component. BAP1 mutations were discordant between epithelioid and sarcomatoid components in 2/2 cases, with mutations identified only in the sarcomatoid component (samples H and I, Table 2B and Supplementary Table 2). SETD2 mutations were discordant in 2/5 cases (samples F and H, Table 2B and Supplementary Table 2), with one of the cases showing a mutation only in the epithelioid component and the other case showing a mutation only in the rhabdoid component. Moreover, an mTOR driver mutation was only identified in the sarcomatoid component of one case, but not in the epithelioid or rhabdoid components (sample I, Table 2B and Supplementary Table 2). The differences in detection of driver mutations among different histologic regions in clear cell renal cell carcinoma are illustrated in Figure 2B, Supplementary Figure 2 and Tables 2A and 2B. We also sequenced tumor foci from different regions of 4 cases of clear cell renal cell carcinoma that lacked any rhabdoid or sarcomatoid features. We found genotypic differences in cancer-related mutations in 3/4 patients (75%) and in driver mutations in 2/4 patients (50%) as shown in Supplementary Table 4.
Figure 2. Distinct occurrence patterns of somatic mutations in rhabdoid and epithelioid components as detected by next generation sequencing.
Representative cases have been shown to highlight the different pattern of somatic mutations shared by the two components as detected by 409-gene NGS panel. (A) A single nucleotide deletion in the VHL gene was observed in both rhabdoid and epithelioid components of the tumor. (B) A 2bp insertion in BAP1 gene was observed only in the rhabdoid component but not in the epithelioid component. The absence of the mutation in the paired normals in both the cases confirm their somatic origin.
Rhabdoid renal cell carcinoma is transcriptionally distinct from high stage, high grade renal cell carcinoma
By gene expression profiling, we evaluated rhabdoid clear cell renal cell carcinoma as a group and compared them to advanced stage clear cell renal cell carcinoma (stage III/IV) and high grade clear cell renal cell carcinoma (Fuhrman grades 3 and 4) without rhabdoid or sarcomatoid foci. We found a distinct contrast between rhabdoid clear cell renal cell carcinoma and both advanced stage and high grade clear cell renal cell carcinoma. This was seen by both RNA-seq and microarray platforms on independent samples as illustrated in Figures 3A–D.
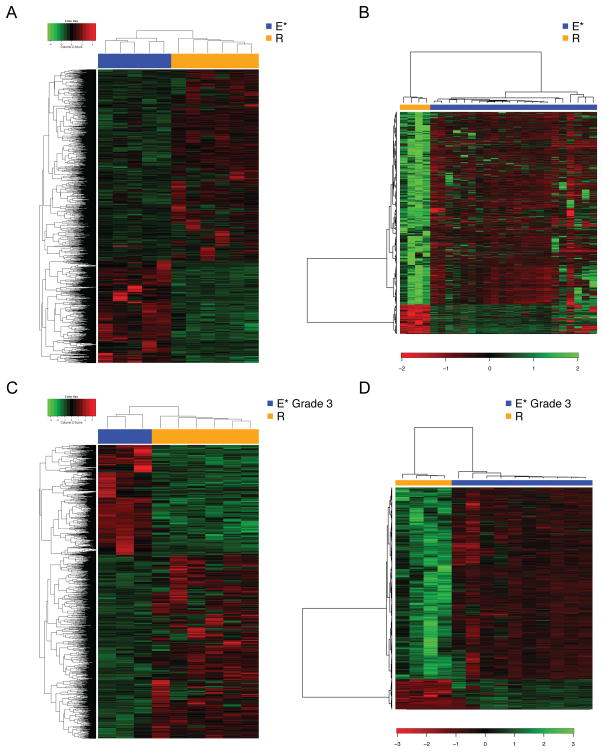
Figure 3. Rhabdoid renal cell carcinoma shows a distinct gene expression signature from that of non-rhabdoid renal cell carcinoma.
The distinctive expression profile of rhabdoid renal cell carcinoma compared with that of advanced-stage (III/IV) non-rhabdoid renal cell carcinoma showed by: (A) an RNA-seq analysis, with a heatmap of the 2976 significant probes in the rhabdoid (E and R) and non-rhabdoid (E*) samples contrasting at a false discovery rate of 0.05; and (B) microarray analysis with a heatmap of the 1669 significant probes in the rhabdoid and non-rhabdoid samples contrasting at a false discovery rate of 0.05. (C) Differential expression profile of rhabdoid renal cell carcinoma compared with that of high grade (Fuhrman grade 3) non-rhabdoid renal cell carcinoma showed by: an RNA-seq analysis, with a heatmap of the 2574 significant probes in the rhabdoid and non-rhabdoid samples contrasting at a false discovery rate of 0.05; and (D) microarray analysis with a heatmap of the 1360 significant probes in the rhabdoid and non-rhabdoid samples contrasting at a false discovery rate of 0.05. Gene expression values were centered before clustering. Samples are ordered by hierarchical clustering (subtype, columns; genes, rows).
When comparing rhabdoid clear cell renal cell carcinoma to renal carcinomas of advanced stage, we identified 2976 genes that were differentially expressed by RNA-seq at a false discovery rate of < 0.05. Rhabdoid clear cell renal cell carcinoma also showed significant differential expression when compared with Fuhrman grade 3 clear cell renal cell carcinoma (2574 genes, false discovery rate < 0.05) and Fuhrman grade 4 clear cell renal cell carcinoma (140 genes, false discovery rate < 0.05). Differentially regulated pathways between rhabdoid and non-rhabdoid clear cell renal cell carcinoma of advanced stage and grade are shown in Supplementary Figure 1.
Gene expression microarray data on independent samples showed a sharp contrast between rhabdoid clear cell renal cell carcinoma versus clear cell renal cell carcinoma of advanced stage (1669 genes, false discovery rate < 0.05) and also compared to Fuhrman grade 3 and grade 4 clear cell renal cell carcinoma (1360 genes and 945 genes, respectively, false discovery rate < 0.05).
Morphologically different components of clear cell renal cell carcinoma share a similar global and prognostic gene expression signature
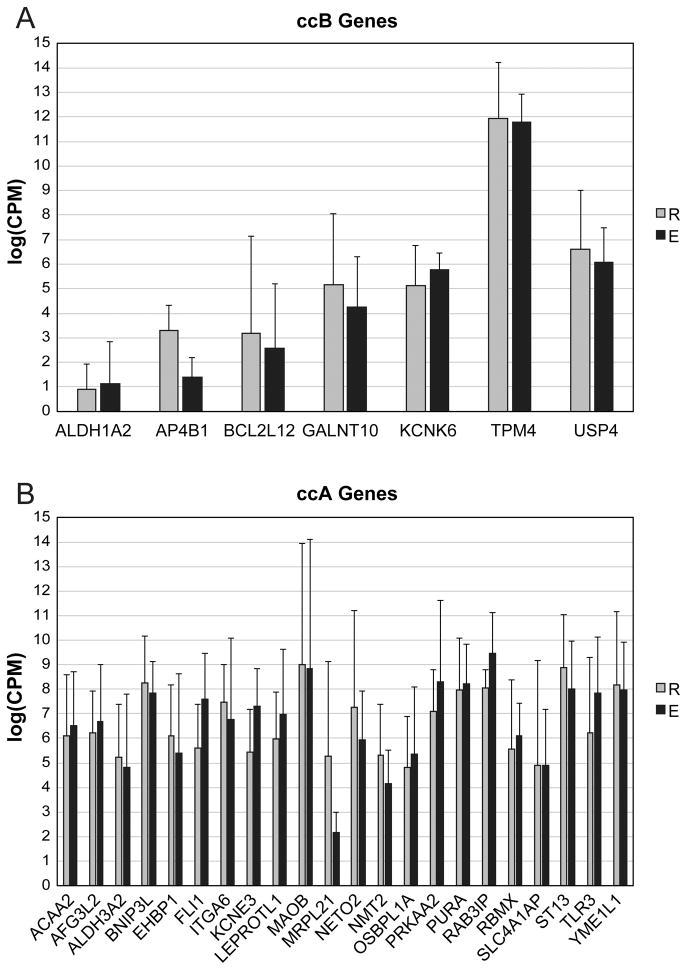
We analyzed RNA-seq based data on 4 cases with respect to the epithelioid and rhabdoid components of rhabdoid clear cell renal cell carcinoma (Table 1 and Supplementary Table 1). Paired analysis of the epithelioid and rhabdoid components yielded relatively few significant genes with low false discovery rate (n=25/8300 at false discovery rate < 0.05). As 3 of these cases also had a defined sarcomatoid component that was macrodissected, we were able to also compare the sarcomatoid and rhabdoid histologies where we again found very few significant differences in gene expression (n=2/8300 genes at false discovery rate < 0.05). These data are illustrated in Supplementary Figure 3. We next performed supervised analysis of the biphasic components of rhabdoid clear cell renal cell carcinoma with respect to good (clear cell type A) and poor (clear cell type B) prognosis genes. By RNA-seq, we found that the epithelioid and rhabdoid components did not show any significant expression differences among the clear cell type A genes. Among the clear cell type B group, only one gene (AP4B1) showed significant elevation (P=0.02, t-test) in the rhabdoid samples (Figure 4). We validated our findings using microarray based gene expression data from 4 independent cases of rhabdoid clear cell renal cell carcinoma in which we did not find any significant differentially expressed genes between the epithelioid and rhabdoid components.
Figure 4. Prognostic gene expression signature of rhabdoid renal cell carcinoma is shared by its clear cell epithelioid and rhabdoid components.
(A) Relative expression of clear cell type B (ccB) poor-prognosis genes in clear cell epithelioid (E) and rhabdoid (R) components demonstrates no significant difference between components for all poor-prognosis genes, except AP4B1 (P<0.02). (B) Relative expression of clear cell type A (ccA) good-prognosis genes in clear cell epithelioid (E) and rhabdoid (R) components demonstrates no significant difference between components in the expression of good-prognosis genes.
DISCUSSION
The incidence of rhabdoid change among all renal cell carcinomas is between 4–7% and rhabdoid change is more commonly observed in clinically advanced renal cell carcinomas. Studies that have specifically addressed this entity are limited with fewer than 150 patients described in the literature (5, 17). This study is the first genome wide examination of rhabdoid clear cell renal cell carcinoma and the first to directly compare the morphologically distinct clear cell epithelioid and rhabdoid components. We did not find any recurrent mutation among our panel of cancer-related genes that was pathognomonic for rhabdoid features in clear cell renal cell carcinoma. Rather, we found frequent VHL mutations that were present in all sampled regions of a given tumor. This confirms previous work (18) where a clonal origin for rhabdoid foci arising from clear cell epithelioid foci was postulated based on an identical pattern of VHL mutations between the rhabdoid and clear cell epithelioid components of clear cell renal cell carcinoma. The uniform mutational pattern of VHL may be exploited clinically in the diagnosis of clear cell renal cell carcinoma if the sampled tumor cells show exclusive rhabdoid morphology that may otherwise suggest a broad differential diagnosis. The overall frequency of SETD2 and BAP1 mutations was higher in this patient cohort than that reported for other Fuhrman grade 4 clear cell renal cell carcinomas (14) in The Cancer Genome Atlas. Higher BAP1 and SETD2 mutation frequencies are consistent with the highly aggressive behavior of rhabdoid clear cell renal cell carcinoma. It must be acknowledged, however, that prior data was based on single biopsy sampling which may have underestimated mutational frequencies. PBRM1, by contrast, was found to be mutated at a significantly lower rate in rhabdoid clear cell renal cell carcinoma compared with Fuhrman grade 4 clear cell renal cell carcinoma from The Cancer Genome Atlas, despite two fold higher sampling per patient in our cohort.
The different pattern of gene expression between rhabdoid clear cell renal cell carcinoma and advanced stage and high grade clear cell renal cell carcinoma lacking rhabdoid foci also underscores the molecular distinctiveness of rhabdoid renal cell carcinoma. Rhabdoid change, traditionally considered as one of the defining features a Fuhrman nuclear grade 4 in renal cell carcinoma, was also recently included as part of the International Society of Urologic Pathologists (ISUP) grade 4 category at the Vancouver consensus conference (19). Our expression data revealed a greater contrast between rhabdoid clear cell renal cell carcinoma and ISUP grade 3 clear cell renal cell carcinoma as compared to grade 4 clear cell renal cell carcinoma, thereby supporting the current grading system.
This is only the second series to evaluate the molecular attributes of different, spatially separate segments of renal cell carcinoma, after the pioneering work of Gerlinger et al. (11, 20) who established the pattern of branched evolution in clear cell renal cell carcinoma by multiregion sequencing of tumor samples. Their studies involved sampling multiple tumor regions from frozen samples of clear cell renal cell carcinoma, acquired in a research setting and profiled using various platforms, including whole exome sequencing. They showed marked intratumoral mutational heterogeneity, with the large majority of mutations not found in all sampled tumor areas. The VHL gene mutation, when present, was the only ubiquitously detected mutation across all tumor regions: this was the so-called truncal mutation that was carried by all clonal subpopulations. These subpopulations had additional mutations that were either “shared” between some—but not all—regions and many mutations that were entirely unique to the region, so-called “private” mutations. The present study complements the work of Gerlinger et al. in that we also found significant intratumoral heterogeneity with only VHL appearing as a truncal type mutation. However, we also explored the mutational landscape of morphologically disparate components of rhabdoid clear cell renal cell carcinoma. Moreover, we better simulated a clinical setting as we used archival formalin-fixed, paraffin-embedded tissues and a Clinical Laboratory Improvement Amendments (CLIA) certified next generation sequencing platform where we assayed only potentially actionable mutations at high sequencing depth. Excepting VHL, we found discordant mutations in relevant driver genes (BAP1, PBRM1, SETD2) across different regions in clear cell renal cell carcinoma. For example, the putatively prognostic BAP1 and PBRM1 mutations would have been missed had we only sampled the clear cell epithelioid tumor region. The sample size in this study is insufficient to establish a definitive association between regional grade and specific mutations. However, our results—drawn from clinical samples and using a CLIA certified platform—suggest that single biopsy approaches for evaluation of mutations are not complete and therefore may not be reliable.
In contrast to the mutational heterogeneity across different regions as well as the reported DNA copy number differences between clear cell and rhabdoid foci (21), we found that the global gene expression pattern between the clear cell epithelioid and rhabdoid histologic components was similar based on unsupervised clustering analysis. Notably, the prognostic gene expression signature of clear cell renal cell carcinoma was stable between the different morphologic areas based on supervised clustering analysis for good (clear cell type A) and poor (clear cell type B) prognosis genes. Rhabdoid clear cell renal cell carcinoma is known to be associated with a very poor outcome, even when the proportion of tumor with rhabdoid histologic features is very small (6, 7). Our results provide an explanation for this observation and argue for a model wherein any percentage of rhabdoid component is potentially ominous since the poor prognosis signature is shared by both clear cell epithelioid and rhabdoid components. Our data also support the idea that the definition of rhabdoid clear cell renal cell carcinoma is not dependent on a minimum percentage of rhabdoid component, since focal rhabdoid change appears to be a marker for the whole tumor.
Clinical and pathologic parameters that are currently used for risk assessment of patients with clear cell renal cell carcinoma are of limited value in the pre-operative setting. For example, radiologic staging and biopsy grading of renal masses is often inaccurate (16, 22). Molecular biomarkers that have shown potential in stratifying patients into prognostic groups include: a) mutations in BAP1 (and possibly SETD2) as markers of tumor aggressiveness; and b) the clear cell type A and clear cell type B gene expression signatures denoting good and poor prognosis tumors, respectively. Although a mutational screen appears straightforward and a potentially attractive ancillary test, our study exposes the limitations of single biopsy approaches in mutational profiling of even a few genes from clinical clear cell renal cell carcinoma tissues. Gene expression, by contrast, was less prone to sampling bias across the different histologic foci. Notably, among 28 evaluated biomarkers in a recent analysis, only the clear cell type B expression signature was significant on multivariate analysis in predicting for worse disease specific survival in clear cell renal cell carcinoma patients (23).
In conclusion, we have described the molecular distinctiveness of rhabdoid clear cell renal cell carcinoma and shown that driver mutations, excepting VHL, are not uniformly distributed across different histologic regions of this tumor. Renal cell carcinoma samples for molecular profiling should be drawn from different regions and should incorporate assessment of the prognostic gene expression signature, which is relatively stable across different morphologic areas in clear cell renal cell carcinoma.
Supplementary Material
Acknowledgments
We would like to acknowledge Kim-Anh Vu, Chi-Wan Chow, Stephanie Garza, and Camille Sanchez for technical and secretarial assistance. This paper is supported in part by the Monteleone foundation and institutional funds from MD Anderson Cancer Center (K.S.) and NIH P30 CA125123 (CJC).
Footnotes
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Delahunt B, McKenney JK, Lohse CM, et al. A novel grading system for clear cell renal cell carcinoma incorporating tumor necrosis. Am J Surg Pathol. 2013;37:311–322. doi: 10.1097/PAS.0b013e318270f71c. [DOI] [PubMed] [Google Scholar]
- 3.Delahunt B. Advances and controversies in grading and staging of renal cell carcinoma. Mod Pathol. 2009;2:S24–36. doi: 10.1038/modpathol.2008.183. [DOI] [PubMed] [Google Scholar]
- 4.Gelb AB, Shibuya RB, Weiss LM, et al. Stage I renal cell carcinoma. A clinicopathologic study of 82 cases. Am J Surg Pathol. 1993;17:275–286. doi: 10.1097/00000478-199303000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Chapman-Fredricks JR, Herrera L, Bracho J, et al. Adult renal cell carcinoma with rhabdoid morphology represents a neoplastic dedifferentiation analogous to sarcomatoid carcinoma. Ann Diagn Pathol. 2011;15:333–337. doi: 10.1016/j.anndiagpath.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Kuroiwa K, Kinoshita Y, Shiratsuchi H, et al. Renal cell carcinoma with rhabdoid features: an aggressive neoplasm. Histopathology. 2002;41:538–548. doi: 10.1046/j.1365-2559.2002.01427.x. [DOI] [PubMed] [Google Scholar]
- 7.Leroy X, Zini L, Buob D, et al. Renal cell carcinoma with rhabdoid features: an aggressive neoplasm with overexpression of p53. Arch Pathol Lab Med. 2007;131:102–106. doi: 10.5858/2007-131-102-RCCWRF. [DOI] [PubMed] [Google Scholar]
- 8.Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. J Clin Oncol. 2014;32:1968–1976. doi: 10.1200/JCO.2012.45.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brannon AR, Reddy A, Seiler M, et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer. 2010;1:152–163. doi: 10.1177/1947601909359929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brannon AR, Haake SM, Hacker KE, et al. Meta-analysis of clear cell renal cell carcinoma gene expression defines a variant subgroup and identifies gender influences on tumor biology. Eur Urol. 2012;61:258–268. doi: 10.1016/j.eururo.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh RR, Patel KP, Routbort MJ, et al. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer. 2014;111:2014–2023. doi: 10.1038/bjc.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 16.Hakimi AA, Ostrovnaya I, Reva B, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19:3259–3267. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Przybycin CG, McKenney JK, Reynolds JP, et al. Rhabdoid differentiation is associated with aggressive behavior in renal cell carcinoma: a clinicopathologic analysis of 76 cases with clinical follow-up. Am J Surg Pathol. 2014;38:1260–5. doi: 10.1097/PAS.0000000000000251. [DOI] [PubMed] [Google Scholar]
- 18.Shannon B, Stan Wisniewski Z, Bentel J, et al. Adult rhabdoid renal cell carcinoma. Arch Pathol Lab Med. 2002;126:1506–10. doi: 10.5858/2002-126-1506-ARRCC. [DOI] [PubMed] [Google Scholar]
- 19.Delahunt B, Cheville JC, Martignoni G, et al. The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am J Surg Pathol. 2013;37:1490–1504. doi: 10.1097/PAS.0b013e318299f0fb. [DOI] [PubMed] [Google Scholar]
- 20.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perrino CM, Hucthagowder V, Evenson M, et al. Genetic alterations in renal cell carcinoma with rhabdoid differentiation. Hum Pathol. 2015;46:9–16. doi: 10.1016/j.humpath.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Volpe A, Finelli A, Gill IS, et al. Rationale for percutaneous biopsy and histologic characterisation of renal tumours. Eur Urol. 2012;62:491–504. doi: 10.1016/j.eururo.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Gulati S, Martinez P, Joshi T, et al. Systematic Evaluation of the Prognostic Impact and Intratumour Heterogeneity of Clear Cell Renal Cell Carcinoma Biomarkers. Eur Urol. 2014;66:936–948. doi: 10.1016/j.eururo.2014.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.