Abstract
The role of nongastric H+-K+-ATPase (HKA) in ion homeostasis of macula densa (MD) cells is an open question. To begin to explore this issue, we developed two mathematical models that describe ion fluxes through a nongastric HKA. One model assumes a 1H+:1K+-per-ATP stoichiometry; the other assumes a 2H+:2K+-per-ATP stoichiometry. Both models include Na+ and NH4+ competitive binding with H+ and K+, respectively, a characteristic observed in vitro and in situ. Model rate constants were obtained by minimizing the distance between model and experimental outcomes. Both 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP models fit the experimental data well. Using both models, we simulated ion net fluxes as a function of cytosolic or luminal ion concentrations typical for the cortical thick ascending limb and MD region. We observed that 1) K+ and NH4+ flowed in the lumen-to-cytosol direction, 2) there was competitive behavior between luminal K+ and NH4+ and between cytosolic Na+ and H+, 3) ion fluxes were highly sensitive to changes in cytosolic Na+ or H+ concentrations, and 4) the transporter does mostly Na+/K+ exchange under physiological conditions. These results support the concept that nongastric HKA may contribute to Na+ and pH homeostasis in MD cells. Furthermore, in both models, H+ flux reversed at a luminal pH that was <5.6. Such reversal led to Na+/H+ exchange for a luminal pH of <2 and 4 in the 1:1-per-ATP and 2:2-per-ATP models, respectively. This suggests a novel role of nongastric HKA in cell Na+ homeostasis in the more acidic regions of the renal tubules.
Keywords: nongastric ATPase, H+-K+-ATPase, mathematical model, Na+ homeostasis, macula densa cell
both the gastric and nongastric or colonic isoforms of H+-K+-ATPase (HKA) are expressed in the kidney (15, 18, 45). Specifically, gastric HKA is mainly expressed in intercalated cells of both the collecting duct and connecting duct in rats (2, 8, 13). This isoform seems to be responsible for normal K+ reabsorption and H+ secretion that contribute to the maintenance of body K+ and acid-base balance.
On the other hand, nongastric HKA has been identified by different means in the rat and rabbit collecting duct, connecting duct, and cortical thick ascending limb (TAL) of Henle (3, 18, 27, 40) and in the distal convoluted tubule of rats (3, 27). In addition, nongastric HKA was immunologically identified in macula densa (MD) cells of rabbits by Verlander et al. (40). This isoform is highly expressed under K+ or Na+ deprivation (3, 27) or under changes in acid-base balance, such as chronic metabolic acidosis (8, 9, 18). Moreover, studies have supported the idea that nongastric HKA plays a role in cellular Na+ homeostasis by extruding Na+ instead of H+ (9, 10, 17, 29).
A study (34) in rabbits has shown that MD cells have low Na+-K+-ATPase activity in the basolateral membrane compared with adjacent TAL cells, which suggests that rabbit MD cells may have an additional mechanism of Na+ extrusion. The fact that these cells have an apical H+(Na+)-K+-ATPase suggests that it may be involved in Na+ homeostasis in rabbit MD cells (29). However, a study (23) in nongastric HKA knockout mice found that tubuloglomerular feedback responses (where MD cells have a NaCl-sensing role) are similar to those in wild-type mice. Moreover, this study also demonstrated that tubuloglomerular feedback responses in mice are critically dependent on MD cell basolateral Na+-K+-ATPase.
Motivated by these findings, and to begin an analysis of Na+ homeostasis in MD cells, we developed two mathematical models that describe ion fluxes through a nongastric HKA. The main difference between the two models is the stoichiometry: 1H+:1K+ per ATP versus 2H+:2K+ per ATP. In addition, the models include Na+ and NH4+ competitive binding with H+ and K+, respectively, a characteristic observed in vitro and in situ (9, 29). Model results showed that both 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP models fit the experimental data well (R2 values were 0.95 and 0.93, respectively). These nongastric HKA models will be essential elements in models of cells that express nongastric HKA, such as MD cells.
MATHEMATICAL MODEL AND METHODS
The H+(Na+)-K+(NH4+)-ATPase kinetic model (Fig. 1) is based on the kinetic diagrams of Weinstein (43) and Cain et al. (8). As in the diagram in Ref. 8, we omit ATP binding to the enzyme before the lumen-to-cytosol translocation. The assumptions made in Ref. 8 regarding ATP binding are based on the crystal structure and modus operandi of Ca2+-ATPase (38, 39) and its homology to H+-K+-ATPase, both of which are part of the P-type ATPase family.
Fig. 1.
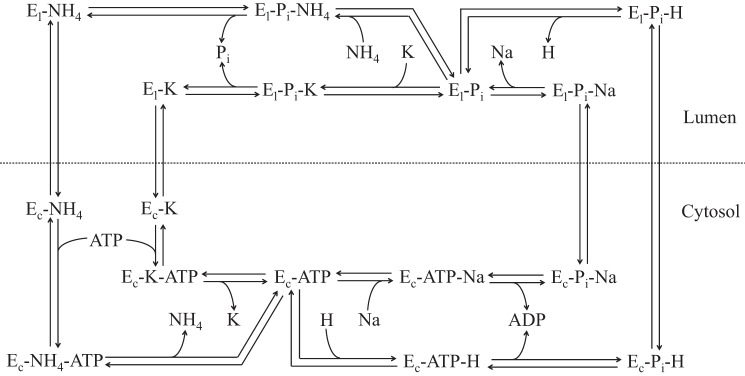
Kinetic model diagram of apical H+(Na+)-K+(NH4+)-ATPase with a 1:1-per-ATP stoichiometry. Ec and El are cytosolic- and luminal-facing enzymes, respectively.
Additional model assumptions are as follows: 1) Na+ and H+ compete for the same site; 2) NH4+ and K+ compete for the same site; 3) conservation of enzyme; 4) ATP, ADP, and Pi association/dissociation kinetics are similar to 3Na+-2K+-ATPase, as Brzezinski et al. (6) and Weinstein (43) have assumed; and 5) thermodynamic restrictions (i.e., the Principle of Detailed Balance) on the parameters are satisfied (see Thermodynamic restrictions on the rate constants in the appendix).
HKA stoichiometry.
There is some uncertainty about the stoichiometry of both gastric and nongastric HKAs. First, a study by Rabon et al. (31) supported a 2H+:2K+-per-ATP stoichiometry with noncooperative behavior between two identical ions for gastric HKA. They also emphasized the possibility that the findings of Renstra and Forte (33) regarding 1H+:1K+-per-ATP stoichiometry in gastric HKA may be due to HCl and KCl leakage as a result of faulty experimental methods. Second, experimental data on nongastric HKA by Grishin et al. (16) suggest noncooperative behavior between two identical ions. However, their work is inconclusive with respect to 2H+:2K+-per-ATP stoichiometry. Third, Burnay et al. (7), who demonstrated the electroneutrality of the ion transport by nongastric HKA, pointed out that their experimental results were insufficient to identify the precise stoichiometry of this isoform. Fourth, a study by Shin et al. (35) on gastric HKA supported the idea of dimerization of two catalytic subunits to activate the ATPase. Moreover, based on the experiments in Ref. 35, Cain et al. (8) presented a gastric HKA structural model and the mechanism of ion translocation (Figs. 2 and 3 in Ref. 8, respectively) based on the notion of a 1H+:1K+-per-ATP stoichiometry per catalytic α-subunit, which results in a 2H+:2K+ ratio per 2 ATP when the subunits are dimerized. Finally, a study by Abe et al. (1) suggested a pH dependence in the H+:K+-per-ATP stoichiometry of gastric HKA, which may be 1H+:1K+ per ATP at low pHs, such as those in the stomach lumen, or 2H+:2K+ per ATP at neutral pH. In this regard, based on electron cryomicroscopic experiments of the structure of gastric HKA at low pHs and on the amount of radioactive Rb+ bound to vesicles rich in this enzyme as a function of bath pH, Abe et al. (1) suggested that gastric HKA has two cation-binding sites: one with low pKa and one with high pKa, resulting in the activation of one site at low pHs and the activation of two sites at higher pHs. Nonetheless, their radioactive Rb+-binding experiments also suggested the dimerization of two gastric HKA α-subunits at near-to-neutral pH, as suggested by Shin and coworkers (35).
Fig. 2.
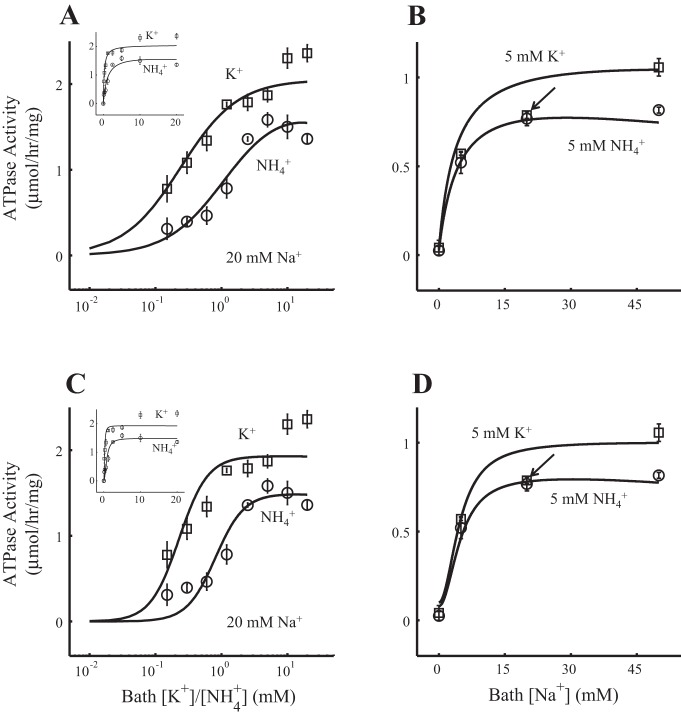
H+(Na+)-K+(NH4+)-ATPase model fit to the experimental data. Each flux is multiplied by the corresponding Jmaxexp shown in Table 4. A and B: the 1H+(1Na+):1K+(1NH4+)-per-ATP stoichiometry model. C and D: the 2H+(2Na+):2K+(2NH4+)-per-ATP stoichiometry model. A and C: ATPase activity as a function of bath K+ concentration ([K+]; squares, Experiment 1 in Table 1) or NH4+ concentration ([NH4+]; circles, Experiment 2 in Table 1), both with bath Na+ concentration ([Na+]) fixed to 20 mM. For a better appreciation of the fittings, the x-axis is presented in the log10 scale and does not include the data corresponding to [K+] and [NH4+] equal to zero. The fittings in the linear scale are shown in the top left corner and include the data corresponding to [K+] and [NH4+] equal to zero. B and D: ATPase activity as a function of bath [Na+] with [K+] fixed to 5 mM (squares, Experiment 3 in Table 1) or [NH4+] fixed to 5 mM (circles, Experiment 4 in Table 1). The arrow identifies the square that corresponds to [Na+] at 20 mM (see results). The R2 value of the model fit to the experimental data was 0.95 for the 1H+(1Na+)-1K+(1NH4+)-ATPase model and 0.93 for the 2H+(2Na+)-2K+(2NH4+)-ATPase model. The resulting H+-to-K+ net flux ratio under the conditions in H+-to-K+ net flux ratio in Table 1 was 0.0099 for the 1H+(1Na+)-1K+(1NH4+)-ATPase model and 0.011 for the 2H+(2Na+)-2K+(2NH4+)-ATPase model.
Fig. 3.
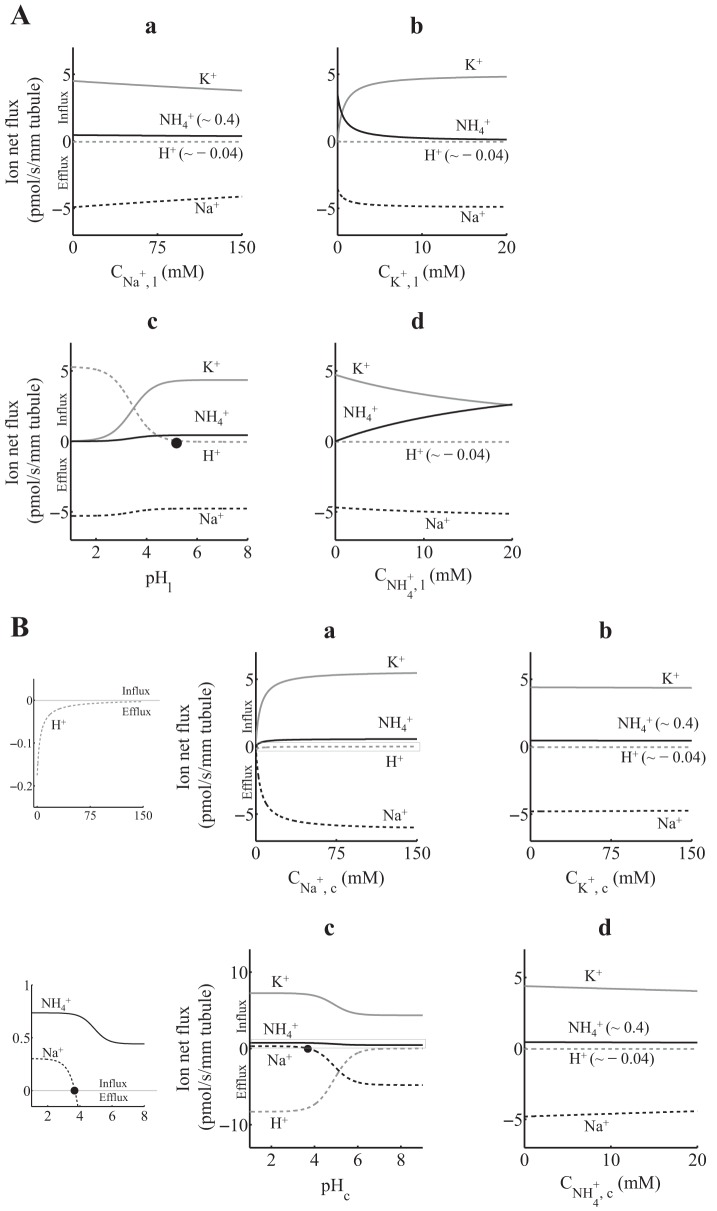
A: 1H+(1Na+)-1K+(1NH4+)-ATPase ion net flux as a function of luminal ion concentration, assuming that the total density of protein ET is 1 pmol/mm tubule. When not varied, lumen and cytosol [Na+], [K+], [NH4+], and pH were set as in Table 1 (Net flux simulation). Positive fluxes are into the cell, and negative fluxes are out of the cell. H+ flux reverses at a luminal pH (pHl) of ∼5.6 (solid circle). B: 1H+(1Na+)-1K+(1NH4+)-ATPase ion net flux as a function of cytosolic ion concentration, assuming that the total density of protein ET is 1 pmol/mm tubule. When not varied, lumen and cytosol [Na+], [K+], [NH4+], and pH were set as in Table 1 (Net flux simulation). Graphs in a and c were zoomed to appreciate small-scale ion net fluxes (left). Positive fluxes are into the cell, and negative fluxes are out of the cell. Na+ flux reverses at a cytosolic pH (pHc) of ∼3.7 (solid circle).
Brzezinski et al. (6) proposed a model of gastric HKA based on a 1H+:1K+-per-ATP stoichiometry. Their model fit the experimental data well in Ref. 42, which showed K+-dependent dephosphorylation rates in a gastric HKA extracted from the mucosa of a hog stomach. On the other hand, based on the gastric HKA model in Ref. 6, Weinstein (43) proposed two mathematical models of a renal ATPase expressed in the inner medullary collecting duct. One model assumed a 1H+:1K+-per-ATP stoichiometry, and the other assumed a 2H+:2K+-per-ATP stoichiometry with noncooperative behavior. Both models accounted for the difference between gastric (very acid) and renal pH ranges (less acid to neutral).
Based on all these findings and previous gastric HKA models, we developed mathematical models for both 1H+:1K+-per-ATP and 2H+:2K+-per-ATP stoichiometries of nongastric HKA with noncooperative behavior for two identical ions in the same compartment.
Ions transported by the ATPase.
Many studies have supported the idea of a Na+-secretory or Na+-reabsorptive property of HKA, making it part of the regulatory mechanisms that maintain body Na+ homeostasis. For example, Zhou et al. (47) observed Na+ reabsorptive activity in gastric HKA. On the other hand, nongastric HKA has also shown Na+ secretory activity, a behavior similar to 3Na+-2K+-ATPase, but in an electroneutral mode (9, 10, 12, 17, 32). In addition, although amino acid comparison and phylogenetic analysis have shown that the α-subunit of nongastric HKA is related to both gastric HKA and Na+-K+-ATPase α-subunits (19), it is even closer to the α-subunit of gastric HKA (11). Furthermore, pharmacological profiles have suggested that rat nongastric HKA shares more similarities with rat Na+-K+-ATPase (10). Moreover, the experiments in Ref. 9 showed that nongastric HKA could secrete Na+, instead of H+, in exchange for K+ (or NH4+). Finally, Swarts et al. (36) observed H+/NH4+ exchange behavior of nongastric HKA. Therefore, we constructed our model based on H+ (or Na+) secretion and K+ (or NH4+) reabsorption.
H+-to-K+ flux ratio when Na+ is present.
Grishin et al. (16) measured 86Rb+ influx and H+ efflux in human embryonic kidney (HEK)-293 cells transfected with a protein complex composed of human (h)ATP1AL1, which is 90% homologous to the nongastric HKA α-subunit (9), and the rabbit gastric β-subunit (rgβHKA). They observed that the H+ net flux was ∼10% of that of K+ when Na+ was present in the medium. In subsequent experiments, Grishin and Caplan (17) measured 86Rb+ influx and Na+ efflux in HEK-293 cells transfected with the hATP1AL1-rgβHKA complex and observed that the Na+ net flux was ∼50% of that of K+, which led us to think that the remaining 50% of the K+ net flux is concurrent to H+ exchange.
It is noteworthy that the combination of two subunits from different species in Refs. 16 and 17 might cause species-specific variations in the enzyme activity profiles. Nonetheless, results obtained by Crambert et al. (12) suggested that the 86Rb+ uptake activity of the hATP1AL1-rgβHKA complex is similar to that of the hATP1AL1-hNKAβ1 complex (hNKAβ1 is the human Na+-K+-ATPase β1-subunit). Moreover, Kraut et al. (21) showed that the nongastric HKA α-subunit is associated with the Na+-K+-ATPase β1-subunit in the rat kidney.
Motivated by the findings in the H+-to-K+ flux ratio in Refs. 16 and 17, we included a constraint in the optimization problem that finds model rate constants (see Optimization problem) such that the model fits the experimental data with parameters that maintain the H+-to-K+ flux ratio within experimental values. This is done by allowing the H+-to-K+ flux ratio to vary within a large range that encompasses the 0.1 and 0.5 net flux ratios reported in Refs. 16 and 17, respectively. In addition, to our knowledge, the NH4+-to-K+ flux ratio in nongastric HKA has not been estimated. Therefore, the optimization problem does not include a constraint for this ratio.
Mathematical model of H+(Na+)-K+(NH4+)-ATPase.
By applying the Principle of Mass Action to the kinetic model shown in Fig. 1, one gets an ordinary differential equation for each enzyme state.1 Each equation computes the net rate of formation of an enzyme population state that is expressed in terms of the ingoing and outgoing rates. We computed the enzyme states by solving the resultant model equations (a system of ordinary differential equations) in steady state. The steady-state equations consist of a system of linear equations of the enzyme-state densities that can be written as follows:
| (1) |
where the matrix A and the vector b, both with elements not all equal to zero, depend on the on and off binding rate constants, translocation rate constants, and solute concentrations, and the vector e contains enzyme-state densities [see Linear equations of the H+(Na+)-K+(NH4+)-ATPase model in the appendix].
Once the linear system in Eq. 1 is solved to obtain the enzyme density at the different states, the net fluxes of Na+, K+, H+, and NH4+ are computed (JXnet, where X is Na+, K+, H+, or NH4+) as follows:
| (2) |
| (3) |
| (4) |
| (5) |
where kXlc and kXcl are the translocation rate constants with X bound from the lumen to cytosol and the cytosol to lumen (where X is Na+, K+, H+, or NH4+), respectively, and eXl and eXc are the densities of the luminal- and cytosolic-facing enzyme, respectively, with X bound (where X is Pi-Na+, K+, Pi-H+, or NH4+). In Eqs. 2–5, the flux is taken positive from the lumen to cytosol direction.
Experimental data.
Codina et al. (9) measured radioactive Pi release in nongastric HKA-rich apical membranes of the rat distal colon submerged in a bath solution at different ion concentrations (Fig. 3, left and middle, in their article). Specifically, they obtained the Pi release by individually varying K+ concentration ([K+]; experiment 1), NH4+ concentration ([NH4+]; experiment 2), or Na+ concentration ([Na+]; experiment 3, [K+] fixed, and experiment 4, [NH4+] fixed) (see Table 1). To fit the model to the experimental data (see Optimization problem below), we defined the model ATPase activity (JXmodel, where X is K+ or NH4+), i.e., net Pi release, as follows:
| (6) |
Table 1.
Experimental settings used to solve the optimization problem in Eqs. 7–9 and to carry out the ion net flux simulations in Figs. 3–5
|
Experiment 1 (bath K+ concentration varies) |
Experiment 2 (bath NH4+ concentration varies) |
Experiment 3 (bath Na+ concentration varies; bath NH4+ concentration = 0) |
||||
|---|---|---|---|---|---|---|
| Solute | Lumen | Cytosol | Lumen | Cytosol | Lumen | Cytosol |
| Na+ | 20 | 20 | 20 | 20 | (0, 5, 20, 50) | |
| K+ | (0, 0.15, 0.3, 0.6, 1.2, 2.5, 5, 10, 20) | 0 | 0 | 5 | 5 | |
| pH | 7.2 | 7.2 | 7.2 | 7.2 | 7.2 | 7.2 |
| NH4+ | 0 | 0 | (0, 0.15, 0.3, 0.6, 1.2, 2.5, 5, 10, 20) | 0 | 0 | |
| ATP | 3 | 3 | 3 | |||
| ADP | 0.04* | 0.04* | 0.04* | |||
| Pi | 5* | 5* | 5* | |||
|
Experiment 4 (bath Na+ concentration varies; bath K+ concentration = 0) |
H+-to-K+ Net Flux Ratio |
Net Flux Simulations |
Fig. 2 in Weinstein's IMCD Model (43) |
|||||
|---|---|---|---|---|---|---|---|---|
| Solute | Lumen | Cytosol | Lumen | Cytosol | Lumen | Cytosol | Lumen | Cytosol |
| Na+ | (0, 5, 20, 50) | 147 | 15† | 25 | 15† | 0 | 0 | |
| K+ | 0 | 0 | 5 | 120† | 5 | 120† | 20 | 150 |
| pH | 7.2 | 7.2 | 7.4 | 7.2† | 7.0 | 7.2† | (0.3–8) | 7.4 |
| NH4+ | 5 | 5 | 0 | 0‡ | 2 | 2 | 0 | 0 |
| ATP | 3 | 2* | 2* | 2* | ||||
| ADP | 0.04* | 0.04* | 0.04* | 0.04* | ||||
| Pi | 5* | 5* | 5* | 5* | ||||
Ion concentrations are shown in mM, pH is dimensionless. If not otherwise specified, ion concentrations in experiments 1–4 are in accordance with the experimental setting in Ref. 9, where apical membranes extracted from the rat distal colon were bathed in different solutions, independently, to perform the ATPase activity assays. For H+-to-K+ net flux ratio and Figure 2 in Weinstein's IMCD model (43) (where IMCD is inner medullary collecting duct), ion concentrations are in accordance with the experimental settings in Refs. 16 and 43, respectively.
where and are the Pi off and on binding rate constants in the cytosol, respectively; is the cytosolic Pi concentration; is the density of the luminal-facing enzyme with both Pi and X bound (where X is K+ or NH4+); and eXl is the density of the luminal-facing enzyme with X bound (where X is K+ or NH4+). Both and eXl are obtained by solving the HKA mathematical model in Eq. 1, which is a function of the varying bath concentrations.
Optimization problem.
To estimate the model parameters, we formulated the optimization problem in Eqs. 7–9. Let J1,jexp and J2,jexp be the jth values of the experimental fluxes in a NH4+-free medium (squares in Fig. 2) and a K+-free medium (circles in Fig. 2), respectively. To compare these values with the model outcomes, we define Ĵ1,jmodel and Ĵ2,jmodel as the jth values of the model fluxes in a NH4+-free medium and a K+-free medium, respectively.
In addition, the model fit to the experimental data is conditioned to a H+-to-K+ net flux ratio range that includes the values calculated in Refs. 16 and 17. To do that, we allowed the absolute value of the H+-to-K+ net flux ratio, JHnet/JKnet, to vary between 0.001 and 1 (or within the interval [10−6, 1] when squared).2 We should mention that we fit the model to the data reported by Codina et al. (9), and, in that study, the authors obtained ATPase activity of nongastric HKA expressed in apical membranes of the rat distal colon, which were submerged in different bath [K+], [NH4+], or [Na+]. On the other hand, Grishin et al. in Refs. 16 and 17, respectively, obtained K+-to-H+ and K+-to-Na+ net flux ratios of nongastric HKA expressed in HEK-293 cells. Thus, to compute JHnet/JKnet, we used the luminal ion concentrations in Ref. 16, which are the same values used in Ref. 17, and cytosolic concentrations of a typical epithelial cell (5) (see H+-to-K+ net flux ratio in Table 1).
Now, the optimization problem can be written as follows:
| (7) |
| (8) |
| (9) |
where k is the vector of unknown rate constants; kL and kU are lower and upper bounds of the parameters in k, respectively; the inequality is taken element by element; and the bounds are prescribed (Table 2). Ĵi,jmodel is computed using Eq. 6 at the corresponding concentrations of the experimental fluxes (i = 1 corresponds to X = K+ and i = 2 corresponds to X = NH4+). Ji,jexp and Ĵi,jmodel are normalized as described below in Normalization methods in the appendix. JHnet/JKnet is obtained using Eqs. 3 and 4 after solving the HKA mathematical model in Eq. 1 with the solute concentrations in H+-to-K+ net flux ratio in Table 1.
Table 2.
Model and literature parameters for the 1:1 and 2:2 H+(Na+)-K+(NH4+)-ATPase models
| Optima† |
Weinstein (43) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | Units | Description | Range* (kL, kU) | 1:1 | 2:2 | Brzezinski et al. (6) 1:1 | 1:1 | 2:2 |
| kNa, coff | s −1 | Na+ off, cytosol | [105, 1010] | 7.01 × 109 | 7.60 × 109 | |||
| kNa, con | mM−1•s −1 | Na+ on, cytosol | [105, 1010] | 8.90 × 108 | 1.23 × 109 | |||
| kADP, coff | s−1 | ADP off, cytosol | 5.00 × 101 | 5.00 × 101 | 5.00 × 101 | 5.00 × 101 | 5.00 × 101 | |
| kADP, con | mM−1•s −1 | ADP on, cytosol | 2.50 × 103 | 2.50 × 103 | 2.50 × 103 | 2.50 × 103 | 2.50 × 103 | |
| kNacl | s−1 | Na+ translocation, cytosol → lumen | [10−2, 103] | 3.34 × 101 | 2.17 × 101 | |||
| kNalc | s−1 | Na+ translocation, lumen → cytosol | ‡ | 3.30 × 102 | 1.00 × 103 | |||
| kNa, loff | s−1 | Na+ off, lumen | [105, 1010] | 9.22 × 109 | 9.54 × 109 | |||
| kNa, lon | mM−1•s−1 | Na+ on, lumen | [105, 1010] | 1.63 × 107 | 6.65 × 107 | |||
| kK, loff | s−1 | K+ off, lumen | [105, 1010] | 4.40 × 109 | 1.03 × 109 | 3.40 × 107 | 3.40 × 107 | 1.50 × 108 |
| kK, lon | mM−1•s−1 | K+ on, lumen | [105, 1010] | 4.30 × 109 | 3.73 × 109 | 2.00 × 107 | 2.00 × 107 | 2.60 × 107 |
| s−1 | Pi off, cytosol | 5.40 × 101 | 5.40 × 101 | 5.40 × 101 | 5.40 × 101 | 5.40 × 101 | ||
| mM−1•s−1 | Pi on, cytosol | 3.20 × 10−2 | 3.20 × 10−2 | 3.20 × 10−2 | 3.20 × 10−2 | 3.20 × 10−2 | ||
| kKlc | s−1 | K+ translocation, lumen → cytosol | [10−2, 103] | 3.66 × 101 | 8.44 × 101 | 1.75 | 1.75 | 1.75 |
| kKcl | s−1 | K+ translocation, cytosol → lumen | [10−2, 103] | 5.53 | 7.91 × 102 | 3.50 × 101 | 3.50 × 101 | 3.50 × 101 |
| kATP, con | mM−1•s−1 | ATP on, cytosol | 1.30 × 104 | 1.30 × 104 | 1.30 × 104 | 1.30 × 104 | 1.30 × 104 | |
| kATP, coff | s−1 | ATP off, cytosol | 6.50 | 6.50 | 6.50 | 6.50 | 6.50 | |
| kK, coff | s−1 | K+ off, cytosol | [101, 108] | 7.97 × 107 | 7.43 × 107 | 4.00 × 103 | 4.00 × 103 | 8.90 × 103 |
| kK, con | mM−1•s−1 | K+ on, cytosol | [101, 108] | 2.52 × 104 | 7.84 × 105 | 4.00 × 101 | 4.00 × 101 | 7.30 × 101 |
| kH, coff | s−1 | H+ off, cytosol | [101, 108] | 1.06 × 107 | 4.03 × 106 | 5.00 × 102 | 5.00 × 102 | 6.60 × 102 |
| kH, con | mM−1•s−1 | H+ on, cytosol | [105, 1010] | 1.84 × 109 | 7.22 × 109 | 1.30 × 106 | 1.30 × 106 | 5.30 × 106 |
| kHcl | s−1 | H+ translocation, cytosol → lumen | [10−2, 103] | 5.47 × 101 | 5.52 × 102 | 4.00 × 101 | 4.00 × 101 | 4.00 × 101 |
| kHlc | s−1 | H+ translocation, lumen → cytosol | ‡ | 6.82 × 101 | 1.74 × 10−1 | 2.00 × 102 | 2.00 × 102 | 2.00 × 102 |
| kH, loff | s−1 | H+ off, lumen | [105, 1010] | 3.21 × 108 | 2.54 × 105 | 5.00 × 107 | 5.00 × 107 | 5.00 × 107 |
| kH, lon | mM−1•s−1 | H+ on, lumen | [105, 1010] | 6.16 × 109 | 7.49 × 109 | 5.00 × 105 | 5.00 × 107 | 8.00 × 109 |
| s−1 | NH4+ off, lumen | [105, 1010] | 5.83 × 109 | 8.74 × 109 | ||||
| mM−1•s−1 | NH4+ on, lumen | [105, 1010] | 1.44 × 109 | 9.11 × 109 | ||||
| s−1 | NH4+ translocation, lumen → cytosol | [10−2, 103] | 8.38 × 102 | 3.72 × 102 | ||||
| s−1 | NH4+ translocation, cytosol → lumen | ‡ | 4.94 × 10−1 | 9.64 × 101 | ||||
| s−1 | NH4+ off, cytosol | [101, 108] | 8.99 × 107 | 7.75 × 107 | ||||
| mM−1•s−1 | NH4+ on, cytosol | [101, 108] | 1.85 × 106 | 1.41 × 106 | ||||
| Keq | mM | Cycle equilibrium constant | 1.00 × 1010 | 1.00 × 1010 | 1.03 × 1010 | 1.03 × 108 | 7.59 × 108 | |
c is the cytosol, l is the lumen, cl is cytosol-to-lumen movement, and lc is lumen-to-cytosol movement.
Numeric methods.
We combined two numerical methods to solve the problem in Eqs. 7–9: the first method, simulated annealing (SA) (28), is a technique that makes a rigorous exploration of a large parameter space and finds satisfactory, rather than optimal, solutions analogously to a physical annealing process (24); the outcome of the first method is then used to start the iteration of a descent method, which is one that seeks a minimum in the direction of the derivative of the function in Eq. 7 under the constraints in Eqs. 8 and 9 (fmincon function in MATLAB). In both methods, for every candidate k, the HKA model in Eq. 1 is, first, solved as a function of the ion concentrations in Experiments 1–4 in Table 1 to obtain model ATPase activities, Ĵi,jmodel, using Eq. 6, and, second, solved using the ion concentrations in H+-to-K+ net flux ratio in Table 1 to obtain the net fluxes, JHnet and JKnet, using Eqs. 3 and 4, respectively.
We implemented and solved the ATPase mathematical model in Eq. 1 and the optimization problem in Eqs. 7–9 in MATLAB using a Mac box with a 2.4-GHz Intel Core 2 Duo processor and 4-GB 1,067-MHz DDR3 memory. In addition, we executed the SA-descent algorithm 100 times, independently, for both the 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP models and selected the best minimum for each model.
RESULTS
Brzezinski et al. (6) obtained model parameters for a gastric 1H+:1K+-per-ATP HKA by fitting model outcomes to experimental data in Ref. 42 that showed enzyme activity as a function of bath [K+]. Weinstein (43) obtained model parameters for a gastric HKA that is expressed in inner medullary collecting duct cells. He specifically modified the 1H+:1K+-per-ATP model of Brzezinski et al. (6) to one with a 1H+:1K+-per-ATP stoichiometry and another with a 2H+:2K+-per-ATP stoichiometry, both based on the differences between gastric and renal lumen pHs. Both Brzezinski et al. and Weinstein's sets of parameters are included in Table 2. Because we are interested in the segment that contains the cortical TAL (cTAL) and MD cells, and nongastric HKA is expressed in that segment in rabbits (40), we obtained model parameters by fitting the mathematical models with either 1H+(1Na+):1K+(1NH4+)-per-ATP or 2H+(2Na+):2K+(2NH4+)-per-ATP stoichiometry to experimental data obtained from nongastric HKA.
Model calibration: 1H+(1Na+):1K+(1NH4+)-per-ATP or 2H+(2Na+):2K+(2NH4+)-per-ATP model fits to the experimental data.
Table 1 shows the solute concentrations for 1) the model fitting to the experimental data in Ref. 9 (Experiments 1–4), 2) computing of the H+ and K+ net fluxes to evaluate the constraint in Eq. 8 (H+-to-K+ net flux ratio; see H+-to-K+ flux ratio when Na+ is present in mathematical model and methods), 3) simulating the net fluxes as a function of ion concentration (Net flux simulations), and 4) emulating Fig. 2 in Ref. 43. Luminal [Na+] values in the net flux simulations correspond to typical values in the cTAL and MD region of the nephron (4).
Figure 2 shows 1:1-per-ATP (A and B) and 2:2-per-ATP (C and D) model fits to the experimental data. The R2 value was 0.95 for the 1:1-per-ATP model and 0.93 for the 2:2-per-ATP model. The H+-to-K+ net flux ratio was ∼0.01 (i.e., the H+ net flux was ∼1% of the K+ net flux) for both models under the conditions shown in H+-to-K+ net flux ratio in Table 1. Table 2 includes the model parameter ranges used to solve the optimization problem in Eqs. 7–9, the 1:1-per-ATP and 2:2-per-ATP model optimal parameter sets, which were obtained by solving the optimization problem in Eqs. 7–9 for each model, and the parameter values reported in Refs. 6 and 43 in their respective HKA models. Table 3 includes the half-maximal concentrations (C50) for the 1:1-per-ATP and 2:2-per-ATP model data sets shown in Fig. 2. To obtain these C50 values, each model data set shown in Fig. 2 was fitted using Eq. 38 as described in Normalization methods in the appendix.
Table 3.
C50 values for the 1:1 and 2:2 H+(Na+)-K+(NH4+)-ATPase model data in Fig. 2
| 1:1-per-ATP model |
2:2-per-ATP model |
|||
|---|---|---|---|---|
| C50, mM | R2 | C50, mM | R2 | |
| Experiment 1 (K+ concentration varies) | 0.240 ± 0.000 | 1.00 | 0.228 ± 0.000 | 1.00 |
| Experiment 2 (NH4+ concentration varies) | 0.915 ± 0.003 | 0.996 | 0.807 ± 0.005 | 1.00 |
| Experiment 3 (Na+ concentration varies; NH4+-free bath) | 3.25 ± 0.00 | 0.999 | 4.25 ± 0.01 | 0.993 |
| Experiment 4 (Na+ concentration varies, K+-free bath) | 2.66 ± 0.01 | 0.974 | 4.12 ± 0.01 | 0.991 |
C50, half-maximal ion concentration. Each data set (i.e., lines that fit circles and squares in Fig. 2, A and B for the 1:1 model and C and D for 2:2 model) was fitted using Eq. 38 [Hill coefficient (n) = 1 for the 1:1 model and n = 2 for the 2:2 model] to obtain C50 (see Normalization methods in the appendix).
As shown in Table 2, there was a large difference between the cytosolic on binding rate constants of NH4+ and K+ for the 1:1-per-ATP model. The binding of NH4+ in the cytosol (kNH4,onc = 1.85 × 106 mM−1·s−1) is ∼73 times faster than the binding of K+ (kK,con = 2.52 × 104 mM−1·s−1). It is difficult to understand the effect of such a large difference in a transport protein that normally carries K+ and NH4+ into the cell. Nonetheless, to our knowledge, there are no experimental measurements of the on and off binding rate constants of these two ions in nongastric HKA.
The model outcomes shown in Fig. 2, B and D, gave a poor approximation for the 5 mM K+ plot datum identified with the arrow (square where bath [Na+] is 20 mM). In both Fig. 2, B and D, notice that this datum is an outlier for both hyperbolic (B) and sigmoidal (D) curves. Even when we extended the range of the parameters in both ATPase models, we obtained similar fits to the ones in Fig. 2, B and D. One possible explanation for why the fit curve is well above this off value is that all experimental data sets were fitted simultaneously and not individually. Regardless of that, the corresponding C50 range of values for the experimental data (5.45 ± 2.24 for n = 1 and 4.26 ± 1.27 mM for n = 2; Table 4) encompassed the models' C50 range of values (3.25 ± 0.00 and 4.25 ± 0.01 mM for the 1:1-per-ATP and 2:2-per-ATP model, respectively; Table 3). Other authors have observed a similar insensitivity of parameter estimates to a single data outlier (25, 26, 44).
Table 4.
Hill parameters for the experimental data
| Jmaxexp, μmol·mg−1·h−1 |
C50, mM |
R2 |
|||||
|---|---|---|---|---|---|---|---|
| n = 1 | n = 2 | n = 1 | n = 2 | Codina et al. (9)* | n = 1 | n = 2 | |
| Experiment 1 (K+ concentration varies) | 2.23 ± 0.09 | 2.01 ± 0.12 | 0.346 ± 0.066 | 0.275 ± 0.059 | 0.2 | 0.968 | 0.894 |
| Experiment 2 (NH4+ concentration varies) | 1.64 ± 0.12 | 1.49 ± 0.10 | 0.974 ± 0.282 | 0.899 ± 0.171 | 2 | 0.942 | 0.930 |
| Experiment 3 (Na+ concentration varies; NH4+-free bath) | 1.11 ± 0.11 | 0.95 ± 0.09 | 5.45 ± 2.24 | 4.26 ± 1.27 | 5 | 0.976 | 0.947 |
| Experiment 4 (Na+ concentration varies, K+-free bath) | 0.88 ± 0.02 | 0.81 ± 0.02 | 3.38 ± 0.48 | 3.73 ± 0.25 | 5 | 0.998 | 0.998 |
See Eq. 38.
C50 values reported by Codina et al. (9); the corresponding Hill coefficient is not reported in Ref. 9.
Model simulations: ion net flux using the 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP models.
Using the 1:1-per-ATP and 2:2-per-ATP models, we simulated ion net fluxes as functions of luminal (Figs. 3A and 4A for the 1:1-per-ATP and 2:2-per-ATP models, respectively) and cytosolic (Figs. 3B and 4B for the 1:1-per-ATP and 2:2-per-ATP models, respectively) ion concentrations. When not varied, luminal ion concentrations were set to typical values in the cTAL region of the nephron (Net flux simulations in Table 1). For all ion variations in the two compartments (lumen and cytosol) and for both models, K+ and NH4+ always flowed in the lumen-to-cytosol direction (i.e., these ions were reabsorbed by HKA). In addition, Na+ and H+ always flowed in the cytosol-to-lumen direction (i.e., these ions were secreted by HKA), except for luminal pH values <5.6 (both models), where H+ is reabsorbed (Figs. 3A,c and 4A,c), and cytosolic pH values <3.7 for the 1:1-per-ATP model and <4.8 for the 2:2-per-ATP model, where Na+ is reabsorbed driven by the high H+ concentration ([H+]) gradient (Figs. 3B,c and 4B,c). Furthermore, the ion net fluxes using both models were minimally affected by changes in the luminal [Na+] (Figs. 3A,a and 4A,a) or by changes in cytosolic [K+] or [NH4+] (Figs. 3B,b and d, and 4B,b and d). In addition to the H+ flux reversal at luminal pH smaller than 5.6, Figs. 3A,c and 4A,c show that, as the luminal pH decreases (luminal pH < 2 for the 1:1-per-ATP model and luminal pH < 4 for the 2:2-per-ATP model), the ATP hydrolysis cycles are turned off (i.e., both K+ and NH4+ fluxes go to zero) and the Na+ efflux matches the H+ influx, which exhibits a cytosolic Na+/lumen H+ exchange favored by the high [H+] gradient. Nonetheless, this behavior should not be expected under normal physiological conditions in the cTAL and MD region of the nephron where pH is >6.
Fig. 4.
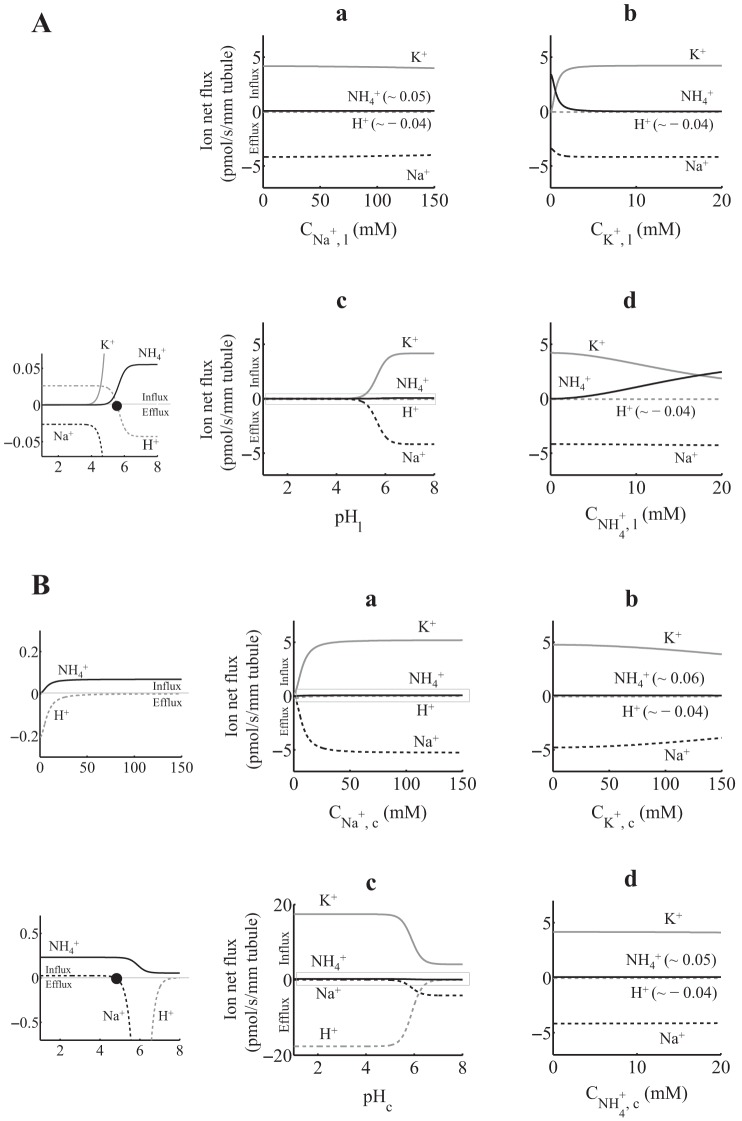
A: 2H+(2Na+)-2K+(2NH4+)-ATPase ion net flux as a function of luminal ion concentration, assuming that the total density of protein ET is 1 pmol/mm tubule. When not varied, lumen and cytosol [Na+], [K+], [NH4+], and pH were set as in Table 1 (Net flux simulation). The graph shown in c was zoomed to appreciate small-scale ion net fluxes (left). Positive fluxes are into the cell, and negative fluxes are out of the cell. H+ flux reverses at pHl of ∼5.6 (solid circle). B: 2H+(2Na+)-2K+(2NH4+)-ATPase ion net flux as a function of cytosolic ion concentration, assuming that the total density of protein ET is 1 pmol/mm tubule. When not varied, lumen and cytosol [Na+], [K+], [NH4+], and pH were set as in Table 1 (Net flux simulation). Graphs in a and c were zoomed to appreciate small-scale ion net fluxes (left). Positive fluxes are into the cell, and negative fluxes are out of the cell. Na+ flux reverses at a pHc of ∼4.8 (solid circle).
Moreover, and as expected, we observed a competitive behavior between luminal K+ and NH4+ (Figs. 3A,b and d, and 4A,b and d) and between cytosolic Na+ and H+ (Figs. 3B,a and c, and 4B,a and c). As shown in Figs. 3B,c and 4B,c, the increase in cytosolic pH (cytosolic pH > 5 for the 1:1-per-ATP model and cytosolic pH > 6 for the 2:2-per-ATP model) led to higher Na+ efflux. In addition, changes in luminal K+ or NH4+ did not have a significant effect on Na+ and H+ effluxes (Figs. 3A,b and d, and 4A,b and d). On the other hand, ion net fluxes were sensitive to changes in cytosolic Na+ or H+ (Figs. 3B,a and c, and 4B,a and c). Therefore, this high sensitivity of ion net fluxes to changes in cytosolic [Na+] and pH suggest a role of nongastric HKA in both the Na+ and pH homeostasis in cells where it is expressed, such as the TAL and MD cells (29).
Differences between the 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP models.
The most notable differences between the 1:1-per-ATP and 2:2-per-ATP models can be observed in the NH4+ net flux profiles shown in Figs. 3,a–c, and 4,a–c, where the NH4+ flux was ∼10-fold larger in the 1:1-per-ATP model. Furthermore, there were notable differences in ion net flux profiles when the luminal or cytosolic pH varied. In this regard, when luminal pH was <2 for the 1:1-per-ATP model and <4 for the 2:2-per-ATP model (Figs. 3A,c and 4A,c, respectively), Na+ effluxes (∼5 pmol·s−1·mm−1 for the 1:1-per-ATP model and ∼0.025 pmol·s−1·mm−1 for the 2:2-per-ATP model) were mainly due to Na+/H+ exchange. However, the Na+ efflux magnitude was ∼200-fold higher in the 1:1-per-ATP model. Moreover, in the 1:1-per-ATP model (Fig. 3A,c), the Na+ efflux that resulted from Na+/H+ exchange (luminal pH < 2) slightly increased compared with the Na+ efflux that resulted from the ATP hydrolysis (i.e., Na+/K+ plus Na+/NH4+ exchange, luminal pH > 2). On the other hand, in the 2:2-per-ATP model (Fig. 4A,c), the Na+ efflux that resulted from Na+/H+ exchange (luminal pH < 4) decreased compared with the Na+ efflux that resulted from ATP hydrolysis (luminal pH > 4). With respect to the ion net flux profiles under changes in cytosolic pH (Figs. 3B and 4B), and in addition to the cytosolic pH value where Na+ flux reversal occurs (∼3.7 for the 1:1-per-ATP model and ∼4.8 for the 2:2-per-ATP model), the K+/H+ exchange was ∼2-fold higher in the 2:2-per-ATP model [i.e., as cytosolic pH decreased, K+ net influx (or H+ net efflux) went to ∼17 pmol·s−1·mm−1 in the 2:2-per-ATP model and to ∼7 in the 1:1-per-ATP model].
The differences between the 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP models may be due to 1) the difference in ion stoichiometries and 2) the different rate constant sets that resulted after solving, independently for each model, the optimization problem in Eqs. 7–9 (e.g., the large difference between the cytosolic on binding rate constants of NH4+ and K+ for the 1:1-per-ATP model; see Model calibration). Despite the differences between both models, the ion net flux profiles of both models were practically the same under normal cytosolic or luminal pH (i.e., pH > 6).
ATPase hydrolysis by nongastric HKA in Na+- and NH4+-free medium.
Figures 3A,c and 4A,c include the ion net fluxes as a function of luminal pH in the presence of both Na+ and NH4+. Weinstein (43) presented a similar simulation in Fig. 2 but with the following differences: [Na+] and [NH4+] were equal to zero in both the lumen and cytosol, cytosolic and luminal [K+] were equal to 150 and 20 mM, respectively, and cytosolic pH was equal to 7.4 [see Fig. 2 in Weinstein's IMCD model (43) in Table 1]. For comparison, we carried out the simulations shown in Fig. 2 in Ref. 43. Specifically, we solved the ATPase mathematical model in Eq. 1 and computed the ATP hydrolysis (i.e., K+ net flux) using Eq. 6 and varying the luminal pH from 0.3 to 8.0 (Fig. 5). The curves (in continuous line) correspond to different model rate constants: gastric 1:1-per-ATP model rate constants of Brzezinski et al. (6) and Weinstein's renal 1:1-per-ATP and 2:2-per-ATP model rate constants (43) (Table 2). The three resulting ATP hydrolysis profiles (continuous lines in Fig. 5) were similar to those shown in Fig. 2 in Ref. 43. We also show the simulations (dashed lines in Fig. 5) using our nongastric 1:1-per-ATP and 2:2-per-ATP model rate constants (Optima in Table 2). Note that the outcomes (dashed lines in Fig. 5) were within Weinstein's renal 1:1-per-ATP and 2:2-per-ATP models.
Fig. 5.
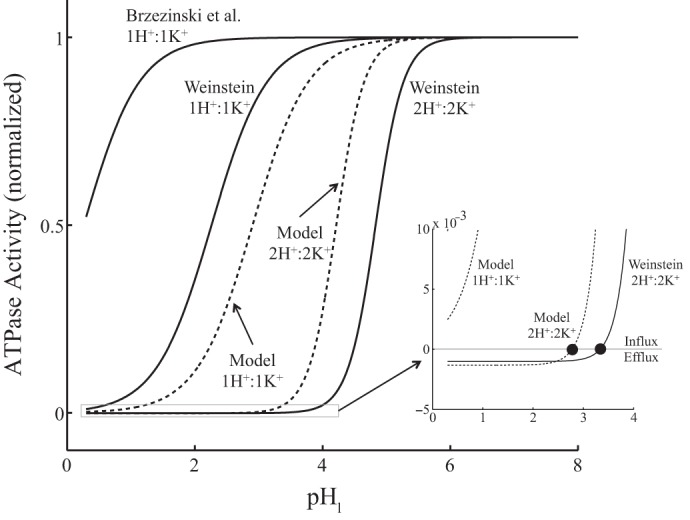
ATPase hydrolysis (i.e., K+ net flux) as a function of luminal pH in Na+- and NH4+-free medium. We simulated Fig. 2 in Ref. 43 using our ATPase mathematical model (Eq. 1) with Weinstein's (43) and Brzezinski et al. (6) model parameter sets in Table 2 (continuous lines). We also show simulations with our nongastric HKA model parameter sets in Table 2 (dashed lines). ATP hydrolysis was computed using Eq. 6. Plots were zoomed to appreciate small-scale ATPase activities. K+ flux reverses at a pHl of ∼2.77 for our 2H+:2K+-per-ATP model and at a pHl of ∼3.33 for Weinstein's 2H+:2K+-per-ATP model (solid circles).
When [Na+] = [NH4+] = 0 in both the lumen and cytosol, the HKA kinetic system results in only one reaction cycle: the H+/K+ exchange cycle. Therefore, we can determine the luminal pH where equilibrium is achieved (i.e., the luminal pH where net ATP hydrolysis is zero) using Eq. 33 in the appendix, the ion concentrations in Table 1 [Fig. 2 in Weinstein's IMCD model (43)], and the equilibrium concentration constant (Keq) for the different HKA models (Table 2). Only two models achieve equilibrium at pHs within the range shown in Fig. 5; these were the Weinstein's 2H+:2K+-per-ATP model at luminal pH ∼ 3.33 and our 2H+:2K+-per-ATP model at luminal pH ∼ 2.77.3
We should emphasize that Brzezinski et al. (6) obtained model parameters using experimental data from hog gastric HKA. In addition, Weinstein, whose models are based on the one from Brzezinski et al., modified the luminal H+ single-ion on binding rate constant (kH,lon in Table 2) to 100-fold larger than that of Brzezinski et al., which resulted in a Keq two orders of magnitude smaller than Brzezinski et al.'s [∼108 mM (Weinstein) vs. ∼1010 mM (Brzezinski et al.); see Table 2]. Weinstein's modification was necessary to adjust Brzezinski et al.'s gastric HKA model to one that is more suitable in the collecting duct. On the other hand, our 1:1-per-ATP and 2:2-per-ATP models are based solely on experimental data from nongastric (colonic) HKA extracted from the rat distal colon, which is located in areas under slightly acidic to neutral pH. In addition, we selected our Keq based on the experimental conditions in Ref. 9 (see Thermodynamic restrictions on the rate constants in the appendix). Consequently, using different experimental data and different assumptions in the model formulation resulted in ATP hydrolysis profile differences that became clear in Fig. 5.
DISCUSSION
The role of nongastric HKA in ion homeostasis of cTAL and MD cells is unresolved. To begin a quantitative analysis of this issue, we developed two mathematical models that predict the ion fluxes through a nongastric HKA. One model assumes a 1H+:1K+-per-ATP stoichiometry, and the other assumes a 2H+:2K+-per-ATP stoichiometry. Both models include competition between both cytosolic Na+ and H+ and luminal K+ and NH4+ (Fig. 1), a behavior that has been observed in vitro and in situ (9, 29). To our knowledge, these are the first mathematical models of a nongastric HKA that include Na+ and NH4+ transport. Our models build on two previous models of gastric HKA [Brzezinski et al. (6) and Weinstein (43)], which is expressed in other renal cells.
Here, we explored the possibility of both a 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP stoichiometry of nongastric HKA. Model outcomes showed that both models provided good fits to the experimental data (Fig. 2). Therefore, based solely on the model fits to the experimental data, we are unable to conclude which is the most suitable model. Moreover, with the exception of NH4+ flux, which is ∼10-fold higher for the 1:1-per-ATP model, both models behaved similarly under cell and luminal pH values >6 (Figs. 3, A,c and B,c, and 4, A,c and B,c).
The results of this study also illustrate the power of nonlinear optimization techniques in the development of transporter models. These methods provide a means to systematically include all applicable experimental studies and constraints in a single simultaneous fitting. Although these methods are more complex than nonlinear regression, they greatly facilitate parameter estimation in models of complex transporters like nongastric HKA.
Our simulations using the 1H+(1Na+):1K+(1NH4+)-per-ATP and 2H+(2Na+):2K+(2NH4+)-per-ATP models showed a model ion net flux sensitivity to both cytosolic [Na+] and [H+] (Figs. 3B,a and c, and 4B,a and c). This suggests a possible role of nongastric HKA in cell pH and Na+ homeostasis in, for example, MD and cTAL cells. To put this into the whole cell perspective, we must consider the ion transport through nongastric HKA and its effect on ion transport through other transport proteins.
For example, regarding cell pH, in the MD/cTAL region of the nephron, luminal Na+ can be very low as a result of flow-dependent tubular fluid dilution by TAL cells. This reduces the trans-apical Na+ gradient in the cortical region and may limit H+ extrusion by the apical Na+/H+ exchanger in MD and cTAL cells. Moreover, this limitation on Na+/H+ exchanger activity should be even greater in MD cells, where cytosolic Na+ essentially tracks luminal Na+ so that the trans-apical [Na+] difference is only ∼10 mM (29). Although the simulations shown in Figs. 3 and 4 suggest that this ATPase will be doing mostly Na+/K+ exchange under physiological conditions, in the cTAL/MD region, [H+] is very small compared with [Na+] (0.0001 to 0.001 mM H+ vs. ∼10 mM Na+). Hence, a ∼100-fold Na+-to-H+ flux ratio, as shown in Figs. 3 and 4, may have an effect on cytosolic pH. Consequently, the expression of apical H+(Na+)-K+(NH4+)-ATPase may well provide a second pathway for acid extrusion in these cells.
Regarding the regulation of cell [Na+], Peti-Peterdi et al. (29) presented evidence suggesting that H+(Na+)-K+(NH4+)-ATPase is the primary Na+ extrusion mechanism in rabbit MD cells, which have low basolateral Na+-K+-ATPase expression (34). In this scenario, Na+ extrusion through apical H+(Na+)-K+(NH4+)-ATPase should exceed that of the basolateral membrane through Na+-K+(NH4+)-ATPases and nonselective cation channels, which are known to be permeable to Na+ (22).
In contrast to Peti-Peterdi et al. (29), Lorenz et al. (23) concluded that the tubuloglomerular feedback response, which is tightly linked to MD cell Na+ homeostasis, is dependent on basolateral Na+-K+-ATPase and is intact in mice lacking nongastric HKA. Although these conflicting conclusions may just reflect a species difference between mice and rabbits, it could also be due in part to methodological issues. In particular, the experiments of Peti-Peterdi et al. (29) were carried out in vitro in the absence of the NH4+/NH3 buffering system,4 whereas the Lorenz et al. (23) study was conducted in intact mice. Therefore, additional in vitro or in situ experiments in the presence of NH4+/NH3 would seem necessary to elucidate the role of nongastric H+(Na+)-K+(NH4+)-ATPases in cell ion homeostasis under conditions closer to those in vivo. Notwithstanding, MD/cTAL cell mathematical models that include nongastric HKA can be of great help to elucidate the overall function of nongastric HKA, its effect on other transport proteins, and its role in cell pH and [Na+] homeostasis under physiological, experimental, or pathophysiological conditions.
GRANTS
This work was supported in part by the National Institutes of Health (NIH) Minority Biomedical Research Support-Research Initiative for Scientific Enhancement through Grant 2-R25-GM061151 (to the University of Puerto Rico) and by NIH Grant SC1-GM-084744 (to M. Marcano).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.N.-Q., L.C.M., and M.M. conception and design of research; M.N.-Q. performed experiments; M.N.-Q. analyzed data; M.N.-Q., L.C.M., and M.M. interpreted results of experiments; M.N.-Q. prepared figures; M.N.-Q. drafted manuscript; M.N.-Q., L.C.M., and M.M. edited and revised manuscript; M.N.-Q., L.C.M., and M.M. approved final version of manuscript.
ACKNOWLEDGMENTS
A portion of this work was presented in poster format at Experimental Biology 2012 (Abstract; FASEB J 26: 867.1, 2012).
Appendix
Linear equations of the H+(Na+)-K+(NH4+)-ATPase model.
We denote the enzyme state density Ej-i in Fig. 1 by the variable eij, where j is the luminal (j = l)- or cytosolic (j = c)-facing enzyme with the ith solute or solutes bound to the enzyme (e.g., i = K+-ATP, Pi-Na+, or NH4+). For example, the enzyme states El-K, Ec-ATP, and El-Pi-Na in Fig. 1 are denoted as eKl, eATPc, and ePi1-Na, respectively. Let Ci,j represent the ith solute concentration (where i = H+, K+, Na+, NH4+, ATP, ADP, and Pi) in the luminal (j = l) or cytosolic (j = c) compartment and kij represent the off (j = off) and on (j = on) binding rate constant of the ith solute in the cytosol or lumen or the translocation rate constant from the cytosol to lumen (j = cl) or from the lumen to cytosol (j = lc) with solute i bound to the enzyme (i = H+, K+, Na+, and NH4+). By applying the Principle of Mass Action in the kinetic model in Fig. 1, one gets an ordinary differential equation for each enzyme state (see Ref. 25 for details). We computed steady-state solutions of the resultant system of ordinary differential equations. The steady-state system of the enzyme state eij is given by the following:
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
| (19) |
| (20) |
| (21) |
| (22) |
| (23) |
| (24) |
| (25) |
This is a homogenous system of linear equations in the variables
| (26) |
where the prime means vector transpose.
Conservation of enzyme implies that
| (27) |
where ET is a constant that represents the total density of ATPase. We arbitrarily selected the variable ePil-H and solved for that variable in Eq. 27 to obtain the following:
| (28) |
Now, Eq. 21 becomes redundant. Substituting the expression for ePil-H from Eq. 28 into Eqs. 14 and 20 results in a system of 15 linear equations in 15 unknown enzyme state densities, which is written as Eq. 1, where the 15 × 1 vector e contains the variables in vector u in Eq. 26, except ePil-H, which is solved by means of Eq. 28.
Thermodynamic restrictions on the rate constants.
The equilibrium constant of a chemical reaction is defined in dimensionless form () as follows (41):
| (29) |
where ΔG is the Gibbs free energy, R is the gas constant (8.3145 J·mol−1·K−1), T is temperature, Π denotes the product, Ci is the concentration of the ith solute, Co is the standard concentration for diluted solutes, which is equal to 1 M, and υi is the stoichiometric number of the ith solute, which is taken as positive when the ith solute is produced or released and negative when it is consumed or binds to the enzyme. Using Eq. 29, based on the experimental conditions in Ref. 9 (where the temperature, pH, and Mg2+ concentration are 37°C, 7.2, and 4 mM, respectively) and from Table 5 in Ref. 30 (which shows the Gibbs free energy as a function of temperature, pH, and Mg2+ concentration), one can get a Gibbs free energy of approximately −10 kcal/mol or approximately −42 kJ/mol, which, using Eq. 29, results in a dimensionless of ∼107.
Moreover, by the Principle of Detailed Balance (20), for the cycles Na+/K+, H+/K+, and Na+/NH4+ in the HKA kinetic model in Fig. 1, the multiplication of the rates in the clockwise direction must equal the multiplication of the rates in the counterclockwise direction. For the first cycle, this is as follows:
| (30) |
This can be written as follows:
| (31) |
where Keq (in mM) is the concentration equilibrium constant. By comparing Eqs. 29 and 31, one obtains the following relation:
| (32) |
which, for Co = 1 M, results in a Keq value of 107 M or 1010 mM.
For the other two cycles (H+/K+ and Na+/NH4+), the thermodynamic restrictions, respectively, are as follows:
| (33) |
| (34) |
To account for the thermodynamic restrictions on the HKA kinetic model, we calculated the rate constants kNalc, kHlc, and kNH4cl by solving for them in Eqs 31, 33, and 34, respectively,
| (35) |
| (36) |
| (37) |
Normalization methods.
We normalized experimental ATPase activity [which is given in μmol·mg−1·h−1 (9)] by dividing it by maximum enzyme activity (Jmax, iexp), which was obtained by fitting the following Hill equation:
| (38) |
independently to each experimental data set i (shown in Table 1) as a function of the variable bath ion concentration (Cion). The Hill model parameters are the maximum enzyme activity (Jmax, iexp), the half-maximal ion concentration (C50, i) and the Hill coefficient (n), which was set to a value of one for the 1H+:1K+-per-ATP model and two for the 2H+:2K+-per-ATP model. The fitting was done with the MATLAB function nlinfit, which solves nonlinear regression problems and gives SEs on the parameters. The Hill model parameters and R2 values for the fittings are shown in Table 4. We need to mention that Eq. 38 depends only on the variable bath ion concentration, which is not the same approach as using a mathematical model that considers the synergism among all solutes in and out of a cell and their interaction with the enzyme.
The enzymatic pathway of an ATPase, such as nongastric HKA, can be expressed in a general form with the following chemical reaction system (14):
| (39) |
where e is the enzyme, eATP is the enzyme-ATP complex (also known as the active intermediate), and kcat (in s−1) is the enzyme turnover rate, i.e., the number of molecules that are converted to product at a given time on a single-enzyme molecule when all its active sites are occupied.
We used ET (from Eq. 27) to normalize the enzyme-state densities and kcat to normalize the reaction rates. Specifically, in the model system, Eq. 1, we computed the fraction of enzyme state instead of the density by dividing each enzyme state (eij) by ET, the total density of ATPase. As a result, each term of the linear system in Eq. 1 and the model fluxes in Eqs. 2–6 are in units of s−1. We normalized the model linear system in Eq. 1 and the model ion fluxes in Eqs. 2–6 by dividing both sides of each equation by the enzyme turnover rate kcat to render them in dimensionless form.
For the 1:1-per-ATP and 2:2-per-ATP models, the rate constants corresponding to the on and off binding of ATP, ADP, and Pi are fixed and assumed equal to those reported by Tanford et al. (37), as Brzezinski et al. (6) and Weinstein (43) assumed in their respective HKA models. Based on these rate constants, Tanford et al. (37) reported a kcat value of 5.8 s−1. Therefore, we set kcat equal to this value.
Considerations for the 2H+(2Na+):2K+(2NH4+)-ATPase model.
So far, our ATPase mathematical model description considers a 1H+(1Na+):1K+(1NH4+) stoichiometry. If we now assume that two H+, K+, Na+, or NH4+ bind (unbind) to (from) two identical enzyme sites with the same off and on binding rate constants (16), then in the lumen
| (40) |
and in the cytosol
| (41) |
where X is H+, K+, Na+, or NH4+ and [X]l and [X]c are the concentrations of ion X in the lumen and cytosol, respectively. The off and on binding rate constants are in units of s−1 and mM−1·s−1. For simplicity, we expressed the chemical reactions in Eqs. 40 and 41 in their respective one-reaction equivalent as follows:
| (42) |
| (43) |
where the on binding rate constants identified with an asterisk are expressed in terms of the single ion on binding rate constants as follows:
| (44) |
| (45) |
Both are expressed in units of mM−2·s−1 units.
Based on the chemical reactions in Eqs. 42–45, for X = H+, K+, Na+, and NH4+, the following modifications are necessary for the HKA model with 2H+(2Na+):2K+(2NH4+) stoichiometry:
1. Enzyme states: substitute X with X2 wherever it appears in the diagram in Fig. 1 and in the subscript i of eij in Eqs. 2–6 and 10–28.
2. On binding rate constants: substitute kX, lon and kX, con with kX, lon* and kX, con*, respectively, in Eqs. 10–25, 29, 31, and 33–37.
3. Ion concentrations: substitute CX, l and CX, c with CX, l2 and CX, c2, respectively, in Eqs. 10–25, 29, 31, 33, and 34.
Footnotes
For this part, we consider a 1H+(1Na+):1K+(1NH4+)-per-ATP stoichiometry to explain the nongastric HKA model. Details for the 2H+(2Na+):2K+(2NH4+)-per-ATP model are included in Considerations for the 2H+(2Na+)-2K+(2NH4+)-ATPase model in the appendix.
Note that, although the results in Ref. 16 and 17 suggest a H+-to-K+ flux ratio near 0.1 and 0.5, respectively (see H+-to-K+ flux ratio when Na+ is present), we increased the variability range of the net flux ratio constraint to account for possible species-specific variations.
The mathematical approach to obtain the luminal pH at which ion net fluxes are zero when both Na+ and NH4+ are present in the solution (as in Figs. 3A,c and 4A,c) is beyond the scope of this study. Nonetheless, zero net fluxes in Figs. 3 and 4 are identified with solid circle.
It is known that NH4+ may occupy the K+-binding site in various transport proteins expressed in cTAL and MD cells, such as the Na+-K+-2Cl− cotransporter and Na+/H+ exchanger (46).
REFERENCES
- 1.Abe K, Tani K, Friedrich T, Fujiyoshi Y. Cryo-EM structure of gastric H+,K+-ATPase with a single occupied cation-binding site. Proc Natl Acad Sci USA 109: 18401–18406, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn KY, Kone BC. Expression and cellular localization of mRNA encoding the “gastric” isoform of the H+-K+-ATPase in rat kidney. Am J Physiol Renal Fluid Electrolyte Physiol 268: F99–F109, 1995. [DOI] [PubMed] [Google Scholar]
- 3.Ahn KY, Park KY, Kim KK, Kone BC. Chronic hypokalemia enhances expression of the H+-K+-ATPase α2-subunit gene in renal medulla. Am J Physiol Renal Fluid Electrolyte Physiol 271: F314–F321, 1996. [DOI] [PubMed] [Google Scholar]
- 4.Bell PD, Lapointe JY, Peti-Peterdi J. Macula densa cell signaling. Annu Rev Physiol 65: 481–500, 2003. [DOI] [PubMed] [Google Scholar]
- 5.Boron WF, Boulpaep EL. Medical Physiology (2nd ed). Philadelphia, PA: Elsevier Health Sciences, 2009. [Google Scholar]
- 6.Brzezinski P, Malmström BG, Lorentzon P, Wallmark B. The catalytic mechanism of gastric H+/K+-ATPase: simulations of pre-steady-state- and steady-state kinetic results. Biochim Biophys Acta 942: 215–219, 1988. [DOI] [PubMed] [Google Scholar]
- 7.Burnay M, Crambert G, Kharoubi-Hess S, Geering K, Horisberger JD. Electrogenicity of Na,K- and H,K-ATPase activity and presence of a positively charged amino acid in the fifth transmembrane segment. J Biol Chem 278: 19237–19244, 2003. [DOI] [PubMed] [Google Scholar]
- 8.Cain BD, Gumz ML, Zies DL, Welch AK. The H+- and H+, K+-ATPases of the collecting duct. In: Epithelial Transport Physiology, edited by Gerencser GA. New York: Humana, 2010, p. 225–243. [Google Scholar]
- 9.Codina J, Pressley TA, DuBose TD. The colonic H+,K+-ATPase functions as a Na+-dependent K+ (NH4+)-ATPase in apical membranes from rat distal colon. J Biol Chem 274: 19693–19698, 1999. [DOI] [PubMed] [Google Scholar]
- 10.Cougnon M, Bouyer P, Planelles G, Jaisser F. Does the non-gastric H,K-ATPase also act as an Na,K-ATPase? Proc Natl Acad Sci USA 95: 6516–6520, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crambert G. H-K-ATPase type 2: relevance for renal physiology and beyond. Am J Physiol Renal Physiol 306: F693–F700, 2014. [DOI] [PubMed] [Google Scholar]
- 12.Crambert G, Horisberger JD, Modyanov NN, Geering K. Human nongastric H+-K+-ATPase: transport properties of ATP1al1 assembled with different β-subunits. Am J Physiol Cell Physiol 283: C305–C314, 2002. [DOI] [PubMed] [Google Scholar]
- 13.DuBose TD Jr, Codina J, Burges A, Pressley TA. Regulation of H+,K+-ATPase expression in kidney. Am J Physiol Renal Fluid Electrolyte Physiol 269: F500–F507, 1995. [DOI] [PubMed] [Google Scholar]
- 14.Fogler HS. Elements of Chemical Reaction Engineering (4th ed). Upper Saddle River, NJ: Pearson Education, 2006. [Google Scholar]
- 15.Greenlee MM, Lynch IJ, Gumz ML, Cain BD, Wingo CS. The renal H,K-ATPases. Curr Opin Nephrol Hypertens 19: 478–482, 2010. [DOI] [PubMed] [Google Scholar]
- 16.Grishin AV, Bevensee MO, Modyanov NN, Rajendran V, Boron WF, Caplan MJ. Functional expression of the cDNA encoded by the human ATP1AL1 gene. Am J Physiol Renal Fluid Electrolyte Physiol 271: F539–F551, 1996. [DOI] [PubMed] [Google Scholar]
- 17.Grishin AV, Caplan MJ. ATP1AL1, a member of the non-gastric H,K-ATPase family, functions as a sodium pump. J Biol Chem 273: 27772–27778, 1998. [DOI] [PubMed] [Google Scholar]
- 18.Gumz ML, Lynch IJ, Greenlee MM, Cain BD, Wingo CS. The renal-K+ ATPases: physiology, regulation, and structure. Am J Physiol Renal Physiol 298: F12–F21, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaisser F, Beggah AT. The nongastric H+-K+-ATPases: molecular and functional properties. Am J Physiol Renal Physiol 276: F812–F824, 1999. [DOI] [PubMed] [Google Scholar]
- 20.Klein MJ. Principle of detailed balance. Phys Rev 97: 1446–1447, 1955. [Google Scholar]
- 21.Kraut JA, Hiura J, Shin JM, Smolka A, Sachs G, Scott D. The Na+-K+-ATPase β1 subunit is associated with the HKα2 protein in the kidney. Kidney Int 53: 958–962, 1998. [DOI] [PubMed] [Google Scholar]
- 22.Lapointe JY, Bell PD, Sabirov RZ, Okada Y. Calcium-activated nonselective cationic channel in macula densa cells. Am J Physiol Renal Physiol 285: F275–F280, 2003. [DOI] [PubMed] [Google Scholar]
- 23.Lorenz JN, Dostanic-Larson I, Shull GE, Lingrel JB. Ouabain inhibits tubuloglomerular feedback in mutant mice with ouabain-sensitive α1 Na,K-ATPase. J Am Soc Nephrol 17: 2457–2463, 2006. [DOI] [PubMed] [Google Scholar]
- 24.Locatelli M. Simulated annealing algorithms for continuous global optimization. In: Handbook of Global Optimization. Dordrecht, The Netherlands: Kluwer Academic, 2002, vol. II, p. 179–229. [Google Scholar]
- 25.Marcano M, Yang HM, Nieves-González A, Clausen C, Moore LC. Parameter estimation for mathematical models of NKCC2 cotransporter isoforms. Am J Physiol Renal Physiol 296: F369–F381, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcano M, Yang H, Nieves-González A, Nadal-Quirós MA, Clausen C, Moore LC. Parameter Estimation for Models of NH4+ Transport by the Renal Na-K-2Cl and K-Cl Cotransporters. Technical Report (online). http://repositorio.upr.edu:8080/jspui/bitstream/10586%20/531/1/NKCC2_KCC_NH4.pdf [1 July 2015]. [Google Scholar]
- 27.Marsy S, Elalouf JM, Doucet A. A quantitative RT-PCR analysis of mRNAs encoding a non-gastric putative H-K-ATPase α subunit along the rat nephron: effect of K+ depletion. Pflügers Arch 432: 494–500, 1996. [DOI] [PubMed] [Google Scholar]
- 28.Nadal-Quirós M. Parameter Estimation for Mathematical Models of Ion Transport in the Renal System (Master's thesis) (online) http://repositorio.upr.edu:8080/jspui/bitstream/10586%20/426/1/MonicaNadalMastersThesis_PrintedVersion-2.pdf [1 July 2015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peti-Peterdi J, Bebok Z, Lapointe LY, Bell PD. Novel regulation of cell [Na+] in macula densa cells: apical Na+ recycling by H-K-ATPase. Am J Physiol Renal Physiol 282: F324–F329, 2002. [DOI] [PubMed] [Google Scholar]
- 30.Phillips RC, George P, Rutman RJ. Thermodynamic data for the hydrolysis of adenosine triphosphate as a function of pH, Mg2+ ion concentration, and ionic strength. J Biol Chem 244: 3330–3342, 1969. [PubMed] [Google Scholar]
- 31.Rabon EC, McFall TL, Sachs G. The gastric [H,K]ATPase: H+/ATP stoichiometry. J Biol Chem 257: 6296–6299, 1982. [PubMed] [Google Scholar]
- 32.Rajendran VM, Sangan P, Geibel J, Binder HJ. Ouabain-sensitive H,K-ATPase functions as Na,K-ATPase in apical membranes of rat distal colon. J Biol Chem 275: 13035–13040, 2000. [DOI] [PubMed] [Google Scholar]
- 33.Renstra WW, Forte JG. H+/K+ ATP stoichiometry for the gastric H+,K+-ATPase. J Membr Biol 61: 55–60, 1981. [DOI] [PubMed] [Google Scholar]
- 34.Schnermann J, Marver D. ATPase activity in macula densa cells of the rabbit kidney. Pflügers Arch 407: 82–86, 1986. [DOI] [PubMed] [Google Scholar]
- 35.Shin JM, Grundler G, Senn-Bilfinger J, Simon WA, Sachs G. Functional consequences of the oligomeric form of the membrane-bound gastric H,K-ATPase. Biochemistry 44: 16321–16332, 2005. [DOI] [PubMed] [Google Scholar]
- 36.Swarts HG, Koenderink JB, Willems PH, De Pont JJ. The non-gastric H,K-ATPase is oligomycin-sensitive and can function as an H+,NH4+-ATPase. J Biol Chem 280: 33115–33122, 2005. [DOI] [PubMed] [Google Scholar]
- 37.Tanford C, Reynolds JA, Johnson EA. Thermodynamic and kinetic cooperativity in ligand binding to multiple sites on a protein: Ca2+ activation of an ATP-driven Ca pump. Proc Natl Acad Sci USA 82: 4688–4692, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405: 647–655, 2000. [DOI] [PubMed] [Google Scholar]
- 39.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418: 605–611, 2002. [DOI] [PubMed] [Google Scholar]
- 40.Verlander JW, Moudy RM, Campbell WG, Cain BD, Wingo CS. Immunohistochemical localization of H-K-ATPase α2c-subunit in rabbit kidney. Am J Physiol Renal Physiol 281: F357–F365, 2001. [DOI] [PubMed] [Google Scholar]
- 41.Vinnakota KC, Wu F, Kushmerick MJ, Beard DA. Multiple ion binding equilibria, reaction kinetics, and thermodynamics in dynamic models of biochemical pathways. Methods Enzymol 454: 29–68, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallmark B, Stewart HB, Rabon E, Saccomani G, Sachs G. The catalytic cycle of gastric (H++K+)-ATPase. J Biol Chem 255: 5313–5319, 1980. [PubMed] [Google Scholar]
- 43.Weinstein AM. A mathematical model of the inner medullary collecting duct of the rat: acid/base transport. Am J Physiol Renal Physiol 274: F856–F867, 1998. [DOI] [PubMed] [Google Scholar]
- 44.Weinstein AM. A mathematical model of rat ascending Henle limb: I. Cotransporter function. Am J Physiol Renal Physiol 298: F512–F524, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wingo CS, Smolka AJ. Function and structure of H-K-ATPase in the kidney. Am J Physiol Renal Fluid Electrolyte Physiol 269: F1–F16, 1995. [DOI] [PubMed] [Google Scholar]
- 46.Weiner ID, Hamm LL. Molecular mechanism of renal ammonia transport. Annu Rev Physiol 69: 317–340, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X, Xia SL, Wingo CS. Chloride transport by the rabbit cortical collecting duct: dependence on H,K-ATPase. J Am Soc Nephrol 9: 2194–2202, 1998. [DOI] [PubMed] [Google Scholar]