Abstract
Objective(s):
Eukaryotic translation initiation factor 4E (eIF4E) is overexpressed in cervical cancer (CC). However, the molecular mechanisms are unclear. This study aimed to investigate the molecular mechanism of eIF4E gene overexpression in CC.
Materials and Methods:
The human papillomavirus (HPV) type 18 E7 and eIF4E mRNAs were measured following knock down or overexpression of E7 gene by RT-PCR and real-time PCR. Cell counting kit-8 assay was used to determine the cell proliferation. Flow cytometry was used to analyze the cell cycle and apoptosis. Transwell system was employed to determine the cell migration.
Results:
Overexpression of E7 gene increased eIF4E mRNA level by 24.3% (P<0.01) in HPV negative C33A cells. Knock down of E7 decreased markedly eIF4E mRNA by 73% (P<0.01) in HPV18 positive HeLa cells. Under the state of high expression of E7, 1) up-regulation of eIF4E drastically promoted the cell proliferation, cell cycle and cell migration, and inhibited the cell apoptosis. 2) down-regulation of eIF4E significantly inhibited the cell proliferation, cell cycle and the ability of cell migration, and also promoted the apoptosis of cervical cancer cells.
Conclusion:
HPV E7 induced eIF4E gene over transcription which might be a new marker for CC. The finding broadens the understanding of the CC carcinogenesis.
Keywords: EIF4E, Cervical cancer, HPV, E7
Introduction
Cervical cancer (CC) is one of the most common female cancers (1, 2), which is closely related to human papillomavirus (HPV) infection. Among the molecules encoded by the high risk HPV DNAs, E7 plays a key role in carcinogenesis. E7 binds the retinoblastoma (Rb) protein family members (pRb, P107, P130) and degrades Rb proteins through the ubiquity 26S protease pathway (3, 4). The Rb degradation leads to the release of elongation 2 factors (E2F) and the activation of proteins which are associated with DNA synthesis and promotes DNA replication, cell division and transformation (5).
Eukaryotic translation initiation factor (eIF4E) is a rate-limiting molecule in the cap-dependent translation initiation (6). Human eIF4E gene is located at chromosome 4q21-q25 and encodes a 24KD protein. EIF4E appears as a cap-binding protein (CBP) that recognizes explicitly the mRNA cap and regulates the mRNA translation. Studies revealed that eIF4E works as a key node of the signal pathway of carcinogenesis and tumor development (7-9). EIF4E was overexpressed in various tumors, including breast cancer, head and neck squamous cell carcinoma, and bile duct cancer (10-12). eIF4E also promotes tumor occurrence, invasion and metastasis by strengthening the translational expression of oncogenes and growth factors such as cellular homolog of the retroviral v-myconcogene (c-Myc), vascular endothelial growth factor (VEGF) and matrix metalloproteinase-9 (MMP9) (10-12). However, the molecular mechanism for eIF4E overexpression remains poorly studied in cervical cancer.
Recent studies reported that eIF4E protein overexpresses in CC (13-15). However, little is known on eIF4E regulation. Van Tranppen et al (13) observed eIF4E over expression in CC tissues by reverse transcription-polymerase chain reaction (RT-PCR) and speculated that eIF4E might take a great part in tumor development. Lee et al (14) further showed through immunohistochemistry (IHC) that eIF4E expression was markedly enhanced following the progression of cervical malignant lesions. So far, studies on eIF4E in CC are still rare, leading to poor understanding of the role and regulation of eIF4E in CC. In our previous study, we found the overexpression of eIF4E was correlated to CC development (15). However, whether HPV E7 could induce eIF4E expression is not known yet, which is an important missing point to clarify the full role of HPV and eIF4E in CC. This paper aimed to explore the mechanism of eIF4E gene overexpression in CC. Here, we provided evidence that E7 can induce eIF4E gene transcription in CC.
Materials and Methods
Cell lines and transfection
Two CC cell lines, HPV positive HeLa and HPV negative C33A were utilized. The cells were cultured in Roswell Park Memorial Institute (RPMI) cell culture medium 1640, which contains 10% Fetal Bovine Serum (FBS), 2 mmol/l l-glutamine, 50 U penicillin and 50 microgram/ml streptomycin) at 37 °C in 5% CO2 in air.
Liposome method was employed for the plasmid transfection. Cells were divided into three groups: Mock group (untreated group, no plasmid was used for the transfection), NC group (negative control group, the plasmid containing a negative DNA fragment was used for the transfection) and treated group (the plasmid containing a tested DNA fragment was used for the transfection). 2.5 µl Lipofectamine™ 2000 (Invitrogen, Guangzhou, China) was used per microgram DNA (15).
eIF4E immunocytochemistry (ICC)
ICC was performed using mAbs specific for eIF4E (Santa Cruz, California, USA). The primary antibody eIF4E in 1:25 dilutions was added to the cell smears and incubated at 4 °C overnight. PBS instead of the eIF4E antibody was used as the negative control staining. Plasmids shE7 RNA interference plasmids were constructed by Genechem Company, Shanghai. Three pairs of shE7 sequences were: ShE7-1 (stem:GCATGGACCTAAGG-CAACA, Loop:AGTGAAGCCACAGATGTA, stem: TGTT- GCCTTAGGTCCATGC), ShE7-2 (stem: GGCAACA-TTGCAAGACATT, Loop:AGTGAAGCCACAGATGTA, stem: AATGTCTTGCAATGTTGCC), ShE7-3(stem:GC- AAGACATTGTATTGCAT, Loop:AGTGAAGCCACAGA-TGTA, Stem:ATGCAATACAATGTCTTGC). E7 and E7 mutant expression plasmids were constructed with pEGFP-C5 vector as described previously (16). E7 contains the wild type E7 of HPV16. E7 mutant contains mutations (TTCGGTTG to TACGTAGG) from nt 191 to nt 198 of E7. The sieIF4E was received as desalted and unprotected oligonucleotides. The sequences of sieIF4E were: 5’-GGAUAUUAU-AAAUAGAUUATT-3’ and 5’-UAAUCUAUUUAUAA-UAUCCTT-3’. Normal control (NC) sequences were: 5’-UUCUCCGAACGUGUCACGUTT-3’ and 5’- ACGUGACACGUUCGGAGAATT-3’.
Detection of E7 and eIF4E mRNA by RT-PCR and real time PCR
E7, eIF4E and GAPDH primer sequences were used for RT-PCR and real-time PCR: 5’-GCGTTAGAGCCCCAAAATGA-3’, 5’-CGTCGGGCTGG-TAAATGTTGA-3’ for E7. 5’-CTGCGGCTGATCTCCAA-G-3’, 5’-CTGCGGCTGATCTCCAAG-3’ for EIF4E. 5’-GAAGGTCGGAGTCAACGGATTT-3’, 5’-CCTGGAAGAT-GGTGATGGGATT-3’ for glyceraldehyde-3-phosph-ate dehydrogenase (GAPDH). RT-PCR was performed using the Access RT-PCR kit (Promega, USA) according to the manufacturer’s protocols. Data was analyzed by sequence detection software from Applied Biosystems. Real-time PCR was performed by Fast Start Universal SYBR Green Master (ROX) assay on the Gene Amp PCR System 9700 (ABI company, USA).
CCK-8 cell proliferation assay
Cell proliferation was determined with Cell counting Kit-8 (CCK-8) (Beyotime Institute of Biotechnology, Shanghai, China) Assay. Cell proliferation ability was represented by the mean absorbance value (AV). The proliferation rate and inhibition rate were calculated:
proliferation rate (%) = (AV treated group/AV mock -1)×100%. Inhibition rate (%)= (1-AV treated group/AV mock) ×100%.
Cell cycle assays and apoptosis assay
Cell cycle and apoptosis were evaluated by flow cytometry. 3×105 cells in 300 μl PBS were stained with 3 μl PI at 4 °C for 30 min in cell cycle assay, and with 3 μl Annexin V-FITC at room temperature for 10 min. The samples were analyzed with MultiCycle software using the flow cytometer BD FACS CantoTM (Becton Dickinson, CA, USA).
Transwell migration assay
Cell migration was determined in a transwell system. 6×104 cells were vaccinated into the upper surface of the transwell membrane and cultured at 37°C in 5% CO2 in air for 24 hr, 48 hr and 72 hr. The number of migrated cells was counted under a microscope (200X).
Statistical analysis
Data were analyzed using SPSS 17.0 software and were reported as means±standard deviation (SD). Independent samples t-test was used for analyzing the two groups and one-way analysis of variance (ANOVA) was used for examining multiple groups. The level of statistical significance was set at P <0.05 or P <0.01.
Results
HPV E7 induced eIF4E expression and promoted the proliferation and migration of HPV negative C33A cells
The E7 expression vector was transfected into C33A cells. For RT-PCR results, two bands with expected sizes for E7 (271bp) (Figure 1A), E7 mutant (271bp) and GAPDH (224bp) were seen. The eIF4E mRNA detection showed two bands for eIF4E (132bp) and GAPDH (224bp) (Figure 1B). By Image J analysis, the relative eIF4E mRNA levels (eIF4E/GAPDH) were 0.682 (mock), 0.613 (NC), 0.808 (E7m group) and 0.762 (E7 group). The eIF4E bands were stronger in E7 and E7m groups than the mock and NC groups. The changes of the eIF4E mRNA were consistent with the changes of E7 mRNA. The lighter changes of the eIF4E mRNA in E7m group suggested that the E7m had a part of function in inducing eIF4E transcription. The results of real-time PCR were similar to that of RT-PCR.
Figure 1.
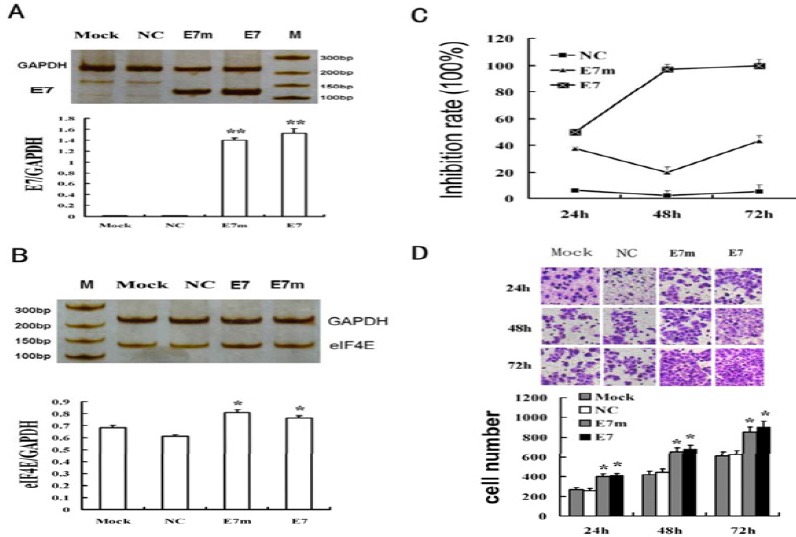
Human papillomavirus e7 gene induced eukaryotic translation initiation factor 4e expression and promoted the proliferation and migration of C33A cells. Cells were divided into 4 groups: untreated C33A cell group (MOCK), p-EGFP blank plasmid group (NC); E7m, E7 mutant group; E7, E7 expression vector group. (A) ecto-E7 gene expression of C33A cells at 20 hr, detected by RT-PCR. (B) eIF4E gene expression of C33A cells at 20 hr, detected by RT-PCR. (C) Transfection of E7 gene promoted the proliferation of C33A cells, detected by CCK-8 assay. (D) Transfection of E7 gene promoted the migration of C33A cells, detected by the transwell migration assay. *: vs Mock, P <0.05; **: vs Mock, P<0.01
The cell proliferation in the mock and NC groups was similar. Compared with the NC group, the proliferation rates increased by 44.3% (24 hr), 97.4% (48 hr) and 96.0% (72 hr) in E7 group, and 32.4% (24 hr), 19.9% (48 hr) and 40.5% (72 hr) in E7m group, respectively (Figure 1C). Here, E7m also influenced the cell proliferation, suggesting E7m maintained partial function of E7.
At any time point, the number of migrating cells in the mock and NC groups was similar. Compared with the NC group, the number of the migrating cells significantly increased by 412.01±21.523 (24 hr), 680.25±40.032 (48 hr) and 900.11±63.22(72 hr), respectively, in the E7 group, and significantly increased by 400.14±29.218 (24 hr), 651.23± 40.036(48 hr) and 850.99±51.001 (72 hr), respectively, in the E7m group (Figure 1D).
E7 mRNA expression was knocked down effectively by the shE7s in HeLa cells
The shE7 vectors carrying GFP were successfully constructed. The E7 mRNA level in the NC group was similar with that in the mock group, determined by both real-time PCR and RT-PCR test. By the real-time PCR detection, the E7 mRNA in shE7-1, 2, 3 treated groups were significantly lower than that of the NC group (Figure 2A). Among shE7-1, 2, 3, shE7-2 showed the most effective inhibition of E7 mRNA, with an inhibition rate of approximately 81%. By the RT-PCR and agarose gel detection, two distinct bands with expected sizes were seen for E7 gene (119bp) and GAPDH gene (224 bp) in all groups (Figure 2B). By Image J band analysis, the E7 mRNA in shE7-1 and shE7-2 but not shE7-3 groups was considerably knocked down, compared with the NC group. The E7 mRNA level in shE7-2 group was lower than that in shE7-1 group (Figure 2B). The changes of the E7 mRNA were comparable in both the real-time PCR and RT-PCR detection (Figure 2).
Figure 2.
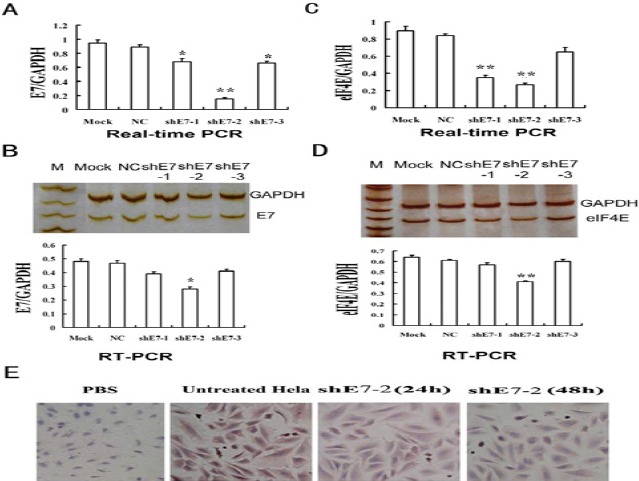
Knockdown of e7 in HeLa cells down regulated eukaryotic translation initiation factor 4e gene expression. (A) E7 mRNA expression was decreased by shE7s at 48 hr detected by real-time PCR. (B) Detection of E7 mRNA expression by RT-PCR; (C) eIF4E mRNA expression decreased after E7 knockdown at 48 hr detected by real-time PCR. (D) Detection of eIF4E mRNA expression by RT-PCR; (E) eIF4E protein expression decreased in HeLa cells at 24 hr and 48 hr after the transfection of shE7. Mock, HeLa cells; NC, blank vector group; *: vs Mock, P <0.05; **: vs Mock, P<0.01
E7 knockdown down regulated eIF4E expression in HPV+ HeLa cells
The eIF4E mRNA level in the NC group was analogous to that in the mock group, by both real-time PCR and RT-PCR detection. EIF4E mRNA levels in shE7-1, 2, 3 treated groups were significantly decreased, compared with the NC group. Among the treated groups, the eIF4E mRNA level in shE7-2 group was reduced most, with an inhibition rate of approximately 73% (Figure 2C). By RT-PCR and agarose gel detection, two discrete bands with expected sizes were seen for eIF4E gene (132 bp) and GAPDH gene (224 bp) (Figure 2D). By Image J band analysis, the eIF4E mRNA levels of shE7-1 and shE7-2 but not shE7-3 groups were substantially decreased, compared with the NC group. The eIF4E mRNA in shE7-2 group was decreased more than that in shE7-1 group. The changes of eIF4E mRNA were comparable in both the real-time PCR and RT-PCR detection. The changes of eIF4E mRNA followed tightly the changes of E7 mRNA in shE7-1, 2, 3 groups.
Using immunocytochemistry, the eIF4E protein expression was detected in HeLa cells (Figure 2E). The eIF4E positive cells were stained yellow brown in cytoplasm and/or nucleus. In the NC group, the rate of eIF4E positive cells was up to 96.8%. When shE7-2 transfection was done for 24 hr and 48 hr, the rates of eIF4E positive cells were decreased to 91.26% and 42.97% (P<0.001), respectively. The intensity of cell staining became considerably weaker in the shE7-2 treated group than in the NC group.
shE7-2 inhibited proliferation, migration and promoted cell apoptosis
After shE7-2 transfection, cell proliferation was significantly inhibited in shE7-2 group. The inhibition rates of cell proliferation were 9.9% (24 hr), 17.3% (48 hr) and 11.3% (72 hr) (Figure 3A).
Figure 3.
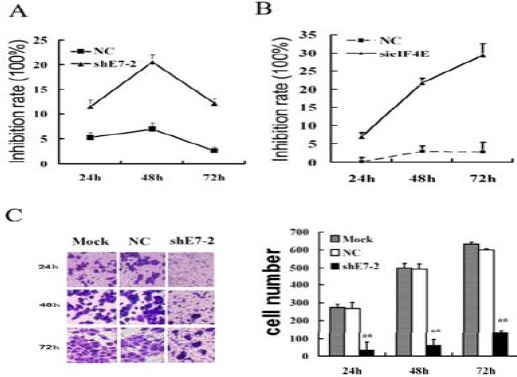
The interference plasmid of e7 or the siRNAs of eukaryotic translation initiation factor 4e transfection influenced HeLa cell biology. (A) shE7-2 inhibited the proliferation of HeLa cells detected by CCK-8 assay; (B) sieIF4E inhibited the proliferation of HeLa cells detected by CCK-8 assay; (C) shE7-2 inhibited the migration of HeLa cells detected by transwell assay. Mock: untreated HeLa cells. NC: blank vector group. shE7-2: shE7-2 group. *: vs Mock, P<0.05;**: vs Mock, P<0.01
Under the condition of high E7 expression, sieIF4E significantly inhibited the proliferation of the HeLa cells, to a degree comparable with the effect of shE7s. Compared with the NC group, the cell proliferation in sieIF4E group was markedly inhibited. The changes of the inhibition rates of cell proliferation in sieIF4E group were 7.635±1.143%(24 hr), 27.505±1.679% (48 hr) and 32.143±3.031% (72 hr), close to that in shE7 group at 24 hr, but significantly higher than that in shE7 group at 48 hr and 72 hr (Figure 3B).
Compared with the NC group, the cell cycle in shE7-2 group was noticeably changed (Table 1). The cell numbers were significantly increased by 14.4% (24 hr), 19.8% (48 hr) and 25.7% (72 hr) in G0/G1 phase, decreased by 15.1% (24 hr), 23.2% (48 hr) and 28.3% (72 hr) in S phase, and changed without significance in G2/M phase. The transfection effect suggested that shE7-2 inhibits the proliferation of HeLa cells through arresting cells at G0/G1 phase.
Table 1.
Cell cycle change of HeLa cells after the interference plasmid of e7 transfection
| Group | Percentage ( ±s, n=3) ±s, n=3) |
||
|---|---|---|---|
| G1% | S% | G2% | |
| HeLa | 32.124±1.425 | 50.263±1.651 | 14.598±1.254 |
| Nc | 36.987±1.251 | 49.541±1.625 | 14.028±1.669 |
| shE7-2 (24 hr) | 47.915±1.805 | 36.255±1.155 | 18.835±1.951 |
| shE7-2 (48 hr) | 53.354±1.552* | 28.101±1.202* | 17.547±1.752 |
| shE7-2 (72 hr) | 59.206±1.401* | 22.987±1.703* | 17.804±1.300 |
VS NC, P<0.01
Annexin V-FITC/PI double staining method was adopted for cell apoptosis detection after the shE7-2 transfection (Table 2). The change of apoptosis between the mock and NC groups was similar. Compared with the NC group, the V-FITC+/PI- cells (early apoptosis cells) in shE7-2 group were significantly increased by 30.1% (24 hr) and 39.2% (48 hr), respectively, while the V-FITC+/PI+ cells (late apoptosis cells) were increased without significance.
Table 2.
Apoptosis change of HeLa cells after the interference plasmid of e7 transfection
| Group | Percentage 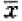 ±s, n=3) ±s, n=3) |
|
|---|---|---|
| V-fitc+/pi- | V-fitc+/pi+ | |
| Hela | 0.783±0.251 | 0.187±0.165 |
| Nc | 11.054±1.254 | 5.817±1.816 |
| shE7-2 (24 hr) | 41.164±3.755* | 10.104±1.020 |
| shE7-2 (48 hr) | 50.205±3.503* | 7.604±1.402 |
VS NC, P<0.01
To analyze the migration of HeLa cells after the shE7 transfection, Transwell Migration Assay was performed (Figure 3C). The cell numbers of migration were comparable between the mock and NC groups. Compared with the NC group, the cell numbers of migration in shE7-2 group were decreased by 33.47±11.563 (24 hr), 61.25±6.629 (48 hr), and 131.68±11.051 (72 hr), respectively (P<0.01) (Figure 3).
Discussion
This study discovered that E7 induced eIF4E expression in CC cells. Here, the ecto-E7 gene expression significantly induced eIF4E transcriptional gene expression in C33A cells (HPV-, eIF4E+). Furthermore, the decreased expression of eIF4E mRNA and protein directly followed the knockdown of E7 in HPV positive HeLa cells; the degree of eIF4E down regulation correspondingly matched the degree of E7 knockdown in the shE7-1, 2, 3 transfection groups of HeLa cells. To the best of our knowledge, this is the first study to suggest that E7 induces eIF4E transcription independently. This could be one of the mechanisms for eIF4E gene overexpression in CC.
However, whether E7 induces eIF4E trans-cription directly or indirectly is not yet known. Additional study will be done in our laboratory to confirm this issue. Evidence revealed that the conserved sequence of E7 could bind pRb, P107 and P130 (17-20). These Rb family members can form the pRb/E2F complex and inactivate the transcription and function of c-Myc, which negatively regulates the progression of G1/S and prohibits the cell cycle. In the process, E7 binds G1-specific pRb to obstruct the formation of the pRb/E2F complex and rescue the transcription and function of c-Myc, leading to the accelerated cell cycle. It was reported that the transcription of eIF4E is induced by c-Myc via a positive eIF4E/c-Myc feedback loop in lymphangiectasis (21). As a result, we concluded that E7 up regulates eIF4E through pRb/c-Myc pathway.
The results suggested that E7 enhanced the cell proliferation, migration and cell cycle progression, and also inhibited cell apoptosis through eIF4E. E7 regulates the transcription of a series of oncogenes by complicated molecular mechanisms (16, 22-24). Since eIF4E directly enhances the translation of many oncogenes whose gene transcription was regulated by E7, the E7/eIF4E pathway might be efficient for E7 to initiate and promote CC.
However, a few studies reported the effects of eIF4E on CC cell biology. Here, ecto-E7 gene expression was performed in C33A cells to investigate the function of E7 on eIF4E. We found that up-regulation of eIF4E gene expression by E7 accelerated the cell proliferation and inhibited the apoptosis (Figure 1). In HPV+ HeLa cells, shE7 down regulated eIF4E gene expression, inhibited the cell proliferation and speeded up the cell apoptosis (Figure 3). These results indicate that eIF4E engaged in the key process of HPV caused carcinogenesis.
In addition, HPV E6 and E7 are proven to produce a bicistronic transcript. Thus, the knockdown of E7 might also lead to the knockdown of E6 gene (25, 26). In this study, we knocked down E7 in HPV+ HeLa cells. The results showed down regulation of eIF4E gene expression and the cell proliferation. This result may be caused by single E7 or E6/E7 because of the presence of the E6/E7 bicistronic transcript. Thus, HPV- C33A cells were chosen to demonstrate the ability of E7 gene affecting eIF4E. After the E7 expression vector was transfected into C33A cells, the expression of E7 up-regulated eIF4E markedly in the absence of E6. The result showed clearly that E7 could induce eIF4E gene transcription. Thus, the down-regulation of eIF4E was caused, at least, mainly by the knockdown of E7. We concluded that E7 could induce eIF4E in HeLa and C33A cells.
Conclusion
EIF4E gene overexpression in CC cell lines was further confirmed in this study. More importantly, we discovered that the transcription of eIF4E could be induced by HPV E7. In addition, the down regulation or up regulation of eIF4E on the condition of sustained high expression of E7 significantly influenced the cell proliferation, cell cycle progression, migration and apoptosis. Our finding suggests that eIF4E is an important target for the treatment and prevention of HPV associated cancers such as CC.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No. 30670860, 81302244), the science and technology planning project of Guangdong Province (No. 2013B021800061, 2013B021800062), by the social technology development project of Dongguan, Guangdong Province, China (No. 2013108101051, 201310810-1060). by the Project of Department of Education of Guangdong Province (No. 2012KJCX0057), by the Science & Technology Innovation Fund of Guangdong Medical College (No. STIF201113), In addition, part of the results described in this paper was part of student thesis.
References
- 1.Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.Hyacinth HI, Adekeye OA, Ibeh JN, Osoba T. Cervical Cancer and Pap Smear Awareness and Utilization of Pap Smear Test among Federal Civil Servants in North Central Nigeria. PLoS One. 2012;7:e46583. doi: 10.1371/journal.pone.0046583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klingelhutz AJ, Roman A. Cellular transformation by human papillomaviruses, lessons learned by comparing high- and low-risk viruses. Virology. 2012;424:77–98. doi: 10.1016/j.virol.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30:F12–F23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 5.Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, et al. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin Suppressor. J Virol. 2007;81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonenberg N. EIF4E, the mRNA cap-binding protein, from basic discovery to translational research. Biochem Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 7.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (EIF4E) in cancer. Clin Cancer Res. 2010;16:4914–4920. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmer SG, DeBenedetti A, Graff JR. Translational control of malignancy, the mRNA cap-binding protein, EIF4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res. 2000;20:1343–1351. [PubMed] [Google Scholar]
- 10.Franklin S, Pho T, Abreo FW, Nassar R, De Benedetti A, Stucker FJ, et al. Detection of the proto-oncogene EIF4E in larynx and hypopharynx cancers. Arch Otolaryngol Head Neck Surg. 1999;125:177–182. doi: 10.1001/archotol.125.2.177. [DOI] [PubMed] [Google Scholar]
- 11.Nathan CA, Sander K, Abreo FW, Nassar R, Glass J. Correlation of p53 and the proto-oncogene EIF4E in larynx cancers, prognostic implications. Cancer Res. 2000;60:3599–3604. [PubMed] [Google Scholar]
- 12.Pavelic Z, Pavelic K, Carter C, Pavelic L. Heterogeneity of c-myc expression in histologically similar infiltrating ductal carcinoma of the breast. J Cancer Res Clin Oncol. 1992;118:16–22. doi: 10.1007/BF01192306. [DOI] [PubMed] [Google Scholar]
- 13.Van Trappen PO, Ryan A, Carroll M, Lecoeur C, Goff L, Gyselman VG, et al. A model for co-expression pattern analysis of genes implicated in angiogenesis and tumour cell invasion in cervical cancer. Br J Cancer 2002. 2002:537–544. doi: 10.1038/sj.bjc.6600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, Choi JJ, Lee KM, Choi CH, Kim TJ, Lee JH, et al. EIF4E expression is associated with histopathologic grades in cervical neoplasia. Hum Pathol 2005. 2005:1197–1203. doi: 10.1016/j.humpath.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Pang T, Gao M, Kang H, Ding W, Sun X, et al. HPV E6 induces eIF4E transcription to promote the proliferation and migration of cervical cancer. FEBS Lett. 2013;587:690–697. doi: 10.1016/j.febslet.2013.01.042. [DOI] [PubMed] [Google Scholar]
- 16.Li G, He L, Zhang E, Shi J, Zhang Q, Le AD, et al. Overexpression of human papillomavirus (HPV) type 16 oncoproteins promotes angiogenesis via enhancing HIF-1αand VEGF expression in non-small cell lung cancer cells. Cancer lett. 2011;311:160–170. doi: 10.1016/j.canlet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Shin MK, Sage J, Lambert PF. Inactivating All Three Rb Family Pocket Proteins Is Insufficient to Initiate Cervical Cancer. Cancer Res. 2012;72:5418–5427. doi: 10.1158/0008-5472.CAN-12-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buitrago-Pérez A, Hachimi MA. Humanized mouse model of HPV-associated pathology driven by E7 expression. PLoS One. 2012;7:e41743. doi: 10.1371/journal.pone.0041743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 20.Gonzalez SL, Stremlau M, He X, Basile JR, Munger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–5334. doi: 10.1158/0008-5472.CAN-07-5876. [DOI] [PubMed] [Google Scholar]
- 22.Sushma M, Vamsikrishna B, Babu M, Mohanraj R. A review on role of Human papilommavirus (HPV) in cervical cancer. Pharma Tutor. 2014;2:21–30. [Google Scholar]
- 23.Bin H, Ruifang W, Ruizhen L, Yiheng L, Zhihong L, Juan L, et al. Detention of HPV L1 capsid protein and hTERC gene in screening of cervical cancer. Iran J Basic Med Sci. 2013;16:797. [PMC free article] [PubMed] [Google Scholar]
- 24.Sadaoui NC, Armaiz-Pena GN, Nagaraja AS, Rupaimoole R, Previs RA, Dalton HJ, et al. Sustained adrenergic signaling promotes cervical cancer progression. Cancer Res. 2014;74:3511–3511. [Google Scholar]
- 25.Stacey SN, Jordan D, Williamson AJ, Brown M, Coote JH, Arrand JR. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J Virol. 2000;74:7284–7297. doi: 10.1128/jvi.74.16.7284-7297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stacey SN, Jordan D, Snijders PJ, Mackett M, Walboomers JM, Arrand JR. Translation of the human papillomavirus type 16 E7 oncoprotein from bicistronic mRNA is independent of splicing events within the E6 open reading frame. J Viral. 1995;69:7023–7031. doi: 10.1128/jvi.69.11.7023-7031.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


