Abstract
The bacterial plant pathogen Pseudomonas syringae pv. phaseolicola (Pph) colonises the surface of common bean plants before moving into the interior of plant tissue, via wounds and stomata. In the intercellular spaces the pathogen proliferates in the apoplastic fluid and forms microcolonies (biofilms) around plant cells. If the pathogen can suppress the plant’s natural resistance response, it will cause halo blight disease. The process of resistance suppression is fairly well understood, but the mechanisms used by the pathogen in colonisation are less clear. We hypothesised that we could apply in vitro genetic screens to look for changes in motility, colony formation, and adhesion, which are proxies for infection, microcolony formation and cell adhesion. We made transposon (Tn) mutant libraries of Pph strains 1448A and 1302A and found 106/1920 mutants exhibited alterations in colony morphology, motility and biofilm formation. Identification of the insertion point of the Tn identified within the genome highlighted, as expected, a number of altered motility mutants bearing mutations in genes encoding various parts of the flagellum. Genes involved in nutrient biosynthesis, membrane associated proteins, and a number of conserved hypothetical protein (CHP) genes were also identified. A mutation of one CHP gene caused a positive increase in in planta bacterial growth. This rapid and inexpensive screening method allows the discovery of genes important for in vitro traits that can be correlated to roles in the plant interaction.
Introduction
Pseudomonas syringae is a Gram-negative plant pathogenic bacterial species with multiple pathovars that cause a number of diseases of a wide range of plants, but can also exist out of the host plant in non-agricultural environments including rivers and snow pack [1]. P. syringae pv. phaseolicola (Pph) causes halo blight disease of the common bean Phaseolus vulgaris [2, 3]. Pph is typically associated with causing disease lesions in the leaf of bean. However, like most P. syringae strains, Pph usually exhibits an epiphytic lifecycle phase, existing on leaf surfaces [4, 5]. Infection involves movement of Pph from these zones to the inside of plant tissue, usually through wounds or natural openings such as stomata. Upon arrival into the apoplast, the change in environment stimulates the activation of genes such as the type III secretion system (T3SS), which is important for subduing the plant resistance mechanisms [6]. If Pph can successfully evade plant detection, and subsequent triggering of resistance, the pathogen will manipulate the plant cells to obtain nutrients allowing it to replicate. The pathogen will rapidly colonise the plant tissue, usually making polysaccharides and forming biofilms [7]. The pathogen then spreads through the apoplast to colonise uninfected tissue. Eventually, the spreading lesions will cause bacteria to remerge onto the external surfaces of the plants where they can be dispersed.
Identification of the genes involved in plant colonisation and virulence has been a long term goal of P. syringae research. Different screening methods have been used to detect genes involved in colonisation and pathogenicity of P. syringae. For example, a modified in vivo expression technology (IVET) approach found in planta-expressed promoter fusions when P. syringae pv. tomato (Pto) infected Arabidopsis thaliana. The study found some known genes such as T3SS effectors, but it also discovered several novel genes [8]. Another approach has been to screen for virulence genes using libraries of P. syringae transposon (Tn) mutants, and subsequently assessing mutants for alterations in virulence. For example, 2000 individual Tn5 mutants of Pph were inoculated into bean pods and four of these mutants lacked the ability to cause disease or induce the hypersensitivity response (HR) [9]. Similarly, when a Tn5 library of Pto was screened on tomato seedlings, nine out of 920 mutants were avirulent or exhibited very mild disease symptoms, of which five also failed to induce HR [10]. In a different study, six Tn mutants were identified in Pph that had lost the ability to cause disease or elicit the HR on bean. These mutants were subsequently identified as insertions into the hypersensitivity response and pathogenicity (hrp) genes which are responsible for the bacteria’s ability to cause disease and a HR on plants [11]. Tn mutants have also been used to identify genes involved in toxin production by the bacteria, for example, 947 Pto DC3000 Tn5 mutants were screened by dip-inoculation of A. thaliana plants and 37 were found to have reduced virulence [12]. Of these Pto Tn mutants, six were found in previously identified phytotoxin coronatine (COR) biosynthesis genes [13]. Large numbers of Tn mutants have also been used to screen for changes in epiphytic growth on the plant surface, for example, Lindow et al. [5] screened 5300 Tn5 mutants of P. syringae B728A for growth on bean leaves, 82 of which had reduced population size.
All of the above screens have been carried out by inoculating the host plant with individual Tn mutants, which is time consuming and labour intensive. A method to overcome the need for large numbers of plants has been developed more recently by screening of P. syringae pv. maculicola ES4326 Tn mutants on A. thaliana seedlings grown in liquid media in 96-well plates [14]. Inoculation of A. thaliana seedlings with the Tn mutants during cultivation resulted in bleaching, which was directly related to virulence of the mutants. Using this approach around 12600 mutants were screened and 40 hits in a number of interesting genes were identified as having an effect on the pathogen’s ability to cause disease, including genes involved in the T3SS, flagella-based motility and periplasmic glucan biosynthesis.
In addition to being time consuming, screening bacteria on plants is also subject to considerable variation in plant responsiveness. Moreover, most of the approaches above focus on alterations in disease symptoms, which often lead only to the identification of T3SS mutants. However, in vitro testing can be a useful approach to identify gene systems involved in plant colonisation. This is based on the knowledge that bacteria can behave similarly in vitro as they do in vivo, so it is possible to use certain in vitro phenotypic tests as proxies for in vivo behaviour. This type of approach is also useful for considering key processes such as pathogen entry into the leaf and spread within the apoplast. It is, of course, important to acknowledge that some of these systems are almost certainly influenced by environmental signals, but we have tested the tractability of this approach. Here we report the use of Tn screens to identify changes in in vitro phenotypes of Pph and to subsequently correlate them to changes in the plant interaction. It is therefore suggested that this would be a reliable and inexpensive method for the identification of genes involved in colonisation and virulence in P. syringae and other similar pathogens of both plants and animals.
Materials and Methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria Bertani (LB, Difco) media supplemented with 15 g/L Bacteriological No.1 agar (Oxoid, UK) and Pseudomonas strains were grown at 25°C on Kings medium B (KB, Difco) [15] or in LB broth. Antibiotics were used at the following concentrations (μg/ml): kanamycin (Km) 50, gentamicin (Gm) 10, and nitrofurantoin (NF) 100.
Table 1. Bacterial strains and plasmids.
| Name | Details | Reference |
|---|---|---|
| P. syringae pv. phaseolicola | ||
| 1302A | Race 4, type strain | [16] |
| 1448A | Race 6, type strain | [16] |
| E. coli | ||
| pCR2.1 TOP10F | Competent cells | Invitrogen, UK |
| S17-1 λpir | Containing plasmid pSCR001 | [17] |
| Plasmids | ||
| pBBR1MCS-5 | Broad host cloning vector, GmR | [18] |
| pSCR001 | Carries IS-Ω -Km/hah, KmR | [17] |
| pCR2.1 cloning vector | KmR AmpR | Invitrogen, UK |
KmR, AmpR and GmR indicate resistance to Kanamycin, Ampicillin and Gentamycin respectively.
Transposon library construction
Tn mutant libraries of Pph strains 1302A and 1448A were generated following bi-parental mating with E. coli S17-1λpir carrying IS-Ω-Km/hah (donor) [17]. Essentially 500 μl of an overnight culture of the donor strain was inoculated into 10 ml of LB broth and incubated for 3 hrs. Following incubation, 30 μl of donor was mixed with 100 μl of an overnight culture of the required Pph strain and the mixture transferred to the centre of an LB plate before incubation at 30°C for 48 hrs. This conjugation mixture was then diluted, plated on KB + Km + NF and incubated at 25°C for 72 hrs. Single transformation mutants were selected and inoculated on KB + Km in a 48 colony grid pattern in a 9 cm petri dish. Tn colonies were numbered, as for example, 14–3.14 (strain Pph 1448A-plate 3.colony 14). The insertion position of the Tns in the Pph genome were identified using Arbitrary Primed (AP)-PCR and DNA sequencing (Eurofins Genomics) [19, 20] (see S1 Table for primer sequences). The resulting sequences were analysed using the BLAST programme from the NCBI and insertion point determined using Artemis: Genome Browser and Annotation Tool [21] (http://www.sanger.ac.uk/resources/software/artemis/).
Screening of transposon insertion libraries
Colony morphology screening
Tn libraries were replica plated onto KB agar plates. The wild type (WT) control strains were spotted onto the plate separately. Each library plate was tested in triplicate and agar plates were incubated at 25°C for three days before visual observation of the colony phenotypes. Selected individual mutants were confirmed either by streak plating on to KB agar or grown overnight in liquid culture, diluted and spread onto KB agar. Plates were incubated at 25°C for 48 hr before microscopic visualization and photography.
Swarming motility screening
Tn libraries were replica plated onto 0.3% LB agar. The WT control strains were spotted onto the agar plate separately. Each library plate was tested in triplicate and agar plates were incubated at 25°C for 72 hr before visual observation of the colonies’ swarming motility. Selected mutants were tested for swarming motility individually. Mutants and WT strains were spotted into the centre of a 0.3% LB agar plate. Each strain was tested in triplicate and plates were incubated and observed for up to six days at 25°C.
Biofilm attachment screening
Two 1302A Tn library plates were replica plated into a 96-well microtitre plate containing 200 μl LB broth + Km. The control 1302A WT was tested without Km by replacing one mutant strain per plate. Plates were incubated at 25°C for seven days without shaking. The biofilm attachment assay of the 96-well plates and individual mutants was a modified protocol based on Spiers et al. [22]. Essentially, after seven days the bacteria attached to the microtitre plates were stained with 1% crystal violet (CV). The density of the eluted CV was determined at OD570 using a microtitre plate reader (FLUOstar OPTIMA, BMG Labtech, Germany). To test individual mutants, strains were incubated in 10 ml of LB broth + Km and kept static at 25°C for seven days. After seven days, attached bacteria were stained as described above with 1 ml of 1% CV.
In vitro growth rate
Three replicates of each bacterial mutant were grown overnight in LB broth + Km and diluted to 8x108 CFU/ml (OD600 1.0). Cells (100 μl) were subcultured into 10 ml fresh LB broth and the optical density measured and recorded. All cultures were incubated at 25°C with shaking (10g) and their optical densities measured and recorded after 16 and 24 h. Log values of growth were calculated and plotted to compare growth of WT versus knockout mutant and ensuing complemented strains.
Plant growth conditions and pathogenicity testing
Pathogenicity tests were carried out on bean leaves (cultivars Canadian Wonder (CW) and Tendergreen (TG) and pods (cultivar unknown) as described previously [23]. Disease was observed as water-soaking lesions around the inoculation site, whereas resistance (HR) was observed as tissue browning. For in planta growth rates, three replicates of each cell suspension were grown overnight in LB broth + Km and diluted to 8x107 CFU/ml (OD600 0.1) for infiltration and 8x108 CFU/ml (OD600 1.0) for spray inoculations. Cells were inoculated into 10 day old bean leaves via a syringe and needle or sprayed onto both surfaces of the leaf using a perfume atomiser until running wet and allowed to dry. Plants were incubated for 48 h (infiltration) or 120 h (spray inoculations) at 23°C, 80% humidity. A 1 cm core borer was used to harvest the inoculated area, this was homogenised in ¼ Ringers solution before being diluted and spread plated onto KB + Km. Total CFU/ml were counted and plotted to compare growth of WT versus disruption mutant and ensuing complemented strains.
Cloning and complementation
In vitro complementation of selected Tn mutants was carried out following PCR amplification of the disrupted genes (see S1 Table for primer sequences) and insertion of the resulting fragment into TOPO pCR2.1 (Invitrogen) by TA cloning following the manufacturer’s instructions. The resulting construct was digested with restriction enzyme EcoR1 and the fragment ligated into broad host range vector pBBR1MCS-5 [18]. These constructs were introduced into their respective mutant strain (TnC) via electroporation, carried out as per the method of Keen et al. [24]. An empty vector was also transformed into the strains to use as a control (TnE).
Results and Discussion
In vitro screening of Psuedomonas syringae pv. phaseolicola transposon mutants
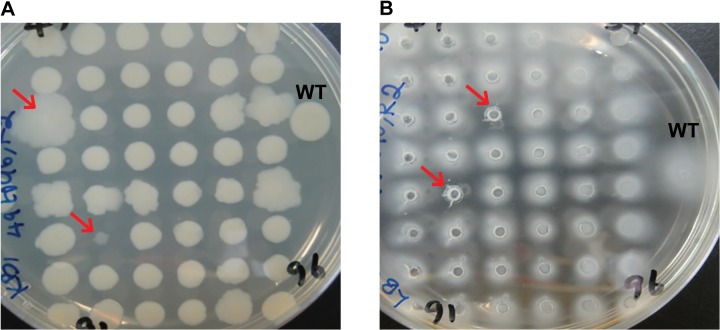
Tn mutant libraries (960 disruption mutants each) were made for Pph strains 1448A (causes disease on CW and TG) and 1302A (causes disease on CW and HR on TG) to screen for mutants that exhibited phenotypic changes. Firstly, colonies were screened for changes in colony morphology which may reflect changes in cell wall structure or motility that are important for colonisation of the plant. Pph1448A::Tn and 1302A::Tn libraries were replica plated (three replicates) onto KB plates, incubated at 25°C for 72 hr and visually compared to their equivalent WT that was also included on each 48 colony plate (Fig 1A). A total of 15 colonies for 1302A::Tn and 42 colonies for 1448A::Tn were observed as being consistently different, that is, either larger or smaller, to their WT equivalents (Table 2). The morphology of selected mutants was also confirmed by subsequently plating out the mutants individually: see Fig 2 for examples.
Fig 1. Screening of Pseudomonas syringae pv. phaseolicola 1448A transposon disruption mutants.
Tn mutant colonies (48) were inoculated onto; A. standard agar plates and changes in colony morphology recorded after 72hr, B. soft agar plates and reduction in swarming ability recorded after 72hr. Selected mutants are highlighted with red arrows. WT, wild type.
Table 2. Number of selected Pseudomonas syringae pv. phaseolicola transposon disruption mutants from 960 screened for each phenotype.
| Phenotypic screening | Pph 1302A | Pph 1448A |
|---|---|---|
| Reduction in swarming motility | 40 | 57 |
| Small colonies | 11 | 17 |
| Large colonies | 4 | 25 |
| Reduced biofilm formation | 6* | NT |
NT—Not tested.
*950 mutants screened.
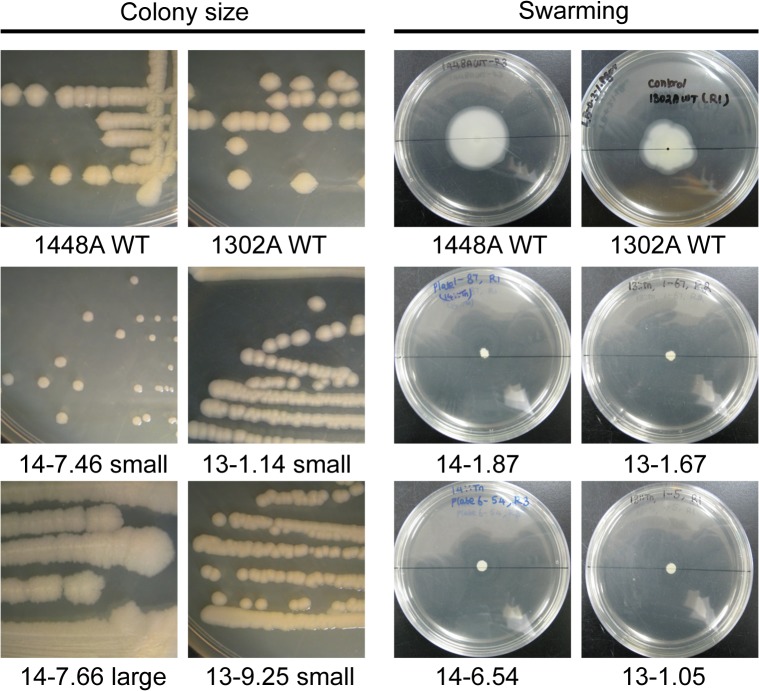
Fig 2. Examples of phenotypes of selected individual Pseudomonas syringae pv. phaseolicola transposon disruption mutants.
Colony size selected mutants were individually streaked onto agar plates and colonies observed after 48 h incubation at the same magnification. Individual swarming mutants were inoculated into the centre of a 9 cm soft agar plate and observed after 144 h incubation. 14-, Pph 1448A; 13-, Pph 1302A; WT, wild type.
The mutant libraries as above were also screened for strains exhibiting altered ability to swarm on soft agar, to identify genes potentially involved in spreading motility, which has been shown to be important for virulence on the plant [25]. Mutant and wildtype strains were replica plated onto soft agar plates to observe swarming motility (movement over the agar surface [26]). Three replicates were carried out for each set of 48 mutant colonies and the plates were incubated at 25°C for 72 hr and visually compared to their equivalent WT (Fig 1B). Mutants that showed a difference to WT on all three replicate plates were individually tested to confirm their altered phenotype (Fig 2). This led to the selection of 97 mutants that showed a reduction in the swarming phenotype (Table 2). No increase in swarming was observed with any of the Tn mutants.
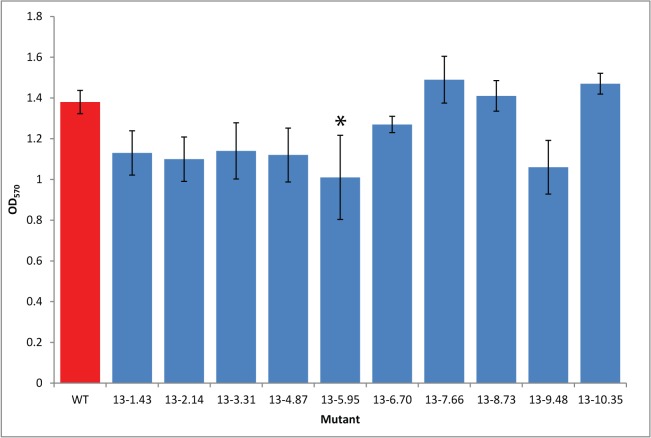
A screen was also used to identify mutants exhibiting reduced adherence to surfaces, a precursor to production of biofilms. Biofilm formation by bacteria is a key strategy in the colonisation of natural environments such as plants [27]. Attachment of bacteria to surfaces is the first stage in the formation of a biofilm and this can be measured by detecting the presence of cells attached to a surface using Crystal Violet (CV) staining [22]. The 1302A::Tn library was replica plated into 96 well plates containing LB media for biofilm assay. One colony from each 96 well plate was removed and replaced with 1302A WT. For the biofilm assay, after seven days, bacteria attached to the sides of the wells were stained with CV, the concentration of which was then measured spectrophotometrically. For each 96 well plate, one mutant that had the highest attachment and one that had lowest attachment compared to WT were selected and tested individually (30 ml vial, 10 ml LB). Individual tests showed that none of the mutants were attaching more than WT which was inconsistent with the initial assay. However, of the 10 mutants selected as having lower attachment in the initial screen, six appeared to be reduced compared to WT although in only one of these was the difference statistically significant at p<0.05 (Fig 3). The six selected biofilm mutants were included in the sequence analysis of the selected mutants (below).
Fig 3. Pseudomonas syringae pv. phaseolicola transposon disruption mutants selected in a biofilm attachment assay.
Transposon mutants and Pph 1302A wild type strain (WT) were cultured static in 10 ml broths. After seven days crystal violet was used to measure (OD570) the attachment of the cells to the culture vessel surface. Error bars represent standard error of the mean of three biological experimental replicates. *above bars indicate a significant difference compared to WT at p<0.05 assessed by a Student’s t-test.
Sequence analysis of selected Pseudomonas syringae pv. phaseolicola transposon mutants
From the phenotypic screens of the Tn mutant libraries, 160 mutants were selected as showing differences to the WT strain. The 160 selected mutants consisted of 106 individual Tn mutants as some mutants were selected in multiple screens. DNA sequences were obtained from 104 of the 106 mutants (we failed on multiple occasions to obtain sequence from two mutants) and the insertion points of the Tn in the P. syringae 1448A genome were determined (S2 Table). The Tn hits were plotted on to a genome map of Pph 1448A using DNAPlotter (Fig 4). The map of the Tn hits shows a diverse distribution of gene knockouts around the genome, with a Tn rich area around 3,900,000 bp, which corresponds to the flagella gene cluster.
Fig 4. Insertion points of transposon hits on a map of the circular chromosome of Pseudomonas syringae pv. phasiolicola 1448A.
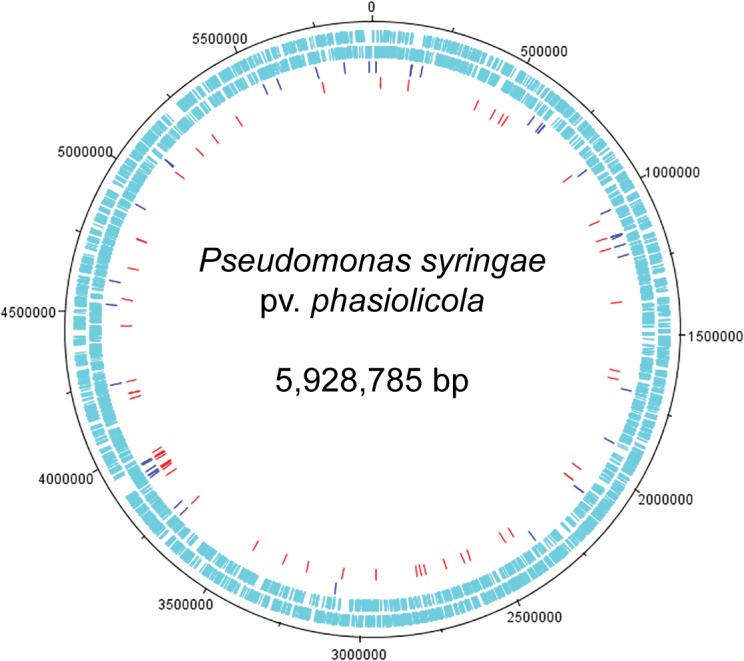
Outer two circles show positions of protein-coding genes on the plus and minus strands. Red dashes, Pph 1448A Tn hits (56); blue dashes, Pph 1302A Tn hits (41). The genome map was generated by using the DNAplotter [28].
Overall most of the Tn hits were in chromosomal genes, but six Tn hits were located on plasmids and one 1302A mutant (13–10.68) had the transposon located in genomic island PPHGI-1 [29]. One of the plasmid Tn hits was to the large plasmid of 1448A and five of the plasmid hits were from the Tn library of Pph strain 1302A, which doesn’t currently have a genome sequence deposited. These five plasmid mutations were found on genes annotated on plasmids in P. syringae B76, savastanoi 3335, maculicola ES4326 and Pto DC3000.
Some genes had Tn insertions identified in the same gene from the different screening methods used here, for example, mutants (13–1.10 and 13–1.12) of 1302A were found from two different screens, for swarming and large colony formation, and the mutated gene is a mannosyltransferase. Also some genes had Tn insertions identified in the same gene from both strains tested, for example, one 1448A mutant and two 1302A mutants (14–6.14, 13–3.09, 13–9.25) had an insertional inactivation of the same OmpA domain protein, but each mutant was found via different phenotypic screens, large colony, swarming ability and small colony respectively. OmpA domain proteins are a family of outer membrane proteins found mainly in Gram-negative bacteria and have been implicated in pathogenicity, for example in bacterial adhesion [30]. However, it is interesting to also note that this OmpA domain protein, PSPPH_0123, has been identified as belonging to the type VI secretion system (T6SS), and labelled as impL in Pph 1448A [31]. The T6Ss secretion systems of Gram-negative bacteria is known to translocate effector proteins into eukaryotic host cells [31] and play a role in bacterial competition [32]. Tn hits in outer membrane proteins could be expected to affect the bacterial cell surface, which may then influence colony size and ability to swarm as was observed in our screen. It was interesting to observe, however, that the mutation in 1302A strain 13–9.25 made the colony smaller while one of the 1448A mutations in the same gene in 1448A 14–6.14 made the colony larger. We also found a number of hits in putative membrane proteins from different screens (14–6.69 and 13–1.01, large colony; 13–1.19, reduced swarming). Some screens, for example small colony size, found a number of hits in related genes. For example four hits were found in pyr genes (14–5.32, pyrB; 14–7.80, pyrD; 13–5.35, pyrF; 13–5.76, pyrF). A number of mutations (11) were identified in genes annotated as ‘transporters’, these came from mutants of both strains and from all the phenotypic screens described.
A number of Tn insertions were identified in genes involved in motility. Seventeen Tn insertions were found in genes involved in the flagella biosynthesis system and associated chemotaxis genes (Table 3), all of which were selected on the basis of reduced swarming ability. Flagella are appendages conferring motility for a number of bacteria [33]. Fourteen Tn mutants described in this study were part of the flagella gene regulon and included mutation of nine genes involved in the flagellum motor/switch (fliN, fliM), hook complex (fliKI, flgE, flgD), the flagellar export pathway (flhB, fliO) and a transcription regulator (fleS) [34]. Schreiber et al. [14] also found a number of Tn hits in flagellar genes in their in vitro screen of P. syringae pv. maculicola Tn mutants against A. thaliana seedlings. They found significant reductions in in planta growth compared to WT when the mutants were inoculated onto plants by spray inoculation [35] but not by pressure infiltration.
Table 3. Pseudomoas syringae pv. phaseolicola transposon insertions mutants identified in the flagellar and associated chemotaxis genes.
| Tn mutant | Locus | Gene | Product |
|---|---|---|---|
| 13–1.42 | PSPPH_0554 | MotA family motility protein | |
| 14–6.54 | PSPPH_3360 | cheA2 | Chemotaxis sensor histidine kinase CheA |
| 13–8.35 | PSPPH_3361 | cheZ | Chemotaxis protein CheZ |
| 14–7.41 14–10.74 13–5.78 | PSPPH_3367 | flhB | Flagellar biosynthetic protein FlhB |
| 13–10.60 | PSPPH_3371 | fliO | Flagellar protein FliO |
| 14–10.63 | PSPPH_3372 | fliN | Flagellar motor switch protein FliN |
| 14–4.52 | PSPPH_3373 | fliM | Flagellar motor switch protein FliM |
| 14–6.51 | PSPPH_3375 | fliK | Flagellar hook-length control protein FliK |
| 13–10.79 | PSPPH_3386 | fleS | Flagellar sensor histidine kinase FleS |
| 14–9.76 | PSPPH_3398 | flgJ | Peptidoglycan hydrolase FlgJ |
| 14–10.54 13–1.67 | PSPPH_3405 | flgE | Flagellar hook protein FlgE |
| 14–2.59 | PSPPH_3406 | flgD | Basal-body rod modification protein FlgD |
| 13–1.05 14–1.87 | PSPPH_3414 | Flagellar basal-body P-ring formation protein FlgA, putative |
13-, Pph 1302A; 14-,Pph 1448A.
Of the three remaining motility-related mutants, two strains had Tn mutations in genes associated with chemotaxis (14–6.54, cheA2; 13–8.35, cheZ). CheA is a histidine protein kinase and CheZ is a signal terminator and both are part of the chemotaxis signalling transduction pathway that connects environmental signals to the flagella and affects the rotation and direction of movement of the bacteria [36, 37]. CheA accepts an activation signal from methyl-accepting chemotaxis proteins (MCPs) via CheW and sets off a signalling cascade that result in the stimulation of the clockwise rotation of the flagella. CheZ allows rapid termination of the signal and therefore gradient sensing. The third mutation was found in motA; MotA (13–1.42) is one of four cytoplasmic membrane proteins that act as motor proteins at the base of the flagellum in P. syringae pathovars. A deletion mutant of all three genes has been shown to not only completely abolish flagella-associated swarming and swimming motilities, but also reduce the ability to cause disease in tobacco leaves by P. syringae pv. tabaci [25]. A number of these 104 mutants are described in more detail below.
Detecting alterations in virulence associated with the mutations
A key test of our approach was to determine whether any of the Tn mutants identified in the in vitro proxy phenotype screens corresponded to alterations in in planta growth and/or symptom defects. We therefore used bought-in bean pods for the tests and confirmed that the 1302A and 1448A WT strains caused HR and disease symptoms, respectively, in pods (S1 Fig). All 106 selected Tn mutants were tested in the bean pods. None of the 1302A mutants exhibited an altered response in causing a HR. However, some 1448A mutants showed a number of differences in symptoms to the WT (S2 Table). For example, of the four strains with mutations in pyr genes (14–5.32, pyrB; 14–7.80, pyrD; 13–5.35, pyrF; 13–5.76, pyrF), two affected symptoms on bean pods (these will be discussed in more detail later). Of the genes annotated as transporters, three reduced the symptoms of disease on bean pods; these were annotated as a major facilitator family transporter (14–6.12), a multidrug efflux transporter (14–10.90) and a putative spermidine/putrescine ABC transporter permease protein (14–6.19). This test confirmed alterations in disease symptoms which could be observed in Tn mutants selected through our in vitro screens.
In vitro growth analysis of selected mutants
A more manageable number of Tn mutants (22) were selected for further investigation (Table 4), based on representation of each phenotypic screen, changes in their response on inoculated bean pods and some mutants with gene knockouts in known virulence factors. We first tested the in vitro growth of the mutants compared to the WT to determine whether the mutations fundamentally altered bacterial growth and to provide the foundation for in planta population growth analysis. The mutants were grown in liquid culture (LB broth) and their OD600 was measured after 24 hours (Fig 5). We observed that a number of the small colony mutants grew to a similar density to the WT indicating that small colony phenotype was not exclusively due to altered growth rate. However, several mutants were significantly different to the WT. Mutants 14–4.44, 14–5.32 and 14–7.09 which have gene disruptions annotated as carA, carB and pyrB all showed very low growth in liquid culture. Genes carA, carB and pyrB are involved in pyrimidine biosynthesis [38, 39]. Mutant 14–7.80 has a disruption in pyrD that is annotated as a dihydroorotate dehydrogenase, which is also involved in pyrimidine biosynthesis, but this does not appear to have affected in vitro growth as much as disruptions in the other genes.
Table 4. Characteristics of selected Pseudomonas syringae pv. phaseolicola transposon disruption mutants.
| Mutant number | Phenotype screen | Tn insertion point (bp) 1 | Gene name/description (% for 1302A hits in 1448A genome) | Locus tag | Bean pod and leaf 2 | In planta growth–TG (% WT) 3 | In planta growth–CW (% WT)3 |
|---|---|---|---|---|---|---|---|
| 13–2.14 | Biofilm | P.s. tomato DC3000 plasmid A, conserved hypothetical protein (88%) | PSPPH_B0022 | HR | 121±11 | 133±6 | |
| 14–7.66 | Large | 3964031..30 | Conserved hypothetical protein | PSPPH_3429 | D | 122±6 | 164±8 |
| 13–1.14 | Small | 13197..98 | plsC, hdtS protein (100%) | PSPPH_0009 | HR | 100±2 | 102±2 |
| 13–9.25 | Small | 140944..45 | impL, OmpA domain protein (98%) | PSPPH_0123 | HR | 76±5 | 84±10 |
| 14–4.22 | Small | 411911..12 | mdoG1, periplasmic glucans biosynthesis protein MdoG | PSPPH_0359 | Null | <1 | <1 |
| 14–4.35 | Small | 4244840..39 | cobyric acid synthase CobQ | PSPPH_3697 | Null | <1 | <1 |
| 14–4.44 | Small | 479168..69 | carA, carbamoyl-phosphate synthase, small subunit | PSPPH_4203 | Null | <1 | <1 |
| 14–5.32 | Small | 537617..18 | pyrB, aspartate carbamoyltransferase | PSPPH_0473 | Null | <1 | <1 |
| 14–6.32 | Small | 2582863..64 | cobalamin synthesis protein/P47K family protein | PSPPH_2224 | Null | 105±7 | 107±2 |
| 14–7.09 | Small | 4791135..36 | carB, carbamoyl-phosphate synthase, large subunit | PSPPH_4202 | Null | <1 | <1 |
| 14–7.46 | Small | 5082664..63 | purH, bifunctional purine biosynthesis protein PurH | PSPPH_4449 | Null | 94±4 | 92±1 |
| 14–7.80 | Small | 2436984..83 | pyrD, dihydroorotate dehydrogenase | PSPPH_2077 | Null | <1 | <1 |
| 13–1.5 | Swarm | 3947618..17 | Flagellar basal-body P-ring formation protein FlgA, putative (99%) | PSPPH_3414 | HR | 80±11 | 88±3 |
| 13–1.67 | Swarm | 3940356..57 | flgE, flagellar hook protein FlgE (80%) | PSPPH_3405 | HR | 73±9 | 85±7 |
| 13–10.60 | Swarm | 389967..66 | fliO, flagellar protein FliO (96%) | PSPPH_3371 | HR | 128±12 | 189±9 |
| 14–1.87 | Swarm | 3947250..60 | Flagellar basal-body P-ring formation protein FlgA, putative | PSPPH_3414 | D | 66±3 | 65±4 |
| 14–2.59 | Swarm | 3941641..42 | flgD, basal-body rod modification protein FlgD | PSPPH_3406 | D | 68±1 | 70±6 |
| 14–4.52 | Swarm | 3901187..86 | fliM, flagellar motor switch protein FliM | PSPPH_3373 | D | 61±2 | 65±2 |
| 14–6.54 | Swarm | 3887619..18 | cheA2, chemotaxis sensor histidine kinase CheA | PSPPH_3360 | D | 69±5 | 65±2 |
| 14–7.41 | Swarm | 3897582..81 | flhB, flagellar biosynthetic protein FlhB | PSPPH_3367 | D | 80±7 | 73±4 |
| 14–10.54 | Swarm | 3940841..42 | flgE, flagellar hook protein FlgE | PSPPH_3405 | D | 68±1 | 71±5 |
| 14–10.63 | Swarm | 3900133..34 | fliN, flagellar motor switch protein FliN | PSPPH_3372 | D | 64±8 | 73±1 |
1Insertion point, where given, is in Pph 1448A genome (accession number CP000058).
2Reaction on bean pods and leaves: HR, hypersensitive response; D, Disease; Null, no symptoms.
3 In planta growth in bean cultivar Tendergreen (TG) and Canadian Wonder (CW) after 2 days compared to cognate WT (1302A growth level was 4.10x106 in TG and 1.80x107 in CW whereas 1448A growth level was 1.40x107 in TG and 2.30x107 in CW cfu/ml).
± represent standard error of the mean of three biological experimental replicates. A value of 100% means that mutant shows equal growth to WT. Phenotypic screens: Swarm, swarming reduction; Small, small colony; Large, large colony; Biofilm, biofilm formation. WT, wild type; 13-, Pph 1302A; 14-, Pph 1448A.
Fig 5. In vitro growth of Pseudomonas syringae pv. phaseolicola transposon mutants.
Transposon mutants and wild type (WT) strains were inoculated in LB broth with shaking for 24 hr and OD600 recorded. Phenotypic screens: Sw, swarming reduction; S, small colony; L, large colony; Bf, biofilm formation. 14-, Pph 1448A; 13-, Pph 1302A. Error bars represent standard error of the mean of three biological experimental replicates. *above bars indicate significant differences compared to WT at p<0.05 assessed with Students t-test.
A number of the selected 1302A mutants had a significantly increased growth rate in vitro compared to 1302A WT. These increased growth rate mutants included a swarming mutant (13–1.67) that was annotated as an insertion in the 1448A flagella gene, flgE (80% identical) and a mutant from the biofilm screen (13–2.14) that is 88% identical to a conserved hypothetical protein from a Pto DC3000 plasmid. A small colony mutant (13–1.14) also had an increased growth rate in vitro. Mutant 13–1.14 is annotated as plsC (100% identical) in 1448A and described as an HdtS protein. HdtS proteins have been described as N-acylhomoserine lactone (AHL) synthases that are involved in production of a quorum-sensing molecule [40]. However, Cullinane et al. [41] found that an hdtS mutant in P. fluorescens made normal levels of HSL and that in this bacterium the hdtS gene encodes a primary lysophosphatidic acid acyltransferase. The P. fluorescens hdtS mutant showed significantly impaired growth rate which we didn’t observe with 13–1.14 in liquid media, however, we did select it on a plate culture as a small mutant.
In planta growth analysis of selected mutants
As we did not know the cultivar of the bean pods used for the initial pathogenicity screening, we analysed in planta growth rate of bacterial strains infiltrated into leaves of bean cv. TG and CW; the symptoms in the leaves were also observed. Table 4 shows the bean pod results (from initial screen), leaf symptoms and the in planta growth rates. We observed that 16 strains exhibited growth rates in planta that were reduced by more than 20% of the WT growth rate and three strains exhibited significantly increased growth; only three mutants exhibited near WT growth levels.
Considering first the reduced growth rate strains, six of the reduced growth rate mutants (all 1448A strains) exhibited less than 1% growth compared to the WT in planta and all six gave rise to a null reaction in bean pods. These corresponded to mutations in carA, carB, pyrB, pyrD, cobQ and mdoG1. Only the carA, carB and pyrB mutations corresponded to dramatic growth loss in vitro (OD of 0.3 cf 2.0 for WT); the pyrD mutation caused a modest in vitro decrease in growth (Fig 5). This suggests these four mutants have auxotrophic phenotypes critical for general cell growth. By contrast, the cobQ (14–4.35) and mdoG1 (14–4.22) mutants exhibited small reduction and no change, respectively, in in vitro growth compared to the WT. This suggests these genes are much more critical for in planta growth and symptom development. The cobQ and mdoG1 mutants were both found as small colony phenotypes. cobQ encodes a cobyric acid synthase which is involved in cobalamin (vitamin B12) biosynthesis [42]. MdoG encodes a periplasmic glucans biosynthesis protein and other work has shown that Salmonella enterica serovar Typhimurium mutants in opgGH (previously referred to as mdoGH) were compromised in virulence in mice compared to WT strain [43].
The remaining ten mutants (a mixture of 1448A and 1302A mutants) exhibited modest reductions in in planta growth compared to their cognate WT. The 1448A strains were still able to cause disease in TG and the 1302A strains still caused an HR, demonstrating these mutations did not alter the basic interaction phenotype. Nine of the strains were mutated in flagellum and chemotaxis genes, all found in the swarming screen and including flgA found in both strains (13–1.5, 14–1.87). The tenth mutant was a 1302A knockout of an OmpA-domain protein gene impL (13–9.25). As discussed earlier the flagellum mutants have previously been shown to be reduced in pathogenicity [34] and OmpA protein is an outer member protein and has been implicated in bacterial pathogenicity [30]. The reduction of growth in planta of the impL mutant may suggest a role for the T6SS system in Pph 1302A. The T6SS has previously been shown to have a role in virulence of Pantoea ananatis in onion plants [32].
Only three mutants exhibited significantly higher in planta growth than their respective WT strain. Of the three faster growing mutants, two were 1302A strains, mutated in a putative plasmid-borne conserved hypothetical protein (CHP) gene (found in biofilm screen; 13–2.14) and, in fliO, a flagellar protein (found in swarming screen; 13–10.60). The single 1448A higher growth mutant (14–7.66) was found because of its large colony phenotype and is mutated in a CHP gene. The CHP mutants both exhibited higher in vitro growth whereas the fliO mutant was not significantly difference from WT in vitro.
Complementation of disruptions in a selection of mutants
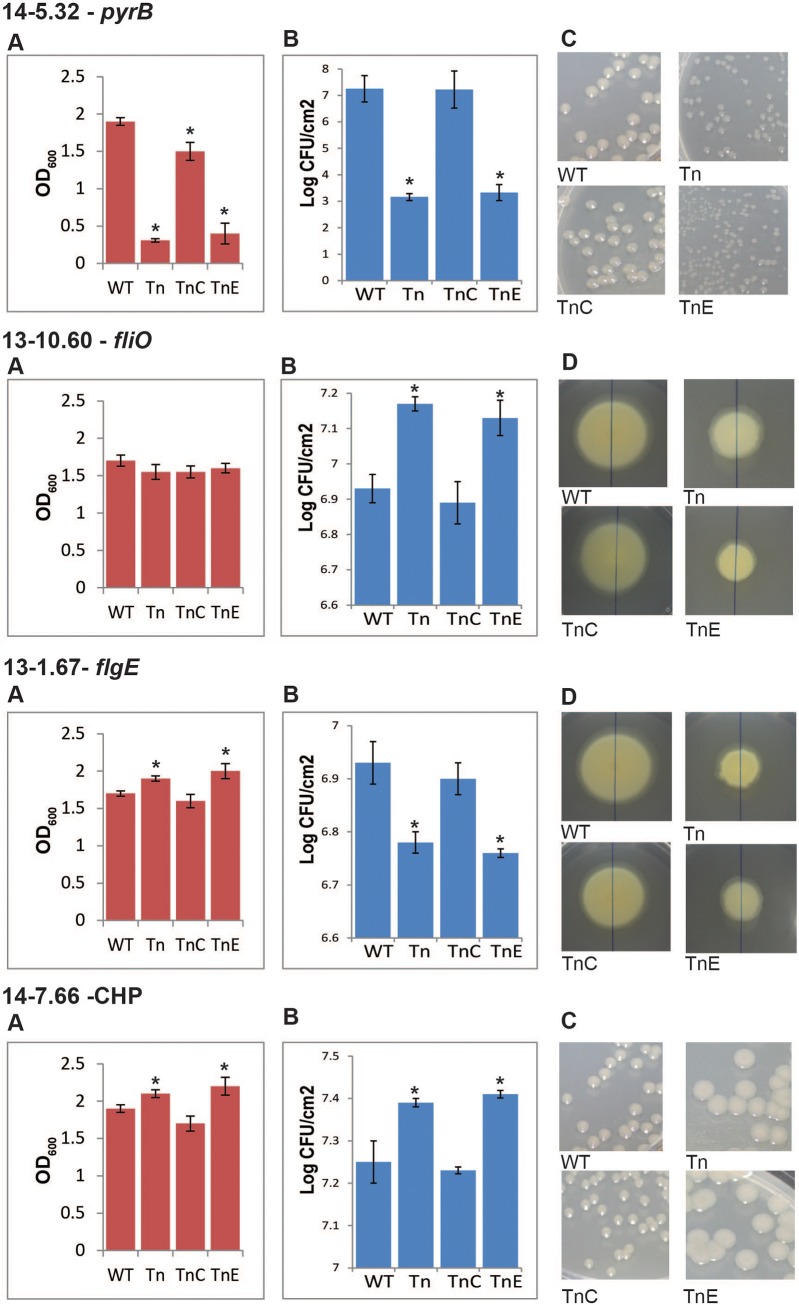
To validate our approach and confirm that the phenotypes observed from the mutants were due to the mutated genes, complementation experiments were undertaken. We focused on four mutants that showed significant differences to WT growth in planta and represented a selection of the phenotypes we had identified. For example one of these, the 1302A::fliO mutant, showed enhanced growth in planta despite still causing an HR. Each gene (open reading frame plus some flanking sequence) was amplified from the WT strain and cloned into broad host range vector pBBR1MCS-5 before electroporation into the mutant strain. An empty vector was also used as a control in all mutants tested. Complementation to WT phenotypes (or near WT) was observed for all four mutants tested, while the empty vector did not affect the mutant phenotypes, confirming that the gene mutated by the Tn was responsible for the change in phenotype (Fig 6; S3 Table).
Fig 6. Complementation of selected Pseudomonas syringae pv. phaseolicola transposon disruption mutants.
Genes identified through transposon (Tn) insertion were cloned into a broad host range vector and transformed into their respective mutant strain (TnC). An empty vector was also transformed into the strains to use as a control (TnE). A number of tests were carried out with these strains: A. in vitro growth in LB broth after 16hrs; B. in planta growth in bean cultivar Canadian Wonder after 2 days; C. colony size after 2 days incubation, shown at the same magnification; D. swarming ability in soft agar after 5 days incubation, shown at the same magnification. 14-, Pph 1448A; 13- Pph 1302A; CHP, conserved hypothetical protein. *above bars indicate significant differences compared to WT at p<0.05 assessed with Students t-test.
The small colony mutant, 14–5.32, had a disrupted pyrB in Pph 1448A. This gene is annotated as an aspartate carbamoyltransferase protein involved in pyrimidine biosynthesis. The mutation in pyrB also leads to a very significant reduction in in planta growth of the bacterium. PyrB is important for bacterial survival, for example, Burns et al. [44] demonstrated that pyrB is essential for cell survival of Helicobactor pylori because a knockout of pyrB was lethal to the bacterium. A group A streptococcus (GAS) Tn library was used to identify genes important for growth and/or survival in whole human blood [36]. A pyrB mutant was found to be important for the fitness of GAS strain 5448 in donor blood. In addition, Tn insertion mutants of Francisella tularensis were screened for their inability to invade and replicate in a hepatic carcinoma cell line [45]. Eighteen mutants were identified as defective in intercellular growth in the hepatic carcinoma cell line, one of which was a mutant of pyrB. In this current study it is clear that the disruption in pyrB is severely reducing the ability of Pph to grow in vitro, so it is not surprising that it lacks the ability to grow in planta. However, it does confirm that mutants selected using this Tn mutagenesis approach reflect their phenotypes in planta.
Another mutant that was identified in the initial swarming screen was 1302A mutant 13–10.60, which has a disruption in a gene with 96% similarity to fliO, a flagella gene from Pph 1448A. Mutant 13–10.60 was selected as having a reduced ability to swarm and showed a significant increase in in planta growth, but no difference to WT in vitro growth was observed and the mutant still causes a visible HR in the resistant bean cv. TG. These results suggest that disruption of fliO removes some of the plants ability to restrict the growth of the bacterium. FilO is predicted to be part of the flagellar export pathway in P. aeruginosa [34]. If this is also true in P. syringae, then the mutation could be reducing the amount of flagellin produced in this mutant. The flg22 domain of flagellin from P. syringae pv. tabaci, which is encoded by fliC, has previously been shown to act as a microbe-associated molecular pattern (MAMP) [46]. MAMPs are conserved microbial molecules that are recognised by the plant and induce defence reactions [47]. Therefore, mutant 13–10.60 may be producing less flagellin and not triggering basal resistance in the plant, thus enabling the increased growth in early stage colonisation that we observed.
In contrast mutant 13–1.67, which was also selected initially for its reduced swarming ability, was mutated in a gene that showed 80% similarity to flgE of Pph 1448A, which is the flagellar hook protein. However this mutant (13–1.67) is reduced in its in planta growth rate (Fig 6) and is also slightly increased in its ability to grow in vitro compared to the 1448A WT. This reduced ability to grow in planta was observed independently in mutant 14–10.54, which also has a hit in flgE and also has a reduced growth rate (68% of WT) in planta (Table 4). Discovery of flagellar genes being involved in virulence to the plant in Pph is not unexpected and disruptions in flagella genes have previously been shown to reduce pathogen virulence in a number of plant pathogens [25, 33, 48, 49]. However, these studies have tended to show reduced in planta growth rate when the mutants were spray inoculated, rather than inoculated using an infiltration method as was carried out here. In order to compare the two inoculation methods we tested mutant 13–10.60 (fliO) and 13–1.67 (flgE) by spray inoculation method and saw no differences in the results compared to our infiltration method results (S4 Table). It may be of interest to further investigate why a mutation in the flagella hook protein is causing a significant reduction in bacterial growth once inside the leaf. Confirming the selection of these flagella mutants does however validate the method used here as a way of identifying genes responsible for flagella formation and indicates that such methodology will be useful to identify genes of interest in other less well characterised molecular systems.
Of particular interest for the discovery of new gene functions, the screens used in this work also identified disruptions in seven genes annotated as conserved hypothetical proteins (CHP) that showed altered phenotypes in colony size and swarming screens. One of these was 14–7.66 which has a disruption in a CHP in Pph 1448A genome and was selected as a large colony mutant in the initial screens. 14–7.66 exhibited a significant increase (110%) in in vitro growth and significantly increased, even higher, growth in planta (122% of WT). Sequence analysis of 14–7.66 showed it to be a hypothetical protein of 224 amino acids and is predicted, using InterProScan 5 [50, 51], to have a transmembrane domain and a SH3 domain. The latter would allow it to bind other proteins at proline rich regions and using String 9.1 [52], with P. syringae 1448A as a target organism, predicts possible binding partners include glycerol-3-phosphate acyltransferase and a histidine kinase, the latter suggesting that it could be involved in modulating signalling. It is possible that this CHP protein has a region external to the cell and therefore may be acting as MAMP. If this CHP can act as a MAMP it would explain why pathogenicity can increase when the protein is not functioning but further work will have to be carried out to investigate this.
Conclusions
In this study we used random mutagenesis in combination with a series of phenotypic screens representing proxies of in planta growth traits to identify mutants with altered phenotypes. Our aim was to test whether the mutants showed a parallel phenotype to the plant response. Previous screening approaches have tended to screen Tn libraries of pathogenic bacteria with a plant-based assay to identify mutants of interest and then obtain DNA sequences to identify the genes involved [9, 10, 11, 12, 14]. Here we took the approach of screening for changes in phenotype that may be associated with the bacteria’s ability to interact with the plant, namely swarming ability, colony size (possibly reflecting changes in the cell wall) and biofilm formation. These assays can be done on a large scale as they are carried out with 48 mutant colonies at a time in a 9 cm Petri dishes or 96 well plates and are relatively cheap, requiring no plant growth facilities for the initial screen. With the decrease in the expense of DNA sequencing it is possible to sequence a large number of Tn hits at a reasonable cost. It was hoped that by using this approach we would find genes of interest that could be used for further investigation of the bacteria-plant interaction.
Overall we screened 1920 Tn mutants and of these 106 were selected for further analysis; of these, Tn-chromosome junction sequence was obtained from 104 strains. After further selection and characterisation we confirmed that some of these Tn hits were indeed important for the bacteria-plant interaction. For example the swarming assay produced a number of hits that were in the flagellum system and those mutants exhibited reduced in planta growth. Other types of genes identified included those annotated as being involved in chemotaxis, membrane proteins, nutrient biosynthesis and transporters.
Interestingly we did not find any hits in hrp genes or effector genes here as have been found previously when mutants have been screened directly on plants [11, 14]. However as the hrp system is very well characterised and not considered to be involved in our particular phenotypes used for screening, this was not unexpected. One of the more interesting results of our screen was the discovery of a CHP that was involved in restricting bacterial growth and which, when mutated, enabled the pathogen to grow to higher levels in the plant. This illustrates how this relatively simple screening technique can be used to identify previously unknown genes that may be important in the bacteria-plant interaction or potentially be targets for adaptation that increase pathogen fitness in the host. This method has generated many more genes of interest that will be useful for investigation in future studies, ultimately helping to identify new potential targets for disease control.
Supporting Information
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council for financial support. This work was carried out under the Department for Environment, Food and Rural Affairs’ Plant Health and Seeds Inspectorate license number PHL 51049/202078-2.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the University of the West of England, Bristol (http://www.uwe.ac.uk/) (BM and DLA) and Biotechnology and Biological Sciences Research Council (http://www.bbsrc.ac.uk/) (BB/J014796/1) (HCN, RWJ and DLA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, et al. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2008. March;2(3): 321–34. 10.1038/ismej.2007.113 [DOI] [PubMed] [Google Scholar]
- 2. Arnold DL, Lovell HC, Jackson RW, Mansfield JW. Pseudomonas syringae pv. phaseolicola: from 'has bean' to supermodel. Mol Plant Pathol. 2011;12(7): 617–27. 10.1111/j.1364-3703.2010.00697.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6): 614–29. 10.1111/j.1364-3703.2012.00804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Upper CD, Hirano SS. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Mol. Biol. Rev. 1999;64: 624–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lindow SE, Andersen G, Beattie GA. Characteristics of Insertional Mutants of Pseudomonas syringae with Reduced Epiphytic Fitness. Appl Environ Microbiol. 1993;59(5): 1593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alfano JR, Collmer A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42: 385–414. [DOI] [PubMed] [Google Scholar]
- 7. Godfrey SA, Mansfield JW, Corry DS, Lovell HC, Jackson RW, Arnold DL. Confocal imaging of Pseudomonas syringae pv. phaseolicola colony development in bean reveals reduced multiplication of strains containing the genomic island PPHGI-1. Mol Plant Microbe Interact. 2010;23(10): 1294–302. 10.1094/MPMI-05-10-0114 [DOI] [PubMed] [Google Scholar]
- 8. Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol Microbiol. 2002;44(1): 73–88. [DOI] [PubMed] [Google Scholar]
- 9. Somlyai G, Hevesi M, Bánfalvi Z. Klement Z, Kondorosi Á. Isolation and characterization of non-pathogenic and reduced virulence mutants of Pseudomonas syringae pv. phaseolicola induced by Tn5 transposon insertions. Physiological and Molecular Plant Pathology. 1986;29: 369–380. [Google Scholar]
- 10. Cuppels DA. Generation and Characterization of Tn5 Insertion Mutations in Pseudomonas syringae pv. tomato . Appl Environ Microbiol. 1986;51(2): 323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lindgren PB, Peet RC, Panopoulos NJ. Gene cluster of Pseudomonas syringae pv phaseolicola controls pathogenicity of bean plants and hypersensitivity of non-host plants. J. Bacteriol. 1986;168: 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, et al. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2004;17(2): 162–74. [DOI] [PubMed] [Google Scholar]
- 13. Bender CL, Alarcón-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63(2): 266–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schreiber KJ, Ye D, Fich E, Jian A, Lo T, Desveaux D. A high-throughput forward genetic screen identifies genes required for virulence of Pseudomonas syringae pv. maculicola ES4326 on Arabidopsis. PLoS One. 2012;7(8): e41461 10.1371/journal.pone.0041461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescen. J Lab Clin Med 1954;44: 301–307. [PubMed] [Google Scholar]
- 16. Taylor JD, Teverson DM, Allen DJ, Pastor-Corrales MA. Identification and origin of races of Pseudomonas syringae pv. phaseolicola from Africa and other bean growing areas. Plant Pathol. 1996a;45: 469–478. [Google Scholar]
- 17. Giddens SR, Jackson RW, Moon CD, Jacobs MA, Zhang XX, Gehrig SM, et al. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc Natl Acad Sci U S A. 2007;104(46): 18247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1): 175–6. [DOI] [PubMed] [Google Scholar]
- 19. Manoil C. Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods Enzymol. 2000;326:35–47. [DOI] [PubMed] [Google Scholar]
- 20. Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2003;100(24): 14339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10): 944–5. [DOI] [PubMed] [Google Scholar]
- 22. Spiers AJ, Bohannon J, Gehrig SM, Rainey PB. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol. 2003;50(1): 15–27. [DOI] [PubMed] [Google Scholar]
- 23. Lovell HC, Jackson RW, Mansfield JW, Godfrey SA, Hancock JT, Desikan R, et al. In planta conditions induce genomic changes in Pseudomonas syringae pv. phaseolicola . Mol Plant Pathol. 2011;12(2): 167–76. 10.1111/j.1364-3703.2010.00658.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keen N.T., Shen H. and Cooksey D.A. Introduction of cloned DNA into plant pathogenic bacteria In: Molecular Plant Pathology. 1992 A practical Approach. Gurr S. J et al. , IRL Press, Oxford. [Google Scholar]
- 25. Kanda E, Tatsuta T, Suzuki T, Taguchi F, Naito K, Inagaki Y, et al. Two flagellar stators and their roles in motility and virulence in Pseudomonas syringae pv. tabaci 6605. Mol Genet Genomics. 2011;285(2): 163–74. 10.1007/s00438-010-0594-8 [DOI] [PubMed] [Google Scholar]
- 26. Harshey RM. Bees aren't the only ones: swarming in gram-negative bacteria. Mol Microbiol. 1994;13(3): 389–94. [DOI] [PubMed] [Google Scholar]
- 27. Koza A, Hallett PD, Moon CD, Spiers AJ. Characterization of a novel air-liquid interface biofilm of Pseudomonas fluorescens SBW25. Microbiology. 2009;155(Pt 5): 1397–406. 10.1099/mic.0.025064-0 [DOI] [PubMed] [Google Scholar]
- 28. Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009;25(1):119–120. 10.1093/bioinformatics/btn578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pitman AR, Jackson RW, Mansfield JW, Kaitell V, Thwaites R, Arnold DL. Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr Biol. 2005;15(24): 2230–5. [DOI] [PubMed] [Google Scholar]
- 30. Confer AW, Ayalew S. The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol. 2013;163(3–4): 207–22. 10.1016/j.vetmic.2012.08.019 [DOI] [PubMed] [Google Scholar]
- 31. Sarris PF, Skandalis N, Kokkinidis M, Panopoulos NJ. In silico analysis reveals multiple putative type VI secretion systems and effector proteins in Pseudomonas syringae pathovars. Mol Plant Pathol. 2010;11(6): 795–804. 10.1111/j.1364-3703.2010.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shyntum DY, Theron J, Venter SN, Moleleki LN, Toth IK, Coutinho TA. Pantoea ananatis Utilizes a Type VI Secretion System for Pathogenesis and Bacterial Competition. Mol Plant Microbe Interact. 2015; 28(4): 420–31. 10.1094/MPMI-07-14-0219-R [DOI] [PubMed] [Google Scholar]
- 33. Jarrell KF, McBride MJ The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6(6): 466–76. 10.1038/nrmicro1900 [DOI] [PubMed] [Google Scholar]
- 34. Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa . Mol Microbiol. 2003;50(3): 809–24. [DOI] [PubMed] [Google Scholar]
- 35. Schreiber KJ, Desveaux D. AlgW regulates multiple Pseudomonas syringae virulence strategies. Mol Microbiol. 2011;80(2): 364–77. 10.1111/j.1365-2958.2011.07571.x [DOI] [PubMed] [Google Scholar]
- 36. Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol. 2011;9(3): 153–65. 10.1038/nrmicro2505 [DOI] [PubMed] [Google Scholar]
- 37. Shah D S H, Perehinec T, Stevens SM, Aizawa SI, Sockett RE. The flagellar filament of Rhodobacter sphaeroides: pH-induced polymorphic transitions analysis of the fliC gene. J. Bacteriol. 2000;182: 5218–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Le Breton Y, Mistry P, Valdes KM, Quigley J, Kumar N, Tettelin H, McIver KS. Genome-wide identification of genes required for fitness of group A Streptococcus in human blood. Infect Immun. 2013;81(3): 862–75. 10.1128/IAI.00837-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gallie J, Libby E, Bertels F, Remigi P, Jendresen CB, Ferguson GC, Desprat N, Buffing MF, Sauer U, Beaumont HJ, Martinussen J, Kilstrup M, Rainey PB. Bistability in a Metabolic Network Underpins the De Novo Evolution of Colony Switching in Pseudomonas fluorescens . PLoS Biol. 2015;13(3): e1002109 10.1371/journal.pbio.1002109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laue BE, Jiang Y, Chhabra SR, Jacob S, Stewart GS, Hardman A, et al. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology. 2000;146 (Pt 10): 2469–80. [DOI] [PubMed] [Google Scholar]
- 41. Cullinane M, Baysse C, Morrissey JP, O'Gara F. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology. 2005;151: 3071–80. [DOI] [PubMed] [Google Scholar]
- 42. Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003. October 17;278(42): 41148–59. [DOI] [PubMed] [Google Scholar]
- 43. Bhagwat AA, Jun W, Liu L, Kannan P, Dharne M, Pheh B, et al. Osmoregulated periplasmic glucans of Salmonella enterica serovar Typhimurium are required for optimal virulence in mice. Microbiology. 2009;155(Pt 1): 229–37. 10.1099/mic.0.023747-0 [DOI] [PubMed] [Google Scholar]
- 44. Burns BP, Hazell SL, Mendz GL, Kolesnikow T, Tillet D, Neilan BA. The Helicobacter pylori pyrB gene encoding aspartate carbamoyltransferase is essential for bacterial survival. Arch Biochem Biophys. 2000;380(1): 78–84. [DOI] [PubMed] [Google Scholar]
- 45. Qin A, Mann BJ. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 2006;6: 69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18(3): 65–76. [DOI] [PubMed] [Google Scholar]
- 47. Bittel P, Robatzek S. Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr Opin Plant Biol. 2007;10(4): 335–41. [DOI] [PubMed] [Google Scholar]
- 48. Tans-Kersten J, Huang H, Allen C. Ralstonia solanacearum needs motility for invasive virulence on tomato. J Bacteriol. 2001;183(12): 3597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Naito K, Taguchi F, Suzuki T, Inagaki Y, Toyoda K, Shiraishi T, et al. Amino acid sequence of bacterial microbe-associated molecular pattern flg22 is required for virulence. Mol Plant Microbe Interact. 2008;21(9): 1165–74. 10.1094/MPMI-21-9-1165 [DOI] [PubMed] [Google Scholar]
- 50. Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9): 847–8. [DOI] [PubMed] [Google Scholar]
- 51. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL-EBI Nucleic acids research. 2010;38 Suppl: W695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue): D808–15. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.