Abstract
Hypertension is a major chronic disease whose molecular mechanisms remain poorly understood. We compared neuroanatomical patterns of microRNAs in the brain stem of the spontaneous hypertensive rat (SHR) to the Wistar Kyoto rat (WKY, control). We quantified 419 well-annotated microRNAs in the nucleus of the solitary tract (NTS) and rostral ventrolateral medulla (RVLM), from SHR and WKY rats, during three main stages of hypertension development. Changes in microRNA expression were stage- and region-dependent, with a majority of SHR vs. WKY differential expression occurring at the hypertension onset stage in NTS versus at the prehypertension stage in RVLM. Our analysis identified 24 microRNAs showing time-dependent differential expression in SHR compared with WKY in at least one brain region. We predicted potential gene regulatory targets corresponding to catecholaminergic processes, neuroinflammation, and neuromodulation using the miRWALK and RNA22 databases, and we tested those bioinformatics predictions using high-throughput quantitative PCR to evaluate correlations of differential expression between the microRNAs and their predicted gene targets. We found a novel regulatory network motif consisting of microRNAs likely downregulating a negative regulator of prohypertensive processes such as angiotensin II signaling and leukotriene-based inflammation. Our results provide new evidence on the dynamics of microRNA expression in the development of hypertension and predictions of microRNA-mediated regulatory networks playing a region-dependent role in potentially altering brain-stem cardiovascular control circuit function leading to the development of hypertension.
Keywords: hypertension, microRNA, neuroinflammation, angiotensin II, brain stem
hypertension is a major chronic disease worldwide, affecting about 50 million adults in the United States alone. It is a multiorgan disease and a major cause of heart failure, coronary heart disease, atrial fibrillation, stroke, and end-stage renal disease, and it has an enormous economic impact. About 90–95% of cases are “essential” with no known medical cause (11). Moreover, only about a third of hypertensive patients have their blood pressure fully controlled (11). A significant and increasing population (∼25%) of hypertensive patients is drug resistant. There remains an unmet need for antihypertensive therapy.
In the context of the recent discovery of novel lines of therapeutic interventions involving approaches to modulate microRNA we decided to elucidate changes in microRNA levels during the development of spontaneous hypertensive rat (SHR) hypertension. The recent explosion in microRNA research has produced new computational and experimental approaches for studying microRNA functions in cell culture and in vivo, which we take advantage of here. MicroRNAs are an abundant class of small (∼22 nt) endogenous noncoding RNAs that direct posttranscriptional regulation of gene expression. There is ample evidence that dysregulation of microRNAs is associated with the pathogenesis of human diseases, including cardiovascular diseases as markers and therapeutic targets (26) specifically in hypertensive patients (26, 29), and shows great promise in application to understanding SHR hypertension (5, 44, 57). Disease-associated microRNAs represent a new class of targets for the development of microRNA-based therapeutic modalities, which may yield patient benefits unobtainable by other therapeutic approaches (7, 16, 36). However, there has been no characterization performed on microRNA expression in the brain stem during hypertension development.
Due to the distinct roles of the nucleus of the solitary tract (NTS) and the rostral ventrolateral medulla (RVLM) in the physiological regulation of blood pressure, it is likely that microRNAs are involved and their expression profiles differ between these two regions. Our goal is to characterize microRNAs that mediate hypertension in the brain stem and to understand their physiological relevance. There are two main types of signaling in the NTS and RVLM that contribute to the development of neurogenic hypertension, including angiotensin II (ANG II) signaling and specific neuroinflammatory pathways involving leukotriene B4 (LTB4) and interleukin 1 (IL-1) signaling (4, 32, 42, 43, 55, 58).
With many signaling pathway changes occurring during the development and onset of hypertension, we wondered whether microRNAs could be involved in such mechanisms and potentially act as master regulators of these physiologically relevant signaling pathways. Given the associations between microRNAs and brain pathologies and their mechanistic potential to contribute to multigenic phenotypes by regulating networks of gene expression, we also wondered if different microRNA expression patterns exist in brain regions associated with blood pressure regulation in hypertension compared with control. Here, we present an extensive characterization of microRNA expression patterns and correlated gene targets in two key structures responsible for regulating cardiovascular function, the NTS and the RVLM. We hypothesized a putative signature exists in each anatomical region of microRNAs and respective targets characterizing the hypertensive state. We aimed to collect data that offer insight into the potential role of microRNAs in an animal model relevant to hypertension susceptibility and that enable further examination into the complexity underlying this disease. Our use of micropunches and probe hybridization and quantitative (q)PCR technologies enabled us to obtain precise neuroanatomical resolution of microRNA and putative gene target expression. Our results reveal different brain regions show distinct microRNA changes with corresponding target-correlation signatures to different molecular signaling pathway targets implicated in hypertension.
MATERIALS AND METHODS
Animal model.
Male Wistar Kyoto (WKY/NHsd) rats and spontaneous hypertensive rats (SHR/NHsd) obtained from Harlan Laboratories were housed one per cage in the Thomas Jefferson University (TJU) animal facility to avoid animal-to-animal stress from dominance that could affect blood pressure. Each condition had an n = 3 or 4 animals. Facilities were maintained at constant temperature and humidity with 12/12 h light cycles (lights on at Zeitgeber time 0). All protocols were approved by the TJU Institutional Animal Care and Use Committee.
Tissue sample punches.
There were three time points of interest to this experiment, referred to as prehypertension, hypertension onset, and chronic hypertension. These time points correspond to the age of the rat at the time of death and mean arterial blood pressure measurements at that time. The prehypertensive stage is 6–7 wk old, hypertension onset is at 10–12 wk, and chronic hypertension occurs around 16 wk old. At the assigned time of death, rat was killed via rapid decapitation and brain stems were excised, placed into ice-cold artificial cerebral spinal fluid (ACSF: 10 mM HEPES, pH 7.4; 140 mM NaCl; 5 mM KCl; 1 mM MgCl2; 1 mM CaCl2; 24 mM d-glucose), and secured with agarose for sectioning [4% UltraPure low-melting-point agarose (Invitrogen) in ACSF]. We made 275 mm transverse sections with a McElwain Tissue Chopper (Gamshall) DVC microdissection with size-matched micropunches (1.25 mm; Stoelting, Wood Dale, IL), as previously reported (27). Bilateral region punches from one animal were treated as a single sample.
RNA extraction and processing.
Total RNA was extracted with the miRNeasy extraction kit (Qiagen, Valencia, CA), which captures all RNA greater than 18 nucleotides in length, DNase-treated (DNA-Free RNA kit; Zymo Research, Orange, CA), and stored at −80°C. Concentration and integrity were assessed with an ND-1000 (NanoDrop, Philadelphia, PA). We processed 100 ng of RNA per experimental condition in the nanoString microRNA assay following manufacturer's protocol. Expression levels of 419 microRNAs were measured from two sections of the brain stem: the NTS and RVLM.
MicroRNA expression data analysis.
nanoString data set has endogenous controls and RNA spike-ins, i.e., additional negative controls from other species purposefully added to the assay to assure that reads were not from nonspecific binding, to assure quality. Total count normalization was performed per the manufacturer's protocol for normalizing nanoString data.
The normalized expression level for ith microRNA in jth sample was calculated as:
Where Countij is the raw count, TotalCountj is the sum of counts for all microRNAs, positive and negative controls in jth sample, and 100,000 is a normalization factor. In essence, this calculation accounts for different amounts of total number of counted molecules across samples (45). Data were log-normalized (base 2), and all microRNAs with maximum normalized expression < 6.0 were removed from subsequent analysis. The threshold of 6.0 corresponded to the maximum value of the negative controls including spike-ins. This analysis yielded 197 microRNAs with expression above background detection limits. The data for each anatomical region (NTS and RVLM) were considered separately in statistical analysis. Data were statistically assessed via a two-way ANOVA using stage of hypertension development and rat strain as independent and interacting factors (P = 0.05); we also confirmed results with Tukey honest significant difference post hoc testing and corrected using a false discovery rate cut-off of 0.05. For visualization purposes, we followed established approaches for normalizing the expression of each microRNA by subtracting the median expression value of that microRNA across all samples from the same neuroanatomical region.
MicroRNA target prediction.
Targets of key microRNAs identified from the nanoString profiling were determined using the miRWalk 1.0 database that provides predicted as well as validated microRNA binding sites for human, mouse, and rat (18). This database utilizes eight established programs for predicting microRNA target genes and combines these results across three genomes, to improve the sensitivity and specificity. Database settings were altered from default to include both the 5′-untranslated region (UTR) and coding sequence for microRNA target matching. Target predictions were filtered based on Gene Ontology and known functional role of in neuronal function. We also used RNA22 to find noncanonical predicted microRNA interactions and combined these lists to compile all possible microRNA-target putative interactions (35).
High-throughput PCR.
Intron-spanning PCR primers and probes for gene target assays were designed using Roche's Universal Probe Library Assay Design Center (http://www.universalprobelibrary.com). The standard BioMark protocol was used to preamplify cDNA samples for 12 cycles using TaqMan PreAmp Master Mix per the manufacturer's protocol (Applied Biosystems, Foster City, CA). qPCR reactions were performed using 3 - 48.48 BioMark Dynamic Arrays (Fluidigm, South San Francisco, CA) enabling quantitative measurement of multiple genes and samples under identical reaction conditions. Runs were 30 cycles (15 s at 95°C, 5 s at 70°C, 60 s at 60°C). The primers are listed in Supplemental Table S6.1
Gene expression data analysis.
Targets assayed were chosen based on their role in neuromodulation, inflammation, and catecholaminergic regulation from literature and previous unpublished data from this lab. Ct values were calculated by the Real-Time PCR Analysis Software (Fluidigm), and software-designated failed reactions were discarded from analysis. Data were median-centered per anatomical region of the brain stem (NTS, RVLM). An independent statistical analysis was conducted by a two-way ANOVA with stage of hypertension development and rat strain as independent and interacting factors (P = 0.05). Pearson correlations between mRNA and microRNA expression were calculated. The microRNA/mRNA pairs with correlation values −0.4 or >0.4 were considered in the downstream network analysis. Hierarchal clustering based on Pearson correlation was performed for each data set using MultiExperiment Viewer part of the TM4 software tool suite (48) and organized graphically with Cytoscape software (12, 49, 50).
RESULTS
Global microRNA expression patterns vary by strain and brain region.
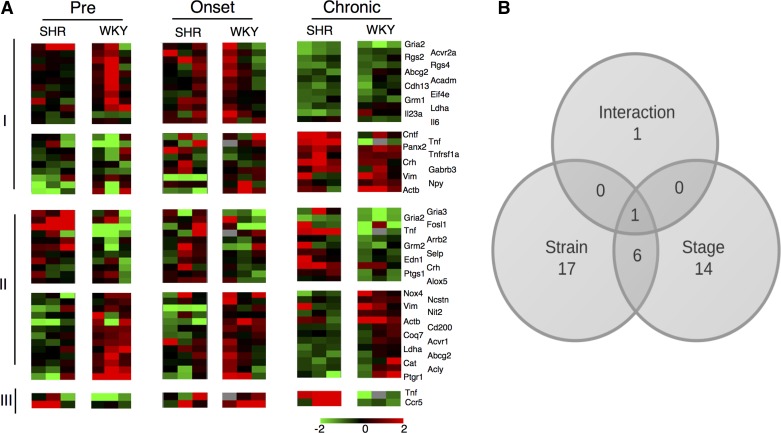
Using the SHR and WKY animals, we measured and compared the expression of 419 microRNAs in NTS and RVLM at three stages: prehypertension, hypertension onset, and chronic hypertension (Fig. 1). In the NTS, 21 microRNAs were differentially expressed between SHR and WKY in a stage-independent manner (Fig. 2A, VI). A set 53 microRNAs were differentially expressed across stage, with a similar temporal pattern between SHR and WKY (Fig. 2A, II–V). Seven microRNAs were differentially expressed across stage in distinct ways between SHR and WKY (Fig. 3A, I). In the RVLM, 12 microRNAs demonstrated differential expression in SHR compared with WKY (Fig. 3B, IV and V), and 20 microRNAs demonstrated differential expression in a stage-dependent manner similarly across animal strains (Fig. 3B, II and III). Sixteen microRNAs demonstrated stage-dependent differential expression in SHR relative to WKY (Fig. 3B, I) in RVLM.
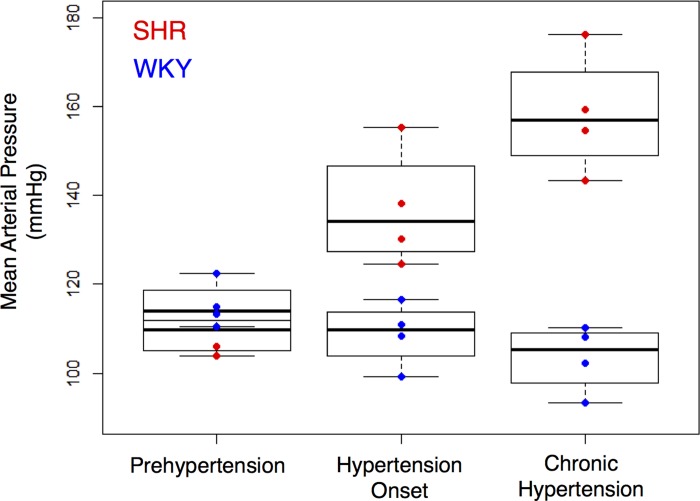
Fig. 1.
Comparison of blood pressure in spontaneously hypertensive rats (SHR) vs. Wistar Kyoto rats (WKY) over the time course of hypertension development. Prehypertension stage corresponds to 6–7 wk of age, hypertension onset stage corresponds to 10–12 wk of age, and chronic hypertension corresponds to 16 wk of age.
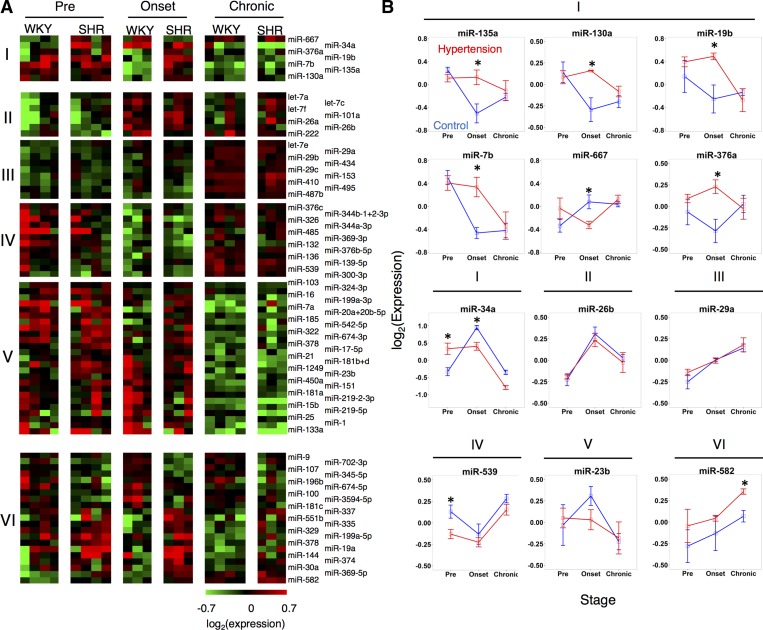
Fig. 2.
Dynamics of microRNA expression in nucleus of the solitary tract (NTS) during the development of hypertension. A: clustered groups of microRNAs showing differential expression. I, across strain (SHR v. WKY) in a stage-dependent manner; II–V, in a stage-dependent and strain-independent manner that shows a gradient of high to low expression from II–V; VI, between strains (SHR v. WKY) without dynamic regulation over time. B: dynamic profiles of all microRNAs in group I and a select subset from other groups. *P < 0.05.
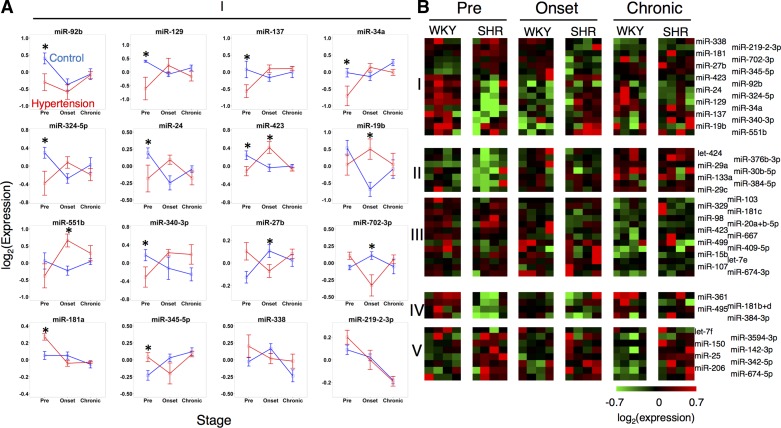
Fig. 3.
Dynamics of microRNA expression in rostral ventrolateral medulla (RVLM) during the development of hypertension. A: dynamic profiles of all microRNAs differentially expressed across strain (SHR v. WKY) in a stage-dependent manner. B: clustered groups of microRNAs showing differential expression across strain (SHR v. WKY) in a stage-dependent manner (I), in a stage-dependent and strain-independent manner (II and III), and between strains (SHR v. WKY) without dynamic regulation over time (IV and V). *P < 0.05.
Our results revealed that the majority of differential expression occurred at distinct stages in the two brain-stem regions studied. In the NTS, the microRNA expression of SHR was higher at the hypertension onset stage compared with controls (Fig. 2B). In the RVLM, the microRNA expression of SHR was lower at the prehypertension stage compared with controls (Fig. 3A). Additionally, 12 microRNAs demonstrated differential expression by stage that was consistent and overlapping between the two brain regions. Only one microRNA, miR-674-5p, was common between the RVLM and NTS with regard differential expression in SHR relative to WKY. Finally, two microRNAs, miR34a and miR-19b, were differentially expressed between SHR and WKY in a stage-dependent manner in both the NTS and the RVLM. All of the dynamically expressed, strain-dependent, differentially expressed microRNAs showed strong correlation at the hypertension onset stage and the chronic hypertension state, whereas they showed an inverse correlation between regions at the prehypertension stage, meaning that when a microRNA had high expression levels, its corresponding putative target has lower expression levels (Fig. 4).
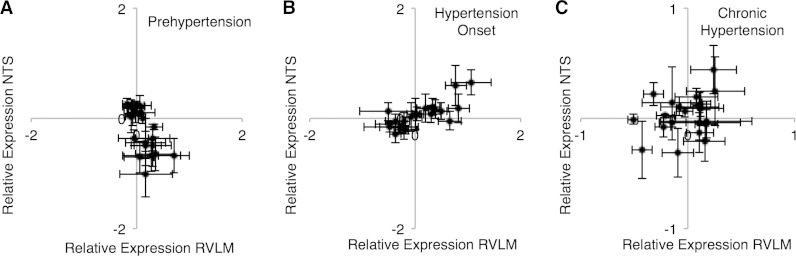
Fig. 4.
Comparison of differential microRNA expression between NTS and RVLM through multiple stages of hypertension development. A: prehypertension stage shows an inverse, inverse correlative relationship. B: onset of hypertension shows a correlative relationship. C: chronic hypertension shows no relationship.
Relating key microRNAs to potential gene regulatory targets with known functional impact on hypertension.
We prioritized a set of microRNAs that showed distinct temporal patterns between SHR and WKY for further analysis to relate these changes to target gene expression (Fig. 2A, I; B, I and Fig. 3A, I; B, I). We focused on potential target genes that are involved processes with known functional impact on development and maintenance of hypertension. We developed a list of 144 potential target transcripts that are associated with AT1R signaling through Pkc, CamkII, Mapk, and Pi3k pathways, immediate early genes (e.g., Fos, Egr1, Egr2, Egr3, Jun, Junb), and ion channels and transporters, guided by our previous studies (27, 34, 39, 40, 42, 51, 53, 54), as well as genes relevant to inflammatory pathways shown to affect blood pressure regulation, including interleukins, chemokines, and leukotrienes (19, 52, 60). Several of these pathways are enriched in astrocytes (e.g., inflammatory processes) compared with neurons (e.g., ion channels) compared with endothelial cells (e.g., junctional proteins), whereas the transcriptional regulators and signaling pathways may be common to all cell types (38).
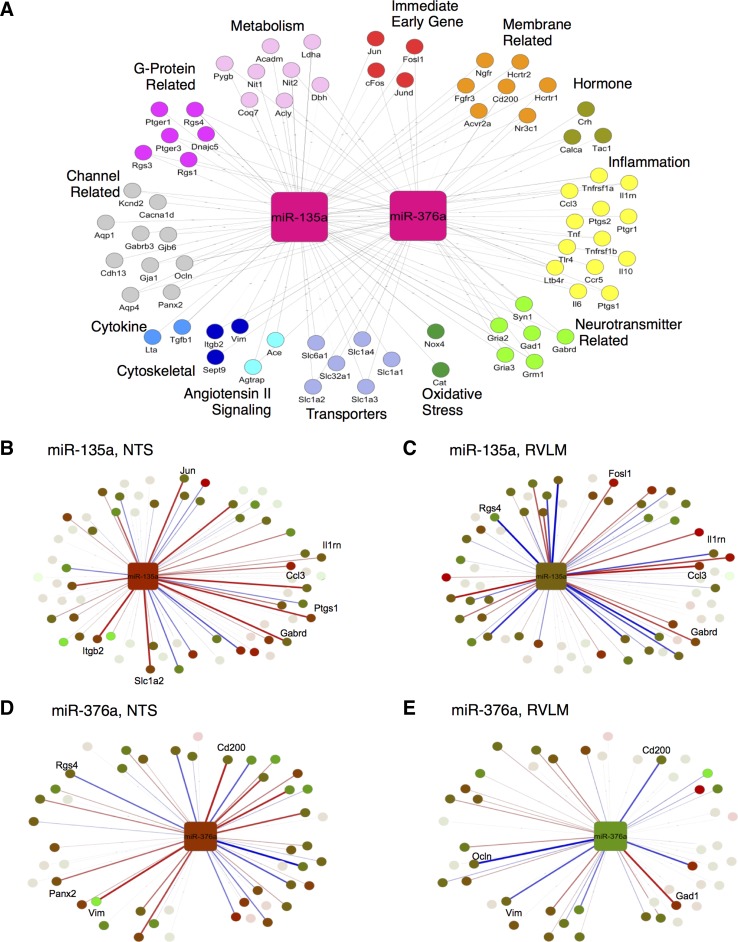
We pursued a combination of approaches for microRNA/mRNA target prediction, including use of miRWALK (18) database, RNA22 (35) tool for pattern matching, and literature-derived known functional interactions (19, 51, 56, 59, 61). Our results revealed that several of the hypertension-relevant functional processes could be targeted by the key differentially expressed microRNAs, yielding a putative microRNA/mRNA regulatory network (Supplemental Table S1). A total of 65 transcripts were predicted to be targets of miR-135a including key transcripts implicated in blood pressure regulation such as angiotensin II receptor antagonist protein (Agtrap), interleukin 1 receptor antagonist (Il1rn), and prostaglandin reductase 1 (Ptgr1). From the potential target transcript list, 45 transcripts were predicted to be targeted by miR-376a, including Agtrap, orexin receptors 1 and 2 (Hcrtr1, Hcrtr2), and chemokine receptor 5 (Ccr5).
High-throughput mRNA target expression level patterns vary by strain and brain region.
We evaluated the expression levels of the 144 prioritized mRNA targets for differential levels that may be consistent with the observed dynamics of microRNA regulation. We employed a high-throughput real-time PCR approach to assay expression of 144 transcripts in 45 samples using the Fluidigm BioMark platform. In the NTS, 24 transcripts were differentially expressed in SHR compared with WKY, and 21 transcripts showed a stage-dependent response. Two transcripts, Tnf and Ccr5, demonstrated stage-dependent differential expression in SHR relative to WKY (Fig. 5). In the RVLM, seven transcripts showed a strain-dependent differential expression, and 41 transcripts were differentially expressed in a stage-dependent manner. Seven transcripts were significantly differentially expressed over time in SHR vs. WKY (Slc1a4, Tlr4, Rgs1, Gabrd, Ace, Nox4, and Gad1) (Fig. 6).
Fig. 5.
NTS differentially expressed transcripts show strain-dependent and stage-dependent differential expression. A: differentially expressed transcripts show stage-dependent differential expression independent of strain (I); strain-dependent differential expression independent of stage, which clusters into 2 groups based on Pearson correlation (II); and strain-dependent dynamic regulation (III). P < 0.05. B: summary of transcripts differentially expressed in strain-, stage-, as well as strain-dependent stage effects.
Fig. 6.
RVLM differentially expressed transcripts show strain-dependent and stage-dependent differential expression. A: differentially expressed transcripts show stage-dependent differential expression independent of strain (I); strain-dependent differential expression independent of stage, which cluster into 2 groups based on Pearson correlation (II); and strain-dependent dynamic regulation (III). P < 0.05. B: summary of transcripts differentially expressed in strain-, stage-, as well as strain-dependent stage effects.
In general, the gene expression patterns were distinct in NTS vs. RVLM, consistent with the observations in the microRNA expression profiles. In NTS, the gene expression levels were lower in SHR at the hypertension onset stage compared with controls (Fig. 5). In contrast, the RVLM gene expression level profiles did not show an obvious grouping based on stage-specific response (Fig. 6).
Key differentially expressed microRNAs and putative transcriptional targets comprise regulatory networks contributing to hypertension.
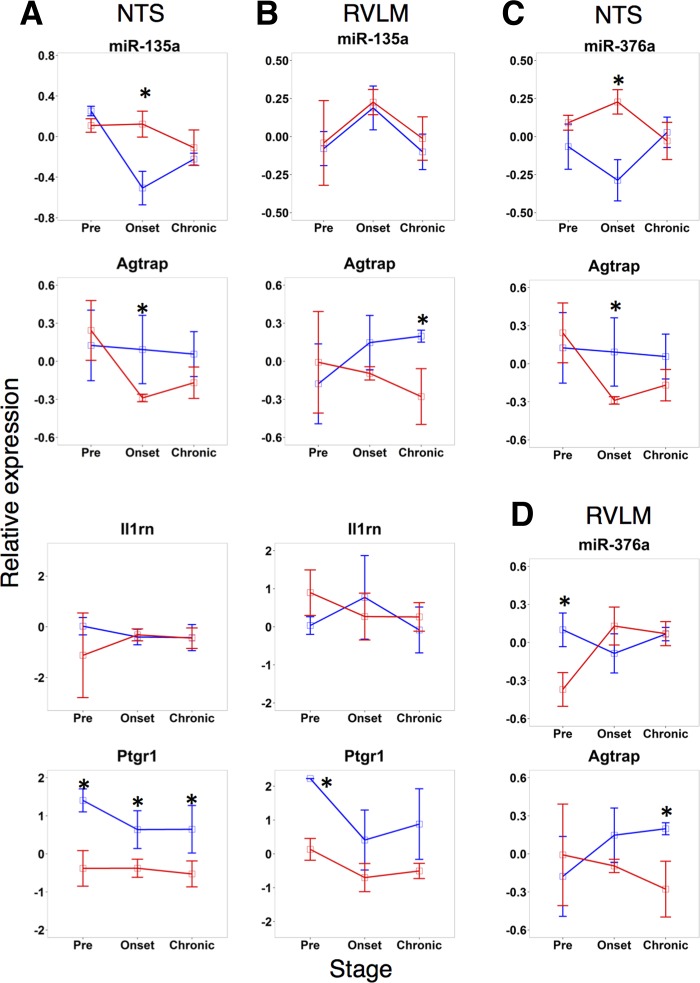
Of the stage and strain-dependent microRNAs (NTS: Fig. 2A, 1; RVLM: Fig. 3B, I), two were common between these regions. Interestingly, these two microRNAs were shown to be enriched in different cell types: miR-135a in astrocytes and miR-376a in neurons (25). We focused the subsequent analysis on this pair of microRNAs and their putative targets. We observed that there were correlational relationships between microRNA and predicted transcript targets when we examined the expression of microRNA-putative target pairs (Fig. 7). Therefore, we assessed all of the microRNA-putative target transcript correlations, and we filtered the microRNA/mRNA network (Supplemental Table S1) to focus on the microRNAs with strain-specific and stage-dependent differential response and their predicted targets (Fig. 8A). We mapped the differential expression data as well as the linear correlation between microRNA expression and target transcript levels on to the postulated regulatory network. We filtered the results to include microRNA/mRNA interactions corresponding to absolute correlation values >0.4. Our analysis revealed a similar extent of positive vs. negative correlation between the microRNA and predicted target transcript patterns. Correlations across SHR animals differed significantly compared with those of WKY animals. Ultimately, we discovered a pair of microRNAs whose regulatory network showed differences in correlations in hypertension compared with control (Fig. 8). Of note are the inflammation transcripts in NTS for microRNA-135a. There appears to be a much stronger inversely correlative relationship between miR-135a and these transcripts in SHR compared with WKY (Fig. 8B).
Fig. 7.
Comparison of microRNA expression patterns with that of the predicted targets by RNA22 and miRWALK algorithms. Key microRNAs plotted with transcripts predicted to be a target of its respective microRNA. A: miR-135a and its predicted target Il1rn show opposite behavior in SHR NTS as the pre and onset stages. B: miR-135a shows anticorrelated behavior with target Agtrap in SHR in NTS and RVLM at the preonset stage and in RVL WKY at the preonset stage. C: miR-376a shows expression levels opposite from target PTGR1. D: miR-376a shows inversely correlated behavior with target Agtrap in SHR NTS at the preonset stage.
Fig. 8.
Network representation of microRNA. A: putative target mapped to pathway annotations derived from literature and gene ontology. Key microRNAs at the hypertension onset stage in NTS of miR-135a regulatory network (B) and miR-376a regulatory network (D). Key microRNAs at the prehypertension stage in RVLM of miR-135a regulatory network (C) and miR-376a regulatory network (E). Edges are mapped to correlation data differences of SHR-WKY. Blue, positive; red, negative. Line connections are present if the transcript is a predicted target of a microRNA with line thickness representing relative strength of correlation subtraction (−2, 2).
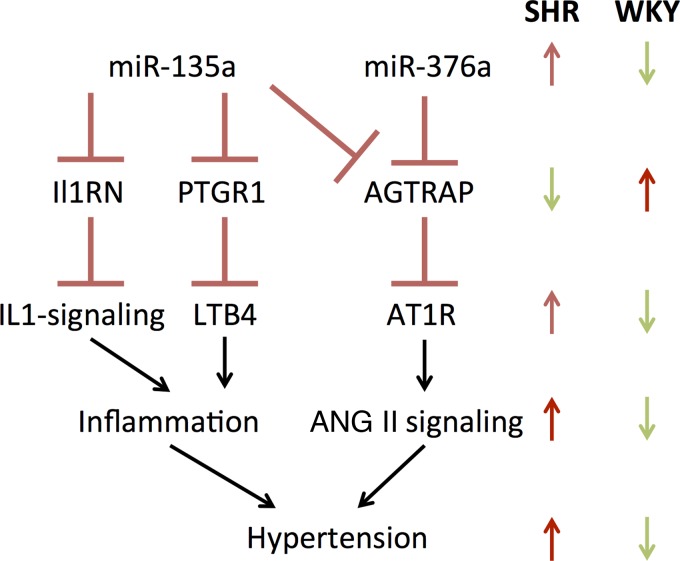
Based on this analysis in both NTS and RVLM, we constructed a model to show how microRNAs affect hypertension. We focused on a pair of microRNAs, miR-135a and miR-376a, which were 1) differentially expressed between SHR and WKY, in both NTS and RVLM, 2) predicted to target transcripts that play a key role in the development of hypertension, and 3) inversely correlated in expression with those key putative targets (Fig. 9). Our model includes miR-135a mediated putative downregulation of Ptgr1 transcript. The corresponding protein, PTGR1, degrades LTB4 and, hence, serves as a negative regulator of leukotriene-mediated inflammation. Similarly, our model includes miR-135a-mediated putative downregulation of Il1rn transcript. The corresponding protein IL-1RA serves as a negative regulator of IL-1-mediated inflammation. In addition, our model incorporates putative downregulation of Agtrap transcript by miR-135a and miR-376a, likely leading to disinhibition of AT1R signaling.
Fig. 9.
Literature-pruned subset of larger data-driven microRNA regulatory network depicted previously captures the interactions relevant to well-established SHR literature focusing on angiotensin II and inflammatory signaling. The network relates key microRNAs either persistently upregulated or showing a larger rise in SHR than in WKY during the developmental phase of hypertension. A common feature of this network is that the 2 key microRNAs are predicted by a consensus of bioinformatics tools to target negative regulators of pathways that are amplified in SHR during the development of hypertension.
DISCUSSION
To our knowledge this is the first study that has examined microRNAs and target gene expression in a high-throughput manner from the NTS and RVLM over the course of development of hypertension. We used the SHR animal model that closely mimics several features of the development and chronic state of human hypertension. Through an unbiased global microRNA expression analysis, we found extensive differential expression of microRNAs in these two key brain-stem regions during the development of hypertension. We correlated these changes to their putative gene expression targets with a focus on how the changes evolve over time (dynamics). We developed a regulatory network model that connects changes in microRNA expression to modulation of key transcript levels participating in inflammation and ANG II signaling to drive hypertension development. The network contains a novel double-negative regulatory motif in which upregulated microRNAs likely downregulate inhibitors of inflammation and ANG II signaling processes.
The recent literature offers abundant evidence for “neurogenic hypertension” in which the central neuronal mechanisms of blood pressure regulation play a key role, in particular autonomic structures in the brainstem, the NTS and the RVLM. Fortunately, current evidence indicates that hypertensive strains of rats and susceptible humans have much in common, both phenotypically and genotypically with respect to neurogenic contributions to hypertension (5, 28, 31, 51), and the SHR is by far the most widely studied animal model of hypertension (44). The SHR has the advantage of hypertension developing from a young age over several weeks, which permits study of transcriptional changes in the prehypertensive age, thereby separating effects of chronic high blood pressure from transcriptional processes that may lead to hypertension during the prehypertension stage. Thus, temporal profiling addresses the cause and effect regarding the association between transcript changes and blood pressure. The animal model supports the time-series study of developmental vs. chronic gene expression changes, which is beneficial for clarifying an optimal therapeutic intervention time point.
Our results highlight the stage-dependent dysregulation of molecular networks in the development of hypertension. These findings are consistent with observations indicating a stage-dependent role for ANG II signaling processes in driving hypertension. Blocking ANG II-mediated AT1R signaling at a young age (4–8 wk of age) in an SHR model, corresponding to intervention at the prehypertension stage, prevented development of hypertension even up to 48 wk later (6). These results, together with our findings, implicate the prehypertensive stage as a key period in the development of hypertension. Our results uncovered a microRNA-mediated regulation as potentially underlying this stage-dependent sensitivity. The microRNAs identified in our study could serve as novel targets for manipulation to influence the ANG II signaling and interfere with the development of hypertension.
Our results suggest that microRNA regulation and downstream effects on putative target transcript levels vary by brain-stem region during development of hypertension. RVLM showed differential microRNA expression prior to the onset of hypertension, at an earlier stage than was observed in the NTS, which showed microRNA expression changes during the hypertension onset stage. The functional consequences of these distinct stage-dependent responses need to be interpreted through the location and role of these brain-stem nuclei in the blood pressure control circuit. The NTS is known to be the primary site of cardiorespiratory regulatory integration (1, 8, 9, 13, 33, 46, 47). The RVLM receives inhibitory projections from the caudal ventrolateral medulla. Disruption of these inhibitory projections leads to the development of hypertension (3, 9, 10, 15, 20, 22, 24). The RVLM is crucial in tonic and reflexive regulation of arterial pressure, and it has been shown to contribute to elevated sympathetic nerve activity and mean arterial pressure in obese Zucker hypertensive rats (24). Studies have shown that RVLM-enhanced angiotensinergic activation and reduced GABAergic inhibition contribute to hypertension in these rats, and the low levels of microRNAs observed in our results are consistent with putative microRNA-mediated regulation of these processes (9, 24, 37). Our results suggest that by the time increases in mean arterial pressure are seen in the hypertension onset stage, it is likely that the aberration in the molecular networks have already occurred in the RVLM. It is interesting to speculate whether the processes that are disrupted earlier in RVLM lead to changes in the microRNA expression and corresponding dysregulation of transcript levels in the NTS at a later stage. In that regard, a question arises as to whether the NTS microRNA network expression changes at hypertension onset are compensatory or are further advancing the pathology leading to aberrant wiring across the baroreflex control circuit.
We interpret the microRNA and putative target gene expression correlations observed in our results as signatures of the underlying mechanistic relationships that are candidates for further experimental testing. Our results show that the microRNA and putative target gene expression correlations differ significantly between the SHR and WKY animals, indicating differences in the underlying regulatory networks between the two genotypes. We prioritized a subset of these relationships based on inverse correlation between microRNAs and putative targets for constructing a regulatory network. The positive correlations observed in our results could arise due to multiple possibilities. For instance, microRNAs have been shown to upregulate certain targets by stabilizing the mRNAs (41). Additionally, the microRNAs could be affecting transcript levels in a positive manner indirectly by downregulating a negative regulator of the transcript.
To examine the effects of microRNA changes on the molecular pathways implicated in hypertension, such as ANG II signaling and inflammation, we employed bioinformatics analyses to predict regulatory targets corresponding to these pathways. From the cohort of differentially expressed microRNAs, we examined two cell type specific microRNAs in additional detail: miR-135a, which is enriched in astrocytes, and miR-376a, which is enriched in neurons (25). Given the expected cell-type enrichment, it is important to localize the correlations between microRNAs and putative targets to specific cell types, as these may be masked or altered when considering tissue-scale samples that we employed in the present study. Literature evidence points to likely cell-type specificity of these pathways, for example, AT1R signaling in neurons and leukotriene metabolism in astrocytes (38, 56). Both microRNAs showed significant differences in expression in NTS and RVLM and are predicted to target transcripts and networks associated with high blood pressure. We developed a regulatory network model containing these two microRNAs and putative target transcripts (Fig. 6).
miR-135a is particularly enriched in astrocytes relative to neurons, oligodendrocytes, and microglia (25). Our target prediction and expression correlation analysis revealed that miR-135a is likely to act by downregulating Ptgr1 transcript, which has been shown to be downregulated in adult SHR rats compared with WKY rats (56), which is consistent with the higher expression of miR-135a observed in our data. Thus, we have hypothesize that miR-135a downregulates the Ptgr1 expression to increase the levels of a key proinflammatory leukotriene, LTB4, likely leading to the development of hypertension. High levels of LTB4 in the NTS have been shown to be prohypertensive, and blocking LTB4 receptor resulted in lowered arterial pressure in SHR (56). In RVLM, proinflammatory cytokines have been shown to be elevated (2), and microinjecting pentoxifylline, an anti-inflammatory drug whose mechanism is partly mediated through leukotriene inhibition, has been shown to lower blood pressure in LPS-induced hypertensive rats (59). Our analyses also pointed to additional routes of influence via miR-135a downregulation of Il1rn, a key anti-inflammatory regulatory player. IL-1RN has been shown to exhibit an anti-inflammatory effect via IL-1 signal attenuation (21). Furthermore, the levels of IL-1, a proinflammatory molecule, are higher in the brain stems of 22 wk old SHR compared with age-matched WKY (51). Additional human studies have also found that Il1rn has single nucleotide polymorphisms that contribute to acute coronary syndrome, which further implicates this gene in cardiovascular disease (19). In our results, Il1rn is expressed at higher levels in WKY than in SHR. We predict that the downregulation of Il1rn transcript via miR-135a contributes to the development of hypertension by disinhibiting an inflammatory signal mediated through IL-1. Another hypothesized route of miR-135a influence predicted by our computational analysis is through modulation of ANG II signaling. NTS contains the highest amount of ANG II receptors in the medulla, and this is evolutionarily conserved (4, 14, 23, 30). RVLM also contains a high amount of ANG II receptors compared with the rest of the medulla (22, 23). miR-135a putative mediation of ANG II signaling occurs via downregulation of Agtrap, angiotensin II receptor-associated mRNA. AGTRAP protein is a key downregulator of the ANG II receptor type 1 (17). Based on this target prediction and expression correlation, we postulate that miR-135a may be downregulating a negative regulator AT1R, therefore increasing ANG II signal transduction, leading to blood pressure elevation.
In contrast to miR-135a, miR-376a is highly expressed in neurons relative to astrocytes, microglia, and oligodendrocytes (25). miR-376a was one of the highly expressed microRNAs in our results with whole tissue samples. Based on the target prediction analysis and transcript and microRNA expression correlation results, we hypothesize that the neuron-enriched miR-376a could act via targeting Agtrap to disinhibit ANG II signaling in NTS. In the rat transcript of Agtrap, there appears to be only one miR-376a predicted binding site in the 3′-UTR beginning at the 5′-end nucleotide 1646 based on RNA22 predictions; however, in the human there is one in the 3′-UTR beginning at 936 from 5′-end of the transcript and an additional predicted binding site in the 5′-UTR beginning at nucleotide 53 from the 5′-end on the transcript (Table 1). miR-135a has more predicted binding sites on the Agtrap transcript than does miR-376a. miR-135a has two predicted target sites on Agtrap, both in the 3′-UTR beginning at nucleotide 1125 and 1509 from 5′-end. In contrast, the human Agtrap transcript has five predicted target sites for miR-135a-1, all of which are located in the 3′-UTR at locations 714, 749, 839, 906, and 961 from 5′-transcript end. Notably, there is no competition between these two microRNAs to bind Agtrap, as none of the predicted binding sites overlap.
Table 1.
Summary of predicted binding sites of key pairs of microRNAs in human and rat
|
Homo sapiens |
Rattus norvegicus |
||||||
|---|---|---|---|---|---|---|---|
| microRNA | Gene Target | Predicted Sites, n | Region | Transcript Location | Predicted Sites, n | Region | Transcript Location |
| miR-376a | Agtrap | 2 | 5′-UTR | 53–74 | 1 | 3′-UTR | 1646–1668 |
| 3′-UTR | 936–959 | ||||||
| miR-135a | Agtrap | 5 | 3′-UTR | 714–726 | 2 | 3′-UTR | 1125–1151 |
| 3′-UTR | 749–770 | ||||||
| 3′-UTR | 839–852 | ||||||
| 3′-UTR | 906–918 | 3′-UTR | 1509–1531 | ||||
| 3′-UTR | 961–980 | ||||||
| miR-135a | Ptgr1 | 2 | CDS | 573–586 | 1 | CDS | 82–104 |
| 3′-UTR | 1215–1232 | ||||||
| miR-135a | II1rn | 2 | 5′-UTR | 14–33 | 1 | 3′-UTR | 1785–1807 |
| CDS | 544–561 | ||||||
UTR, untranslated region; CDS, coding sequence.
Our results also highlight several additional microRNAs differentially expressed between the SHR and WKY rats and correlated with their downstream transcript targets, providing a cadre of putative microRNA regulated pathways underlying the development of hypertension. Our correlation analysis pointed to several putative influences of microRNA changes aside from the canonical direct targeting via seed region base pairing in the 3′-UTR. For instance, miR-135a expression was inversely correlated with that of Ace, which was not predicted to be a direct target of miR-135a in our analysis. This suggests a regulatory network interaction involving multiple intermediate steps, or alternatively, potential novel direct targeting of Ace by miR-135a.
Additional experiments involving manipulation of microRNAs in disease models are required to further develop the putative target transcript correlations into a mechanistic causative role for microRNAs in the development of hypertension. Our results identify several microRNAs, including those with a cell type-specific role, for prioritization in such follow-on experimental studies. We would propose miR-135a and 376a as an initial point for future functional studies as we predict they are positioned to act as “hubs” or “master regulators” for several relevant transcripts that may drive development of cardiovascular disease in SHRs. Additional opportunities for investigation include analysis of other neuroanatomical regions participating in blood pressure regulation, such as the caudal ventrolateral medulla and the paraventricular nucleus of the hypothalamus. Understanding the dynamics of microRNA-mediated changes across the nuclei in the blood pressure control circuit would yield new targets for testing for effects on prevention as well as potential rescue from the hypertensive condition.
GRANTS
The work presented here was funded through National Institutes of Health Grants R01 GM-083108 and R01HL-111621.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: D.D., H.Z., and A.B. performed experiments; D.D. and R.V. analyzed data; D.D., J.S.S., and R.V. interpreted results of experiments; D.D. and R.V. prepared figures; D.D. drafted manuscript; D.D., J.S.S., and R.V. edited and revised manuscript; D.D., H.Z., A.B., J.S.S., and R.V. approved final version of manuscript; J.S.S. and R.V. conception and design of research.
Supplementary Material
ACKNOWLEDGMENTS
D. DeCicco acknowledges Warren A. Anderson for guidance with comparative network analysis and Sirisha Achanta for assistance in conducting initial high-throughput qPCR experiments.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Affleck VS, Coote JH, Pyner S. The projection and synaptic organisation of NTS afferent connections with presympathetic neurons, GABA and nNOS neurons in the paraventricular nucleus of the hypothalamus. Neuroscience 219: 48–61, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal D, Welsch MA, Keller JN, Francis J. Chronic exercise modulates RAS components and improves balance between pro- and anti-inflammatory cytokines in the brain of SHR. Basic Res Cardiol 106: 1069–85, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal SK, Calaresu FR. Reciprocal connections between nucleus tractus solitarii and rostral ventrolateral medulla. Brain Res 523: 305–308, 1990. [DOI] [PubMed] [Google Scholar]
- 4.Allen AM, McKinley MJ, Oldfield BJ, Dampney RA, Mendelsohn FA. Angiotensin II receptor binding and the baroreflex pathway. Clin Exp Hypertens A 10, Suppl 1: 63–78, 1988. [DOI] [PubMed] [Google Scholar]
- 5.Atanur SS, Birol I, Guryev V, Hirst M, Hummel O, Morrissey C, Behmoaras J, Fernandez-Suarez XM, Johnson MD, McLaren WM, Patone G, Petretto E, Plessy C, Rockland KS, Rockland C, Saar K, Zhao Y, Carninci P, Flicek P, Kurtz T, Cuppen E, Pravenec M, Hubner N, Jones SJM, Birney E, Aitman TJ. The genome sequence of the spontaneously hypertensive rat: analysis and functional significance. Genome Res 20: 791–803, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann M, Janssen BJA, Hermans JJR, Peutz-Kootstra C, Witzke O, Smits JFM, Struijker Boudier HAJ. Transient AT1 receptor-inhibition in prehypertensive spontaneously hypertensive rats results in maintained cardiac protection until advanced age. J Hypertens 25: 207–215, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Bhajun R, Guyon L, Pitaval A, Sulpice E, Combe S, Obeid P, Haguet V, Ghorbel I, Lajaunie C, Gidrol X. A statistically inferred microRNA network identifies breast cancer target miR-940 as an actin cytoskeleton regulator. Sci Rep 5: 8336, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campagnole-Santos MJ, Diz DI, Ferrario CM. Baroreceptor reflex modulation by angiotensin II at the nucleus tractus solitarii. Hypertension 11: I167–I171, 1988. [DOI] [PubMed] [Google Scholar]
- 9.Chan RK, Sawchenko PE. Organization and transmitter specificity of medullary neurons activated by sustained hypertension: implications for understanding baroreceptor reflex circuitry. J Neurosci 18: 371–387, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan YS, Wong TM. Relationship of rostral ventrolateral medullary neurons and angiotensin in the central control of blood pressure. Biol Signals 4: 133–141, 2014. [DOI] [PubMed] [Google Scholar]
- 11.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jones DW, Materson BJ, Oparil S, Wright JT, Roccella EJ. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension 42: 1206–1252, 2003. [DOI] [PubMed] [Google Scholar]
- 12.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colombari E, Sato MA, Cravo SL, Bergamaschi CT, Campos RR, Lopes OU. Role of the medulla oblongata in hypertension. Hypertension 38: 549–554, 2001. [DOI] [PubMed] [Google Scholar]
- 14.Dampney R, Fontes M, Hirooka Y, Horiuchi J, Potts P, Tagawa T. Role Of angiotensin II receptors in the regulation of vasomotor neurons in the ventrolateral medulla. Clin Exp Pharmacol Physiol 29: 467–472, 2002. [DOI] [PubMed] [Google Scholar]
- 15.Dampney RA, Polson JW, Hirooka Y, Horiuchi J. Functional organization of brain pathways subserving the baroreceptor reflex: studies in conscious animals using immediate early gene expression. Cell Mol Neurobiol 23: 597–616, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dávalos A, Chroni A. Antisense oligonucleotides, microRNAs, and antibodies. Hand Exp Pharmacol 224: 649–689, 2015. [DOI] [PubMed] [Google Scholar]
- 17.Daviet L, Lehtonen JY, Tamura K, Griese DP, Horiuchi M, Dzau VJ. Cloning and characterization of ATRAP, a novel protein that interacts with the angiotensin II type 1 receptor. J Biol Chem 274: 17058–17062, 1999. [DOI] [PubMed] [Google Scholar]
- 18.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk - Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44: 839–847, 2011. [DOI] [PubMed] [Google Scholar]
- 19.Fragoso JM, Delgadillo H, Llorente L, Chuquiure E, Juárez-Cedillo T, Vallejo M, Lima G, Furuzawa-Carballeda J, Peña-Duque MA, Martínez-Ríos MA, Vargas-Alarcón G. Interleukin 1 receptor antagonist polymorphisms are associated with the risk of developing acute coronary syndrome in Mexicans. Immunol Lett 133: 106–111, 2010. [DOI] [PubMed] [Google Scholar]
- 20.Geraldes V, Goncalves-Rosa N, Liu B, Paton JFR, Rocha I. Essential role of RVL medullary neuronal activity in the long term maintenance of hypertension in conscious SHR. Auton Neurosci 186: 22–31, 2014. [DOI] [PubMed] [Google Scholar]
- 21.Goehler LE, Relton JK, Dripps D, Kiechle R, Tartaglia N, Maier SF, Watkins LR. Vagal paraganglia bind biotinylated interleukin-1 receptor antagonist: a possible mechanism for immune-to-brain communication. Brain Res Bull 43: 357–364, 1997. [DOI] [PubMed] [Google Scholar]
- 22.Granata AR, Ruggiero DA, Park DH, Joh TH, Reis DJ. Brain stem area with C1 epinephrine neurons mediates baroreflex vasodepressor responses. Am J Physiol Heart Circ Physiol 248: H547–H567, 1985. [DOI] [PubMed] [Google Scholar]
- 23.Head GA, Saigusa T, Mayorov DN. Angiotensin and baroreflex control of the circulation. Braz J Med Biol Res 35: 1047–59, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Huber DA, Schreihofer AM. Altered regulation of the rostral ventrolateral medulla in hypertensive obese Zucker rats. Am J Physiol Heart Circ Physiol 301: H230–H240, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jovičić A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, Luthi-Carter R. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci 33: 5127–5137, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karapetsas A, Tokamani M, Kolettas E, Sandaltzopoulos R. Novel microRNAs as putative therapeutic targets in cardiovascular diseases. Curr Vasc Pharmacol [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 27.Khan RL, Vadigepalli R, McDonald MK, Rogers RF, Gao GR, Schwaber JS. Dynamic transcriptomic response to acute hypertension in the nucleus tractus solitarius. Am J Physiol Regul Integr Comp Physiol 295: R15–R27, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight J, Munroe PB, Pembroke JC, Caulfield MJ. Human chromosome 17 in essential hypertension. Ann Hum Genet 67: 193–206, 2003. [DOI] [PubMed] [Google Scholar]
- 29.Kontaraki JE, Marketou ME, Zacharis EA, Parthenakis FI, Vardas PE. MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: potential markers of target-organ damage. J Am Soc Hypertens 8: 368–3 75, 2014. [DOI] [PubMed] [Google Scholar]
- 30.Li YW, Polson JW, Dampney RA. Angiotensin II excites vasomotor neurons but not respiratory neurons in the rostral and caudal ventrolateral medulla. Brain Res 577: 161–164, 1992. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto K, Yamada T, Natori T, Ikeda K, Yamada J, Yamori Y. Genetic variability in SHR (SHRSR), SHRSP and WKY strains. Clin Exp Hypertens A 13: 925–938, 1991. [DOI] [PubMed] [Google Scholar]
- 32.Mauno V, Hannu K, Esko K. Proinflammation and hypertension: a population-based study. Mediators Inflamm 2008: 619704, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelini LC. The NTS and integration of cardiovascular control during exercise in normotensive and hypertensive individuals. Curr Hypertens Rep 9: 214–221, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Miller GM, Ogunnaike a B, Schwaber JS, Vadigepalli R. Robust dynamic balance of AP-1 transcription factors in a neuronal gene regulatory network. BMC Syst Biol 4: 171, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217, 2006. [DOI] [PubMed] [Google Scholar]
- 36.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 3: e194, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muratani H, Averill DB, Ferrario CM. Effect of angiotensin II in ventrolateral medulla of spontaneously hypertensive rats. Am J Physiol Regul Integr Comp Physiol 260: R977–R984, 1991. [DOI] [PubMed] [Google Scholar]
- 38.Okaty BW, Sugino K, Nelson SB. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS One 6: e16493, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, Brureau A, Kernan K, Starks A, Gulati S, Ogunnaike B, Schwaber J, Vadigepalli R. Inputs drive cell phenotype variability. Genome Res 24: 930–941, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J, Ogunnaike B, Schwaber J, Vadigepalli R. Identifying functional gene regulatory network phenotypes underlying single cell transcriptional variability. Prog Biophys Mol Biol 117: 87–98, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13: 271–282, 2012. [DOI] [PubMed] [Google Scholar]
- 42.Paton JFR, Waki H. Is neurogenic hypertension related to vascular inflammation of the brainstem? Neurosci Biobehav Rev 33: 89–94, 2009. [DOI] [PubMed] [Google Scholar]
- 43.Paul M, Poyan Mehr A, Kreutz R. Physiology of local renin-angiotensin systems. Physiol Rev 86: 747–803, 2006. [DOI] [PubMed] [Google Scholar]
- 44.Pravenec M, Křen V, Landa V, Mlejnek P, Musilová A, Šilhavý J, Šimáková M, Zídek V. Recent progress in the genetics of spontaneously hypertensive rats. Physiol Res 63, Suppl 1: S1–S8, 2014. [DOI] [PubMed] [Google Scholar]
- 45.Prokopec SD, Watson JD, Waggott DM, Smith AB, Wu AH, Okey AB, Pohjanvirta R, Boutros PC. Systematic evaluation of medium-throughput mRNA abundance platforms. RNA 19: 51–62, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers RF, Paton JF, Schwaber JS. NTS neuronal responses to arterial pressure and pressure changes in the rat NTS neuronal responses to arterial and pressure changes in the rat. Am J Physiol Regul Integr Comp Physiol 265: R1355–R1368, 1993. [DOI] [PubMed] [Google Scholar]
- 47.Rogers RF, Rose WC, Schwaber JS, Rogers RF, Rose C, Schwaber JS. Simultaneous encoding of carotid sinus pressure and dP/dt by NTS target neurons of myelinated baroreceptors Simultaneous Encoding of Carotid Sinus Pressure and dP/dt by NTS target neurons of myelinated aroreceptors. J Neurophysiol 76: 2644–2660, 1996. [DOI] [PubMed] [Google Scholar]
- 48.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378, 2003. [DOI] [PubMed] [Google Scholar]
- 49.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Meth 9: 1069–1076, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun L, Gao YH, Tian DK, Zheng JP, Zhu CY, Ke Y, Bian K. Inflammation of different tissues in spontaneously hypertensive rats. Sheng Li Xue Bao 58: 318–323, 2006. [PubMed] [Google Scholar]
- 52.Takagishi M, Waki H, Bhuiyan M, Gouraud S, Kohsaka A, Cui H, Yamazaki T, Paton JFR, Maeda M. IL-6 microinjected in the nucleus tractus solitarii attenuates cardiac baroreceptor reflex function in rats. Am J Physiol Regul Integr Comp Physiol 298: R183–R190, 2010. [DOI] [PubMed] [Google Scholar]
- 53.Vadigepalli R, Gonye GE, Paton JFR, Schwaber JS. Adaptive transcriptional dynamics of A2 neurons and central cardiovascular control pathways. Exp Physiol 97: 462–468, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waki H, Gouraud SS, Maeda M, Paton JFR. Specific inflammatory condition in nucleus tractus solitarii of the SHR: novel insight for neurogenic hypertension? Auton Neurosci 142: 25–31, 2008. [DOI] [PubMed] [Google Scholar]
- 55.Waki H, Gouraud SS, Maeda M, Raizada MK, Paton JFR. Contributions of vascular inflammation in the brainstem for neurogenic hypertension. Respir Physiol Neurobiol 178: 422–428, 2011. [DOI] [PubMed] [Google Scholar]
- 56.Waki H, Hendy EB, Hindmarch CCT, Gouraud S, Toward M, Kasparov S, Murphy D, Paton JFR. Excessive leukotriene B4 in nucleus tractus solitarii is prohypertensive in spontaneously hypertensive rats. Hypertension 61: 194–201, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Gong L, Tan Y, Hui R, Wang Y. Hypertensive epigenetics: from DNA methylation to microRNAs. J Hum Hypertens [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 58.Winklewski PJ, Radkowski M, Wszedybyl-Winklewska M, Demkow U. Brain inflammation and hypertension: the chicken or the egg? J Neuroinflamm 12: 85, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu KLH, Chan SHH, Chan JYH. Neuroinflammation and oxidative stress in rostral ventrolateral medulla contribute to neurogenic hypertension induced by systemic inflammation. J Neuroinflamm 9: 212, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida M, Watanabe Y, Yamanishi K, Yamashita A, Yamamoto H, Okuzaki D, Shimada K, Nojima H, Yasunaga T, Okamura H, Matsunaga H, Yamanishi H. Analysis of genes causing hypertension and stroke in spontaneously hypertensive rats: gene expression profiles in the brain. Int J Mol Med 33: 887–896, 2014. [DOI] [PubMed] [Google Scholar]
- 61.Zubcevic J, Waki H, Raizada MK, Paton JFR. Autonomic-immune-vascular interaction: an emerging concept for neurogenic hypertension. Hypertension 57: 1026–1033, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.