Highlights
-
•
NDRG2 promotes C2C12 myoblast proliferation but not overall differentiation.
-
•
NDRG2 increases caspase 3/7 activities in differentiating C2C12 muscle cells.
-
•
NDRG2 reduces hydrogen peroxide-induced oxidative and ER stress but not lipotoxicity.
-
•
Ser and Thr residues in the C-terminus of mouse NDRG2 contribute to its function.
Abbreviations: NDRG2, N-myc downstream-regulated gene 2; MRFs, myogenic regulatory factors; Cdk, cyclin-dependent kinase; Rb, retinoblastoma; Myf5, myogenic factor 5; MyoD, myogenic differentiation; p27, p27 kip1; p21, p21 waf1/cip1; Akt, thymoma viral proto-oncogene; PKCθ, protein kinase C theta; SGK1, serum- and glucocorticoid-inducible kinase 1; Bcl-2, B cell leukemia/lymphoma 2; Bax, Bcl-2-associated X protein; Bcl-xL, Bcl-2-like 1; Mcl-1, myeloid cell leukemia 1; PARP, poly (ADP-ribose) polymerase family, member; Caspase, apoptosis-related cysteine peptidase; GRP78, glucose-regulated protein 78; Myh7, myosin, heavy polypeptide 7; Ckm, muscle creatine kinase; Acta1, skeletal muscle alpha-actin; PA, palmitate; H2O2, hydrogen peroxide; ER, endoplasmic reticulum
Keywords: NDRG2, Myoblast, Myotube, Proliferation, Differentiation, Apoptosis, Oxidative stress, Lipotoxicity, ER stress
Abstract
The function of the stress-responsive N-myc downstream-regulated gene 2 (NDRG2) in the control of myoblast growth, and the amino acids contributing to its function, are not well characterized. Here, we investigated the effect of increased NDRG2 levels on the proliferation, differentiation and apoptosis in skeletal muscle cells under basal and stress conditions. NDRG2 overexpression increased C2C12 myoblast proliferation and the expression of positive cell cycle regulators, cdk2, cyclin B and cyclin D, and phosphorylation of Rb, while the serine/threonine-deficient NDRG2, 3A-NDRG2, had less effect. The onset of differentiation was enhanced by NDRG2 as determined through the myogenic regulatory factor expression profiles and myocyte fusion index. However, the overall level of differentiation in myotubes was not different. While NDRG2 up-regulated caspase 3/7 activities during differentiation, no increase in apoptosis was measured by TUNEL assay or through cleavage of caspase 3 and PARP proteins. During H2O2 treatment to induce oxidative stress, NDRG2 helped protect against the loss of proliferation and ER stress as measured by GRP78 expression with 3A-NDRG2 displaying less protection. NDRG2 also attenuated apoptosis by reducing cleavage of PARP and caspase 3 and expression of pro-apoptotic Bax while enhancing the pro-survival Bcl-2 and Bcl-xL levels. In contrast, Mcl-1 was not altered, and NDRG2 did not protect against palmitate-induced lipotoxicity. Our findings show that NDRG2 overexpression increases myoblast proliferation and caspase 3/7 activities without increasing overall differentiation. Furthermore, NDRG2 attenuates H2O2-induced oxidative stress and specific serine and threonine amino acid residues appear to contribute to its function in muscle cells.
1. Introduction
The progression of dividing myoblasts to commence fusion into multinucleated muscle cells is a tightly coordinated process involving cell cycle regulators interacting with myogenic regulatory factors (MRFs) as well as apoptosis. The MRFs, myogenic factor 5 (Myf5) and myogenic differentiation (MyoD) are required for normal myoblast proliferation and are regulated by positive cell cycle regulatory complexes such as cyclin E-cyclin-dependent kinase 2 (Cdk2) [1] and cyclin B-Cdk1 [2]. The activation of Cdks and cyclins in turn phosphorylate the tumor suppressor retinoblastoma (Rb) protein releasing its inhibitory effect on cell cycle progression (reviewed in [3]). In addition, MyoD promotes the onset of cell cycle arrest and commitment to differentiate through activation of the cell cycle inhibitor p21 waf1/cip1 (p21) [4]. The MRF myogenin is also responsible for cell cycle exiting and assists myoblasts to fuse in conjunction with p21 to enter the post-mitotic state [5]. During this process, a proportion of myoblasts undergo apoptosis that are thought to enhance myoblast fusion [6], whereas p21 protects fusing myocytes against apoptosis once they are committed to differentiate, particularly during in vitro myogenesis [7]. During differentiation, the activities of the pro-apoptotic enzymes caspase 9 and caspase 3 increase and play essential roles in myoblast differentiation [8,9] while the anti-apoptotic B cell leukemia/lymphoma 2 (Bcl-2) gene also contributes to skeletal muscle postnatal development, particularly in fast twitch muscle fiber formation [10]. In addition to normal apoptosis during myogenesis, apoptosis in skeletal muscle may occur during disease conditions [11]. During stress conditions, Bcl-2 members, such as the pro-apoptotic Bcl-2-associated X protein (Bax), may help mediate apoptosis in myotubes [11] as observed following palmitate (PA)-induced lipotoxicity [12]. However, our understanding of the molecular factors controlling and coordinating muscle cell proliferation and differentiation, in conjunction with apoptosis and increased caspase activity, is far from complete and requires continued investigation.
The N-myc downstream-regulated gene 2 (NDRG2) is highly expressed in skeletal muscle tissue both at the gene level [13] and as a phosphoprotein [14]. NDRG2 expression increases with myoblast differentiation and contributes to muscle cell growth as its knockdown attenuates C2C12 myoblast proliferation and differentiation [15]. In addition, NDRG2 is considered a stress-response molecule as its expression is induced under hypoxic conditions [16,17] and radiation [17] in cancer cells, for example; and following catabolic treatment in skeletal muscle cells [15]. Conversely, anabolic factors including testosterone, insulin and insulin-like growth factor suppress NDRG2 gene expression in muscle cells [15]. The increased endogenous expression of NDRG2 could indicate a natural protective response against stress effects. Indeed, a loss of NDRG2 attenuated the protective effect of thymoma viral proto-oncogene (Akt) overexpression in pancreatic beta cells following PA treatment [18], and reduced Bcl-2 levels in cervical cancer cells promoting their sensitivity to cisplatin treatment [19]. Furthermore, NDRG2 overexpression protected against radiation-induced apoptosis and the up-regulation of Bax in Hela cells [17]. In contrast to these studies, the overexpression of NDRG2 is reported to reduce the neuroprotective effect of sevoflurane pre-treatment in astrocytes following oxygen–glucose deprivation [20]. Currently, it is unclear as to whether enhanced levels of NDRG2 will have an overall beneficial outcome during different stress conditions or whether the benefit is cell-type specific. As skeletal muscle cells are sensitive to multiple stress conditions such as lipotoxicity [21] and oxidative stress through hydrogen peroxide treatment [22], which is also linked to the activation of endoplasmic reticulum (ER) stress [23], the effects of such stresses in the presence of elevated NDRG2 will further our understanding of NDRG2’s role in skeletal muscle function.
The specific protein regions contributing to the function of NDRG2 are not well characterized. In skeletal muscle cells, previous studies have identified serine (Ser) and threonine (Thr) amino acid-rich regions in the C-terminus of human and mouse NDRG2 [24,25]. These residues in mouse NDRG2, including the Ser332, Thr348 and Ser350 amino acids, may be phosphorylated by insulin and by Ser/Thr kinases including Akt and protein kinase C theta (PKCθ) [24,25], factors that affect myoblast proliferation, fusion and myotube development [26–28]. Interestingly, PKCθ’s phosphorylation of Ser332 reduces insulin’s ability to phosphorylate Thr348 [24] although the functional consequence of this for NDRG2 is unknown. In cancer cells, Ser and Thr amino acids appear to contribute, either individually or collectively, to NDRG2’s ability to inhibit colon carcinoma cell proliferation [29] indicating that the phosphorylation status of NDRG2 is important for its function. Therefore, the aims of this study were to investigate the effect of NDRG2 overexpression on proliferation, differentiation and apoptosis under both normal growth and stress-induced conditions in C2C12 muscle cells. Furthermore, we determined whether the combined removal of the Ser332, Thr348 and Ser350 residues, making NDRG2 phospho-deficient, impacted the role of NDRG2 on these processes.
2. Materials and methods
2.1. Cell culture
Mouse C2C12 myoblasts (ATCC, Manassas, VA, USA) were cultured in growth medium containing 25 mM glucose Dulbecco’s Modified Eagle Medium (DMEM) and 10% fetal bovine serum (FBS) (Life Technologies, Mulgrave, VIC, AUS) at 37 °C in 5% CO2. To differentiate cells, differentiation medium consisting of 2% horse serum and 25 mM glucose DMEM was added when cells were confluent and refreshed every 48 h. Platinum-E (Plat-E) cells (Jomar Bioscience, Welland, SA, AUS) were used for the production of retrovirus and were cultured in growth medium at 37 °C in 5% CO2. To induce oxidative stress, myoblasts were treated with 0.5 mM hydrogen peroxide (H2O2; Sigma–Aldrich, St. Louis, MO, USA) in 10% FBS DMEM for 4 or 8 h. For lipotoxic treatments, PA (Sigma–Aldrich, P0500) was dissolved in 100% ethanol and conjugated for 1 h at 37 °C with gentle rotation with 10% fatty acid-free bovine serum albumin (FAF-BSA, Sigma–Aldrich) in serum-free DMEM at a final concentration of 4.5 mM PA and 1.5 mM FAF-BSA (3:1 ratio). The equivalent amount of 100% ethanol was added to 10% FAF-BSA/DMEM as a vehicle control treatment. C2C12 myoblasts were treated with 0.5 mM PA conjugated with FAF-BSA or with vehicle control for 8 or 16 h in 10% FBS DMEM.
2.2. Protein stability study
The production of the FLAG-tagged wild type and phospho-deficient mouse NDRG2 expression constructs, pCMV/SV-FLAG1-NDRG2 and -3A-NDRG2, were described previously [24]. The amino acids Ser332, Thr348 and Ser350 in the phospho-deficient form of mouse NDRG2 (3A-NDRG2) had been replaced with alanine residues. C2C12 myoblasts plated at 8 × 104 cells/ml and 24 h later plasmids were transiently transfected into Plat-E cells using Lipofectamine 2000 transfection reagent (Life Technologies) and OptiMem medium (Life Technologies) as recommended by the manufacturer. Cells were treated with 10 μg/ml cycloheximide for up to 24 h prior to harvesting of protein lysates.
2.3. Retroviral infection
NDRG2 and 3A-NDRG2 were amplified from pCMV/SV-FLAG1 plasmids using primers: forward 5′ AGTGAGATCTACCATGGCAGAACTTCAGGAGGT and reverse 5′ GTTCGTCGACTCAACAGGAGACTTCCATGG. The cDNAs were cloned into the SalI and BglII sites of the pMSCV-IRES-hygromycin (pMIH) retroviral vector as previously described [30] to generate wild type and phospho-deficient, pMIH-WT-NDRG2 and pMIH-3A-NDRG2 plasmids, respectively. For the production of the retroviruses, empty pMIH vector (vector), pMIH-WT-NDRG2 (NDRG2) and pMIH-3A-NDRG2 (3A-NDRG2) plasmids were transiently transfected into Plat-E cells plated at 4 × 105 cells/ml essentially as described before [15]. Virus was allowed to accumulate in the media over 48 h prior to harvesting. The supernatant containing each virus was centrifuged to remove dead cells and then diluted 1:6 in growth medium containing 6 μg/ml polybrene (Sigma–Aldrich) before adding to C2C12 myoblasts plated at 1.8 × 104/ml 24 h earlier. Cells with virus media were centrifuged at 1800 rpm for 45 min and left to incubate at 37 °C in 5% CO2 for 5 h before being replaced with fresh growth medium. The following day, cells were selected with 800 μg/ml of hygromycin for 36 h prior to re-plating at 1.5 × 104/ml for all assays. Cells were harvested across a time-course encompassing both proliferating and differentiating myoblasts. Myoblasts were analzsed during proliferation at two and three days (P2 and P3) post-plating and then at day four (D0) when myoblasts were confluent and differentiation medium was added. Differentiating myoblasts were differentiated for up to six days and harvested at days 1, 2, 4 or 6 of differentiation (D1, D2, D4 or D6).
2.4. Gene expression analysis
Total RNA was extracted using Tri-Reagent (Ambion Inc, Austin, TX, USA), treated with DNAse I and RNase H (Life Technologies) and quantitated using the NanoDrop 1000 spectrophotometer (ThermoFisher Scientific, Scoresby, VIC, AUS). Half a microgram of RNA was reverse-transcribed to form cDNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s instructions. Semi-quantitative polymerase chain reaction (qPCR) was performed using the Mx3000 PCR system (Stratagene, La Jolla, CA, USA) with SYBR Green Master Mix (Applied Biosystems). The cycling conditions included: 95 °C for 10 min (1 cycle), 30 s at 95 °C and 60 °C for one min (40 cycles). Primers were synthesized by GeneWorks (Adelaide, SA, AUS) and their sequences are listed in Table 1. Samples were measured in triplicate and the relative gene expression expressed as arbitrary units (A.U.) or fold-change, which was calculated using 2−ΔCt and normalized to 36B4 gene expression (for proliferation) or cDNA input as determined by Quant-iT™ OliGreen® ssDNA Assay Kit (for differentiation) (Life Technologies).
Table 1.
Primers used for qPCR analysis.
| Gene ID | GenBank accession No. | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| 36B4 | NM_007475.5 | ttgtgggagcagacaatgtg | agtcctccttggtgaacacg |
| Ndrg2 | NM_013864.2 | gagttagctgcccgcatcc | gtgaccgagccataaggtgtc |
| p21/Cdkn1a | NM_001111099.1 | gccttgtcgctgtcttgc | cgcttggagtgatagaaatctgtc |
| p27/Cdkn1b | NM_009875.4 | gagcagtgtccagggatgagg | tccacagtgccagcgttcg |
| Ccnb1/Cyclin B1 | NM_172301.3 | agccatggcgctcagggtcac | gggtcagccccatcatctgcg |
| Ccnd1/Cyclin D1 | NM_007631.2 | gcccgaggagctgctgcaaa | tcaggccttgcatcgcagcc |
| Cdk2 | NM_183417.3 | tacccagtactgccatccga | ccggaagagttggtcaatct |
| Myod1 | NM_010866.2 | ctggttcttcacgcccaaa | ctggaagaacggcttcgaagg |
| Myogenin | NM_031189.2 | tccatcgtggacagcatcac | caatctcagttgggcatggttt |
| Myf5 | NM_008656.5 | caccacaaccaaccctaacca | actctcaatgtagcggattgca |
| Acta1 | NM_001272041.1 | gagcgtggctattccttcgt | ctttgatgtcgcgcacaatc |
| Ckm | NM_007710.2 | cccacagacaagcataagac | cccgtcaggctgttgagag |
| Myh7 | NM_080728.2 | accctcaggtggctccgaga | tgcagccccaaatgcagcca |
2.5. Western blotting
Cells were lysed in 1× modified RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA) (Merck Millipore, North Ryde, NSW, AUS) containing dilution of 1:1000 protease inhibitor cocktail (Sigma–Aldrich) and 1:100 Halt phosphatase inhibitor cocktail (ThermoFisher Scientific). Protein lysate concentrations were determined using Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific). For de-phosphorylation analysis, cells were harvested in 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1% Triton-X100 and protease inhibitor cocktail. Following determination of protein concentration, lysates were treated with 1000 U lambda phosphatase for 2 h at 30 °C (New England Biolabs, MA, USA). Thirty or sixty micrograms of protein lysates were electrophoresed on either 12% or 8% SDS–PAGE gels and transferred to Immobilon®-FL polyvinylidene difluoride membrane (Merck Millipore, Kilsyth, VIC, AUS). Membranes were blocked in 5% BSA/Tris-Buffered saline with Tween-20 (TBST; 50 mM Tris, 150 mM NaCl, 0.1% Tween-20) for 1 h before incubation at 4 °C overnight in primary antibodies diluted in 5% BSA/TBST. Rabbit polyclonal anti-cyclin B1 (#4138; 1:500), anti-p27 Kip1 (#2552; 1:300), anti-caspase 3 (#9662; 1:2500), anti-phospho Rb (Ser780) (#3590; 1:500), anti-phospho Akt (Ser473) (#9271; 1:1000), anti-PARP (#9532; 1:300), rabbit monoclonal anti-Cdk2 (#2546; 1:250) and mouse monoclonal anti-cyclin D3 (#2936; 1:1000) were obtained from Cell Signaling (Beverly, MA, USA). Rabbit polyclonal antibodies anti-Myf5 (sc-302; 1:200) and anti-MyoD (sc-760; 1:350), and mouse monoclonal antibodies anti-myogenin (sc-12732; 1:200) and anti-GRP78 (sc-376768; 1:200) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Mouse monoclonal antibodies, anti-myosin, type 1 slow (M8421; 1:5000), anti-Bax (B9054; 1:100) and anti-FLAG (M2; 1:1000), and rabbit polyclonal anti-NDRG2 (HPA002896; 1:5000) were obtained from Sigma–Aldrich. Mouse monoclonal antibodies, anti-Bcl-2 (#610539; 1:100) and anti-Bcl-xL (#610747; 1:100) were from BD Transduction Laboratories (BD Biosciences, San Jose, CA, USA) and the rabbit polycolonal anti-Mcl-1 (1:8000) was from Rockland Immunochemicals Inc. (Limerick, PA USA). Rabbit monoclonal anti-p21 waf/cip1 (ab109199; 1:1000) and mouse monoclonal antibodies, anti-α-tubulin (clone DM1A; 1:5000) and anti-β-Actin (ab6276; 1:10,000), were obtained from Abcam (Cambridge, MA, USA). The detection of proteins was performed using goat anti-rabbit AlexaFluor®680 or donkey anti-mouse AlexaFluor®800 IgG antibodies (Life Technologies) diluted at 1:10,000 in 50% Odyssey blocking buffer (Li-COR Biosciences, Lincoln, NE, USA), 50% TBST and 0.01% SDS. Membranes were imaged using the infrared imaging system (Li-COR Biosciences). Proteins were normalized against heat shock protein 90 (HSP90), α-tubulin or β-actin protein levels using the Li-COR software.
2.6. BrdU labeling assay
The proliferation rate of myoblasts was assessed using the colorimetric 5-bromo-2′-deoxy-uridine (BrdU) Labeling and Detection Kit III assay (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s protocol. The BrdU label reagent was added at a 1:90 dilution to the medium 16 h prior to analysis. BrdU incorporation into the DNA was detected using horseradish peroxidize conjugated BrdU antibodies and measured using a Synergy 2 Multi-Mode microplate reader (BioTek, Winooski, VT, USA) at 405 nm with a reference wavelength of 490 nm following the addition of the peroxidase substrate, ABTS (2,2′-azino-bis, 3-ethylbenzthiazoline-6-sulfonic acid).
2.7. Immunohistochemistry and fusion index
Cells were washed in PBS before being fixed in cold 4% paraformaldehyde (PFA) (Sigma–Aldrich) in PBS for 20 min. Following washing, cells were permeabilized in 0.5% Triton X-100/PBS for 10 min, washed again and blocked overnight in 3% BSA/PBS at 4 °C. Cells were incubated in 1:1000 dilution of mouse monoclonal anti-myosin, type 1 slow (M8421; Sigma–Aldrich) for 24 h at 4 °C. Following washing, cells were incubated for 1 h with anti-mouse AlexaFluor®546 IgG antibody (1:500; Life Technologies), washed again and incubated 15 min with diamidino-2-phenylindole (DAPI) (1:1000; Sigma–Aldrich). Cells were imaged using the Olympus IX70 fluorescent microscope (Olympus, Notting Hill, VIC, AUS) at 10× magnification and 10 fields of view were imaged per condition. The fusion index of differentiating myocytes was analyzed as described previously [31]. Fusion index was calculated as the number of myosin-positive cells displaying at least three nuclei divided by the total number of nuclei per field of view as determined using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.8. Caspase 3/7 assays
Caspase 3/7 activities were measured using Caspase-Glo 3/7 Assay (Promega, Annandale, NSW, AUS) according to manufacturer’s instructions. Briefly, cells were grown in white-walled 96-well plates and the Caspase-Glo 3/7 reagent was added to the cells (1:1) and control wells (growth media only). Incubation time was optimized at 30 min and caspase 3/7 luminescence as relative light units (RLU) was measured using the Synergy 2 Multi-Mode microplate reader. The average background reading of control wells were subtracted from all values and final values were normalized to protein content.
2.9. TUNEL analysis
Myoblasts were prepared using the APO-BrdU TUNEL Assay Kit (Invitrogen) according to manufacturer’s instructions. Briefly, cells were fixed in 1% PFA/PBS and placed on ice for 15 min. Cells were washed in PBS and resuspended in 70% ethanol and stored at −20 °C. Cells were then labeled with BrdU 5′-triphosphate and terminal deoxynucleotidyl transferase at room temperature overnight with gentle agitation. Cells were washed and the Alexa Fluor 488 dye-labeled anti-BrdU antibody was added and incubated for 30 min away from light. Cells were then diluted in PBS and imaged using the Olympus IX70 fluorescent microscope. Ten fields of view were analyzed per condition. The number of apoptotic cells were counted and divided by the total number of cells per field of view using ImageJ software (National Institutes of Health).
2.10. Apoptotic DNA laddering
Genomic DNA was extracted from cells using a Genomic DNA Extraction kit (Life Technologies). DNA concentrations were determined using the NanoDrop 1000 spectrophotometer (ThermoFisher Scientific). An apoptotic positive control (lyophilized apoptotic U937 cells; Roche, Basel, Switzerland) and 2 μg of each DNA sample were electrophoresed on a 1% agarose-Tris–acetate–EDTA gel. Gel images were obtained using the Gel Doc™ XR+ (Bio-Rad, Hercules, CA, USA).
2.11. Statistics
Data presented as the mean ± S.E.M. Statistical significance was determined using one-way ANOVA with Bonferroni post hoc comparisons using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). A P-value of <0.05 was considered statistically significant.
3. Results
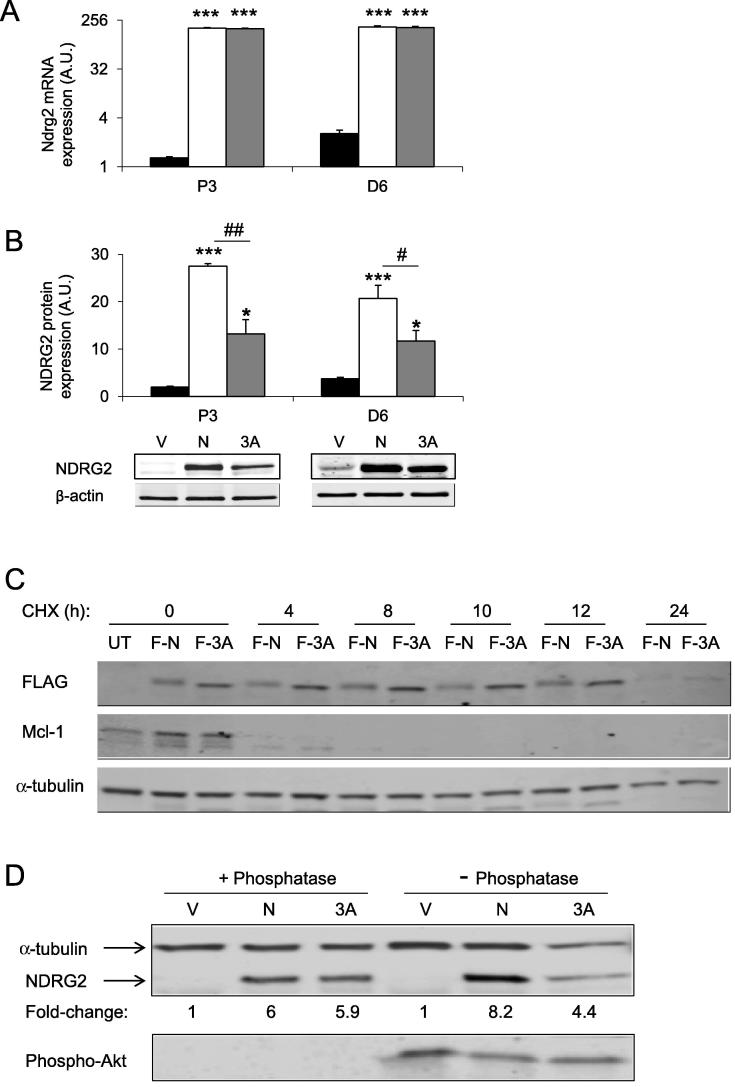
Initially, the expression levels of NDRG2 and 3A-NDRG2 following their overexpression were established in both proliferating and differentiated myoblasts. NDRG2 and 3A-NDRG2 mRNA levels increased approximately 250-fold in myoblasts at day 3 of proliferation (P3) (P < 0.001) and 90-fold at day 6 of differentiation (D6) in myotubes (P < 0.001) when compared to endogenous NDRG2 gene expression represented by the vector control (Fig. 1A). Accordingly, NDRG2 and 3A-NDRG2 overexpression increased their respective protein levels significantly above endogenous NDRG2 protein expression (Fig. 1B), which increases with C2C12 myoblast differentiation as previously reported [15]. However, overexpressed NDRG2 protein remained 2-fold higher than 3A-NDRG2 protein levels at each stage of proliferation and differentiation (P < 0.05; Fig. 1B). To address if there was a difference in protein stability, the half-life of the overexpressed NDRG2 and 3A-NDRG2 proteins tagged with a FLAG epitope were measured. The half-life of NDRG2 protein was reported previously as less than 8 h in pancreatic beta-cells [18]. Here, however, we found that both NDRG2 and 3A-NDRG2 displayed a half-life of more than 12 h and were both essentially degraded by 24 h suggesting similar stabilities (Fig. 1C). The short-lived pro-survival protein, myeloid cell leukemia 1 (Mcl-1), had also decayed by 4 h as expected [32]. Moreover, detection of the two NDRG2 proteins with the FLAG antibody revealed comparable levels of expression. Finally, treatment of the protein lysates with lambda protein phosphatase, a Ser, Thr and tyrosine phosphatase, revealed equivalent levels of de-phosphorylated NDRG2 protein expression in both NDRG2 and 3A-NDRG2 compared with untreated samples (Fig. 1D). Phospho-Akt levels were also measured to confirm the de-phosphorylation effect of the phosphatase. Therefore, we attribute the difference between the overexpressed NDRG2 and 3A-NDRG2 protein levels to greater reactivity displayed by the NDRG2-specific antibody for the phosphorylated form of NDRG2, compared to 3A-NDRG2, rather than a stability difference between the two proteins.
Fig. 1.
NDRG2 and 3A-NDRG2 overexpression characterization. NDRG2 mRNA (A) and protein (B) expression levels following infection with empty vector (black bars; V), NDRG2 (white bars; N) or 3A-NDRG2 (gray bars; 3A) at day 3 of proliferation (P3) and day 6 of differentiation (D6) (n = 3 per treatment). β-Actin expression indicates protein levels loaded. ***P < 0.001 and *P < 0.05 compared to vector; #P < 0.05 and ##P < 0.01 compared to NDRG2. (C) Immunoblots representing a time-course (h) of FLAG-NDRG2 (F-N) and FLAG-3A-NDRG2 (F-3A) protein stability following 10 μg/ml cycloheximide (CHX) treatment. An untransfected control (UT) is included and FLAG, Mcl-1 and α-tubulin protein levels are indicated. (D) Phosphorylated and de-phosphorylated levels of NDRG2, α-tubulin and phospho-Akt proteins in untreated (− phosphatase) and lambda protein phosphatase-treated (+ phosphatase) protein lysates following infection with vector (V), NDRG2 (N) or 3A-NDRG2 (3A) at P3. Normalized to α-tubulin protein, fold-change of NDRG2 expression to vector controls is indicated.
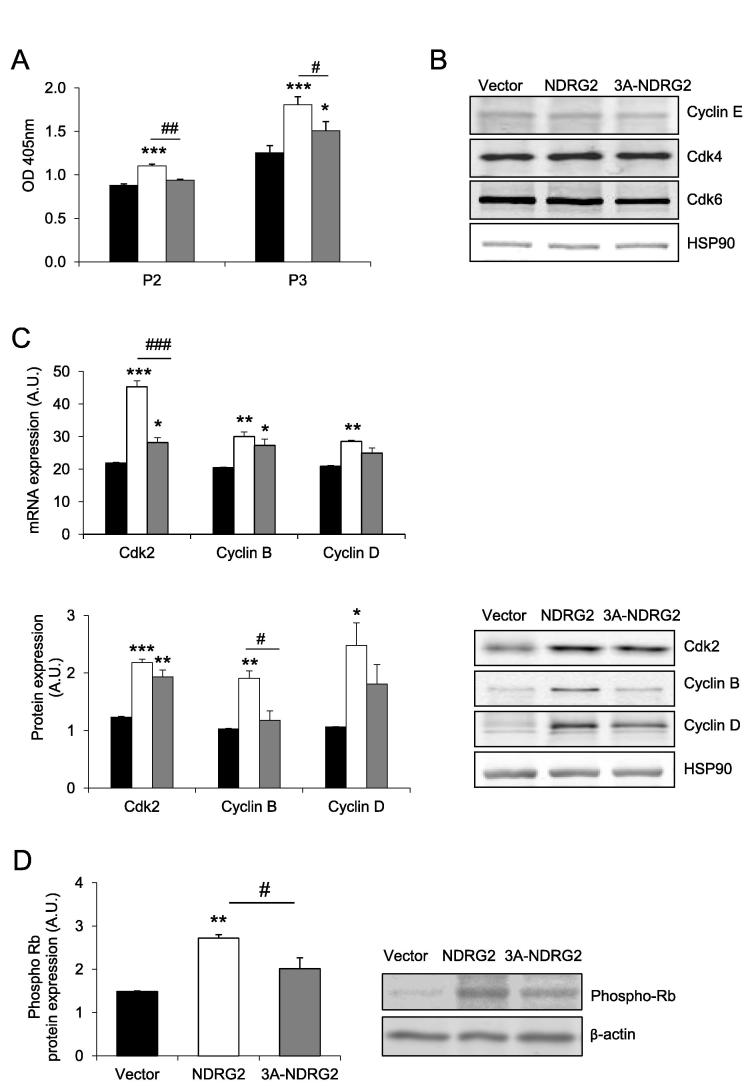
Having established comparable overexpression levels between NDRG2 and 3A-NDRG2, C2C12 myoblast proliferation rates were assessed at P2 and P3. To note, in this study, our aim was not to compare between time-points, but rather to focus on the effect of the NDRG2 proteins at each given time-point. At P2, NDRG2 increased proliferation 1.25-fold (P < 0.001) when compared with the vector. There was no effect of 3A-NDRG2 on proliferation. At P3, both NDRG2 and 3A-NDRG2 increased proliferation by 1.4-fold (P < 0.001) and 1.2-fold (P < 0.05), respectively (Fig. 2A). The increase in proliferation with NDRG2 overexpression remained higher than the effect of 3A-NDRG2 (P < 0.05). NDRG2 or 3A-NDRG2 did not affect cyclin E, Cdk4 or Cdk6 protein levels (Fig. 2B) at P3. However, NDRG2 increased both the mRNA and protein expression of the positive cell cycle regulators Cdk2 (P < 0.001), cyclin B (P < 0.01) and cyclin D (P < 0.05; Fig. 2C). Similarly, 3A-NDRG2 increased the levels of Cdk2 mRNA and protein, and cyclin B mRNA (P < 0.05). The phosphorylated levels of Rb also increased significantly following NDRG2 overexpression above both vector and 3A-NDRG2 (P < 0.05; Fig. 2D) indicating increased inhibition of Rb and enhanced cell cycle progression.
Fig. 2.
The effect of NDRG2 and 3A-NDRG2 overexpression on C2C12 myoblast proliferation and expression of positive cell cycle regulators at day 2 (P2) or day 3 (P3) of proliferation. (A) Proliferation rate of C2C12 myoblasts following infection with vector (black bars), NDRG2 (white bars) or 3A-NDRG2 (gray bars) (n = 6 per treatment) at P2 and P3. (B) Protein expression of cell cycle regulators cyclin E, Cdk4 and Cdk6. (C) mRNA (upper panel) and protein (lower panel) expression of Cdk2, cyclin B, cyclin D, and (D), Rb phosphorylation levels following vector, NDRG2 and 3A-NDRG2 overexpression at P3. Data are mean of three independent experiments (n = 3 per treatment). HSP90 or β-actin expression indicates protein levels loaded. ***P < 0.001, **P < 0.01, *P < 0.05 compared to vector; ###P < 0.001, ##P < 0.01 and #P < 0.05 compared to NDRG2.
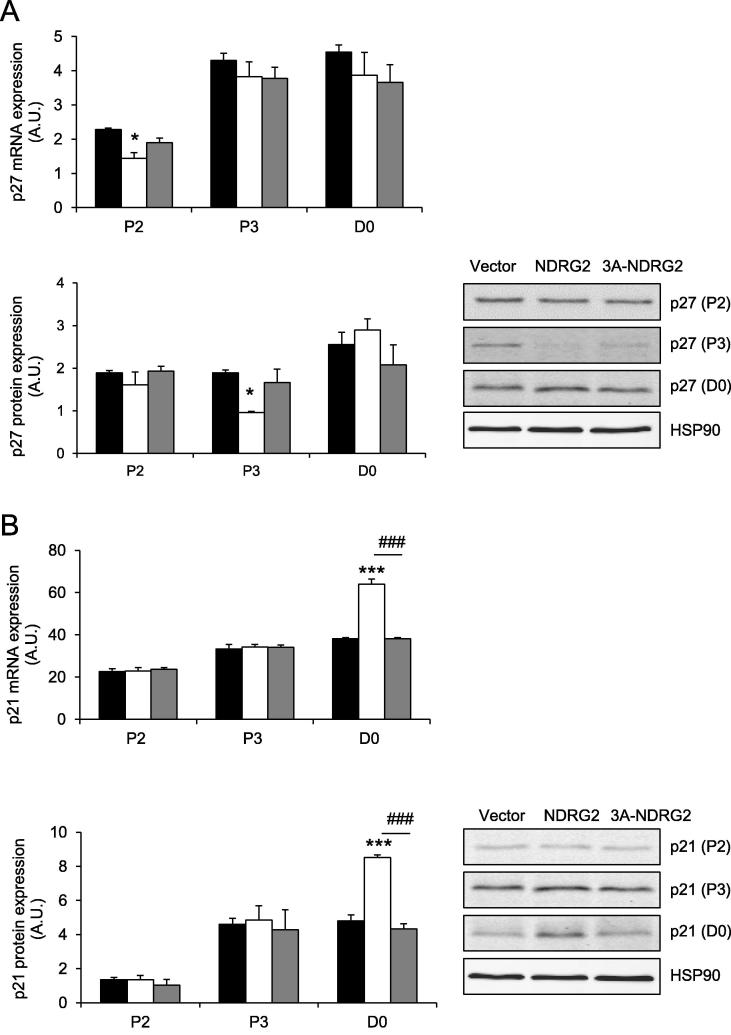
The expression of the cell cycle inhibitors p27 Kip1 (p27) and p21 were also investigated. Following NDRG2 overexpression p27 mRNA levels were reduced 1.4-fold (P < 0.05) at P2 with a corresponding 2-fold suppression of its protein levels at P3 (P < 0.05, Fig. 3A). In contrast, no difference in p21 mRNA was observed at P2 or P3. However, when myoblasts reached confluence (D0), p21 mRNA and protein levels increased 1.7-fold with NDRG2 treatment (P < 0.001; Fig. 3B). No effect of 3A-NDRG2 was observed for any of the mRNA or protein targets measured.
Fig. 3.
The expression of negative cell cycle regulators following NDRG2 and 3A-NDRG2 overexpression in myoblasts at days 2 and 3 of proliferation (P2 and P3) and at confluence (D0). (A) p27 mRNA (upper panel) and protein (lower panel) expression, and (B) p21 mRNA (upper panel) and protein (lower panel) expression with representative blots indicated. Data are mean of three independent experiments (n = 3 per treatment). HSP90 expression indicates protein levels loaded. ***P < 0.001 and *P < 0.05 compared to vector; ###P < 0.001 compared to NDRG2.
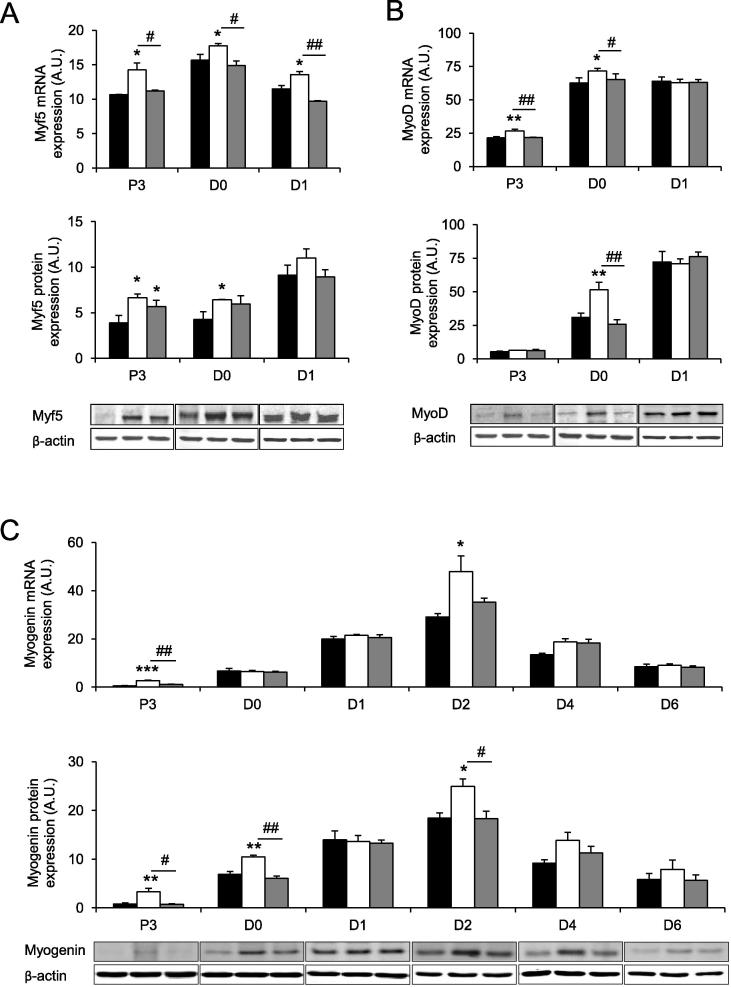
The effects of NDRG2 and 3A-NDRG2 overexpression during the early stages of differentiation were next investigated through the expression analyses of the MRFs, Myf5, MyoD and myogenin. NDRG2 overexpression increased Myf5 mRNA and protein expression by 1.5- and 1.3-fold at P3 and D0, respectively, while mRNA levels remained significantly elevated by 1.2-fold at D1 (P < 0.05) while 3A-NDRG2 increased Myf5 protein levels at P3 (P < 0.05; Fig. 4A). For MyoD expression levels, NDRG2 overexpression increased mRNA expression by 1.2-fold at P3 and D0, and protein levels by 1.7-fold at D0 (P < 0.05; Fig. 4B). No difference in MyoD mRNA or protein expression was observed at D1 and no effect by 3A-NDRG2 was found. Finally, myogenin levels were examined over the course of differentiation and NDRG2 overexpression increased myogenin mRNA and protein expression by 4.4-fold at P3 (P < 0.05) as well as increasing myogenin protein expression by 1.5-fold at D0 (P < 0.01) and both mRNA and protein levels at D2 when maximal myogenin expression was reached (P < 0.05; Fig. 4C). No difference in myogenin mRNA or protein expression was observed at D1 or with 3A-NDRG2 overexpression.
Fig. 4.
NDRG2 and 3A-NDRG2 overexpression effect on MRFs expression levels. (A) Myf5, (B) MyoD and (C) Myogenin mRNA (upper panels) and protein (lower panels) expression levels with representative blots indicated in C2C12 cells at P3, D0, D1 and up to D6 for myogenin. Myoblasts are infected with vector alone (black bars), NDRG2 (white bars) and 3A-NDRG2 (gray bars). All data are averages of three independent experiments (n = 3 per condition). ***P < 0.001, **P < 0.01 and *P < 0.05 compared to vector; ##P < 0.01, #P < 0.05 compared to NDRG2.
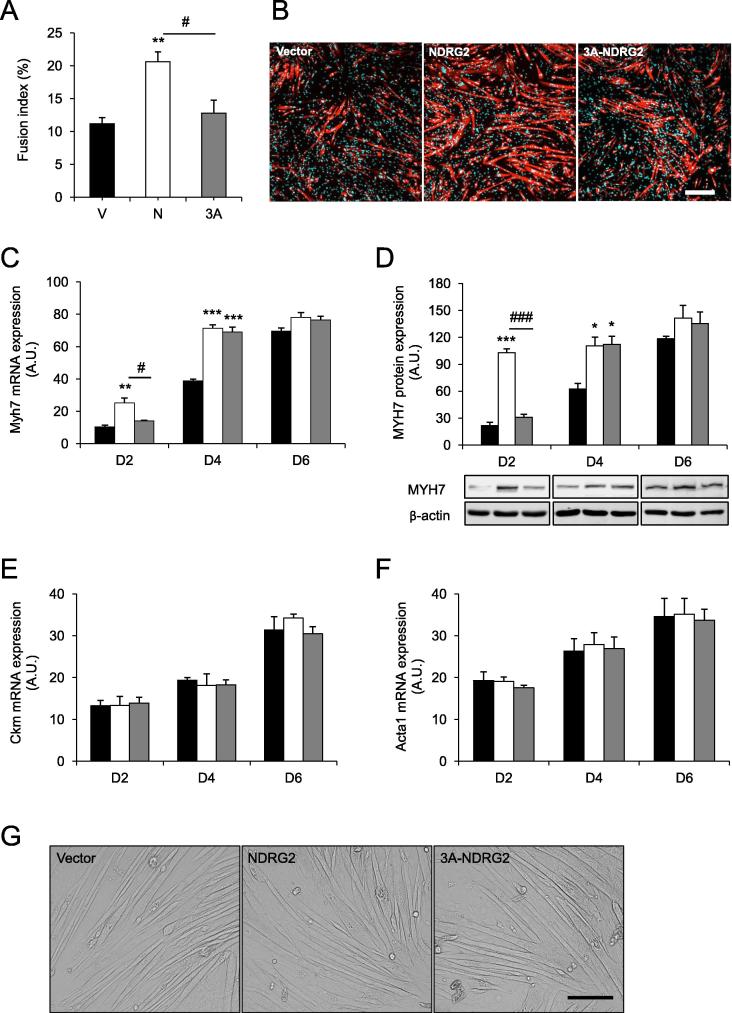
Myocyte fusion was next assessed by measuring the number of nuclei in differentiating myocytes as identified through their expression of myosin, type 1 slow (myosin, heavy polypeptide 7; MYH7) protein at D2. MYH7-positive myocytes with more than three nuclei per myocyte were counted in the analyses (Fig. 5A). When compared to the vector control and 3A-NDRG2, NDRG2 overexpression increased the fusion index by 46% (P < 0.01) and 38% (P < 0.05), respectively (Fig. 5B). 3A-NDRG2 did not significantly increase myocyte fusion. Myh7 mRNA and MYH7 protein were then measured at D2, D4 and D6 with NDRG2 significantly increasing expression at D2 and D4 but not at D6 (Fig. 5C and D) when cells are well differentiated (Fig. 5G). 3A-NDRG2 also increased Myh7 mRNA and MYH7 protein expression at D4. No difference, however, was measured between treatments on any day for markers of differentiated muscle cells [33,34] including muscle creatine kinase (Ckm) and skeletal muscle alpha-actin (Acta1) (Fig. 5E and F) and no clear phenotypic differences in the differentiated myotubes were evident at D6 between treatments (Fig. 5G).
Fig. 5.
NDRG2 and 3A-NDRG2 overexpression effect on myocyte fusion index and gene markers of differentiation. (A) Fusion index (n = 10 fields of view per condition) following vector (V), NDRG2 (N) and 3A-NDRG2 (3A) overexpression. (B) Immunofluorescent images of differentiating C2C12 myocytes at D2. Nuclei are stained with DAPI (blue labeling) and fusing myocytes are stained for MYH7 expression (red labeling). Scale bar = 200 μM. (C) mRNA and (D) protein expression levels of Myh7, and mRNA levels of (E) Ckm and (F) Acta1 at D2, D4 and D6 (n = 3 per condition). (G) Phenotypic appearance of C2C12 myotubes at D6 following vector, NDRG2 and 3A-NDRG2 overexpression. Scale bar = 50 μM. ***P < 0.001, **P < 0.01 and *P < 0.05 compared to vector; ###P < 0.001, #P < 0.05 compared to NDRG2.
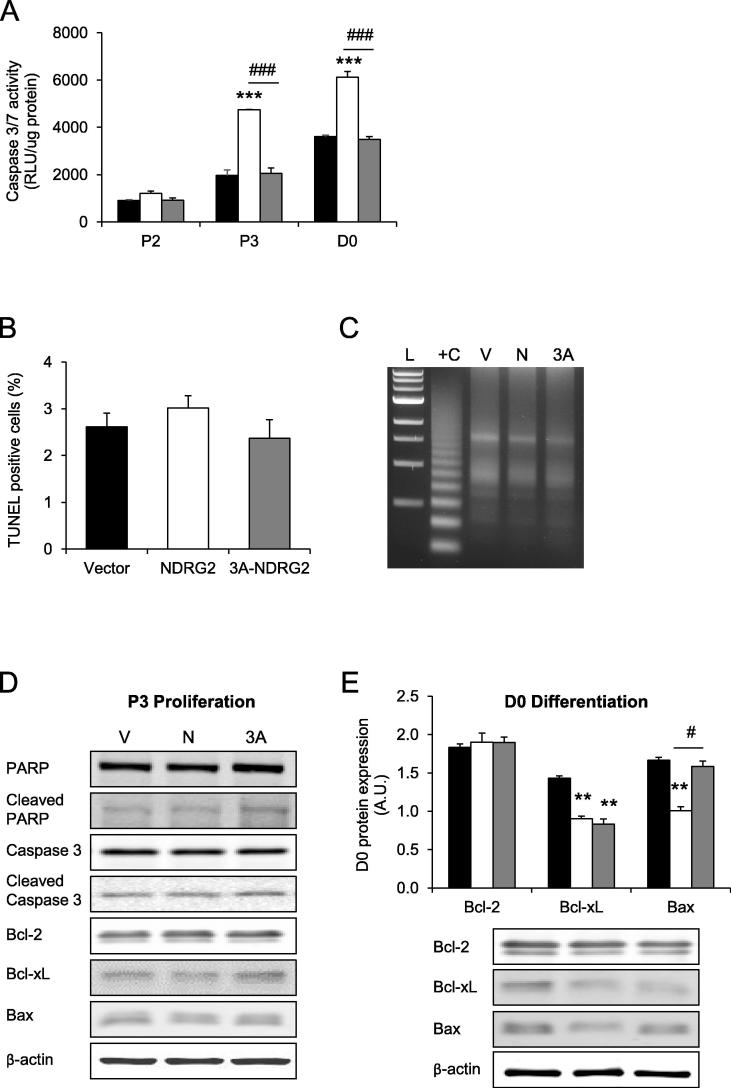
To determine whether increased expression of NDRG2 promoted apoptosis in C2C12 myoblasts during normal growth conditions, indices of apoptosis were measured in the proliferating and differentiating cells. Initially, the activities of caspases 3/7, effector caspases of apoptosis initiation, were investigated. NDRG2 overexpression increased caspase 3/7 activities by 1.3-, 2.4- and 1.7-fold at P2, P3 and D0, respectively (P < 0.001 at P3 and D0; Fig. 6A), whereas 3A-NDRG2 had no effect. However, no difference in the number of TUNEL positive cells was observed at P3 (Fig. 6B). Additionally, no increase in apoptotic DNA laddering at P3 (Fig. 6C) or during D1–2 (data not shown) was found with any of the treatments. Furthermore, the protein levels of cleaved poly (ADP ribose) polymerase (PARP) and cleaved caspase 3 proteins at P3 (Fig. 6D) and during D0–2 (data not shown) were not altered. The anti-apoptotic regulators Bcl-2 and Bcl-2-like 1 (Bcl-xL), as well as the pro-apoptotic protein Bax remained unchanged at P3 (Fig. 6D). However, at D0 the protein expression of Bax and Bcl-xL were both decreased by 1.4-fold following NDRG2 overexpression (P < 0.01; Fig. 6E). 3A-NDRG2 also decreased Bcl-xL protein expression by 1.5-fold (P < 0.01), but had no effect on Bax expression.
Fig. 6.
Caspase 3/7 activities and apoptosis levels in proliferating and differentiating C2C12 myoblasts following NDRG2 and 3A-NDRG2 overexpression. (A) Caspase 3/7 activities in myoblasts at P2, P3 and D0. Cells were infected with vector alone (black bars), NDRG2 (white bars) or 3A-NDRG2 (gray bars). Luminescence as a reflection of activity is represented as relative light units (RLU), n = 6 per sample. (B) Percentage of TUNEL positive myoblasts (10 fields of view counted per treatment), and (C) apoptotic DNA laddering gel at P3. Lanes: L, DNA 1 kb ladder; +C, positive control apoptotic U937 cells; V, vector; N, NDRG2; and 3A, 3A-NDRG2. (D) Immunoblots of total and cleaved PARP, total and cleaved caspase 3, Bcl-2, Bcl-xL and Bax proteins at P3, and (E), at or D0 for Bcl-2, Bcl-xL and Bax proteins (n = 3 per treatment). β-Actin protein indicates protein levels loaded. Data are mean of three independent experiments. ***P < 0.001 and **P < 0.01 compared to vector; ###P < 0.001 and #P < 0.05 compared to NDRG2.
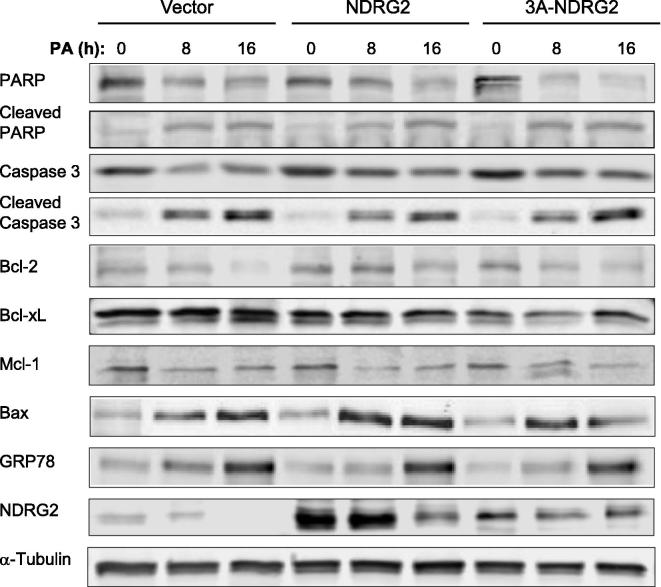
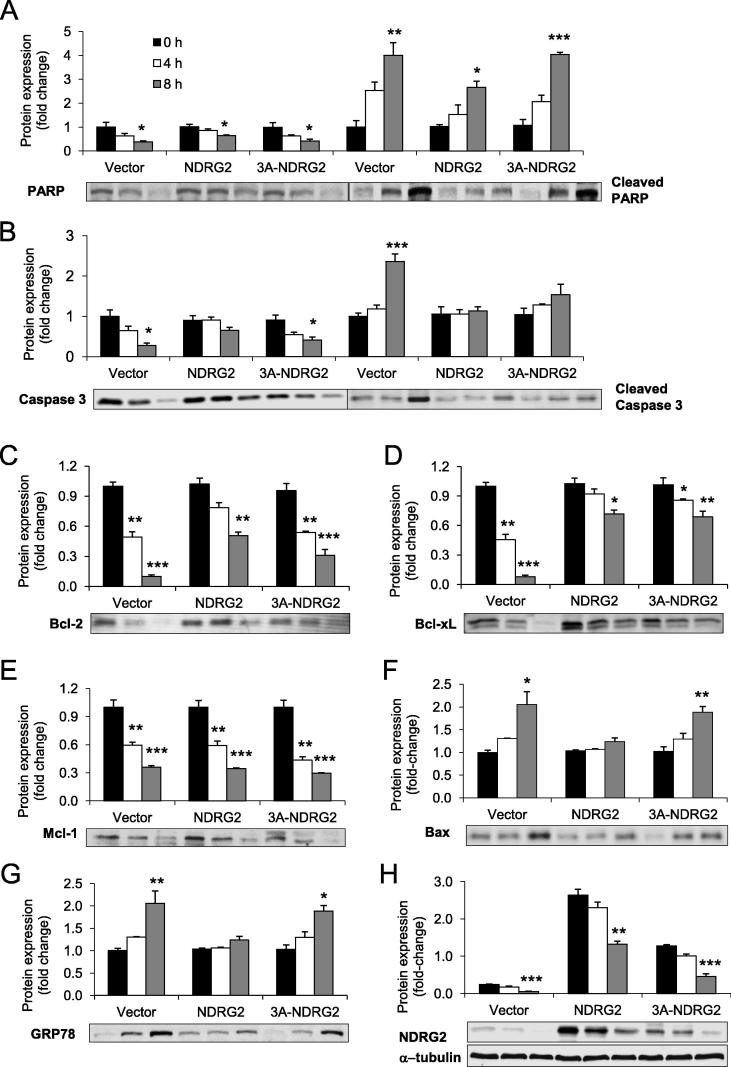
We next assessed the role for NDRG2 and 3A-NDRG2 under different stress treatments during myoblast proliferation. Both PA and H2O2 treatments cause significant apoptosis with oxidative and ER stress observed in C2C12 muscle cells [21–23,35]. Accordingly, we determined that 0.5 mM PA and 0.5 mM H2O2 treatments reduced myoblasts proliferation by nearly 50% after 16 and 8 h treatments, respectively (P < 0.001), while shorter exposure times also significantly decreased proliferation (P < 0.05; Table 2). Both NDRG2 and 3A-NDRG2 overexpression were unable to alter the proliferation rates of the myoblasts in the presence of PA treatment (Table 2). Nor did they alter the protein levels of apoptosis or ER stress markers including PARP and caspase 3 cleavage, Bcl-2 family members Bcl-2, Bcl-xL, Bax and Mcl-1, and glucose-regulated protein 78 (GRP78) (Fig. 7). In contrast, NDRG2 overexpression inhibited the decrease in cell proliferation following 4 h H2O2 treatment and attenuated the effect of H2O2 at 8 h with only a 17% proliferation decrease measured (P < 0.05) when compared to the 48% decrease with the vector alone (P < 0.001; Table 2). Moreover, NDRG2 reduced the amount of PARP and caspase 3 cleavage at 8 h (Fig. 8A and B) and reduced the loss of Bcl-2 and Bcl-xL (Fig. 8C and D) but Mcl-1 expression did not change (Fig. 8E). The induction of both Bax and GRP78 expression levels by H2O2 treatment was blocked by NDRG2 at 4 and 8 h time-points (Fig. 8F and G). 3A-NDRG2 only had moderate effects in preventing the loss in cell proliferation following H2O2 treatment (Table 2) and in regulating the expression of the apoptotic and ER stress protein markers (Fig. 8). Interestingly, both endogenous and overexpressed NDRG2 protein levels were significantly reduced by 8 h of H2O2 exposure with 80%, 50% and 60% decrease in NDRG2 expression measured in vector, NDRG2- and 3A-NDRG2-treated myoblasts, respectively (Fig. 8H).
Table 2.
The % decrease in myoblast proliferation at P3 following vector, NDRG2 and 3A-NDRG2 overexpression and 0.5 mM PA or 0.5 mM H2O2 treatments.
| PA | Vector | NDRG2 | 3A-NDRG2 |
|---|---|---|---|
| 0 h | 0% | 0% | 0% |
| 8 h | ↓ 32%⁎⁎ | ↓ 30%⁎⁎ | ↓ 29%⁎ |
| 16 h | ↓ 44%⁎⁎⁎ | ↓ 49%⁎⁎⁎ | ↓ 44%⁎⁎⁎ |
| H2O2 | |||
| 0 h | 0% | 0% | 0% |
| 4 h | ↓ 23%⁎ | ↓ 3% | ↓ 15%⁎ |
| 8 h | ↓ 48%⁎⁎⁎ | ↓ 17%⁎ | ↓ 36%⁎⁎⁎ |
P < 0.05.
P < 0.01.
P < 0.001 to each 0 h control group; n = 6 per treatment.
Fig. 7.
Protein expression levels of NDRG2, apoptotic and ER stress markers following 0, 8 and 16 h palmitate (PA) treatment of vector, NDRG2 and 3A-NDRG2-infected C2C12 myoblasts at P3. Protein loading is indicated by the α-Tubulin immunoblots.
Fig. 8.
Protein expression levels of apoptotic and ER stress markers following H2O2 treatment of vector, NDRG2 and 3A-NDRG2-infected C2C12 myoblasts at P3. Protein expression levels of (A) total and cleaved PARP, (B) total and cleaved caspase 3, (C) Bcl-2, (D) Bcl-xL, (E) Mcl-1, (F) Bax, (G) GRP78 and (H) NDRG2 proteins. Alpha-tubulin (α-Tubulin) protein indicates protein levels loaded. Hydrogen peroxide treatment times were 0 h (black bars), 4 h (white bars) or 8 h (gray bars). Data are the mean of three independent experiments (n = 3 per treatment). ***P < 0.001, **P < 0.01 and *P < 0.05 compared to each 0 h control.
4. Discussion
This study aimed to investigate the effect of NDRG2 on myogenic processes and apoptosis under both normal growth and stress-induced conditions in C2C12 muscle cells. We identified that increased levels of NDRG2 promoted the proliferation, and hence, onset of differentiation of C2C12 myoblasts although the overall level of differentiation in mature myotubes was not different. In addition, NDRG2 overexpression attenuated the stress induced by H2O2 treatment while a phospho-deficient form of NDRG2 displayed reduced function. These findings extend our previous observations identifying that the knockdown of NDRG2 impaired myogenesis in vitro [15] and corroborate a unique role for NDRG2 in myoblast growth and response to stress in skeletal muscle cells.
Here, we demonstrated that NDRG2 enhanced the expression of the positive cell cycle proteins Cdk2, cyclin B and cyclin D and down-regulated the negative cell cycle regulator p27 during proliferation. In addition, increased phosphorylation of the tumor suppressor Rb protein was measured, most likely due to enhanced activity of cyclins and Cdks (reviewed in [3]). This was followed by the up-regulation of p21 at myoblast confluence corresponding to cell cycle exiting and commitment to differentiation [5,36]. Cyclins A and B target the phases of growth (G2) and mitosis (M) phases, whereas Cdk2 complexes with cyclins A and E acting on the later G1 and synthesis phases, and cyclin D targets the resting G0 to early G1 phase (reviewed in [37]). Therefore, it appears that NDRG2 overexpression generates a positive broad cell-cycle effect in C2C12 myoblasts rather than targeting a single, specific phase of the cell cycle. Indeed, a recent study that overexpressed NDRG2 in proliferating C2C12 myoblasts found that NDRG2 had no specific effect on any particular phase of the cell cycle [38]. However, this study also reported that NDRG2 overexpression increased the pool of dividing myoblasts and the G2/M phase with less cells in the G1/G0 resting phase within 24 h of adding differentiation media to the myoblasts [38]. The authors interpreted this as a delay in cell cycle withdrawal during differentiation, which may equate to increased proliferation but this was not measured under these conditions. In contrast, we did not observe any delay in cell cycle exiting with our myoblasts in growth medium. We observed both a higher proliferation rate and significantly elevated p21, MyoD and myogenic expression levels from NDRG2 overexpression at D0 indicating cell cycle exiting and commitment to differentiate earlier than myoblasts infected with vector alone or with 3A-NDRG2 overexpression. While a higher myocyte fusion index was measured at D2, the overall level of differentiation was not greater by D6 suggesting that NDRG2 primarily affected myoblast proliferation. The broad effects of NDRG2 on cell cycle control are further highlighted in reports in cancer cells. NDRG2 overexpression down-regulated cyclin D1 in human colon carcinoma cells [29] whereas it suppressed cyclin E, but not cyclin D or Cdk2, resulting in cell cycle arrest at the G1/S phase in rat liver cells [39]. Similarly to our findings here, a Ser/Thr phospho-deficient form of human NDRG2 did not significantly alter cyclin D or p21 expression levels in the colon carcinoma cells. This NDRG2 mutant also displayed a reduced inhibitory effect on cancer growth [29] compared with human NDRG2 overexpression suggesting that the specific Ser and Thr residues in both the human and mouse NDRG2 protein contribute to its activity in cancer and muscle cells. How NDRG2 affects cellular proliferation and why changes in NDRG2 expression have opposing effects on cell proliferation in C2C12 myoblasts compared with human cancer cells are currently unknown. It is interesting to note that the knockdown of the ortholog NDRG4 reduces cardiac myocyte proliferation [40] despite being described as a tumor suppressor while NDRG3 appears to play a more cancer growth-promoting role [41]. Clearly, further investigation into the mechanisms of how the NDRG proteins control cell growth in cancer versus non-cancer cells is warranted.
While NDRG2 has been described to enhance apoptosis in non-cancer [39,42] and cancer [43,44] cell types, we did not observe an increase in myoblast apoptosis under basal growth conditions in C2C12 myoblasts nor change in Bcl-2 levels unlike the decreased Bcl-2 expression reported in Hela cells following NDRG2 overexpression [19]. A decrease in the expression of both the anti-apoptotic Bcl-xL and the pro-apoptotic Bcl-2 family member Bax were measured following overexpression NDRG2 but are in contrast to the increased Bax expression observed in liver cells [39]. The significance of reduced Bax and Bcl-xL levels in muscle cells is unknown although overexpression of Bcl-xL can inhibit myotube formation [9]. Indeed, non-apoptotic roles for the Bcl-2 family are being realized including regulation of calcium homeostasis and ER function [45]. The role of caspase 3 in muscle cell differentiation is not entirely clear at present. Enhanced activities of caspase 3, and potentially its upstream initiator, caspase 9, are required for myoblast fusion and differentiation [8,9,46,47]. In C2C12 myoblasts, caspase 3 activation along with transient activation of caspase 2 in differentiating myoblasts were measured although caspase 8 or 9 activities were not increased [48]. Interestingly, this study also determined that neither X-linked inhibitor of apoptosis protein nor mitochondrial-based apoptosis were involved in the transient caspase 3 activation measured [48]. Indeed, increased caspase 3 has been linked to the cleavage of actomyosin and ubiquitin–proteasome muscle proteolysis [49] and instead may be required for organelle remodeling during normal cell function [50]. Caspase 3 is also thought to contribute to myogenesis through the regulation of mechanosensitive cation channels [46] and through an effector molecule, the Mammalian Sterile Twenty-like kinase [8].
During H2O2-induced stress conditions, we observed that NDRG2 helped attenuate a loss in cell proliferation and the induction of apoptosis. This was in parallel with a reduced loss in the pro-survival factors Bcl-2 and Bcl-xL, but not Mcl-1, suggesting that NDRG2 targets specific Bcl family members. With time, however, decreased levels of endogenous and exogenous NDRG2 were observed also with the induced stress. The cause of this loss in NDRG2 is not known currently but NDRG2 may be a target for ubiquitin-mediated degradation during stress as NDRG2 has been reported a substrate of the E3 Ligase Trim32 ubiquitination in skeletal muscle [38]. Indeed, Cyclin D1 is a targeted for ubiquitin-dependent proteasomal degradation during oxidative stress mediated by H2O2 exposure [51]. Interestingly, no protective effect was identified during PA treatment despite H2O2 and PA treatments inducing both oxidative and ER stress and apoptosis [21–23,35]. A potential explanation for this difference may lie within the regulation of NDRG2’s C-terminal amino acids, Ser332, Thr348 and Ser350. Our data confirm that the phosphorylation status is important for NDRG2 function as the removal of these residues did ablate the NDRG2 increase in caspase 3/7 activities, myocyte fusion and protection against H2O2 treatment. Whether Ser332, Thr348 and Ser350 contribute significantly individually or in combination to the function of mouse NDRG2 is yet to be determined. The Ser/Thr kinases that potentially target these residues in skeletal muscle include Akt, PKCθ and serum- and glucocorticoid-inducible kinase 1 (SGK1) [24,25]. The possible impact of Akt and SGK1 on NDRG2 phosphorylation and regulation in the context of myoblast proliferation and differentiation, particularly by Akt1 and Akt2, has been discussed previously [15]. The role for PKCθ in muscle growth and homeostasis is less clear. While PKCθ promotes myoblast fusion [27], reduced levels of PKCθ appear to enhance myogenesis [28] and ameliorate muscle dystrophy [52]. Of particular interest to this study here is the potential inhibitory regulation of NDRG2 by PKCθ [24], which itself becomes activated upon PA treatment in C2C12 cells [53,54]. As PKCθ phosphorylates Ser332 in the mouse NDRG2 protein sequence, it has been shown it can block phosphorylation of the Thr348 site by insulin [24]. Potentially, this inhibition of Thr348 phosphorylation during PA treatment, and presumably PKCθ activation, may prevent NDRG2’s protective function and provide an explanation for NDRG2’s inability to attenuate lipotoxicity in C2C12 muscle cells. The positive effects of Akt and insulin on NDRG2 function are highlighted where the knocked down of NDRG2 removed the ability of a constitutively active Akt to block apoptosis induced by PA in pancreatic beta cells [18]. Whether NDRG2 overexpression alone could protect against lipotoxicity in the pancreatic beta cells was not reported.
In conclusion, this study demonstrates that in C2C12 myoblasts NDRG2 promotes their proliferation and earlier onset of myogenic differentiation, which may be contributed to by at least one of its Ser332, Thr348 or Ser350 phosphorylation sites. Furthermore the pro-myogenic role of NDRG2 was elicited partially through enhanced caspase 3/7 activities. NDRG2 overexpression attenuated oxidative and ER stress and apoptosis induced by treatment with H2O2, but not from PA-induced lipotoxicity. Confirmation of these findings in vivo and the amino acid residues specifically targeted by Ser/Thr kinases during skeletal muscle development and under different stress conditions remain to be determined. Such studies will further enhance our understanding of the physiological role and regulation of NDRG2 in skeletal muscle.
Conflicts of interest
None are declared by the authors.
Acknowledgments
This work was supported by Deakin University and the Centre for Physical Activity and Nutrition Research (C-PAN), Australia. The authors would like to thank Dr D. Segal for helpful discussions and advice, Prof D. Huang and Dr T. Okamoto from the Walter and Eliza Hall Institute of Medical Research, Parkville, Australia, for their kind gift of the pMSCV-IRES-hygromycin retroviral vector, and Dr C. Schmitz-Peiffer, Garvan Institute, Sydney, Australia, for his generous provision of the pCMV/SV-FLAG1 plasmids expressing mouse wild type and phospho-deficient NDRG2 cDNAs. VF conceived and designed the study, KA and VF acquired the data, KA analyzed the data, VF, KA and AR interpreted the data and wrote the paper.
References
- 1.Tintignac L.A., Leibovitch M.P., Kitzmann M., Fernandez A., Ducommun B., Meijer L., Leibovitch S.A. Cyclin E-cdk2 phosphorylation promotes late G1-phase degradation of MyoD in muscle cells. Exp. Cell Res. 2000;259:300–307. doi: 10.1006/excr.2000.4973. [DOI] [PubMed] [Google Scholar]
- 2.Batonnet-Pichon S., Tintignac L.J., Castro A., Sirri V., Leibovitch M.P., Lorca T., Leibovitch S.A. MyoD undergoes a distinct G2/M-specific regulation in muscle cells. Exp. Cell Res. 2006;312:3999–4010. doi: 10.1016/j.yexcr.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Adams P.D. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim. Biophys. Acta. 2001;1471:M123–M133. doi: 10.1016/s0304-419x(01)00019-1. [DOI] [PubMed] [Google Scholar]
- 4.Halevy O., Novitch B.G., Spicer D.B., Skapek S.X., Rhee J., Hannon G.J., Beach D., Lassar A.B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 5.Andrés V., Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochreiter-Hufford A.E., Lee C.S., Kinchen J.M., Sokolowski J.D., Arandjelovic S., Call J.A., Klibanov A.L., Yan Z., Mandell J.W., Ravichandran K.S. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;497:263–267. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez P.C., Frank S.R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernando P., Kelly J.F., Balazsi K., Slack R.S., Megeney L.A. Caspase 3 activity is required for skeletal muscle differentiation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray T.V., McMahon J.M., Howley B.A., Stanley A., Ritter T., Mohr A., Zwacka R., Fearnhead H.O. A non-apoptotic role for caspase-9 in muscle differentiation. J. Cell Sci. 2008;121:3786–3793. doi: 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- 10.Dominov J.A., Houlihan-Kawamoto C.A., Swap C.J., Miller J.B. Pro- and anti-apoptotic members of the Bcl-2 family in skeletal muscle: a distinct role for Bcl-2 in later stages of myogenesis. Dev. Dyn. 2001;220:18–26. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1088>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 11.Sandri M., Carraro U. Apoptosis of skeletal muscles during development and disease. Int. J. Biochem. Cell Biol. 1999;31:1373–1390. doi: 10.1016/s1357-2725(99)00063-1. [DOI] [PubMed] [Google Scholar]
- 12.Peterson J.M., Wang Y., Bryner R.W., Williamson D.L., Alway S.E. Bax signaling regulates palmitate-mediated apoptosis in C(2)C(12) myotubes. AJP Endocrinol. Metab. 2008;295:E1307–E1314. doi: 10.1152/ajpendo.00738.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu X., Zhai Y., Wei H., Zhang C., Xing G., Yu Y., He F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol. Cell. Biochem. 2002;229:35–44. doi: 10.1023/a:1017934810825. [DOI] [PubMed] [Google Scholar]
- 14.Hojlund K., Bowen B.P., Hwang H., Flynn C.R., Madireddy L., Geetha T., Langlais P., Meyer C., Mandarino L.J., Yi Z. In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC–ESI-MS/MS. J. Proteome Res. 2009;8:4954–4965. doi: 10.1021/pr9007267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foletta V.C., Prior M.J., Stupka N., Carey K., Segal D.H., Jones S., Swinton C., Martin S., Cameron-Smith D., Walder K.R. NDRG2, a novel regulator of myoblast proliferation, is regulated by anabolic and catabolic factors. J. Physiol. 2009;587:1619–1634. doi: 10.1113/jphysiol.2008.167882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., Liu N., Yao L., Li F., Zhang J., Deng Y., Liu J., Ji S., Yang A., Han H., Zhang Y., Zhang J., Han W., Liu X. NDRG2 is a new HIF-1 target gene necessary for hypoxia-induced apoptosis in A549 cells. Cell. Physiol. Biochem. 2008;21:239–250. doi: 10.1159/000113765. [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Zhang J., Wang X., Li Y., Chen Y., Li K., Zhang J., Yao L., Guo G. HIF-1 and NDRG2 contribute to hypoxia-induced radioresistance of cervical cancer Hela cells. Exp. Cell Res. 2010;316:1985–1993. doi: 10.1016/j.yexcr.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 18.Shen L., Liu X., Hou W., Yang G., Wu Y., Zhang R., Li X., Che H., Lu Z., Zhang Y., Liu X., Yao L. NDRG2 is highly expressed in pancreatic beta cells and involved in protection against lipotoxicity. Cell. Mol. Life Sci. 2010;67:1371–1381. doi: 10.1007/s00018-010-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Yang L., Zhang J., Zhang J., Chen Y., Li K., Li Y., Li Y., Yao L., Guo G. Knock-down of NDRG2 sensitizes cervical cancer Hela cells to cisplatin through suppressing Bcl-2 expression. BMC Cancer. 2012;12:370–378. doi: 10.1186/1471-2407-12-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Luo P., Wang F., Yang Q., Li Y., Zhao M., Wang S., Wang Q., Xiong L. Inhibition of N-myc downstream-regulated gene-2 is involved in an astrocyte-specific neuroprotection induced by sevoflurane preconditioning. Anesthesiology. 2014;121:549–562. doi: 10.1097/ALN.0000000000000314. [DOI] [PubMed] [Google Scholar]
- 21.Patkova J., Andel M., Trnka J. Palmitate-induced cell death and mitochondrial respiratory dysfunction in myoblasts are not prevented by mitochondria-targeted antioxidants. Cell. Physiol. Biochem. 2014;33:1439–1451. doi: 10.1159/000358709. [DOI] [PubMed] [Google Scholar]
- 22.Siu P.M., Wang Y., Alway S.E. Apoptotic signaling induced by H2O2-mediated oxidative stress in differentiated C2C12 myotubes. Life Sci. 2009;84:468–481. doi: 10.1016/j.lfs.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierre N., Barbe C., Gilson H., Deldicque L., Raymackers J.M., Francaux M. Activation of ER stress by hydrogen peroxide in C2C12 myotubes. Biochem. Biophys. Res. Commun. 2014;450:459–463. doi: 10.1016/j.bbrc.2014.05.143. [DOI] [PubMed] [Google Scholar]
- 24.Burchfield J.G., Lennard A.J., Narasimhan S., Hughes W.E., Wasinger V.C., Corthals G.L., Okuda T., Kondoh H., Biden T.J., Schmitz-Peiffer C. Akt mediates insulin-stimulated phosphorylation of Ndrg2: evidence for cross-talk with protein kinase C theta. J. Biol. Chem. 2004;279:18623–18632. doi: 10.1074/jbc.M401504200. [DOI] [PubMed] [Google Scholar]
- 25.Murray J.T., Campbell D.G., Morrice N., Auld G.C., Shpiro N., Marquez R., Peggie M., Bain J., Bloomberg G.B., Grahammer F., Lang F., Wulff P., Kuhl D., Cohen P. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalfant C.E., Ciaraldi T.P., Watson J.E., Nikoulina S., Henry R.R., Cooper D.R. Protein kinase C theta expression is increased upon differentiation of human skeletal muscle cells: dysregulation in type 2 diabetic patients and a possible role for protein kinase C theta in insulin-stimulated glycogen synthase activity. Endocrinology. 2000;141:2773–2778. doi: 10.1210/endo.141.8.7591. [DOI] [PubMed] [Google Scholar]
- 27.Madaro L., Marrocco V., Fiore P., Aulino P., Smeriglio P., Adamo S., Molinaro M., Bouche M. PKC theta signaling is required for myoblast fusion by regulating the expression of caveolin-3 and beta1D integrin upstream focal adhesion kinase. Mol. Biol. Cell. 2011;22:1409–1419. doi: 10.1091/mbc.E10-10-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marino J.S., Hinds T.D., Jr., Potter R.A., Ondrus E., Onion J.L., Dowling A., McLoughlin T.J., Sanchez E.R., Hill J.W. Suppression of protein kinase C theta contributes to enhanced myogenesis in vitro via IRS1 and ERK1/2 phosphorylation. BMC Cell Biol. 2013;14:39. doi: 10.1186/1471-2121-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y.J., Yoon S.Y., Kim J.-T., Choi S.C., Lim J.-S., Kim J.H., Song E.Y., Lee H.G., Choi I., Kim J.W. NDRG2 suppresses cell proliferation through down-regulation of AP-1 activity in human colon carcinoma cells. Int. J. Cancer. 2009;124:7–15. doi: 10.1002/ijc.23945. [DOI] [PubMed] [Google Scholar]
- 30.Foletta V.C., Brown E.L., Cho Y., Snow R.J., Kralli A., Russell A.P. Ndrg2 is a PGC-1alpha/ERR alpha target gene that controls protein synthesis and expression of contractile-type genes in C2C12 myotubes. Biochim. Biophys. Acta. 2013;1833:3112–3123. doi: 10.1016/j.bbamcr.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Filigheddu N., Gnocchi V.F., Coscia M., Cappelli M., Porporato P.E., Taulli R., Traini S., Baldanzi G., Chianale F., Cutrupi S., Arnoletti E., Ghe C., Fubini A., Surico N., Sinigaglia F., Ponzetto C., Muccioli G., Crepaldi T., Graziani A. Ghrelin and des-acyl ghrelin promote differentiation and fusion of C2C12 skeletal muscle cells. Mol. Biol. Cell. 2007;18:986–994. doi: 10.1091/mbc.E06-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akgul C., Moulding D.A., White M.R., Edwards S.W. In vivo localisation and stability of human Mcl-1 using green fluorescent protein (GFP) fusion proteins. FEBS Lett. 2000;478:72–76. doi: 10.1016/s0014-5793(00)01809-3. [DOI] [PubMed] [Google Scholar]
- 33.Devlin R.B., Emerson C.P., Jr. Coordinate accumulation of contractile protein mRNAs during myoblast differentiation. Dev. Biol. 1979;69:202–216. doi: 10.1016/0012-1606(79)90286-0. [DOI] [PubMed] [Google Scholar]
- 34.Dym H., Yaffe D. Expression of creatine kinase isoenzymes in myogenic cell lines. Dev. Biol. 1979;68:592–599. doi: 10.1016/0012-1606(79)90229-x. [DOI] [PubMed] [Google Scholar]
- 35.Rieusset J., Chauvin M.A., Durand A., Bravard A., Laugerette F., Michalski M.C., Vidal H. Reduction of endoplasmic reticulum stress using chemical chaperones or Grp78 overexpression does not protect muscle cells from palmitate-induced insulin resistance. Biochem. Biophys. Res. Commun. 2012;417:439–445. doi: 10.1016/j.bbrc.2011.11.135. [DOI] [PubMed] [Google Scholar]
- 36.Zabludoff S.D., Csete M., Wagner R., Yu X., Wold B.J. P27Kip1 is expressed transiently in developing myotomes and enhances myogenesis. Cell Growth Differ. 1998;9:1–11. [PubMed] [Google Scholar]
- 37.Arellano M., Moreno S. Regulation of CDK/cyclin complexes during the cell cycle. Int. J. Biochem. Cell Biol. 1997;29:559–573. doi: 10.1016/s1357-2725(96)00178-1. [DOI] [PubMed] [Google Scholar]
- 38.Mokhonova E.I., Avliyakulov N.K., Kramerova I., Kudryashova E., Haykinson M.J., Spencer M.J. The E3 ubiquitin ligase TRIM32 regulates myoblast proliferation by controlling turnover of NDRG2. Hum. Mol. Genet. 2015;24:2873–2883. doi: 10.1093/hmg/ddv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Li Y., Wu L., Zhang Z., Han T., Guo H., Jiang N., Tao K., Ti Z., Liu X., Yao L., Dou K. NDRG2 in rat liver regeneration: role in proliferation and apoptosis. Wound Repair Regen. 2010;18:524–531. doi: 10.1111/j.1524-475X.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- 40.Qu X., Jia H., Garrity D.M., Tompkins K., Batts L., Appel B., Zhong T.P., Baldwin H.S. Ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev. Biol. 2008;317:486–496. doi: 10.1016/j.ydbio.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., Li Y., Li Y., Hong A., Wang J., Lin B., Li R. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int. J. Cancer. 2009;124:521–530. doi: 10.1002/ijc.23961. [DOI] [PubMed] [Google Scholar]
- 42.Li T., Hu J., He G.H., Li Y., Zhu C.C., Hou W.G., Zhang S., Li W., Zhang J.S., Wang Z., Liu X.P., Yao L.B., Zhang Y.Q. Up-regulation of NDRG2 through nuclear factor-kappa B is required for Leydig cell apoptosis in both human and murine infertile testes. Biochem. Biophys. Acta. 2012;1822:301–313. doi: 10.1016/j.bbadis.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Liu N., Wang L., Li X., Yang Q., Liu X., Zhang J., Zhang J., Wu Y., Ji S., Zhang Y., Yang A., Han H., Yao L. N-Myc downstream-regulated gene 2 is involved in p53-mediated apoptosis. Nucleic Acids Res. 2008;36:5335–5349. doi: 10.1093/nar/gkn504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi H., Li N., Li S., Chen C., Wang W., Xu C., Zhang J., Jin H., Zhang H., Zhao H., Song W., Feng Q., Feng X., Shen X., Yao L., Zhao Q. Expression of NDRG2 in esophageal squamous cell carcinoma. Cancer Sci. 2010;101:1292–1299. doi: 10.1111/j.1349-7006.2010.01529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonneau B., Prudent J., Popgeorgiev N., Gillet G. Non-apoptotic roles of Bcl-2 family: the calcium connection. Biochim. Biophys. Acta. 2013;1833:1755–1765. doi: 10.1016/j.bbamcr.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 46.Wedhas N., Klamut H.J., Dogra C., Srivastava A.K., Mohan S., Kumar A. Inhibition of mechanosensitive cation channels inhibits myogenic differentiation by suppressing the expression of myogenic regulatory factors and caspase-3 activity. FASEB J. 2005;19:1986–1997. doi: 10.1096/fj.05-4198com. [DOI] [PubMed] [Google Scholar]
- 47.Hunt L.C., Upadhyay A., Jazayeri J.A., Tudor E.M., White J.D. Caspase-3, myogenic transcription factors and cell cycle inhibitors are regulated by leukemia inhibitory factor to mediate inhibition of myogenic differentiation. Skelet. Muscle. 2011;1:17. doi: 10.1186/2044-5040-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloemberg D., Quadrilatero J. Mitochondrial pro-apoptotic indices do not precede the transient caspase activation associated with myogenesis. Biochim. Biophys. Acta. 2014;1843:2926–2936. doi: 10.1016/j.bbamcr.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Wang X.H., Mitch W.E. Muscle wasting from kidney failure-a model for catabolic conditions. Int. J. Biochem. Cell Biol. 2013;45:2230–2238. doi: 10.1016/j.biocel.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandri M. Apoptotic signaling in skeletal muscle fibers during atrophy. Curr. Opin. Clin. Nutr. Metab. Care. 2002;5:249–253. doi: 10.1097/00075197-200205000-00003. [DOI] [PubMed] [Google Scholar]
- 51.Pyo C.W., Choi J.H., Oh S.M., Choi S.Y. Oxidative stress-induced cyclin D1 depletion and its role in cell cycle processing. Biochim. Biophys. Acta. 2013;1830:5316–5325. doi: 10.1016/j.bbagen.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 52.Madaro L., Pelle A., Nicoletti C., Crupi A., Marrocco V., Bossi G., Soddu S., Bouche M. PKC theta ablation improves healing in a mouse model of muscular dystrophy. PLoS One. 2012;7:e31515. doi: 10.1371/journal.pone.0031515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jove M., Planavila A., Sanchez R.M., Merlos M., Laguna J.C., Vazquez-Carrera M. Palmitate induces tumor necrosis factor-alpha expression in C2C12 skeletal muscle cells by a mechanism involving protein kinase C and nuclear factor-kappaB activation. Endocrinology. 2006;147:552–561. doi: 10.1210/en.2005-0440. [DOI] [PubMed] [Google Scholar]
- 54.Wang X., Yu W., Nawaz A., Guan F., Sun S., Wang C. Palmitate induced insulin resistance by PKCtheta-dependent activation of mTOR/S6K pathway in C2C12 myotubes. Exp. Clin. Endocrinol. Diabetes. 2010;118:657–661. doi: 10.1055/s-0030-1252069. [DOI] [PubMed] [Google Scholar]