Abstract
Foot-and-mouth disease virus (FMDV) causes morbidity and mortality in a range of animals and threatens local economies by acting as a barrier to international trade. The outbreak in the United Kingdom in 2001 that cost billions to control highlighted the risk that the pathogen poses to agriculture. In response, several mathematical models have been developed to parameterize and predict both transmission dynamics and optimal disease control. However, a lack of understanding of the multi-strain etiology prevents characterization of multi-strain dynamics. Here, we use data from FMDV serology in an endemic setting to probe strain-specific transmission and immunodynamics. Five serotypes of FMDV affect cattle in the Far North Region of Cameroon. We fit both catalytic and reverse catalytic models to serological data to estimate the force of infection and the rate of waning immunity, and to detect periods of sustained transmission. For serotypes SAT2, SAT3, and type A, a model assuming life-long immunity fit better. For serotypes SAT1 and type O, the better-fit model suggests that immunity may wane over time. Our analysis further indicates that type O has the greatest force of infection and the longest duration of immunity. Estimates for the force of infection were time-varying and indicated that serotypes SAT1 and O displayed endemic dynamics, serotype A displayed epidemic dynamics, and SAT2 and SAT3 did not sustain local chains of transmission. Since these results were obtained from the same population at the same time, they highlight important differences in transmission specific to each serotype. They also show that immunity wanes at rates specific to each serotype, which influences patterns of local persistence. Overall, this work shows that viral serotypes can differ significantly in their epidemiological and immunological characteristics. Patterns and processes that drive transmission in endemic settings must consider complex viral dynamics for accurate representation and interpretation.
Introduction
Foot-and-mouth disease virus (FMDV) is currently ranked as one of the most significant foreign animal disease threats in most major global economies [1, 2]. Perhaps surprisingly, this is not because the virus causes exceptionally severe disease or because there is no preventative vaccine; in fact, morbidity is moderate, disease-induced mortality is only slightly elevated in animals up to one year of age [1], and vaccines exist against multiple FMDV serotypes [3, 4]. The high threat raking is due, at least partially, to the intersection between epidemiology and economics. The World Organisation for Animal Health (OIE) classifies countries into three epidemiological categories with regards to foot-and-mouth (FMD) disease status: countries with active FMDV outbreaks, countries that are free of disease in the presence of vaccination, and countries that are free of the disease in the absence of vaccination [5]. Countries experiencing active FMDV outbreaks have the most restricted export market, while countries that are FMDV free without vaccination have the most accessible export market. Following the outbreak of FMD in the UK in 2001, which cost £4 billion to successfully eliminate the Pan-Asia O serotype [6], computational efforts have been initiated in many countries that are free of FMDV in the absence of vaccination like the United States [7–10], Australia [11, 12], and throughout Europe [13–15]. These projects analyze threats and plan disease management strategies if an epidemic of any of the multiple circulating FMDV serotypes were to be introduced.
We contend that for any of these efforts to be successful in making robust predictions about future epidemic scenarios, we need to understand the epidemiology of FMDV in much greater detail by quantifying strain-specific transmission and immunodynamics in the endemic setting. A quantity of particular interest is the force of infection (FOI), which measures the rate at which susceptible individuals acquire infection. The FOI can inform epidemic behavior, identify heterogeneities in transmission, and specify targets for disease control [16–19]. Standard approaches to estimating the FOI fit accurate counts of both susceptible and infected individuals to dynamical catalytic models. However, this information is difficult to obtain, especially for endemic disease when there is little or inconsistent surveillance, or when the disease can be subclinical. In these cases, the lack of necessary information prohibits the design and implementation of disease control programs.
One solution to quantifying disease dynamics in the absence of counts of infected and susceptible individuals is to fit dynamical models to serology data. These data classify an individual based on the presence of host antibodies against a pathogen, which indicates past exposure to the pathogen. Here, we show that serology data matched with host age can be fitted to dynamical models to provide a record of historical exposure, even when animals are sampled only in a single event such as a cross-sectional study. The serology data also allow us to estimate the duration of immunity for different strains—an important epidemiological parameter that is poorly known for FMD. We use serology data that indicate past exposure to FMDV for cattle in the Far North Region, Cameroon, where there is evidence that five of the seven serotypes are present either endemically or intermittently [20].
Approximately 650,000 cattle are herded in the Far North Region, Cameroon [21]. Herd management is highly individual and non-intensive, but can be broadly classified in one of two ways. The majority of cattle herds (~93%) are sedentary, residing near villages and grazing around them. The minority of cattle herds (~7%) are mobile, participating in transhumance which allows them to graze on pastures in different geographic areas throughout the year depending on seasonal availability. Wild bovids are absent from this site. Herders report frequent FMD outbreaks; however, there was a complete absence of formal control and no access to FMDV vaccines at the time of the study [20, 22].
To address uncertainty in the duration of a host immune response against FMDV in the absence of vaccination, we fit contrasting models of FMDV transmission and immunity to the serology data: the catalytic model and reversible catalytic model [23, 24]. The catalytic model describes transmission followed by lifelong seroconversion, suggesting that the antibody response lasts for the lifespan of the animal. Alternatively, the reversible catalytic model assumes that infection leads to temporary seroconversion, and that the immune memory wanes over time. We also discriminate between two hypotheses about variation in the FOI. Previous studies have shown that the FOI is age-specific, due to the fact that animals have contact rates that vary with age, in an endemic steady state [16, 25]. Other studies have also shown that the FOI may be time-varying, due to the fact that incidence often exhibits cycles or local extinction due to depletion of susceptible individuals which do not support the idea of a disease steady state [17]. We distinguish between age-specific and time-varying differences in the force of infection by estimating the force of infection for five FMDV serotypes independent of each other and then comparing results across serotypes. In this way, we test between two sets of alternative hypothesizes: first, between lifelong seroconversion versus waning immunity and second, between age-specific FOI versus time-varying FOI.
Within this framework, we characterize and quantify three aspects of FMDV dynamics in the endemic setting. First, we determine the duration of seropositivity as a result of natural infection in the absence of vaccination. Second, we determine if the FOI is age-specific or time-varying and quantify the FOI for each serotype. Third, we use the FOI patterns inferred to estimate key epidemiological quantities that describe endemic FMDV dynamics. By estimating these key epidemiological parameters, we are able to reconstruct the historical burden of FMDV in an endemic area and quantify control efforts necessary to stop transmission.
Materials and Methods
Data collection
Data were obtained in four phases from cattle over one year of age in the Far North Region, Cameroon between February 5, 2010 and December 29, 2010 and again between January 30, 2012 and February 8, 2012. The cattle were from 15 mobile herds, 15 sedentary herds. First, cattle ages were obtained by interviews with herders. Second, serum samples were taken from the same 30 herds using a sampling scheme detailed in Ludi et al. [20]. The herds were visited twice during the study period and the same animals were located and sampled on subsequent visits whenever possible. In 2010, 531 samples were obtained from 498 animals. Four hundred sixty six animals were sampled once, thirty-one animals were sampled twice, and one animal was sampled three times during the study period (A Text in S1 File). In 2012, 124 samples were obtained. Animal ages collected in the first phase were confirmed upon sampling conducted in the second phase and the samples and age data were then linked by an animal identification number. Third, serum samples were shipped to the USA and subjected to serum neutralization tests (SNTs) at Plum Island Animal Diagnostic Center (PIADC), Agricultural Research Service (ARS), United States Department of Agriculture (USDA). Each serum sample was tested for serotypes O, A, SAT1, SAT2, and SAT3 as reported in Ludi et al. [20]. Fourth, the remainders of the serum samples were irradiated and subjected to antibody testing, in which animal autoantibody profiles were determined at the National Veterinary Services Laboratory (NVSL), USDA. This analysis confirmed the identities of serially sampled animals.
Initial data analysis
A subset of animals (n = 32) were sampled two or three times in 2010. For this subset, the serology status of serially sampled animals was compared across sampling events, with an interval between sampling events that ranged from 94 to 190 days (A Text in S1 File). Paired samples taken from a single animal at two different time points were classified into one of four groups: animals that retained a positive serology status between the two sampling events; animals that had a positive serology status at the first sampling event and a negative serology status at the second sampling event; animals that had a negative serology status at the first sampling event and a positive serology status at the second sampling event; and animals that retained a negative serology status between the two sampling events. Paired samples were separately classified for each of the five FMDV serotypes. We used this information only as a preliminary investigation to determine plausibility of investigating the hypothesis of waning immunity for each serotype.
Modeling transmission and immunity
We compared two models to determine which better fit the age and serology data collected in Cameroon. The first model was a catalytic model, which assumes that immunity is lifelong. The second model was a reverse catalytic model, which assumes that immunity is transient and wanes over time. We fit both models to the dataset of all animals (n = 498) sampled once, twice, and three times, which amounted to 531 samples and was much larger than the subset used in the initial data analysis. We selected the model that best fit the dataset of all animals using AIC. In this way, we could determine if lifelong or waning immunity best described immunological processes in our study population for each FMDV serotype.
To define the two models mathematically, let a represent the age of each animal in years and let t represent a calendar year. Let λ(t) represent the force of infection—the rate per capita per year at which seronegative individuals become seropositive due to infection. According to the catalytic model [16, 18, 23, 24, 26], assuming lifelong immunity, the age-specific seroprevalence is given by
| (1) |
In our analysis, we can ignore any maternally derived antibodies since all animals were at least one year of age. We can use the reverse catalytic model to estimate the age-specific seroprevalence, given by
| (2) |
assuming seroconversion lasts for an average duration of and then wanes [23, 24]. In both models, we accounted for gradual changes in the force of infection by fitting the force of infection as a b-spline that varied by year. The b-spline, fit with the bs command in the splines package in R, was a cubic spline with four uniformly spaced internal knots.
Model selection, parameterization, and sensitivity
The catalytic and reverse catalytic models were compared and the best-fit model was selected using Akaike’s Information Criterion (AIC) [27]. To find estimates for the force of infection and rate of waning immunity, we estimated values for the b-splined force of infection and rate of waning immunity by maximizing the binomial likelihood of seropositivity by age according to
| (3) |
where N a is the total number of animals sampled at age a and p a is the proportion of animals that are seropositive. Likelihood estimations were performed using the optim command in R.
Plots of the time-varying forces of infection were compared across serotypes and local minima and maxima were visually determined. If local minima and maxima occurred at the same ages regardless of serotype, we can assume that the FOI is age-related and FMD dynamics are at or near a steady state. If local minima and maxima occurred at different ages across serotypes, we can assume that the FOI is time-varying and FMD dynamics are not at a steady state. (See C Text in S1 File for discussion.)
Ages were calculated from birth months and years reported in interviews. Therefore, the accuracy of reported ages may affect model selection and parameterization. We investigated the impact of inaccuracy in age reporting using the Morris Method for parameter sensitivity [28] to find the deviation from the reported age at which the model selected would change.
Estimating key epidemiological quantities
The number of secondary cases produced by a primary infectious case, or R t, can be calculated from the parameterized FOI and the average life expectancy (L). Assuming Type I mortality, in which all animals live until a certain age and then die, the average life expectancy L = 16 years (see S3 Text in S1 File for discussion). It follows that we can estimate R t from the L and the mean FOI (), according to [29]
| (4) |
Assuming a fully efficacious vaccine, the critical proportion (p c) of the population that needs to be vaccinated to prevent an outbreak depends on R t according to
| (5) |
All analyses were performed using R version 2.15.0 [30] in RStudio [31].
Ethics statement
The animal use protocol for this study was reviewed and approved by The Ohio State University’s Animal Care and Use Program (Animal Welfare Assurance Number A3261-01). The protocol number in the Ohio State University system is 2010A0018, which superseded 2009A0113 (approval for the pilot project and first round of sampling in January and February 2010). The project was also approved through the Ministry of Scientific Research and Innovation of Cameroon with permit numbers 0000114/MINRESI/B00/C00/C10/C13 (pilot project and 2010 data) and 004/MINRESI/CRRI-EN/BAG/2011 (for 2011–2012). Herd owners formally consented to participation in the study at the beginning of the study. At each visit to the herd, herd owners and/or herders (whoever was present) assented to sampling of the animals and to responding to a survey, which included the age data presented here. Assent was recorded through use of a verbal consent script by the technicians collecting samples and administering the survey. The data used in this manuscript did not require IRB approval; though, the survey contained other information that did require IRB approval. The larger survey was approved by the Ohio State University Institutional Review Board/ Human Research Protection Program (Federal-wide Assurance #00006378 from the Office for Human Research Protections in the Department of Health and Human Services: protocol 2010B0004). Within this ethical review for the survey the protocol was approved for a waiver of signed consent forms due to the low literacy of the population and cultural inappropriateness of obtaining signatures to record consent.
Results
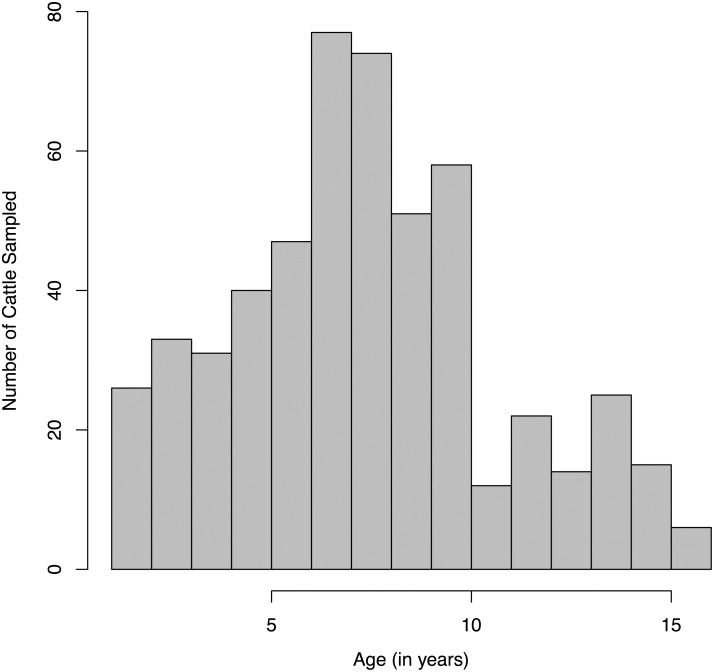
Sampled animals ranged in age from one-year-old to sixteen-years-old with a mean of 7.8 years (Fig 1). Ages of animals sampled were similar to previous demographic surveys of cattle in the Far North Region [32]; however, the oldest animals in our study were two years older than the oldest animals previously recorded.
Fig 1. Age distribution of sampled cattle.
Four hundred sixty nine cattle, ranging in age from one year old to sixteen years old, were sampled for serotype-specific FMDV antibodies.
Serotype-specific waning immunity
As a preliminary investigation to check for any evidence of waning immunity after natural infection with FMDV, we categorized the subset of cattle that were serially sampled cattle (n = 31) and showed seropositivity at the first sampling event and seronegativity at subsequent sampling events. Among the 31 animals that had been sampled twice, seropositivity waned for SAT1 (n = 3), SAT2 (n = 9), SAT3 (n = 4), and type A (n = 4; Table 1). For the single animal that had been sampled three times, seropositivity waned for SAT1, SAT3, and type O. Thus, observations indicate that it is possible that immunity wanes for all five FMDV serotypes but the significance of this process at the population level still needs to be clarified.
Table 1. Changes in seropositive status among 31 animals sampled twice in 2010.
| FMDV Serotype | Lost | Gained | No change |
|---|---|---|---|
| SAT1 | 3 | 1 | 27 |
| SAT2 | 9 | 2 | 20 |
| SAT3 | 4 | 3 | 24 |
| Type O | 0 | 5 | 26 |
| Type A | 4 | 5 | 22 |
To determine if seropositivity after natural infection was lifelong or waned, we fit the entire dataset consisting of all samples (Table 2) to both the catalytic model (Eq 1) and to the reverse catalytic model (Eq 2).
Table 2. Number of samples positive, by age, for all animals sampled in 2010.
| Age | SAT1 | SAT2 | SAT3 | A | O | total |
|---|---|---|---|---|---|---|
| 1 | 0 | 1 | 0 | 0 | 3 | 8 |
| 2 | 1 | 6 | 1 | 2 | 8 | 18 |
| 3 | 8 | 13 | 9 | 15 | 16 | 33 |
| 4 | 4 | 12 | 2 | 8 | 16 | 31 |
| 5 | 4 | 13 | 8 | 18 | 29 | 40 |
| 6 | 8 | 21 | 9 | 15 | 30 | 47 |
| 7 | 11 | 27 | 14 | 24 | 62 | 77 |
| 8 | 18 | 37 | 16 | 26 | 54 | 74 |
| 9 | 18 | 37 | 12 | 22 | 41 | 51 |
| 10 | 12 | 24 | 13 | 29 | 42 | 58 |
| 11 | 6 | 6 | 6 | 10 | 10 | 12 |
| 12 | 8 | 9 | 5 | 9 | 11 | 22 |
| 13 | 3 | 6 | 2 | 7 | 7 | 14 |
| 14 | 9 | 10 | 8 | 12 | 16 | 25 |
| 15 | 3 | 10 | 1 | 7 | 13 | 15 |
| 16 | 2 | 3 | 0 | 2 | 6 | 6 |
Model selection, based on AIC scores, indicates if the seropositivity significantly wanes or not at the population level; point estimates for parameter values (ω; Eq 2) further quantify the duration of seropositivity. Data for serotypes SAT1 and type O best fit the reverse catalytic model, indicating that seropositivity does not last for the lifespan of cattle in Cameroon (Table 3). Estimated parameters for Type O showed longer duration of seropositivity, lasting for 3.8 years (Table 3). In contrast, estimated parameters for SAT1 showed a shorter duration of seropositivity, inducing antibody reactions that last for less than one year (Table 3). Data for serotypes SAT2, SAT3, and A best fit the catalytic model, indicating that seropostivitiy broadly lasts for the lifespan of cattle in Cameroon (Table 3). Sensitivity of age to model selection results was low, as these results held through analyses in which the animal’s age was increased by 1 or 3 years. In this way, there are serotype-specific host responses that manifest in waning seropositivity for serotypes SAT1 and type O and lifelong seroprevalence for serotypes SAT2, SAT3, and type A.
Table 3. Model selection and parameterization.
| FMDV Serotype | Model selected | Δ AIC | Duration of natural seropositivity (1/ω) | Mean force of infection (λ) |
|---|---|---|---|---|
| SAT1 | Reverse catalytic | 8.64 | 0.45 years | 0.68 years-1 |
| SAT2 | Catalytic | 6.39 | Lifelong | 0.021 years-1 |
| SAT3 | Catalytic | 1.53 | Lifelong | 0.014 years-1 |
| Type O | Reverse catalytic | 4.02 | 3.80 years | 0.69 years-1 |
| Type A | Catalytic | 8.29 | Lifelong | 0.052 years-1 |
Time-varying serotype-specific FOI
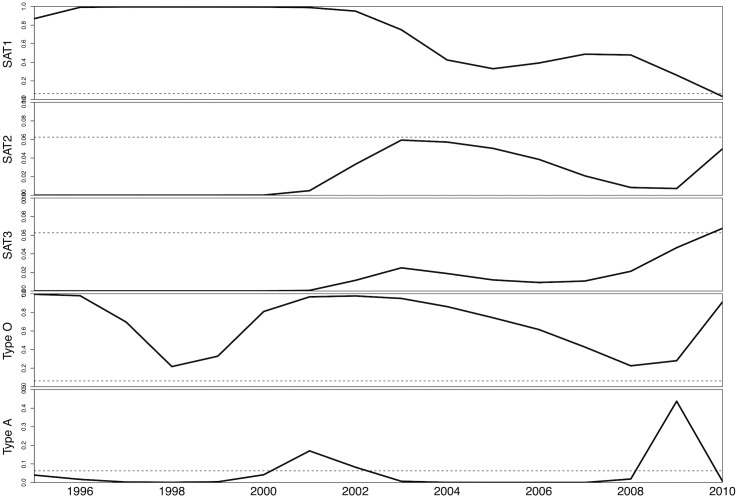
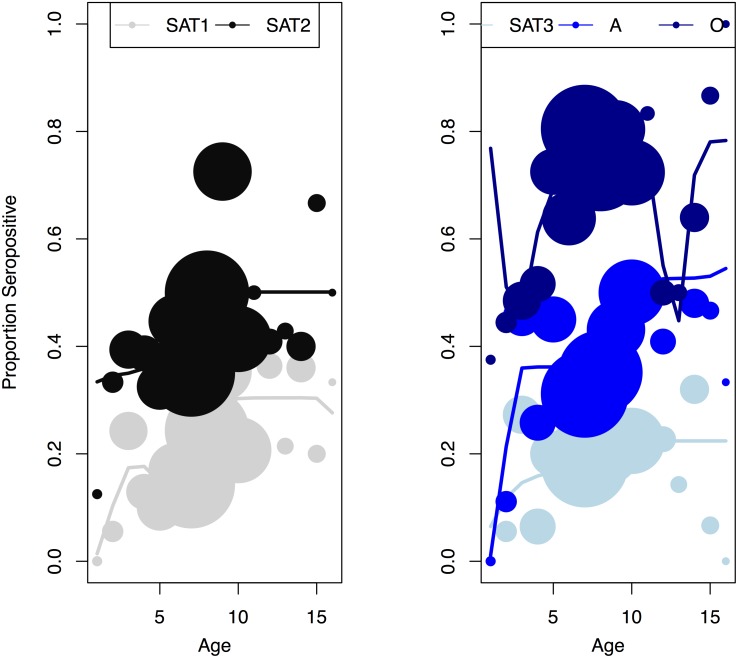
To quantify the force of infection, we fit the entire dataset for SAT1 and type O to reverse catalytic models and data for SAT2, SAT3, and A to catalytic models, all of which quantified the time-variation in the force of infection using a b-spline. Since local minima and maxima in the force of infection did not occur at the same ages across serotypes, we infer that the force of infection is time-varying. Between 1996 and 2010, estimates for the FOI for SAT1 decrease through time while estimates for the FOI for SAT2 and SAT3 increase through time (Fig 2). Type O shows cyclic variation in the estimated FOI though time, and type A shows two distinct peaks of estimated FOI: one between 2000 and 2002 and the other between 2008 and 2010. Simulations using the better-fit version of the catalytic model with estimated values for the duration of seropositivity after natural infection and serotype-specific time-varying FOI produced a good fit with the data (Fig 3). The mean estimated FOI ranged from the maximum of 0.69 per year for type O to a minimum of 0.021 per year for SAT 2 (Table 3). Therefore, there are clear serotype specific differences in the FOI with regards to magnitude and temporal patterns.
Fig 2. Yearly force of infection by serotype.
Estimates derived by model fitting for the time-varying force of infection (FOI) for SAT1, SAT2, SAT2, type O, and type A inferred for the 16-year period preceding data collection represented by a solid line. The dotted line represents the epidemic threshold R 0 = 1; the shaded areas represent estimated FOI that correspond to R 0 > 1 and therefore, sustained chains of transmission within the study population. Note that the scale of the y-axis varies across panels.
Fig 3. Model selection and parameterization results for five FMDV serotypes.
Best-fit model results using the catalytic model (for SAT2, SAT3, and type A) or reverse catalytic mode (for SAT1 and type O) with values for the duration of immunity and serotype-specific time-varying forces of infection estimated from serology data.
Serotype-specific R t and critical vaccine coverage estimates
To calculate the number of secondary cases produced by each infectious case (R t), we considered both the mean and the maximum FOI estimated from the best-fit model for each serotype and the cattle lifespan (Eq 4). For SAT1 and type O, R t could be as high as 16 (Table 4). For SAT2 and SAT3, R t remained near unity or just below (Table 4). While the average R t for type A was below unity, the maximum showed almost 7 secondary cases for each infectious animal. These values resulted in critical proportions (p c) that needed to be vaccinated been 85–95% for SAT1, type O and type A to block circulation (Table 4). Therefore, there were serotype-specific differences in whether or not the estimated FOI breached the epidemic threshold and the targeted vaccination rates to prevent such a breach.
Table 4. Epidemiological quantities.
| FMDV serotype | mean Rt | max Rt | pc |
|---|---|---|---|
| SAT1 | 10.88 | 15.94 | 0.94 |
| SAT2 | < 1 | 0.95 | n/a |
| SAT3 | < 1 | 1.08 | 0.07 |
| Type O | 11.04 | 15.93 | 0.94 |
| Type A | < 1 | 6.99 | 0.86 |
To determine temporal patterns in epidemic potential, we calculated the FOI that corresponded to the threshold value of R t = 1 and plotted it on Fig 2. When the serotype-specific estimated FOI is greater than the threshold value, the population can sustain chains-of-transmission among cattle; when the serotype-specific estimated FOI is less than the threshold value, the population cannot sustain chains-of-transmission. Chains were sustained between 1996 and 2010 by SAT1 and type O and were not sustained by SAT2 and SAT3 (Fig 2). Chains were sustained only for two periods for type A (Fig 2). Thus, there were important time-varying patterns in whether or not the estimated FOI breached the epidemic threshold.
Variation in transmission and immunity by herd management
To determine if there was variation in the duration of seropositivity or force of infection by herd management type, we stratified the data into three subsets: market cattle, mobile cattle, and sedentary cattle. We fit each subset independently to both the catalytic and reverse catalytic models. We found that the duration of seropositivity was dependent on both serotype and herd management practice: model selection supported waning immunity for 2 serotypes when using the all cattle dataset (Table 3), 1 serotype when using subset of sedentary cattle data, 2 serotypes when using the subset of mobile cattle data, and 4 serotypes when using the subset of market cattle data (A Table in S1 File). Therefore, FMD seroprevalence may be variable and differ among market cattle, mobile cattle, and sedentary cattle.
To determine the serotype-specific FOI, we estimated the parameter from the best-fit model as listed above. We found that the FOI differed between mobile and sedentary cattle and by serotype. For SAT1, SAT2, and SAT3, the FOI was greater in sedentary cattle. For type O and type A, the FOI was greater in mobile cattle (Table 5).
Table 5. Mean force of infection (λ) for different herd management practices.
| FMDV serotype | Mobile cattle | Sedentary cattle |
|---|---|---|
| SAT1 | 0.032 yr-1 | 0.044 yr-1 |
| SAT2 | 0.093 yr-1 | 0.72 yr-1 |
| SAT3 | 0.013 yr-1 | 0.020 yr-1 |
| Type O | 0.58 yr-1 | 0.19 yr-1 |
| Type A | 0.60 yr-1 | 0.12 yr-1 |
These results suggest that FMDV transmission differs between mobile and sedentary herds in a complex manner.
Detection of circulating FMDV serotypes
To determine if changes in circulating FMDV serotypes detected by local surveillance were detected by the models, we quantified the force of infection for 2011 and 2012 by fitting models to samples collected in 2012. For SAT2, the FOI increased over these years, while for serotypes O and A, the FOI decreased. For SAT1 and SAT3, the FOI remained the same over these years. These results correspond with detected increased circulation of SAT2 and decreased circulation of type O between 2010 and 2012 (Table 6).
Table 6. Force of infection (λ) for all serotypes in 2011 and 2012.
| Serotype | 2011 FOI | 2012 FOI |
|---|---|---|
| SAT1 | 0.0363 | 0.0336 |
| SAT2 | 0.0482 | 0.0655 |
| SAT3 | 0.0305 | 0.0305 |
| O | 0.127 | 0.0451 |
| A | 9.36 e-6 | 3.80 e-4 |
Discussion
Our results show that the five FMDV serotypes detected in the Far North Region, Cameroon—SAT1, SAT2, SAT3, type O, and type A—differ in their magnitude of transmission as measured by the FOI, duration of host immunity, and their ability to sustain chains-of-transmission. The mean FOI was greatest for serotypes SAT1 and type O, and much lower for serotypes SAT2, SAT3, and type A. Only SAT1 and type O showed waning seropositivity; type O showed a duration of nearly four years while SAT1 seropositivity lasted approximately for half of a year. Time-variation was observed in the FOI, with some serotypes always above the outbreak threshold (SAT1 and type O), other serotypes always below the outbreak threshold (SAT2 and SAT3), and one serotype alternating above and below the threshold (type A). Changes in circulating FMDV serotypes detected by local surveillance in 2011 and 2012 were also detected by our analyses. These results illustrate the complex dynamics that occur in endemic transmission and the numerous differences that exist even between pathogens that are genetically related.
Previous studies investigate the duration of immunity in both experimental and field settings, but show variation in results. Among studies that estimated the duration of immunity in small groups of cattle experimentally infected with FMDV by injection, a single animal was protected against secondary infection 4.5 years after the primary FMDV infection [33] and eight other cattle were protected 5.5 years after the first infection [34], but there was no investigation of immune response at longer durations. On the other hand, a previous survey of cattle in Adamawa Region, Cameroon indicated that antibody prevalence might wane for serotype O [35]. Taken together, these studies indicate no clear pattern for FMDV immunity among cattle. Here, we present two possible explanations for the lack of clear patterns in FMDV immunity. First, the duration of FMDV immunity is serotype specific: some serotypes show lifelong immunity, while others might induce a serological response for less than one year. Second, these results are relative to the lifespan of the host. Waning immunity in an endemic FMDV system where animals have longer lifespans might manifest as lifelong immunity in epidemic FMDV systems where animals have shorter lifespans. In this way, the duration of immunity is specific to both serotype and context of FMDV dynamics.
In Cameroon, where FMDV is endemic, we find periods in which the cattle population can sustain chains-of-transmission internally and other periods in which the cattle population cannot sustain chains-of-transmission but still show a large proportion of seropositive animals. Two serotypes (O and SAT1) show evidence of sustained chains-of-transmission over the last 16 years, indicating an endemic disease regime. One serotype (A) shows periods with chains-of-transmission and other periods without chains-of-transmission, indicating an epidemic disease regime. One period in which our analysis detected sustained chains of transmission for serotype A (2008–2009) corresponds to an outbreak of the same serotype in Nigeria, which borders our study site [36]. Two other serotypes (SAT2 and SAT3) display a lack of chains-of-transmission, indicating that transmission occurs via stuttering chains or repeated contact with some other animal reservoir. The disease regime affects control strategies, and vaccinating 85–95% of the population would help control serotypes that display endemic or epidemic behavior. However, for those serotypes that exhibit other disease mechanisms, controlling access to a source population may be more beneficial. These results indicate that complex dynamics govern endemic FMD and must be considered when planning appropriate control programs.
Certain features of the analysis permitted the characterization of serotype specific epidemiological parameters and dynamics. First, the comparison of multiple pathogens sampled from the same animals at the same time allowed the FOI to be categorized as either age-specific or time-varying. Because the FOI did not show the same patterns with age—the maximum or minimum values did not occur at the same ages across serotypes—then we can conclude that the FOI must be time-varying. Because age-specificity in the FOI has been well-documented across pathogens, it is likely that there are also some age-specific factors at play, including between-host contacts or within-host immunological responses that differ by age. Second, the data collection scheme permitted separation of two fundamental phenomena that drive transmission: pathogen characteristics and patterns of contact within a population. These samples were obtained from the same animals at the same time, effectively holding patterns of contact constant. Factors uncontrolled for in this analysis would have affected all serotypes equally, since identical animals were sampled for all five serotypes. Any differences among the serotypes can be attributed to differences in pathogen characteristics that are specific to each serotype. Thus, time-variation and serotype-specific differences driven by viral biology can be detected.
Serology from serially sampled animals did not show waning antibody prevalence for serotype O; however, the model selected indicated that immunity lasted for almost four years and then waned. This combination might arise for pathogens that display cyclic multiannual dynamics sampled between the onset of the outbreak and the peak number of cases. If our sampling occurred as an outbreak was in an early phase, most animals would be gaining immunity and no information about waning immunity or immune duration would be provided by only analyzing paired samples. It would be interesting to sample serology over multiple years and during different seasons to determine if the signature of cyclic outbreaks or waning immunity could be detected.
Serology from serially sampled animals indicated waning antibody prevalence for serotypes SAT2, SAT3, and type A; however, the model selected indicated lifelong antibody prevalence. This discrepancy might arise if the duration of antibody prevalence is so short that the waning cannot be detected in models that have a timestep of one year. The serotypes that show the discrepancy are those with the highest counts of immunity loss in the paired sample data, so it is possible that they exhibit waning immunity. It would be interesting to sample serology at shorter intervals over multiple years and fit catalytic and reverse catalytic models with shorter timesteps to see if waning immunity can be detected at an enhanced temporal resolution.
Our analyses suggested variation in the FOI and duration of immunity among market cattle, mobile cattle, and sedentary cattle. Variation might be caused by greater stress in market and mobile herds, which has been shown to negatively impact immune function. Alternatively, variation might be driven by heterogeneity in disease exposures that differ between herd types, with repeated exposure in sedentary cattle causing immune boosting that lengthens immune durations for multiple serotypes. In either case, if control were to be implemented in Cameroon, mobile and market herds would be ideal candidates for vaccination since waning immunity suggested for multiple serotypes.
Conclusion
FMDV endemicity is complex due to the presence of multiple co-circulating serotypes that differ in characteristics relating to transmission and immunity. Although epidemic patterns and processes are well defined [37–41], they do not accurately represent the true global FMD burden, which occurs primarily in endemic settings [42, 43]. In order to understand the epidemiology of this pathogen, we must consider patterns and processes in infection and immunity that differ from the well-studied patterns evident in acute, immunizing diseases.
Supporting Information
Supplementary Material for Serotype-specific transmission and waning immunity of endemic foot-and-mouth disease virus including dates of serology sampling (A Text and Fig A), model derivation (B Text), derivation for age-specific FOI versus time-varying FOI (C Text), additional results from stratifying data by herd management type (D Text and Table A), and additional results from analyzing 2012 serology data (E Text and Table B).
(PDF)
Acknowledgments
LWP is grateful for insight provided by Niel Hens, Mario Peruggia, and Karla Moreno Torres, and for helpful comments from both the Disease Ecology and Computer Modeling Laboratory (DECML) at Ohio State and the Ohio State University Center for the Study and Teaching of Writing Postdoc Group. We are grateful to our collaborators at the United States Department of Agriculture including Luis Rodriguez, Anna Ludi, and Zaheer Ahmed at the Plum Island Animal Diagnostic Center (PIADC), Agricultural Research Service (ARS) and Amy Winter at the National Veterinary Services Laboratory. We are also grateful to collaborators in Cameroon, including the Center for the Support of Research and Pastoralism (CARPA), the University of Ngaoundere, the University of Maroua, the National Veterinary Laboratory (LANAVET), the Ministry of Livestock, Fisheries, and Animal Industries (MINEPIA), and the Ministry of Scientific Research and Innovation (MINRESI). Finally, we thank the Global Foot-and-mouth disease Research Alliance (GFRA). This work was supported by an allocation of computing time from the Ohio Supercomputer Center.
Data Availability
All relevant data are within the paper (in Tables 1 and 2) or the supplementary material.
Funding Statement
This work was supported by award R24-HD058484 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (http://www.nichd.nih.gov/Pages/index.aspx), awarded to the Ohio State University Initiative in Population Research and RG, and by award DEB-1015908 from the National Science Foundation (http://www.nsf.gov), awarded to RG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Grubman MJ, Baxt B. Foot-and-mouth disease. Clinical Microbiology Reviews. 2004;17(2):465–+. 10.1128/cmr.17.2.465-493.2004 WOS:000220972100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leforban Y. Prevention measures against foot-and-mouth disease in Europe in recent years. Vaccine. 1999;17(13–14):1755–9. 10.1016/S0264-410X(98)00445-9 [DOI] [PubMed] [Google Scholar]
- 3. Doel TR. FMD vaccines. Virus Research. 2003;91(1):81–99. 10.1016/S0168-1702(02)00261-7 [DOI] [PubMed] [Google Scholar]
- 4. Rodriguez LL, Gay CG. Development of vaccines toward the global control and eradication of foot-and-mouth disease. Expert Review of Vaccines. 2011;10(3):377–87. 10.1586/erv.11.4 [DOI] [PubMed] [Google Scholar]
- 5. World Organisation for Animal Health (OIE). Terrestrial Animal Health Code Paris: OIE; 2014. [cited 2014 December 9]. 23rd:[Available from: http://www.oie.int/en/international-standard-setting/terrestrial-code/access-online/. [Google Scholar]
- 6. Thompson D, Muriel P, Russell D, Osborne P, Bromley A, Rowland M, et al. Economic costs of the foot and mouth disease outbreak in the United Kingdom in 2001. Revue scientifique et technique de l'Office international des épizooties. 2002;21(3):675–87. [DOI] [PubMed] [Google Scholar]
- 7. Tildesley MJ, Smith G, Keeling MJ. Modeling the spread and control of foot-and-mouth disease in Pennsylvania following its discovery and options for control. Preventive Veterinary Medicine. 2012;104(3–4):224–39. 10.1016/j.prevetmed.2011.11.007 WOS:000303032900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carpenter TE, O'Brien JM, Hagerman AD, McCarl BA. Epidemic and economic impacts of delayed detection of foot-and-mouth disease: a case study of a simulated outbreak in California. Journal of Veterinary Diagnostic Investigation. 2011;23(1):26–33. WOS:000286953200003. [DOI] [PubMed] [Google Scholar]
- 9. Ward MP, Laffan SW, Highfield LD. Modelling spread of foot-and-mouth disease in wild white-tailed deer and feral pig populations using a geographic-automata model and animal distributions. Preventive Veterinary Medicine. 2009;91(1):55–63. 10.1016/j.prevetmed.2009.05.005 WOS:000269033900008. [DOI] [PubMed] [Google Scholar]
- 10. Buhnerkempe MG, Tildesley MJ, Lindstrom T, Grear DA, Portacci K, Miller RS, et al. The Impact of Movements and Animal Density on Continental Scale Cattle Disease Outbreaks in the United States. PLoS ONE. 2014;9(3):e91724 10.1371/journal.pone.0091724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doran RJ, Laffan SW. Simulating the spatial dynamics of foot and mouth disease outbreaks in feral pigs and livestock in Queensland, Australia, using a susceptible-infected-recovered cellular automata model. Preventive Veterinary Medicine. 2005;70(1–2):133–52. 10.1016/j.prevetmed.2005.03.002 WOS:000230371100010. [DOI] [PubMed] [Google Scholar]
- 12. Garner MG, Beckett SD. Modelling the spread of foot-and-mouth disease in Australia. Australian Veterinary Journal. 2005;83(12):758–66. 10.1111/j.1751-0813.2005.tb11589.x WOS:000234174200022. [DOI] [PubMed] [Google Scholar]
- 13. Lindstrom T, Lewerin SS, Wennergren U. Influence on disease spread dynamics of herd characteristics in a structured livestock industry. Journal of the Royal Society Interface. 2012;9(71):1287–94. 10.1098/rsif.2011.0625 WOS:000303108400018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vernon MC, Keeling MJ. Impact of regulatory perturbations to disease spread through cattle movements in Great Britain. Preventive Veterinary Medicine. 2012;105(1–2):110–7. 10.1016/j.prevetmed.2011.12.016 WOS:000303950200012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bajardi P, Barrat A, Savini L, Colizza V. Optimizing surveillance for livestock disease spreading through animal movements. Journal of the Royal Society Interface. 2012;9(76):2814–25. 10.1098/rsif.2012.0289 WOS:000309269100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Long GH, Sinha D, Read AF, Pritt S, Kline B, Harvill ET, et al. Identifying the Age Cohort Responsible for Transmission in a Natural Outbreak of Bordetella bronchiseptica . PLoS Pathog. 2010;6(12):e1001224 10.1371/journal.ppat.1001224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reiner RC, Stoddard ST, Forshey BM, King AA, Ellis AM, Lloyd AL, et al. Time-varying, serotype-specific force of infection of dengue virus. Proceedings of the National Academy of Sciences. 2014;111(26):E2694–E702. 10.1073/pnas.1314933111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferrari MJ, Djibo A, Grais RF, Grenfell BT, Bjornstad ON. Episodic outbreaks bias estimates of age-specific force of infection: a corrected method using measles as an example. Epidemiology & Infection. 2010;138(01):108–16. 10.1017/S0950268809990173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grenfell BT, Anderson RM. The estimation of age-related rates of infection from case notifications and serological data. Epidemiology & Infection. 1985;95(02):419–36. 10.1017/S0022172400062859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ludi A, Ahmed Z, Pomeroy LW, Pauszek SJ, Smoliga GR, Moritz M, et al. Serotype Diversity of Foot-and-Mouth-Disease Virus in Livestock without History of Vaccination in the Far North Region of Cameroon. Transboundary and Emerging Diseases. 2014:n/a–n/a. 10.1111/tbed.12227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seignobos C. In: Iyebi-Mandjek CSO, editor. Atlas de la Province Extrême-Nord Cameroun: IRD Éditions, MINREST/INC; 2000. [Google Scholar]
- 22. Healy Profitos JM, Moritz M, Garabed RB. What to do with chronically sick animals? Pastoralists' management strategies in the Far North Region of Cameroon. Pastoralism. 2013;3(1):1–11. 10.1186/2041-7136-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muench H. Catalytic Models in Epidemiology. Cambridge, Massachusetts: Harvard University Press; 1959. 110 p. [Google Scholar]
- 24. Hens N, Aerts M, Faes C, Shkedy Z, LeJeune O, Van Damme P, et al. Seventy-five years of estimating the force of infection from current status data. Epidemiology & Infection. 2010;138(06):802–12. 10.1017/S0950268809990781 [DOI] [PubMed] [Google Scholar]
- 25. Lavine JS, Bjørnstad ON, de Blasio BF, Storsaeter J. Short-lived immunity against pertussis, age-specific routes of transmission, and the utility of a teenage booster vaccine. Vaccine. 2012;30(3):544–51. 10.1016/j.vaccine.2011.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griffiths DA. A Catalytic Model of Infection for Measles. Journal of the Royal Statistical Society Series C (Applied Statistics). 1974;23(3):330–9. 10.2307/2347126 [DOI] [Google Scholar]
- 27. Akaike H. Factor analysis and AIC. Psychometrika. 1987;52(3):317–32. 10.1007/bf02294359 [DOI] [Google Scholar]
- 28. Wu J, Dhingra R, Gambhir M, Remais JV. Sensitivity analysis of infectious disease models: methods, advances and their application. Journal of The Royal Society Interface. 2013;10(86). 10.1098/rsif.2012.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford: Oxford University Press; 1991. 757 p. [Google Scholar]
- 30. R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0, URL http://www.R-project.org/. 2012. [Google Scholar]
- 31. RStudio. RStudio: Integrated development environment for R (Version 0.98.994). Boston, MA: Retrieved August 2, 2014. Available from http://www.rstudio.org/. 2014. [Google Scholar]
- 32. Njoya A, Bouchel D, Ngo Tama AC, Moussa C, Martrenchar A, Letenneur L. Systèmes d'élevage et productivité des bovins en milieu paysan au Nord-Cameroun. World Animal Review 1997;89:12–23. [Google Scholar]
- 33. Cunliffe HR. Observations on the duration of immunity in cattle after experimental infection with foot-and-mouth disease. Cornell Veterinarian. 1964;54:501–10. [PubMed] [Google Scholar]
- 34. Garland AJM. Inhibitory activity of secretions in cattle against foot-and-mouth disease virus.: University of London; 1974. [Google Scholar]
- 35. Bronsvoort BMdC, Anderson J, Corteyn A, Hamblin P, Kitching RP, Nfon C, et al. Geographical and age-stratified distributions of foot-and-mouth disease virus-seropositive and probing-positive cattle herds in the Adamawa province of Cameroon. Veterinary Record. 2006;159(10):299–+. WOS:000240673400005. [DOI] [PubMed] [Google Scholar]
- 36. Ehizibolo DO, Perez AM, Carrillo C, Pauszek S, AlKhamis M, Ajogi I, et al. Epidemiological Analysis, Serological Prevalence and Genotypic Analysis of Foot-and-Mouth Disease in Nigeria 2008–2009. Transboundary and Emerging Diseases. 2014;61(6):500–10. 10.1111/tbed.12054 [DOI] [PubMed] [Google Scholar]
- 37. Alexandersen S, Zhang Z, Donaldson AI, Garland AJM. The Pathogenesis and Diagnosis of Foot-and-Mouth Disease. Journal of Comparative Pathology. 2003;129(1):1–36. 10.1016/S0021-9975(03)00041-0 [DOI] [PubMed] [Google Scholar]
- 38. Chis Ster I, Dodd PJ, Ferguson NM. Within-farm transmission dynamics of foot and mouth disease as revealed by the 2001 epidemic in Great Britain. Epidemics. 2012;4(3):158–69. MEDLINE:22939313. [DOI] [PubMed] [Google Scholar]
- 39. Keeling MJ, Woolhouse MEJ, Shaw DJ, Matthews L, Chase-Topping M, Haydon DT, et al. Dynamics of the 2001 UK foot and mouth epidemic: Stochastic dispersal in a heterogeneous landscape. Science. 2001;294(5543):813–7. 10.1126/science.1065973 WOS:000171851800036. [DOI] [PubMed] [Google Scholar]
- 40. Tildesley MJ, Deardon R, Savill NJ, Bessell PR, Brooks SP, Woolhouse MEJ, et al. Accuracy of models for the 2001 foot-and-mouth epidemic. Proceedings of the Royal Society B-Biological Sciences. 2008;275(1641):1459–68. 10.1098/rspb.2008.0006 WOS:000255503300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boender GJ, van Roermund HJW, de Jong MCM, Hagenaars TJ. Transmission risks and control of foot-and-mouth disease in The Netherlands: Spatial patterns. Epidemics. 2010;2(1):36–47. 10.1016/j.epidem.2010.03.001 WOS:000208233300005. [DOI] [PubMed] [Google Scholar]
- 42. Rweyemamu M, Roeder P, Mackay D, Sumption K, Brownlie J, Leforban Y, et al. Epidemiological Patterns of Foot-and-Mouth Disease Worldwide. Transboundary and Emerging Diseases. 2008;55(1):57–72. 10.1111/j.1865-1682.2007.01013.x [DOI] [PubMed] [Google Scholar]
- 43. Knight-Jones TJD, Rushton J. The economic impacts of foot and mouth disease—What are they, how big are they and where do they occur? Preventive Veterinary Medicine. 2013;112(3–4):161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material for Serotype-specific transmission and waning immunity of endemic foot-and-mouth disease virus including dates of serology sampling (A Text and Fig A), model derivation (B Text), derivation for age-specific FOI versus time-varying FOI (C Text), additional results from stratifying data by herd management type (D Text and Table A), and additional results from analyzing 2012 serology data (E Text and Table B).
(PDF)
Data Availability Statement
All relevant data are within the paper (in Tables 1 and 2) or the supplementary material.