Graphical abstract
Keywords: X-ray crystallography, Crystal structure, Angiotensin-I converting enzyme, Metalloprotease, Zinc binding protein, Inhibitor binding
Highlights
-
•
We present a new crystal structure of Drosophila AnCE in a different crystal form.
-
•
Effects of crystal packing on the new structure are considered in detail.
-
•
This structure may be more suitable for the development of new ACE inhibitors.
Abstract
Angiotensin converting enzyme (ACE) is a zinc-dependent dipeptidyl carboxypeptidase with an essential role in blood pressure homeostasis in mammals. ACE has long been targeted in the treatment of hypertension through ACE inhibitors, however current inhibitors are known to cause severe side effects. Therefore, there is a requirement for a new generation of ACE inhibitors and structural information will be invaluable in their development. ACE is a challenging enzyme to work with due to its extensive glycosylation. As such, the Drosophila melanogaster ACE homologue, AnCE, which shares ∼60% sequence similarity with human ACE, can be used as a model for studying inhibitor binding. The presence of ligands originating from the crystallisation condition at the AnCE active site has proved an obstacle to studying the binding of new inhibitor precursors. Here we present the crystal structure of AnCE (in a new crystal form) at 1.85 Å resolution, using crystals grown under different conditions. This new structure may be more suitable for studying the binding of new compounds, with the potential of developing a new generation of improved ACE inhibitors.
1. Introduction
Angiotensin-I converting enzyme, ACE (EC 3.4.15.1) is a zinc dependent dipeptidyl carboxypeptidase with an essential role in mammalian blood pressure regulation. ACE is an integral component of the renin–angiotensin aldosterone system (RAAS), a complex hormone system that is the main mechanism for blood pressure homeostasis in mammals. The primary function of ACE in the RAAS is to produce the potent vasoconstrictor angiotensin II from its inactive precursor angiotensin I, thereby increasing blood pressure. ACE also acts to increase blood pressure by destroying the peptide bradykinin, a potent vasodilator [1–4]. As such, ACE has long been targeted in the treatment of hypertension and other cardiovascular ailments [5–11]. Whilst ACE inhibitors have proven to be highly effective, there are a number of side effects associated with their use. These range from relatively mild: a persistent dry cough, to potentially fatal: angioedema [12].
Two isoforms of ACE are found in humans: somatic (sACE) and testis (tACE) ACE [13]. Both are transcribed from the same twenty-six exon gene. sACE consists of two homologous domains arranged in tandem, the N- and C-terminal domains, each with its own functional active site. In contrast, tACE transcription is initiated from an intragenic promoter, resulting in a single domain enzyme identical to the C-domain of sACE [14–17].
Although the two domains of sACE share approximately 60% amino acid sequence identity, they have distinct biophysical and biochemical properties [18,19]. Crucially, the C-domain hydrolyses angiotensin I to angiotensin II much more efficiently than the N-domain and appears to be sufficient for in vivo blood pressure regulation [20].
The two domains of sACE are likely to be the result of a gene duplication event, which would have enabled the N-domain to evolve and acquire new functions [16]. There is evidence for the N-domain being involved in a number of other physiological processes, including inflammation and immunity [21,22]. As current inhibitors are unable to discriminate between the N- and C-domains, it is these newly acquired functions of the N-domain that are thought to be the major cause of the side effects associated with the use of ACE inhibitors.
It is therefore clear that there is a requirement for a new generation of domain specific ACE inhibitors and structural information will no doubt be invaluable in achieving this. The first ACE structure, of human tACE, was reported in 2003 [23] however due to its extensive glycosylation, estimated at 50% by weight [1], human ACE is a challenging enzyme to work with.
An ACE homologue from Drosophila melanogaster, AnCE, shares approximately 60% overall sequence similarity with each domain of human ACE (considerably higher in key catalytic regions). Hence, AnCE, has previously been used successfully as a model for studying inhibitor binding to ACE [24–26].
The crystal structure of AnCE was first determined in 2003 at 2.40 Å resolution using crystals in the P21 space group (form I structure) [27]. This was followed in 2010 by a series of higher resolution structures of AnCE in complex with various inhibitors that were obtained from crystals in the R3 space group (form II structure) [24]. Over the five years that have followed, a number of further AnCE-inhibitor complex structures have been reported using the form II structure [24–26,28].
All of the current ACE inhibitors are competitive inhibitors, which use either a carboxyl or sulphydryl group to co-ordinate the catalytic zinc ion. The structure based design of a new generation of ACE inhibitors will need to begin by studying the mechanism of binding of novel compounds which are much less potent inhibitors than those currently available. By using structural information to study their mode of inhibition it would then be possible to exploit features of their interactions with the ACE active site to develop them into better inhibitors.
The crystallisation condition that gives rise to the R3 crystals used to determine the form II AnCE structure contains a high concentration of citrate ion [24]. In the absence of an inhibitor a citrate ion co-ordinates the active site catalytic zinc ion. New, weaker inhibitors are unable to displace the citrate ion; hence it has been very difficult to study their binding.
Here we present a new structure of AnCE at 1.85 Å resolution from crystals in the P212121 space group grown in the absence of citrate ions. The atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession code PDB: 5A2R. Comparison with the form I and form II structures obtained previously shows that overall the three are very similar, thus we can confidently use the new crystallisation condition for future inhibitor binding studies. There are some small differences resulting from crystal packing interactions, some of which are explored in detail in this report. The main focus of the comparison is the form II structure, as this has previously been used extensively for ligand binding studies.
2. Results and discussion
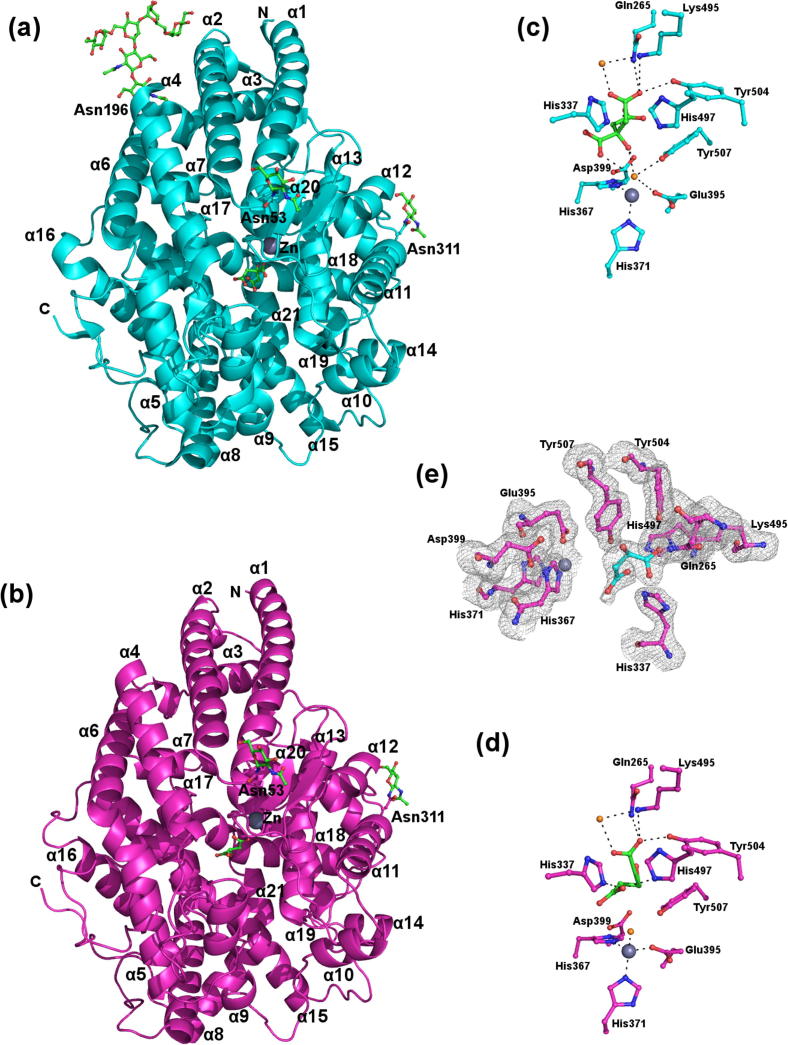
In contrast to the crystals used to determine the form I and form II structures, which belonged to the P21 and R3 space groups, respectively, the crystals obtained here were found to belong to the P212121 space group. As for the form II structure there is one molecule per asymmetric unit. Electron density was observed for residues 18-614 (20-614 in the form II structure). The catalytic zinc ion found in all ACE homologues was modelled in the active site along with a malate ion from the crystallisation media (Fig. 1d).
Fig. 1.
Comparison of the overall appearance of the form II AnCE structure with the new structure presented here. (a) The overall structure of the form II AnCE structure (PDB 2X8Y) with the catalytic zinc ion visible as a grey sphere alongside a citrate ion from the crystallisation media (green sticks) at the centre of the substrate binding channel. Helices α1–α3 form a lid capping the substrate binding channel. N-linked glycosylation visible at Asn53, Asn196 and Asn311 is shown as green sticks. (b) The overall structure of the new AnCE structure is identical to the form II structure, apart from the absence of N-linked glycosylation at Asn196. (c) Key active site residues from the form II structure are shown as cyan sticks co-ordinating the catalytic zinc ion (grey sphere) and a citrate ion from the crystallisation condition, green sticks. Two water molecules involved in co-ordination are shown as orange spheres, one of which completes the co-ordination of the zinc ion by mediating an interaction with the citrate. Hydrogen bonds between the citrate ion and active site residues are shown as black dashes. (d) The same selection of residues as in (c) shown as magenta sticks for the new structure. The positions of these residues are unchanged between the two structures. The active site of the new structure contains a malate ion (green sticks) from the crystallisation solution, which makes additional interactions with His397 and His337 not replicated by the citrate. The zinc ion and water molecules involved in co-ordination are shown as grey and orange spheres, respectively. Although the zinc co-ordinating water molecule from the form II structure is conserved it does not interact with the malate ion. (e) Electron density at the active site of the new structure showing key side chains as pink sticks and the malate ion as cyan sticks.
The overall structure is predominantly helical, consisting of twenty-one α-helices, nine 310 helices and two anti-parallel β-strands. Key features of the enzyme that are characteristic of ACE homologues include a large central channel with the active site at its centre which is capped by a lid formed by helices α1, α2 and α3 (Fig. 1b). The overall topology of this structure is identical to that of the form I and form II structures reported previously; Cα RMS deviation 0.27 and 0.33 Å, respectively. There are no Ramachandran outliers.
As illustrated in Fig. 1e, a malate ion was modelled in the active site electron density. The malate ion binds in a similar manner to the citrate ion in the form II structure and it is therefore not surprising that the positions of the active site residues are unchanged in the two structures.
Both the malate ion in this structure and the citrate ion in the form II structure interact with active site residues Tyr504, Gln265 and Lys495 through a carboxylate group. The second carboxylate group of the malate ion also contacts His494 and His337, interactions that are not replicated by the citrate (Fig. 1).
The remaining two carboxylate groups of the citrate ion make water mediated interactions with active site residues Asp399, Tyr507 and Glu395, the latter of which is also involved in zinc co-ordination. Crucially, the citrate ion also forms a water-mediated interaction with the catalytic zinc ion. Although the water molecule involved in this is conserved in the form III structure, the orientation of the malate ion means that it is unable to replicate this interaction. A summary of hydrogen bond interactions made by the citrate and malate ions is presented in Table 2.
Table 2.
List of hydrogen bond interactions formed by the citrate and malate ions in the active sites of the form II and form III AnCE structures, respectively.
| Interacting atoms | Distance (Å) | |
|---|---|---|
| Citrate atom | ||
| OA1 | Water | 2.61 |
| OA2 | Y504OH | 2.87 |
| OA2 | Q265NE2 | 3.28 |
| OA2 | K495NZ | 3.22 |
| OHB | Water | 2.86 |
| OG2 | Water | 3.20 |
| OB1 | Water | 2.96 |
| Malate atom | ||
| O1 | Y504OH | 2.52 |
| O1 | Q265NE2 | 3.02 |
| O1 | K495NZ | 2.70 |
| O2 | Water | 2.79 |
| O4 | H497NE2 | 2.95 |
| O4 | H337NE2 | 2.67 |
The lack of zinc co-ordination by the malate ion indicates that it may not be held so tightly in the active site and so would be more easily displaced by weaker inhibitors. This could make this crystallisation condition more suitable for developing new inhibitors.
A noticeable difference between the three structures is in the glycosylation profiles of the enzymes. No glycosylation was reported for the form I structure, whereas electron density is clearly visible at three N-linked glycosylation sites (Asn53, Asn196 and Asn311) in the form II structure (Fig. 1a). The form II structure and the new structure presented here were both determined using AnCE expressed by Pichia pastoris. In spite of this, electron density is only observed at two of these sites, Asn53 and Asn311, in the new structure (Fig. 1b). This is most likely due to differences in crystal packing.
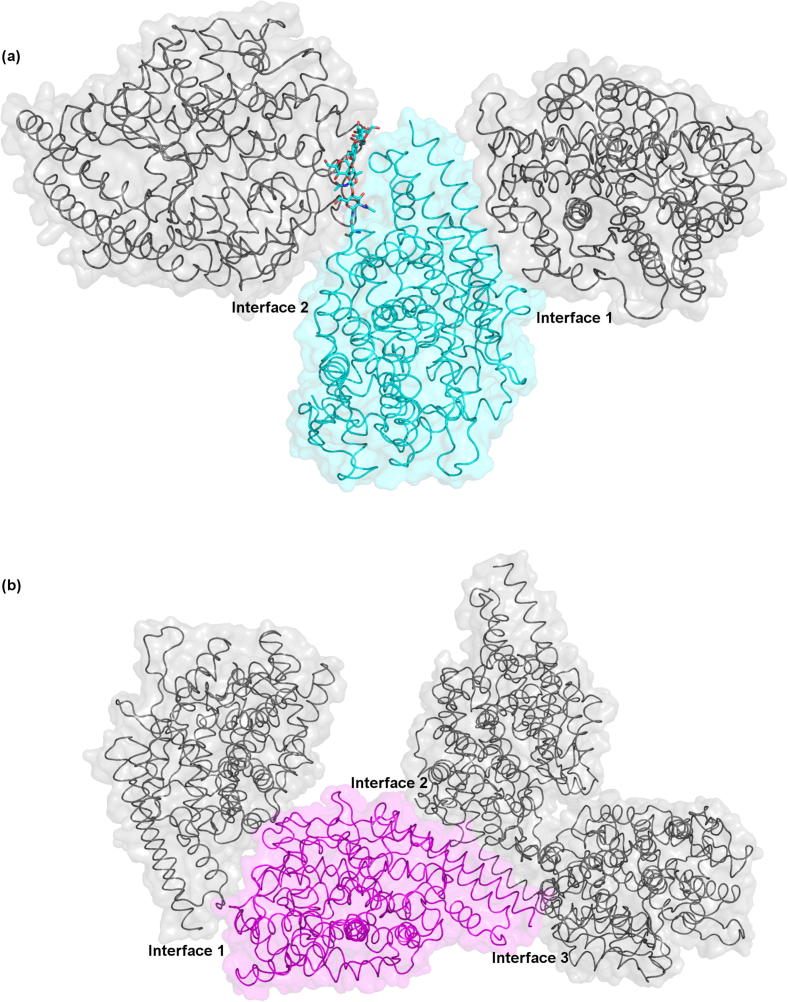
In the form II structure each molecule contacts four other molecules, there are therefore two different interfaces between symmetry related molecules, as illustrated in Fig. 2a. The first involves helices α1 and α2 of the N-terminal lid of one molecule and regions of α10, α11, α14 and α18 of the symmetry molecule. The second interface is significantly larger than the first. It involves helices α3, α6, α8 and the N-linked glycosylation at Asn196 of the first molecule interacting with helices α5, α8 and α12 of the symmetry molecule.
Fig. 2.
Crystal contacts in the two AnCE structures. (a) In the form II structure there are two different interfaces between symmetry molecules. To illustrate this three AnCE molecules are shown as cartoon and surface representations. The central molecule is shown as cyan and the two symmetry molecules in grey. The two interfaces are labelled as interface 1 and interface 2. The N-linked glycosylation at Asn196 is shown as cyan sticks and is an integral part of interface 2. (b) The three different interfaces between symmetry molecules in the new structure. The interfaces are labelled 1–3. The molecules are shown as surface and cartoons with the central molecule in magenta and the three symmetry molecules in grey.
In the new structure each molecule contacts six symmetry molecules, involving three different interfaces, as illustrated in Fig. 2b. The first interface involves α5, α8 and α9 of the first molecule interacting with regions of α1, α3, α8, α18 and α20 of the symmetry molecule. The second interface is similar to the first interface in the form II structure, involving α1, α2 and α4 of the N-terminal lid of the first molecule contacting α5 and α8 of the symmetry related molecule. The third interface is much smaller than any of the other interfaces, involving Lys20 of α1 of the first molecule and four residues from α18 of the symmetry molecule.
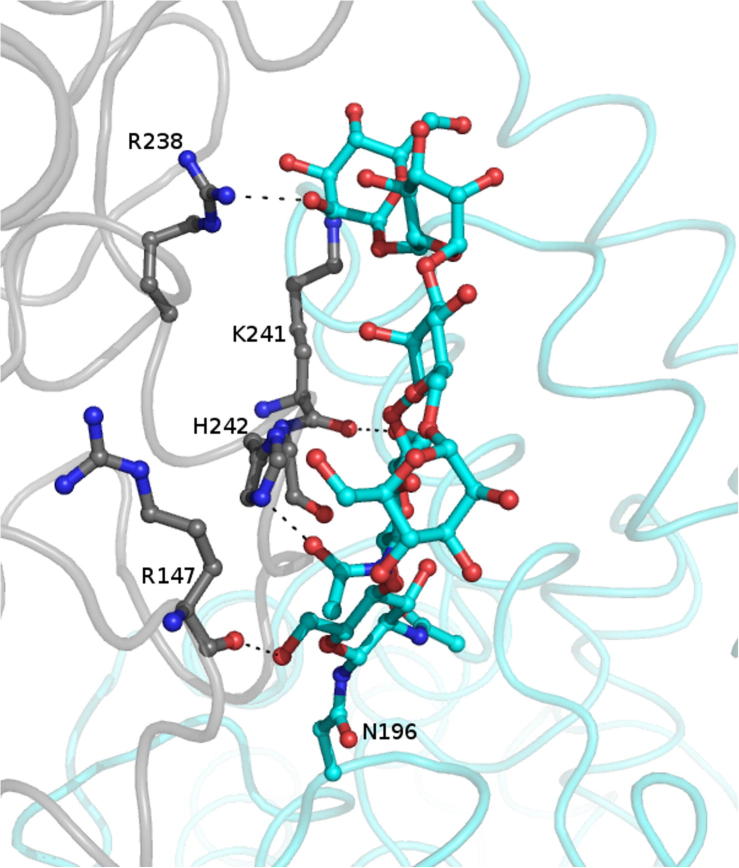
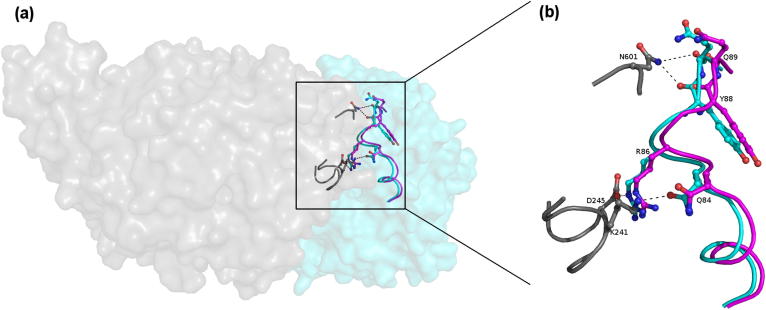
The absence of N-linked glycosylation at Asn196 in the new structure can be explained by examining these interactions with symmetry related molecules. As illustrated in Fig. 2a, this glycosylation is an integral part of the second interface in the form II structure, forming hydrogen bond interactions with Arg157, Arg238, Lys241 and His242 of the symmetry related molecule (Fig. 3). These interactions hold the glycan chain in a single conformation, making it visible in the structure. Conversely, in the new structure there is no symmetry related molecule in this region. The glycan chain is therefore likely to be highly mobile hence no electron density is visible.
Fig. 3.
Crystal contacts at interface 2 in the form II structure stabilize the N-linked glycosylation at Asn196. The glycosylation is shown as blue sticks with hydrogen bonding residues from the symmetry molecule in grey. Hydrogen bonds are shown as black dashes.
There are further differences between the new structure and the form II structure associated with this interface. As illustrated in Fig. 4, the top of helix α2 and the loop region between α2 and α3 is shifted by up to 1.3 Å towards the symmetry molecule in the form II structure. This is driven by hydrogen bonding between the main chains of Tyr88 and Gln89 with the side chain of Asn601 of the symmetry molecule, and with the side chains of Gln84 and Arg86 with the side chains of Lys241 and Asp245, respectively of the symmetry related molecule.
Fig. 4.
Interactions between the symmetry molecules at interface 2 in the form II structure results in movement of α2 by 1.3 Å. (a) A surface representation of the interface with one molecule shown in cyan and the symmetry molecule in grey and (b) close-up view of the interface. Interacting residues from the symmetry molecule are shown in grey and those from the central molecule in cyan. Corresponding residues from the new structure are superposed and shown in magenta to illustrate the movement of the peptide backbone in this region. Hydrogen bonding interactions with Asn601, Asp245 and Lys241 of the symmetry molecule in the form II structure pull part of α2 and the loop between α2 and α3 towards the symmetry molecule, resulting in movement of the peptide backbone by up to 1.3 Å compared to in the new structure.
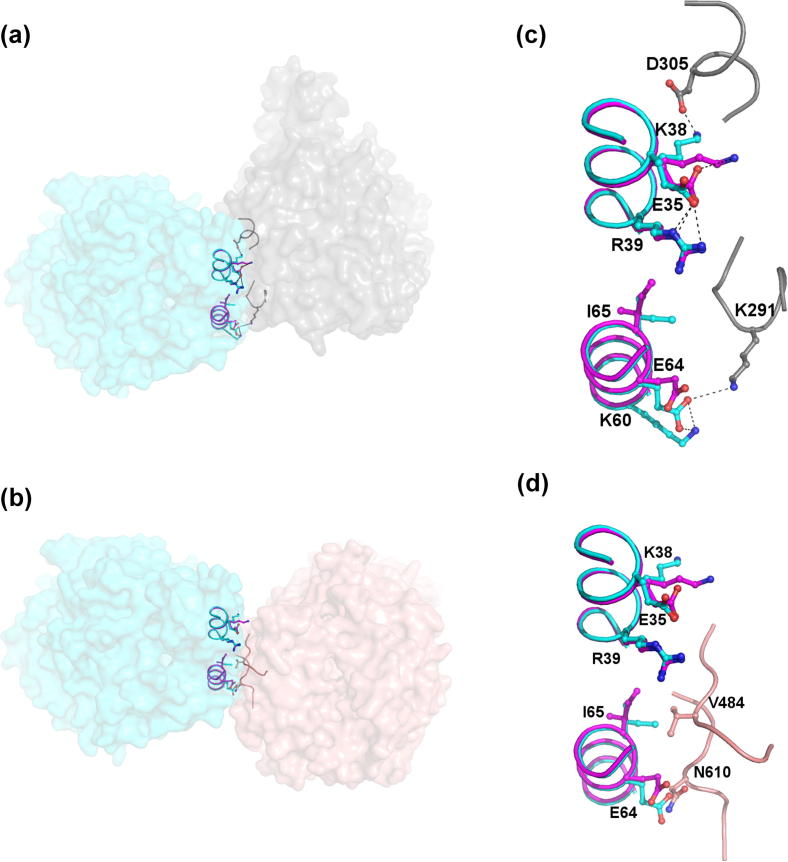
More subtle differences between the two structures are concentrated around the first interface in the form II structure and the second interface of the new structure. This is not surprising as there is considerable overlap between these two interfaces. In the form II structure, the side chain of Lys38 is positioned to hydrogen bond with Asp305 of the symmetry molecule. In the new structure there are no symmetry residues within hydrogen bonding distance of Lys38. A 5 Å movement of the side chain is observed that enables it to form a hydrogen bond with neighbouring Glu35, as illustrated in Fig. 5. In order to accommodate this interaction there is a small change in the position of the Glu35 side chain in the new structure, which results in one of the two interactions with Arg39 being lost, Fig. 5b.
Fig. 5.
Comparison of positions adopted by side-chains at interface 2 in the form II structure and interface I in the new structure. (a) Surface representation of interface 2 of the form II structure with AnCE shown in cyan and the symmetry molecule in grey. Key interacting residues at this interface are shown as sticks with a close-up view in (c). For comparison, equivalent residues from the new structure are shown in (c) as magenta sticks. The Lys38 side chain moves by approximately 5 Å in the two structures to hydrogen bond with Asp305 of the symmetry molecule in the form II structure and Glu35 of the same molecule in the new structure. (b) Surface representation of interface 1 in the new structure, which overlaps significantly with interface 2 in the form II structure. The AnCE surface is shown in cyan with the symmetry molecule in coral. Key interacting residues are shown as sticks with a close-up view in (d), where symmetry molecule residues are in coral and AnCE residues from the new structure in magenta with residues from the original structure shown in cyan for comparison. To avoid a clash with Asn610 of the symmetry molecule, Glu64 in the new structure adopts a different conformation to in the form II structure where it forms a hydrogen bond with Lys291 of the symmetry molecule. Similarly, to avoid a clash with Val484 of the symmetry molecule Ile65 exists as a different rotamer in the two structures.
Moving further along the helix to the area where there is most overlap between the two interfaces more small changes are observed. In the form II structure Glu64 hydrogen bonds to Lys291 of the symmetry related molecule and Lys60 within the molecule. In the new structure Glu64 adopts a different conformation so as to avoid a clash with Asn610 of the symmetry molecule. This results in the intra-molecular interaction with Lys60 being lost (Fig. 5d). Similarly, Ile65 exists as different rotamers in the two structures. As illustrated in Fig. 5d, this is to avoid a clash with Val484 of the symmetry related molecule in the new structure.
3. Conclusions
We present here a new structure of the D. melanogaster ACE homologue, AnCE in a new space group to the two previous structures. Although there are some minor differences between the structures caused by interactions with symmetry related molecules, the overall folds are identical, indicating that this is a valid structure. The absence of a citrate ion co-ordinating the catalytic zinc ion may make this crystallisation condition more suitable for studying the binding of new, weaker inhibitors, an important starting point in the development of a new generation of ACE inhibitors.
4. Materials and methods
4.1. Protein expression and purification
AnCE was expressed using P. pastoris and purified by hydrophobic interaction chromatography and size exclusion chromatography as described previously [24,29]. Briefly, the culture supernatant was treated with 1.5 M ammonium sulphate, 20 mM Tris, pH 8 and clarified by centrifugation at 4000g, 4 °C. The clarified supernatant was loaded onto a phenyl sepharose fast flow 6 column (GE Healthcare) previously equilibrated with 1.5 M ammonium sulphate, 20 mM Tris pH 8. Elution was performed with a gradient of decreasing salt and fractions containing AnCE were pooled and concentrated to a final volume of approximately 1.5 ml. Concentrated fractions were dialysed into 150 mM NaCl, 20 mM Tris pH 8 and loaded onto a superdex 200 16/600 column (GE healthcare) equilibrated with this buffer. Fractions containing AnCE were pooled together and dialysed into 5 mM HEPES pH 7.5, 0.1 mM PMSF before being concentrated to 15 mg ml−1 for crystallisation.
4.2. X-ray crystallography
A range of crystallisation conditions were screened using 96-well pre-prepared screens (Molecular Dimensions) and the Phoenix protein crystallisation robot (Art Robbins Instruments) using the sitting drop vapour diffusion method. 300 nl drops were set-up with a 1:1 ratio of protein:reservoir and plates were incubated at 21 °C. Crystals were grown in a condition containing 0.1 M MMT buffer pH 4.0 and 25% w/v PEG 1500. Crystals were taken directly from this drop for data collection at Diamond Light Source (Oxon, UK) on beamline I24.
Data were collected to 1.85 Å resolution. Raw data images were processed using MOSFLM [30,31] and Aimless [32] from the CCP4 suite [33]. This indicated that the crystals belonged to the space group P212121 with one molecule per asymmetric unit. Initial phases were calculated by molecular replacement using Phaser [34] with the previously determined structure of AnCE (PDB code 2X8Y) as the search model. The resulting model was refined using REFMAC5 [35,36] and 5% of reflections were separated as the Rfree set for use in cross-validation. After an initial round of rigid body refinement rounds of restrained refinement with manual adjustments of the model using COOT [37] were performed. Water molecules were added where Fo–Fc electron density peaks exceed 3σ and potential hydrogen bonds could be made. Ligands were added to the active site based on interpretation of the electron density and further rounds of refinement were performed. Validation was performed with the aid of the program MOLPROBITY [38]. There were no residues in the disallowed regions of the Ramachandran plot. Crystallographic data statistics are summarised in Table 1. All figures were drawn using PyMOL [39].
Table 1.
X-ray data collection, processing and refinement statistics.
| Beamline | I24 – Diamond Light Source, Didcot, UK |
| Detector | PILATUS 6M |
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 68.52–1.85 |
| Space group | P212121 |
| Cell dimensions (Å, °) | a = 86.12, b = 94.90, c = 99.03, α = 90.00, β = 90.00, γ = 90.00 |
| Molecules per asymmetric unit | 1 |
| Solvent content (%) | 56.87 |
| Unique/Total reflections | 219797/68121 |
| Completeness (%) | 97.8 (89.5) |
| Multiplicity | 3.2 (2.2) |
| #Rmerge (%) | 6.1 (58.8) |
| +Rpim (%) | 5.0 (49.2) |
| Mean I/σ(I) | 10.6 (2.2) |
| §Rcryst/##Rfree (%) | 16.31/18.56 |
| Wilson B factor (Å2) | 25.0 |
| Average B factor (Å2) | |
| Overall | 25.59 |
| Protein | 24.35 |
| Ligands: | |
| Zinc | 21.00 |
| Tris | 41.90 |
| Malate | 28.33 |
| Glycosylated carbohydrates | 49.35 |
| Solvent | 34.73 |
| R.m.s.d from ideal values in bond lengths (Å) | 0.007 |
| R.m.s.d from ideal values in bond angles (°) | 1.28 |
| Ramachandran plot statistics (%) | |
| Favoured | 99.16 |
| Disallowed | 0 |
| PDB code | 5A2R |
, where Ii(hkl) is the intensity of ith measurement and <I(hkl)> is the average of symmetry-related observations of a unique reflection.
.
, where Fo and Fc are observed and calculated structure factor amplitudes of reflection h, respectively.
Rfree is equal to Rcryst for a randomly selected 5% subset of reflections.
##Ramachandran plots were prepared for all residues other than Gly and Pro.
Author contribution
CH performed protein expression, purification and structural biology experiments, analysed the structure and wrote the manuscript. KRA supervised the study, analysed the data and edited the manuscript. Both authors reviewed the final manuscript.
Acknowledgements
We thank Diamond Light Source (Didcot, Oxon, UK) for access to beam line I24, which contributed to the results presented here. CH is supported by a postgraduate studentship by the Biotechnology and Biological Sciences Research Council (BBSRC, UK). The research on ACE in KRA’s laboratory is supported by the Medical Research Council (MRC, UK) through a project grant (number G1001685).
References
- 1.Acharya K.R., Sturrock E.D., Riordan J.F., Ehlers M.R.W. ACE revisited: a new target for structure-based drug design. Nat. Rev. Drug Discov. 2003;2:891–902. doi: 10.1038/nrd1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cushman D.W., Cheung H.S. The spectrophotometric assay and properties of the angiotensin-converting enzyme of rabbit lung. Biochem. Pharmacol. 1971;20:1637–1648. doi: 10.1016/0006-2952(71)90292-9. [DOI] [PubMed] [Google Scholar]
- 3.Das M., Soffer R.L. Pulmonary angiotensin-converting enzyme structural and catalytic properties. J. Biol. Chem. 1975;250:6762–6768. [PubMed] [Google Scholar]
- 4.Skeggs L.T., Marsh W.H., Kahn J.R., Shumway N.P. The existence of two forms of hypertension. J. Exp. Med. 1954;100:275–282. doi: 10.1084/jem.99.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cushman D.W., Cheung H.S., Sabo E.F., Ondetti M.A. Design of potent competitive inhibitors of angiotensin-converting enzyme. Carboxyalkanoyl and mercaptoalkanoyl amino acids. Biochemistry. 1977;16:5484–5491. doi: 10.1021/bi00644a014. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira S.H. A bradykinin-potentiating factor (BPF) present in the venom of Bothrops jararaca. Br. J. Pharmacol. Chemother. 1965;24:163–169. doi: 10.1111/j.1476-5381.1965.tb02091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira S.H., Greene L.J., Alabaster V.A., Bakhle Y.S., Vane J.R. Activity of various fractions of bradykinin potentiating factor against angiotensin I converting enzyme. Nature. 1970;225:379–380. doi: 10.1038/225379a0. [DOI] [PubMed] [Google Scholar]
- 8.Ondetti M.A., Williams N.J., Sabo E.F., Plusec J., Weaver E.R., Kocy O. Angiotensin converting enzyme inhibitors from the venom of Bothrops jararaca. Isolation, elucidation of structure, and synthesis. Biochemistry. 1971;19:4033–4039. doi: 10.1021/bi00798a004. [DOI] [PubMed] [Google Scholar]
- 9.Patchett A.A., Harris E., Tristram E.W., Wyvratt M.J., Wu M.T., Taub D., Peterson E.R., Ikeler T.J., Broeke J., Payne L.G., Ondeyka D.L., Thorsett E.D., Greenlee W.J., Lohr N.S., Hoffsommer R.D., Joshua H., Ruyle W.V., Rothrock J.W., Aster S.D., Maycock A.L., Robinson F.M., Hirschmann R., Sweet C.S., Ulm E.H., Gross D.M., Vassil T.C., Stone C.A. A new class of angiotensin-converting enzyme inhibitors. Nature. 1980;288:280–283. doi: 10.1038/288280a0. [DOI] [PubMed] [Google Scholar]
- 10.Turner A.J., Hooper N.M. The angiotensin converting enzyme gene family: genomics and pharmacology. Trends Pharmacol. Sci. 2002;23:177–183. doi: 10.1016/s0165-6147(00)01994-5. [DOI] [PubMed] [Google Scholar]
- 11.Zaman M.A., Oparil S., Calhoun D.A. Drugs targeting the renin–angiotensin–aldosterone system. Nat. Rev. Drug Discov. 2002;1:621–636. doi: 10.1038/nrd873. [DOI] [PubMed] [Google Scholar]
- 12.Steckelings U.M., Artuc M., Wollschlager T., Wiestutz S., Henz B.M. Angiotensin-converting enzyme inhibitors as inducers of adverse cutaneous reactions. Acta Derm. Venereol. 2001;81:321–325. doi: 10.1080/000155501317140007. [DOI] [PubMed] [Google Scholar]
- 13.Soubrier F., Alhenc-Gelas F., Hubert C., Allegrini J., John M., Tregear G., Corvol P. Two putative active centers in human angiotensin I-converting enzyme revealed by molecular cloning. Proc. Natl. Acad. Sci. USA. 1988;85:9386–9390. doi: 10.1073/pnas.85.24.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corvol P., Williams T.A., Soubrier F. Peptidyl dipeptidase A: angiotensin I-converting enzyme. Methods Enzymol. 1995;248:283–305. doi: 10.1016/0076-6879(95)48020-x. [DOI] [PubMed] [Google Scholar]
- 15.Ehlers M.R., Fox E.A., Strydom D.J., Riordan J.F. Molecular cloning of human testicular angiotensin-converting enzyme: the testis isozyme is identical to the C-terminal half of endothelial angiotensin-converting enzyme. Proc. Natl. Acad. Sci. USA. 1989;86:7741–7745. doi: 10.1073/pnas.86.20.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubert C., Houot A., Corvol P., Soubrier F. Structure of the angiotensin I-converting enzyme gene. J. Biol. Chem. 1991;266:15377–15383. [PubMed] [Google Scholar]
- 17.Wei L., Clauser E., Alhenc-Gelas F., Corvol P. The two homologous domains of human angiotensin I-converting enzyme interact differently with competitive inhibitors. J. Biol. Chem. 1992;267:13398–13405. [PubMed] [Google Scholar]
- 18.Sturrock E.D., Danilov S.M., Riordan J.F. Limited proteolysis of human kidney angiotensin-converting enzyme and generation of catalytically active N- and C-terminal domains. Biochem. Biophys. Res. Commun. 1997;236:16–19. doi: 10.1006/bbrc.1997.6841. [DOI] [PubMed] [Google Scholar]
- 19.Voronov S., Zueva N., Orlov V., Arutyunyan A., Kost O. Temperature-induced selective death of the C-domain within angiotensin-converting enzyme molecule. FEBS Lett. 2002;522:77–82. doi: 10.1016/s0014-5793(02)02888-0. [DOI] [PubMed] [Google Scholar]
- 20.Georgiadis D., Beau F., Czarny B., Cotton J., Yiotakis A., Dive V. Role of the two active sites of somatic angiotensin-converting enzyme in the cleavage of angiotensin I and bradykinin: insights from selective inhibitors. Circ. Res. 2003;93:148–154. doi: 10.1161/01.RES.0000081593.33848.FC. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein K.E., Shen X.Z., Gonzalez-Villalobos R.A., Billet S., Okwan-Duodu D., Ong F.S., Fuch S. Different in vivo functions of the two catalytic domains of angiotensin-converting enzyme (ACE) Curr. Opin. Pharmacol. 2011;11:105–111. doi: 10.1016/j.coph.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Villalobos R.A., Shen X.Z., Bernstein E.A., Janjulia T., Taylor B., Giani J.F., Blackwell W.-L.B., Shah K.H., Shi P.D., Fuchs S., Bernstein K.E. Rediscovering ACE: novel insights into the many roles of the angiotensin-converting enzyme. J. Mol. Med. (Berl) 2013;91:1143–1154. doi: 10.1007/s00109-013-1051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natesh R., Schwager S.L.U., Sturrock E.D., Acharya K.R. Crystal sturcture of the human angiotensin-converting enzyme–lisinopril complex. Nature. 2003;421:1427–1429. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 24.Akif M., Georgiadis D., Mahajan A., Dive V., Sturrock E.D., Isaac R.E., Acharya K.R. High-resolution crystal structures of Drosophila melanogaster angiotensin-converting enzyme in complex with novel inhibitors and antihypertensive drugs. J. Mol. Biol. 2010;400:502–517. doi: 10.1016/j.jmb.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 25.Akif M., Ntai L., Sturrock E.D., Isaac R.E., Bachmann B.O., Acharya K.R. Crystal structure of a phosphonotripeptide K-26 in complex with angiotensin converting enzyme homologue (AnCE) from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2010;398:532–536. doi: 10.1016/j.bbrc.2010.06.113. [DOI] [PubMed] [Google Scholar]
- 26.Masuyer G., Akif M., Czarny B., Beau F., Schwager S.L.U., Sturrock E.D., Isaac R.E., Dive V., Acharya K.R. Crystal structures of highly specific phosphinic tripeptide enantiomers in complex with the angiotensin-I converting enzyme. FEBS J. 2013;281:943–956. doi: 10.1111/febs.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H.M., Shin D.R., Yoo O.J., Lee H., Lee J.-O. Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril. FEBS Lett. 2003;538:65–70. doi: 10.1016/s0014-5793(03)00128-5. [DOI] [PubMed] [Google Scholar]
- 28.Akif M., Masuyer G., Bingham R.J., Sturrock E.D., Isaac R.E., Acharya K.R. Structural basis of peptide recognition by the angiotensin-1 converting enzyme homologue AnCE from Drosophila melanogaster. FEBS J. 2012;279:4525–4534. doi: 10.1111/febs.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houard X., Williams T.A., Michaud A., Dani P., Isaac R.E., Shirras A.D., Coates D., Corvol P. The Drosophila melanogaster-related angiotensin-I-converting enzymes Acer and Ance distinct enzymic characteristics and alternative expression during pupal development. Eur. J. Biochem. 1998;257:599–606. doi: 10.1046/j.1432-1327.1998.2570599.x. [DOI] [PubMed] [Google Scholar]
- 30.Leslie A.G. The integration of macromolecular diffraction data. Acta Crystallogr. 2006;D62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 31.Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. 2011;D67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. 2013;D69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., Read R.J., Vagin A., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.S., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. 2011;D53:240–255. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-stom structure validation for macromolecular crystallography. Acta Crystallogr. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger LLC.