Abstract
Objective
We sought to employ an innovative tool based on common biological pathways to identify specific phenotypes among women with spontaneous preterm birth (SPTB), in order to enhance investigators' ability to identify to highlight common mechanisms and underlying genetic factors responsible for SPTB.
Study Design
A secondary analysis of a prospective case-control multicenter study of SPTB. All cases delivered a preterm singleton at SPTB ≤34.0 weeks gestation. Each woman was assessed for the presence of underlying SPTB etiologies. A hierarchical cluster analysis was used to identify groups of women with homogeneous phenotypic profiles. One of the phenotypic clusters was selected for candidate gene association analysis using VEGAS software.
Results
1028 women with SPTB were assigned phenotypes. Hierarchical clustering of the phenotypes revealed five major clusters. Cluster 1 (N=445) was characterized by maternal stress, cluster 2 (N=294) by premature membrane rupture, cluster 3 (N=120) by familial factors, and cluster 4 (N=63) by maternal comorbidities. Cluster 5 (N=106) was multifactorial, characterized by infection (INF), decidual hemorrhage (DH) and placental dysfunction (PD). These three phenotypes were highly correlated by Chi-square analysis [PD and DH (p<2.2e-6); PD and INF (p=6.2e-10); INF and DH (p=0.0036)]. Gene-based testing identified the INS (insulin) gene as significantly associated with cluster 3 of SPTB.
Conclusion
We identified 5 major clusters of SPTB based on a phenotype tool and hierarchal clustering. There was significant correlation between several of the phenotypes. The INS gene was associated with familial factors underlying SPTB.
Keywords: Spontaneous preterm birth, phenotype, cluster analysis, gene-based analysis
Introduction
Spontaneous preterm birth (SPTB) remains the leading cause of morbidity and mortality1 in non-anomalous newborns, yet our understanding of the causes of SPTB is limited. This is, in part, because SPTB is a multifactorial condition with multiple etiologies and likely results from specific interactions between the environment and genetic factors.2-6 There is support of a genetic component to SPTB suggested by the presence of racial disparities that persist despite controlling for multiple risk factors.7 In addition, there is a strong risk for recurrence of SPTB in women with a personal history of SPTB in a previous pregnancy.8,9 In addition, a clear familial predisposition has been demonstrated.10 Finally, twin studies support the role of genetic risk factors in preterm birth by estimating the heritability at 20 to 40 percent.11
Efforts to identify the genetic causes of SPTB have produced overall disappointing results. A recent large genome wide association (GWA) study of SPTB identified specific single nucleotide polymorphisms (SNPs) that were associated with SPTB but these could not be subsequently validated.12 One attempt to summarize the genetic contribution to SPTB concluded that no robustly validated genetic variants contributing to this complex disease process have been identified.13 This lack of success is likely due, at least in part, to inadequate phenotyping of SPTB cases, the heterogeneity of the disease process, differences amongst patient populations, or a combination of these factors.
The Genomic and Proteomic Network for Preterm Birth Research (“GPN-PBR”, abbreviated GPN) was established by the Eunice Kennedy Shriver National Institute for Child Health and Human Development to study the genetic and environmental etiologies, and with a goal of deciphering mechanisms underlying SPTB. Accurate and precise phenotypes were needed to accomplish this goal. We have previously created a unique phenotyping tool using clinical features present at the time of delivery to define nine phenotypes suggestive of underlying etiologies of SPTB. We applied the phenotype tool to more than 1000 women with SPTB, were able to classify over 95% of women into one or more phenotype categories, and demonstrated that most cases of SPTB have evidence of two or more phenotypes present and that phenotypes vary by gestational age at delivery and by race.14 Assigning a phenotype that suggests similar underlying etiology or etiologies for SPTB among a group of women will likely result in an enhanced ability to identify genes or pathways associated with that phenotype.
We hypothesized that associations exist between SPTB phenotypes that highlight common mechanisms responsible for SPTB will enhance our ability to identify the underlying genetic factors responsible for this complication. We further hypothesized that cluster analysis using sub-categories within phenotypes might identify subsets of women with a similar genetic risk for SPTB. We sought to test this by evaluating candidate genes that might be associated with SPTB among one of the subsets identified.
Materials and Methods
This is a secondary analysis of a multicenter, prospectively cohort women enrolled in the GPN case-control study.
Patient Recruitment
Women with SPTB and matched uncomplicated term controls were prospectively recruited from November 2007 through January 2011 across eight clinical sites including the University of Utah / Intermountain Healthcare, University of Texas Medical Branch – Galveston, University of Alabama at Birmingham, Columbia University, Northwestern University, University of Texas – Houston, University of North Carolina – Chapel Hill, and Brown University. This study was approved by the Institutional Review Board at each center, and a written informed consent was obtained from all participants.
Women were included in the study if they experienced a preterm birth of a singleton pregnancy between 20 0/7 and 33 6/7 weeks gestation following spontaneous labor. The inclusion criteria for the study have been published previously.12
Women were excluded from the study if they were diagnosed with a stillbirth prior to presentation to labor and delivery or if they needed an indicated delivery for maternal or fetal complications. Women who experienced an intrapartum stillbirth or who had spontaneous labor in addition to maternal or fetal complications were not excluded.
A control group was also collected consisting of women who experienced a singleton live birth after spontaneous labor at 39 weeks or greater. Controls were excluded if they had a history of a prior pregnancy complicated by SPTB. Controls were used only for the analysis of candidate genes.
Data Collection
Clinical and demographic data were collected for cases and controls by trained research nurses using in-person interviews prior to hospital discharge whenever possible. All interviews and abstraction of medical records were performed within 14 days of delivery. Data collected included demographics, medical, social, family, and obstetric history, obstetric course and complications during the current pregnancy. Patients also completed validated questionnaires to assess factors such as anxiety (Beck anxiety index), depression (Beck depression inventory), perceived stress (Perceived stress scale), and attitude of the subject and partner with respect to pregnancy.
Cluster Analysis
A phenotyping tool was designed by the authors (MSE, TAM, MWV) that grouped maternal social, demographic, family history, and obstetric factors into SPTB categories.14 (See Table 1) Category clinical factors were classified into levels of evidence as providing “strong”, “moderate”, and “possible” evidence of the phenotype. Cluster analysis was used to classify 1028 unique SPTB cases. The data included binary indicator variables for several phenotypes relating to SPTB including Infection/inflammation, maternal stress (Hypothalmus-Pituitary Axis (HPA) activation), decidual hemorrhage, uterine distension, cervical insufficiency, preterm premature rupture of membranes (PPROM), placental dysfunction, maternal comorbidities, and familial phenotypes. There were two or three levels of evidence for each of the phenotypes. Identification of one level of evidence of a specific phenotype was not mutually exclusive for the other levels of evidence for the same phenotype. For example, one subject might have strong, moderate and possible evidence for one or more phenotypes. It is possible that the true presence of a phenotype may be more likely in women who had more than one indicator of the phenotype. Thus, this information was used to calculate a “weighted” score for each factor. Three points were given for “strong” evidence, 2 points for “moderate” evidence, and 1 point for “possible” evidence for each phenotype. Thus a subject with evidence from each of the categories “strong”, “moderate” and “possible” for a particular phenotype would receive six points for that phenotype. The maximum score any individual could receive for each phenotype was therefore 6 points. There was no limit to the number of phenotypes or levels of evidence that each woman could be assigned, provided she met criteria.
Table 1.
Phenotyping tool used for analysis. Characteristics are labeled as “Strong”, “Moderate” and “Possible” evidence of each phenotype. The tool was applied to all women with SPTB and each subject could thus have Strong, Moderate and/or Possible evidence of more than one phenotype.
| Phenotype | Strong Evidence | Moderate Evidence | Possible Evidence |
|---|---|---|---|
| Infection / Inflammationa |
|
|
|
| Decidual Hemorrhagea |
|
|
|
| Maternal Stress |
|
|
|
| Cervical Insufficiency |
At least one pregnancy loss prior to 24 weeks gestation due to painless cervical dilation |
|
|
| Uterine Distensiona | n/a |
|
|
| Placental Dysfunctiona |
|
|
|
| Preterm premature rupture of membranes |
|
|
|
| Maternal Comorbidities |
|
|
n/a |
| Familial |
|
|
|
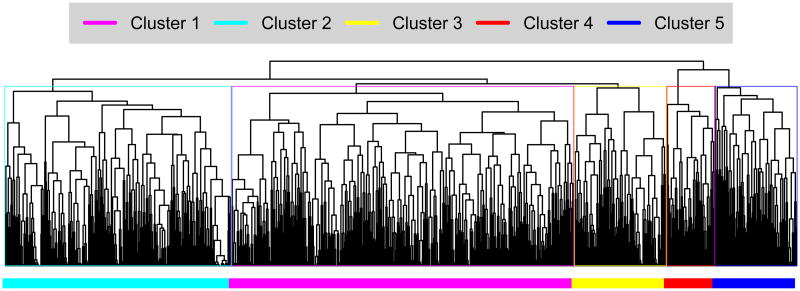
Cluster analysis incorporated demographic variables (including maternal age, race, Hispanic ethnicity, educational attainment, marital status and nulliparity), binary indicators for each level of phenotypic evidence and the weighted score for each phenotype category. Using these variables as input, a sample dissimilarity matrix was generated using the “Daisy” method in the R “Cluster” package. Chi-square analysis was performed to evaluate potential correlation among specific phenotypes.15,16 Figure 1. Illustrates the clustering of each individual included in the analysis.
Figure 1.
Hierarchical clustering of 1028 women with SPTB. The samples were divided into five main clusters for further analysis, as indicated by colors in the figure.
Candidate Gene Analysis
Once cluster analysis was complete, one cluster (cluster 3) was selected to use for gene-based analysis. We chose this cluster because it contained women with a strong familial phenotype and we thought it likely that they might have a genetic contribution to their SPTB. The women within the sample cluster were compared to 717 term controls.
All cluster cases and term controls had biologic samples collected at the time of their delivery and DNA was subsequently extracted for all study subjects. Genotypes for 905,682 SNPs were generated with the Affymetrix SNP 6.0 genotyping array as previously described.12 For the present study, genotype data were downloaded from dbGaP in binary PLINK format. The files contained genotypes for 1419 individual mothers including 702 with at least one SPTB and 717 women with no history of PTB. Quality assurance testing was done to identify an appropriate set of samples and SNPs for use in association testing. The samples were screened for sex discrepancies, sample duplications, and high Mendelian error rates. Principal components analysis (PCA) was performed to ascertain population stratification within the data and to confirm the reported ancestry of individuals in the study. Identity-by-descent (IBD) estimates were calculated to assess relationships between all pairs of samples. Samples were removed from the analysis if the mean pair-wise IBD value compared to all other samples was greater than 0.04. Samples with autosomal SNP call rates less than 0.95 were also excluded from further analysis. SNPs with call rates less than 0.95, minor allele frequency less than 0.005, or significant departure from Hardy-Weinberg equilibrium (p<5 × 10-8 in non-Hispanic Caucasian controls) were removed from analysis. A total of 841,350 SNPs on chromosomes 1-22 and the X chromosome passed all criteria used for association testing.
Gene-based testing
Genotype association tests were performed to compare SPTB cases in cluster 3 to the non-SPTB controls. All tests were performed with Golden Helix SNP and Variation Suite software version 8.1. Significance was tested with logistic regression, assuming an additive genetic model. Results were adjusted for three principal components to account for population structure and ethnic stratification within the data. The output of the SNP tests was processed with the VEGAS program to generate gene-level association test results.17 The VEGAS program compiles the significance of all SNPs in or near each gene to determine the significance of the entire gene region using a simulation procedure. The test for each gene includes all SNPs within 50kb of the gene, thereby capturing most cis regulatory regions and other important features in the region of the gene. VEGAS combines the significance of individual SNPs using a linkage disequilibrium (LD) model to determine the expected correlation patterns within the gene. Several independent SNP associations within a gene may thus be combined to assess the overall significance of the gene region.
966 genes from previously identified inflammatory pathways were selected for evaluation in this candidate gene analysis.18 We chose to use genes from inflammatory pathways because inflammation is a common underlying mechanism for multiple etiologies of SPTB. Based on the number of tests required to evaluate 966 genes, a p-value of about 5e-5 was required to declare significance in the analysis.
Results
We applied the phenotyping tool to 1,028 women with SPTB. Hierarchical clustering of the dissimilarity matrix using R revealed five major data clusters. Clusters can be visualized in Figure 1. Cluster 1 (n=445) is characterized by “maternal stress” (HPA activation), cluster 2 (n=294) by premature membrane rupture, cluster 3 (n=120) by familial factors, and cluster 4 (n=63) by maternal comorbidities. Cluster 5 (n=106) is multifactorial, characterized by infection, decidual hemorrhage and placental dysfunction. Significant co-occurrence was observed between these three phenotypic categories. Chi-square analysis shows correlation between placental dysfunction and decidual hemorrhage (p<2.2e-6), placental dysfunction and inflammation/infection (p=6.2e-10), and between inflammation/infection and decidual hemorrhage (p=0.0036).
We chose to use cluster 3 for additional gene based testing since this cluster was characterized primarily by women with a family history of SPTB. The seventy-eight women with genotyping data available from dbGaP from cluster 3 were compared to 717 term controls. Demographic information for the cases and controls is found in Table 2. There are notable differences in maternal race, education level and marital status likely due to the fact that these variables were part of the hierarchical clustering. Gene test results were adjusted for race using PCA.
Table 2.
Comparison of demographic information between cases from Cluster 3 and control subjects. There is a significant difference in races between the two groups. All gene association tests are adjusted for principal components in order to account for racial differences.
| Cases in Cluster 3 | Controls | p-value | |
|---|---|---|---|
| N | 78 | 717 | |
| Age (mean, SD) | 25.6, 5.5 | 25.5, 5.7 | 0.897 |
| Caucasians | 66 (84.6%) | 486 (67.9%) | 0.003 |
| African Americans | 8 (10.3%) | 169 (23.6%) | 0.011 |
| Hispanic (any race) | 10 (12.8%) | 133 (18.6%) | 0.273 |
| Education=4 (13-16 years) | 41 (52.6%) | 313 (43.7%) | 0.166 |
| Education=3 (9-12 years) | 34 (43.6%) | 347 (48.5%) | 0.492 |
| Married/living with partner | 47 (60.3%) | 400 (55.9%) | 0.525 |
| Never married, living with partner | 14 (17.9%) | 118 (16.5%) | 0.860 |
| Never married, not living with partner | 12 (15.4%) | 174 (24.3%) | 0.105 |
The insulin (INS) gene (p=3.8e-5) was significant in the cluster-3 analysis. The result is based on the combined evidence from 34 SNPs located in or near the INS gene. None of the SNPs were individually significant after multiple test adjustment.
Comment
We have identified five major clusters of women with SPTB using precise phenotyping and clustering tools. There was significant correlation between several of the phenotypes indicating the possibility of common mechanisms underlying these groups. We hypothesized that use of this approach would identify a SPTB cluster that may be associated with a genotype and indeed we found a significant difference in the INS gene, which had a group of 34 SNPs that were significantly different between cluster cases in which women either had recurrent SPTB or a strong family history of PTB and controls.
We found significant co-occurrence among placental dysfunction, decidual hemorrhage and the inflammation/infection pathways. The decidual hemorrhage and inflammation/infection pathways have previously been identified in the same population of Caucasian women with SPTB.19 Thrombosis within the decidua and placenta is often associated with an acute inflammatory process, and therefore it is not surprising that thrombosis is seen in conjunction with inflammation more frequently in SPTB.20 In addition, one study reported that findings of inflammation and/or hemorrhage in placental pathology at the time of SPTB are associated with an increased risk of recurrent SPTB in subsequent pregnancies.21 It is therefore logical to find a cluster that includes all three clinical presentations.
Previous attempts to identify the genetic causes of SPTB have met with only modest success.13 In fact, a recent GWA study performed on the women included in this analysis identified only a few maternal and fetal SNPs associated with SPTB, despite relatively large numbers of prospectively collected cases and controls.12 This may be due to insufficient numbers of cases and controls and thus inadequate power to detect the more subtle differences that exist between the groups. An alternative hypothesis is that the phenotypes that are currently used to define cases and controls are too broad, do not identify groups of women who share similar etiologies for their SPTB, and thus are not likely to identify common genetic causes. Several authors have recently proposed more sophisticated approaches to improve the assignment of phenotype to cases of preterm birth.22,23 We have previously described a phenotyping system that was used to evaluate the SPTBs that occurred in the GPN case control study.14 Our phenotyping tool improves on other previously proposed PTB classification systems by providing more specific classification yet was applied with the use of readily available data. Use of this system demonstrated that most women had evidence of 2 or more phenotypes confirming the complex nature of SPTB. Previous reports of increased risk of recurrent SPTB even after a prior indicated PTB highlight the potential for overlap in the pathophysiology responsible different type of PTB.9,24
In order to further refine the phenotype of SPTB, in the current study we used hierarchal clustering to identify women with common clinical characteristics that are not limited to the pre-defined phenotypes of the original tool. The hierarchal cluster analysis found five separate groups of women with one or more common clinical features and thus provides another opportunity to identify genetic factors associated with specific subsets of SPTB.
We chose the cluster with family history of SPTB as a predominant feature (cluster 3) for genetic analyses. We hypothesized that women with a strong personal history (recurrent SPTB) or family history of SPTB would be more likely to have inherited a genetic factor that would increase their risk of SPTB.8,25 We chose a previously reported comprehensive set of inflammatory genes as the candidates for our candidate gene analysis.18 The INS gene, which is found on chromosome 11 and is known to be involved in the MAPK and in the NF-kB signaling pathways,18 was significantly associated with SPTB among women in cluster 3. Both the MAPK signaling and NFkB signaling pathways play important roles in the inflammatory response and have been previously implicated in the pathogenesis of SPTB.26,27 However, the exact contribution of the INS gene to SPTB remains to be evaluated.
There are several limitations to this study. First, some of the clinical information used by the phenotyping tool was absent in some of the women with SPTB. For example, placental pathology was not available for every subject. Placental pathology after a SPTB is a powerful tool for elucidating the underlying etiology of the delivery. There is a well-characterized association between SPTB and the histologic presence of chorioamnionitis, with funisitis and vasculitis being particularly associated with deliveries at increasingly early gestational ages.28 In addition, the identification of phenotypes of decidual hemorrhage and placental dysfunction would be enhanced by routine pathologic evaluation of the placenta in cases of SPTB and these findings may be associated with the risk of recurrent SPTB.21 It is clear that this phenotyping approach would be enhanced if all pertinent data were available for all subjects, however, the absence of some data would lead to under-classification of subjects and would make the identification of genetic contributors more difficult. There are likely some number of confounders, such as environmental exposures, that were not assessed. Separation of women into clusters was a second limitation in that it resulted in a reduction of the number of subjects available for the candidate gene analysis and therefore reduced the power of this study to identify genes with less robust association. Our study was also limited to the maternal genetic contribution to SPTB. Finally, we have chosen to focus on inflammatory genes and it is possible that other genes from other pathways are associated with SPTB. Future studies should assess the role of fetal genotype, as well as other pathways and other clusters of SPTB phenotypes in the genetic causes of SPTB.
Despite the limitations, this study has taken a novel, granular approach to the phenotyping of women who experience SPTB and who are thus more likely to share common etiologies and common genetic predispositions. We have identified one gene that was associated with women who have SPTB with a strong familial history. We recognize that the interaction between a pregnant woman's environment and her personal biological responses is complex and that advances in our understanding of these relationships is an ongoing, iterative process. However, we believe that the use of hierarchical clustering generates unbiased phenotyping information, which may enhance our understanding of the pathways leading to SPTB. Future studies may address the genetic similarities of the other clusters identified in this study providing new insight into the phenotypes that cluster together based on clinical characteristics. In addition, this type of evaluation may be used to assess recurrence risks in future pregnancies or neonatal outcomes among women with similar clinical phenotypes.
Figure 2.
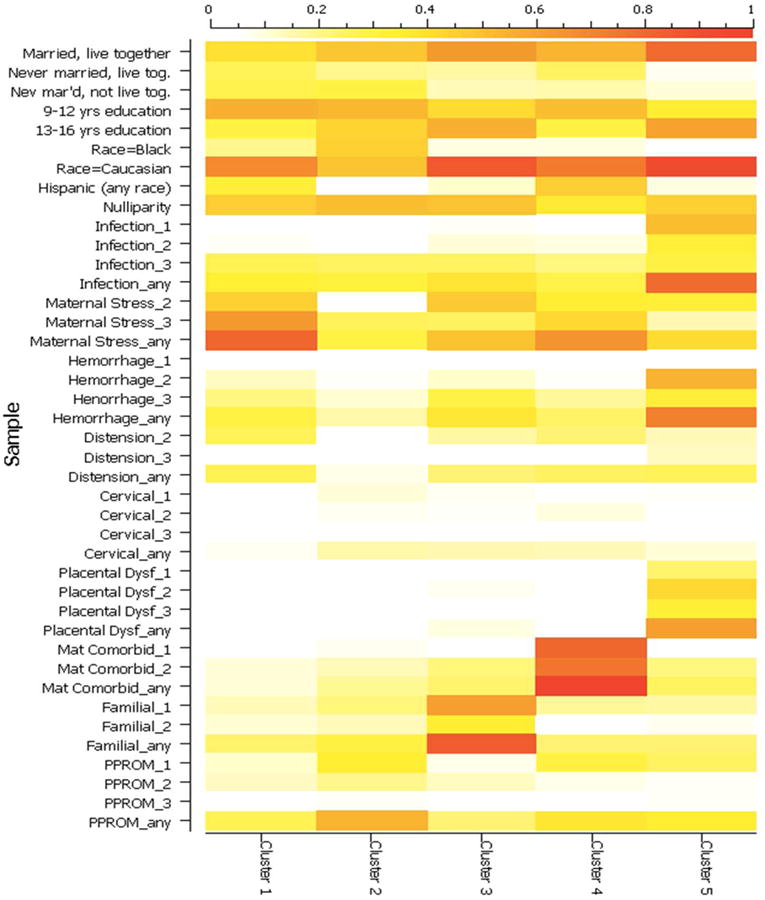
Graphic representation of phenotype distributions for samples in each cluster. The coloring of each cell in the figure indicates the proportion of samples in the cluster that are positive for the specified variable. For example, the mean of “maternal stress_any” for Cluster 1 is 0.81, indicating that 81% of samples in the cluster are positive for some level of HPA activation. The color gradient is defined in the legend on the top edge of the figure. The labels on the left side represent the different levels of evidence from each of the phenotype categories that were included in the final cluster analysis. The first word indicates the phenotype category and the number represents the level of evidence within that category (1=Strong, 2= Moderate, and 3=Possible).
Table 3.
This table demonstrates the raw values for each of the variables assessed in each cluster. The number in each space represents the percentage of women in each cluster that were found to have each specific characteristic. For example, 40.2% of women in cluster 1 were married and living with their partner (See Column one, Row 1)
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | |
|---|---|---|---|---|---|
| Married, live together | 40.2% | 48.6% | 62.5% | 54.0% | 78.3% |
| Never married, live tog. | 26.3% | 19.0% | 16.7% | 25.4% | 7.5% |
| Never mar'd, not live tog. | 28.1% | 29.9% | 14.2% | 15.9% | 10.4% |
| 9-12 yrs education | 54.8% | 53.1% | 40.8% | 50.8% | 34.0% |
| 13-16 yrs education | 30.3% | 43.2% | 55.8% | 30.2% | 60.4% |
| Race=Black | 19.1% | 44.9% | 9.2% | 9.5% | 0.9% |
| Race=Caucasian | 68.8% | 49.3% | 85.0% | 74.6% | 92.5% |
| Hispanic (any race) | 31.7% | 5.8% | 11.7% | 46.0% | 9.4% |
| Nulliparity | 45.4% | 51.0% | 49.2% | 36.5% | 44.3% |
| Infection_1 | 3.8% | 2.0% | 6.7% | 0.0% | 50.9% |
| Infection_2 | 6.3% | 5.8% | 10.0% | 9.5% | 32.1% |
| Infection_3 | 27.2% | 25.5% | 25.0% | 20.6% | 30.2% |
| Infection_any | 33.3% | 31.0% | 38.3% | 28.6% | 78.3% |
| Maternal Stress_2 | 44.7% | 5.1% | 47.5% | 34.9% | 32.1% |
| Maternal Stress_3 | 63.1% | 25.9% | 25.0% | 42.9% | 15.1% |
| Maternal Stress_any | 80.7% | 29.9% | 49.2% | 65.1% | 41.5% |
| Hemorrhage_1 | 0.4% | 0.0% | 0.0% | 0.0% | 0.9% |
| Hemorrhage_2 | 12.8% | 6.1% | 11.7% | 6.3% | 53.8% |
| Henorrhage_3 | 20.0% | 11.2% | 29.2% | 17.5% | 32.1% |
| Hemorrhage_any | 30.3% | 15.3% | 37.5% | 23.8% | 71.7% |
| Distension_2 | 26.1% | 3.4% | 16.7% | 22.2% | 14.2% |
| Distension_3 | 1.6% | 4.4% | 5.8% | 4.8% | 13.2% |
| Distension_any | 26.7% | 7.8% | 22.5% | 25.4% | 26.4% |
| Cervical_1 | 4.0% | 10.2% | 7.5% | 4.8% | 6.6% |
| Cervical_2 | 2.9% | 7.1% | 6.7% | 9.5% | 2.8% |
| Cervical_3 | 1.6% | 1.4% | 3.3% | 3.2% | 4.7% |
| Cervical_any | 7.4% | 16.0% | 15.0% | 14.3% | 10.4% |
| Placental Dysf_1 | 0.7% | 0.3% | 4.2% | 3.2% | 23.6% |
| Placental Dysf_2 | 2.5% | 1.4% | 7.5% | 4.8% | 42.5% |
| Placental Dysf_3 | 3.1% | 0.7% | 3.3% | 0.0% | 33.0% |
| Placental Dysf_any | 4.9% | 2.0% | 9.2% | 4.8% | 61.3% |
| Mat Comorbid_1 | 1.1% | 7.1% | 4.2% | 81.0% | 5.7% |
| Mat Comorbid_2 | 10.3% | 13.9% | 21.7% | 76.2% | 20.8% |
| Mat Comorbid_any | 10.6% | 18.4% | 23.3% | 100.0% | 24.5% |
| Familial_1 | 13.9% | 21.4% | 60.8% | 17.5% | 17.0% |
| Familial_2 | 10.8% | 13.9% | 35.8% | 4.8% | 7.5% |
| Familial_any | 23.4% | 29.9% | 84.2% | 22.2% | 22.6% |
| PPROM_1 | 12.1% | 34.4% | 8.3% | 30.2% | 25.5% |
| PPROM_2 | 13.3% | 19.0% | 12.5% | 7.9% | 6.6% |
| PPROM_3 | 3.6% | 6.8% | 3.3% | 3.2% | 6.6% |
| PPROM_any | 26.5% | 53.7% | 22.5% | 38.1% | 34.0% |
Acknowledgments
The National Institutes of Health, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and the National Center for Advancing Translational Sciences provided grant support for the NICHD Genomics and Proteomics Network for Preterm Birth (GPN). While NICHD staff did have input into the study design, conduct, analysis, and manuscript drafting, the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data collected at participating sites of the GPN were transmitted to Yale University, the data coordinating center (DCC) for the network, which stored, managed and analyzed the data for this study. On behalf of the GPN, Drs. Heping Zhang (DCC Principal Investigator) and Yaji Xu (DCC Statistician) had full access to all the data in the study and take responsibility for the integrity of the data and accuracy of the data analysis.
We are indebted to our medical and nursing colleagues and the infants and their parents who agreed to take part in this study. The following investigators, in addition to those listed as authors, participated in this study:
Steering Committee Chair: Yoel Sadovsky, MD, Magee-Womens Research Institute, University of Pittsburgh, Pittsburgh, PA.
Eunice Kennedy Shriver National Institute of Child Health & Human Development – Stephanie Wilson Archer, MA.
University of Alabama at Birmingham Health System (U01 HD50094, UL1 RR25777) – Rachel L. Copper, MSN CRNP; Pamela B. Files, MSN CRNP; Stacy L. Harris, BSN RN.
University of Pennsylvania (U01 HD5088) – Don A. Baldwin, PhD; Rita Leite, MD.
University of Texas Medical Branch at Galveston (U01 HD50078) – Margaret L. Zimmerle, BSN; Janet L. Brandon, RN MSN; Sonia Jordan, RN BSN; Angela Jones, RN BSN.
University of Utah Medical Center, Intermountain Medical Center, LDS Hospital, McKay-Dee Hospital, and Utah Valley Regional Medical Center (U01 HD50080) – Kelly Vorwaller, RN BSN; Sharon Quinn, RN; Valerie S. Morby, RN CCRP; Kathleen N. Jolley, RN BSN; Julie A. Postma, RN BSN CCRP.
Yale University School of Public Health, Collaborative Center for Statistics in Science (U01 HD50062) – Kei-Hoi Cheung, PhD; Donna Losi DelBasso; Buqu Hu, MS; Lina Jin, PhD; Analisa L. Lin, MPH; Charles C. Lu, MS; Lauren Perley, MA; Laura Jeanne Simone, BA; Chi Song, PhD; Feifei Xiao, PhD; Yaji Xu, PhD.
Alpert Medical School of Brown University, Women & Infants Hospital of Rhode Island – Dwight J. Rouse, MD MSPH; Donna Allard, RNC.
Columbia University Hospital, Drexel University, Christiana Care Health Systems, and St. Peter's University Hospital – Ronald Wapner, MD; Michelle Divito, RN MSN; Sabine Bousleiman, RN MSN MsPH; Vilmarie Carmona, MA; Rosely Alcon, RN BSN; Katty Saravia, MA; Luiza Kalemi, MA; Mary Talucci, RN MSN; Lauren Plante, MD MPH; Zandra Reid, RN BSN; Cheryl Tocci, RN BSN; Marge Sherwood; Matthew Hoffman, MD; Stephanie Lynch, RN; Angela Bayless, RN; Jenny Benson, RN; Jennifer Mann, RN; Tina Grossman, RN; Stephanie Lort, RN; Ashley Vanneman; Elisha Lockhart; Carrie Kitto; Edwin Guzman, MD; Marian Lake, RN; Shoan Davis; Michele Falk; Clara Perez, RN.
Northwestern University – Alan M Peaceman MD, Lara Stein RN, Katura Arego, Mercedes Ramos-Brinson B.S., Gail Mallett RN BSN.
University of North Carolina – John M. Thorp, Jr, MD MPH; Karen Dorman, RN MS; Seth Brody, MD MPH.
University of Texas Health Science Center at Houston and Lyndon Baines Johnson General Hospital/Harris County Hospital District – Sean C. Blackwell, MD; Maria Hutchinson, MPH.
Funding: This study was funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Genomic and Proteomic Network for Preterm Birth Research (U01-HD-050062; U01-HD-050078; U01-HD-050080; U01-HD-050088; U01-HD-050094), all authors. This study was also funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development 5K23HD067224 (Dr. Manuck).
Footnotes
Author Affiliation Notes: Since the study was conducted, Dr. Manuck has moved to the University of North Carolina-Chapel Hill Department of Obstetrics and Gynecology Division of Maternal Fetal Medicine (Chapel Hill, NC). Dr. Bukowski has since moved to Yale University School of Medicine, Division of Maternal Fetal Medicine (New Haven, CT).
Conflict of Interest/Disclosure Statement: Dr. Esplin serves on the Scientific Advisory Boards of Sera Prognostics and Clinical Innovations. Neither of these companies was involved in any way with the present study.
The remaining authors report no conflict of interest
Presentation: This study was presented in part at the 35th Annual Society of Maternal Fetal Medicine Meeting (February 2015, San Diego, CA) as a poster presentation (#186)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. The New England journal of medicine. 2010;362:529–35. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 2.Preterm birth: crisis and opportunity. Lancet. 2006;368:339. doi: 10.1016/S0140-6736(06)69080-6. [DOI] [PubMed] [Google Scholar]
- 3.Esplin MS. Overview of spontaneous preterm birth: a complex and multifactorial phenotype. Clin Obstet Gynecol. 2014;57:518–30. doi: 10.1097/GRF.0000000000000037. [DOI] [PubMed] [Google Scholar]
- 4.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moutquin JM. Classification and heterogeneity of preterm birth. BJOG: an international journal of obstetrics and gynaecology. 2003;110(Suppl 20):30–3. doi: 10.1016/s1470-0328(03)00021-1. [DOI] [PubMed] [Google Scholar]
- 6.Lockwood CJ, Kuczynski E. Risk stratification and pathological mechanisms in preterm delivery. Paediatric and perinatal epidemiology. 2001;15(Suppl 2):78–89. doi: 10.1046/j.1365-3016.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 7.York TP, Strauss JF, 3rd, Neale MC, Eaves LJ. Racial differences in genetic and environmental risk to preterm birth. PloS one. 2010;5:e12391. doi: 10.1371/journal.pone.0012391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esplin MS, O'Brien E, Fraser A, et al. Estimating recurrence of spontaneous preterm delivery. Obstetrics and gynecology. 2008;112:516–23. doi: 10.1097/AOG.0b013e318184181a. [DOI] [PubMed] [Google Scholar]
- 9.Simonsen SE, Lyon JL, Stanford JB, Porucznik CA, Esplin MS, Varner MW. Risk factors for recurrent preterm birth in multiparous Utah women: a historical cohort study. BJOG: an international journal of obstetrics and gynaecology. 2013;120:863–72. doi: 10.1111/1471-0528.12182. [DOI] [PubMed] [Google Scholar]
- 10.Porter TF, Fraser AM, Hunter CY, Ward RH, Varner MW. The risk of preterm birth across generations. Obstetrics and gynecology. 1997;90:63–7. doi: 10.1016/S0029-7844(97)00215-9. [DOI] [PubMed] [Google Scholar]
- 11.Clausson B, Lichtenstein P, Cnattingius S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG: an international journal of obstetrics and gynaecology. 2000;107:375–81. doi: 10.1111/j.1471-0528.2000.tb13234.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Baldwin DA, Bukowski RK, et al. A Genome-Wide Association Study of Early Spontaneous Preterm Delivery. Genetic epidemiology. 2015 doi: 10.1002/gepi.21887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolan SM, Hollegaard MV, Merialdi M, et al. Synopsis of preterm birth genetic association studies: the preterm birth genetics knowledge base (PTBGene) Public health genomics. 2010;13:514–23. doi: 10.1159/000294202. [DOI] [PubMed] [Google Scholar]
- 14.Manuck TA, Esplin MS, Biggio J, et al. The Phenotype of Spontaneous Preterm Birth: Application of a Clinical Phenotyping Tool. American journal of obstetrics and gynecology. 2015 doi: 10.1016/j.ajog.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gower JC. A general coefficient of similarity and some of its properties. Biometrics. 1971;27:857–74. [Google Scholar]
- 16.Kaufman La, R PJ. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley; New York: 1990. [Google Scholar]
- 17.Liu JZ, McRae AF, Nyholt DR, et al. A versatile gene-based test for genome-wide association studies. American journal of human genetics. 2010;87:139–45. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loza MJ, McCall CE, Li L, Isaacs WB, Xu J, Chang BL. Assembly of inflammation-related genes for pathway-focused genetic analysis. PloS one. 2007;2:e1035. doi: 10.1371/journal.pone.0001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velez DR, Fortunato SJ, Thorsen P, Lombardi SJ, Williams SM, Menon R. Preterm birth in Caucasians is associated with coagulation and inflammation pathway gene variants. PloS one. 2008;3:e3283. doi: 10.1371/journal.pone.0003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldenberg RL, Andrews WW, Faye-Petersen O, Cliver S, Goepfert AR, Hauth JC. The Alabama Preterm Birth Project: placental histology in recurrent spontaneous and indicated preterm birth. American journal of obstetrics and gynecology. 2006;195:792–6. doi: 10.1016/j.ajog.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 21.Hackney DN, Tirumala R, Salamone LJ, Miller RK, Katzman PJ. Do placental histologic findings of chorion-decidual hemorrhage or inflammation in spontaneous preterm birth influence outcomes in the subsequent pregnancy? Placenta. 2014;35:58–63. doi: 10.1016/j.placenta.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Goldenberg RL, Gravett MG, Iams J, et al. The preterm birth syndrome: issues to consider in creating a classification system. American journal of obstetrics and gynecology. 2012;206:113–8. doi: 10.1016/j.ajog.2011.10.865. [DOI] [PubMed] [Google Scholar]
- 23.Villar J, Papageorghiou AT, Knight HE, et al. The preterm birth syndrome: a prototype phenotypic classification. American journal of obstetrics and gynecology. 2012;206:119–23. doi: 10.1016/j.ajog.2011.10.866. [DOI] [PubMed] [Google Scholar]
- 24.Laughon SK, Albert PS, Leishear K, Mendola P. The NICHD Consecutive Pregnancies Study: recurrent preterm delivery by subtype. American journal of obstetrics and gynecology. 2014;210:131 e1–8. doi: 10.1016/j.ajog.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manuck TA, Major HD, Varner MW, Chettier R, Nelson L, Esplin MS. Progesterone receptor genotype, family history, and spontaneous preterm birth. Obstetrics and gynecology. 2010;115:765–70. doi: 10.1097/AOG.0b013e3181d53b83. [DOI] [PubMed] [Google Scholar]
- 26.Uzun A, Dewan AT, Istrail S, Padbury JF. Pathway-based genetic analysis of preterm birth. Genomics. 2013;101:163–70. doi: 10.1016/j.ygeno.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menon R, Fortunato SJ, Thorsen P, Williams S. Genetic associations in preterm birth: a primer of marker selection, study design, and data analysis. Journal of the Society for Gynecologic Investigation. 2006;13:531–41. doi: 10.1016/j.jsgi.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Hillier SL, Martius J, Krohn M, Kiviat N, Holmes KK, Eschenbach DA. A case-control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. The New England journal of medicine. 1988;319:972–8. doi: 10.1056/NEJM198810133191503. [DOI] [PubMed] [Google Scholar]