Abstract
Septa from sheep hearts at 130 days gestation, term, and 14-day-old lambs were used to model the changes in gene expression patterns during the perinatal period using Agilent 15k ovine microarrays. We used Bioconductor for R to model five major patterns of coexpressed genes. Gene ontology and transcription factor analyses using Webgestalt modeled the biological significances and transcription factors of the gene expression patterns. Modeling indicated a decreased expression of genes associated with anatomical development and differentiation during this period, whereas those associated with increased protein synthesis and growth associated with maturation of the endoplasmic reticulum rose to term but did not further increase from the near term expression. Expression of genes associated with cell responsiveness, for example, immune responses, decreased at term but expression returned by postnatal day 14. Changes in genes related to metabolism showed differential substrate-associated patterns: those related to carbohydrate metabolism rose to term and remained stable thereafter, whereas those associated with fatty acid oxidation facility rose throughout the period. The timing of many of these maturational processes was earlier in relation to birth than in the rodent. The importance of the transcription factors, estrogen-related receptors, and v-myc avian myelocytomatosis viral oncogene homolog was also highlighted in the pattern of gene expression during development of the perinatal sheep heart.
Keywords: transcriptomics, heart, perinatal ontogeny, lamb
the period of development from the last 2 wk of gestation through the first 2 wk of life spans a period of great functional and metabolic challenge to the fetal and neonatal lamb heart. Substantial maturation in form and function occur to meet these challenges. In late gestation cardiomyocytes cease replication, becoming progressively binucleated and enlarged, with increased width (8, 29). The mitochondria in cardiomyocytes undergo a maturation process, becoming more closely associated with sarcomeres, and form a network in association with cellular membranes. By term there are two clear populations of mitochondria, those around the poles of the nuclei and those associated with the myofibrils, although the process of mitochondrial network formation continues postnatally (7). The extracellular matrix (ECM) between cardiomyocytes becomes fine-tuned; the cardiomyocytes, already individually sheathed by collagen, are bundled into functional groups of eight cardiomyocytes and form different contacts with the ECM (7). The heart switches from utilization of glucose and lactate as the primary energy sources in utero, to utilization of lactate and fatty acids in the first few weeks after birth (4). This change in the efficiency of metabolism complements the changing prevailing tissue oxygen status and substrate supply, as well as functional changes in cardiac work. At birth as the shunts in the fetal circulation close, the biventricular pumping by the fetal heart, to provide systemic cardiac output against a relatively low arterial pressure, transitions to the two chambers acting in parallel, with increased afterload in the left heart, but decreased afterload for the right heart (61).
It has been appreciated for some time that cortisol, thyroid hormone (T3), the insulin-like growth factor family (IGF1 and IGF2), and estrogen-related receptors (ERRs) among others are important contributors driving the maturation of the heart during this time period (for review see Ref. 23). For example, plasma T3 concentration increases at about 135 days gestation. It is unclear whether other mechanisms also alter the heart over this time period. The increased plasma T3 has been implicated in the differentiation of the cardiomyocytes, slowing proliferation and stimulating binucleation (11) from 110 days gestation through term (29). Cortisol and IGFs have also been implicated in this process (11, 25, 68, 72), and both global glucocorticoid receptor (GR) knockout mice and cardiomyocyte and smooth muscle-specific knockout mice show significant deficits in cardiac function and anatomy (59).
It has been assumed that cortisol and T3 act primarily at glucocorticoid and thyroid hormone response elements, respectively. But it is not known if these hormones are the only maturing factors for the heart, nor whether they act at classical hormone receptors to mediate the maturation. Recently it has been shown that both glucocorticoids and thyroid hormones can act in several other ways, for example with membrane receptors that signal through second messenger systems (41) in hybrid receptor dimers with other nuclear-acting receptors directly binding to chromatin (18), by affecting the activity of coactivators and corepressors either by attracting and tethering to them or blocking their binding at functional sites on the genome (30, 62) or by binding to negative response elements (69). In the amphibian, the unliganded thyroid hormone receptor alpha has been shown to affect growth and maturation when thyroid hormone levels are low or absent (14, 73).
In this study gene expression microarray analyses were used to test two hypotheses: first, that distinct temporal patterns of gene expression patterns in the fetal and newborn heart will reflect the temporal patterns of maturation events related to proliferation, structural maturation, and metabolic changes in the heart over the last weeks of gestation and the first weeks of life; and second, that cortisol and/or thyroid hormones would be the most important transcription factors driving these patterns. We analyzed RNA isolated from the septum of developing sheep at 130 days, term, and 14 days postpartum, intervals of ∼14 days spanning late gestation and early postnatal life. The microarray analysis methods used allowed examination of temporal patterns of gene expression over this period of cardiac transition, analysis of the function of the groups of genes with each particular pattern of expression, and the transcription factors underpinning the changes in gene expression.
MATERIALS AND METHODS
Animal treatment.
All animal use in this study was approved by the Institutional Animal Care and Use Committee of the University of Florida. The animals used for the experiments at 130 days gestation (n = 6) and at term (n = 8) were the control animals in previous publications (31, 56). The ewes were euthanized (Euthasol; Fort Dodge, IA); the fetuses removed; and sections of the septum dissected, frozen in liquid nitrogen, and stored at −80°C until RNA extraction was performed (21, 56). Lambs (n = 8, 4 male and 4 female) were born naturally and lived with their mothers in a temperature- and light-controlled room in the animal care facility for 14 days. The lambs were monitored every 12 h for 2 days after birth and then daily for weight, temperature, and plasma hormones, and every 3 days for electrolyte and hormone concentrations via a jugular catheter placed on the day of birth. Lambs were also subjected to a glucose tolerance test or insulin tolerance tests on days 5, 7, and 10 after birth (data reported elsewhere). The lambs were treated with antibiotics postnatally (Polyflex; Fort Dodge, IA at 12.5–15 mg/kg bid and/or Naxcel, 1 mg/kg sid). At 14 days of age the lambs were euthanized, the heart removed, the septum dissected, and samples of tissue frozen and stored as described for the fetal heart tissues. We chose to use samples of septum in this study so as to include genes related to both myocyte and Purkinje fiber maturation.
RNA extraction, labeling, and microarray hybridization.
These protocols have been described in detail for the 130-day gestation (n = 6) and term hearts (n = 8) (56); the same protocol was used for the 14-day-old samples (n = 8). RNA was extracted with Trizol (Invitrogen), further purified and DNase-treated with Qiagen RNeasy+ kits, and labeled with Cy3-dCTP dye using Agilent one-color labeling kits. The labeled cRNA was fragmented, hybridized to Agilent 8 × 15k microarray slides (ovine 019221 arrays; G4813A, GPL14112 platform), washed, and scanned at the Interdisciplinary Center for Biotechnology Research core at the University of Florida. The annotation of this array platform was described in a previous publication from our laboratory group (54). The raw normalized array data for this experiment have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE66725 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66725).
Microarray analyses.
The microarray data were imported into R using the limma package for background correction and data normalization using the quantile normalization method (67). Approximately 4,000 probes were excluded from analysis because of low intensity on the arrays, reflective of low gene expression levels. Gene collapsing using the weighted gene coexpression network analysis (WGCNA) package (34) was performed to obtain the mean intensity of probes that were repeated on the array and/or shared the same gene symbol, after these adjustments 7,345 genes were uniquely identified. Unsupervised analyses using principal component analysis (PCA, prcomp function-R) and hierarchical clustering analysis [Cluster 3.0 software, (16)] were performed to detect major patterns of change in gene expression within these data. The normalized, background-corrected, unique gene intensity data were statistically analyzed using the Bayesian estimation of temporal regulation (BETR) method with an false discovery rate of 0.05 to detect differentially regulated (DR) genes (3). This returned the probabilities of differential gene expression for each gene in the data set; those genes with a probability of >99.999 were subjected to further analysis.
Then patterns of changed coexpressed genes over time within the DR genes were detected using the WGCNA package for R. The automatic method was employed for block-wise network construction and detection of clusters. The resulting clusters were related with gestational age to identify clusters of coexpressed genes with different expression patterns over time (34).
Each cluster of genes with a similar pattern of gene expression was modeled in Webgestalt (71, 75) using gene ontology terms to identify significantly enriched biological processes, molecular functions, and cellular compartments associated with them as previously described (56). For the present analysis the identified genes on the ovine microarray platform was used as the reference set. Some of the known factors contributing to the maturation of the heart, such as T3 receptors and ERRs, are transcription factors. Therefore, analysis of microarray results with Webgestalt was used to discover transcription factor targets among the DR genes in each expression pattern. Webgestalt uses the Broad Institute's molecular signatures database that uses TRANSFAC gene sets. These gene sets comprise genes that share a transcription factor binding site in the −2 to +2 kb around the transcription start site. There are ∼615 transcription factor target gene sets in the database. We used the human genome as the reference set in these analyses. This allowed identification of transcription factors targets overrepresented in each temporal expression pattern to suggest transcription factors that might be important in regulating the DR genes. The transcription factors associated with each pattern of gene expression were modeled to discern both the timing of the actions of these known factors and the involvement and timing of other transcription factors. Finally protein-protein interaction modeling was performed with Webgestalt to indicate biologically important associations and to refine the modeling of the biological processes associated with the five temporal patterns of coexpressed DR gene expression. For analysis Homo sapiens gene symbols were used for the corresponding ovine genes; therefore, some DR gene sequences were not included in the subsequent analyses, as no human homolog has been identified for them. In this study they represented <2.2% of the genes.
Histology.
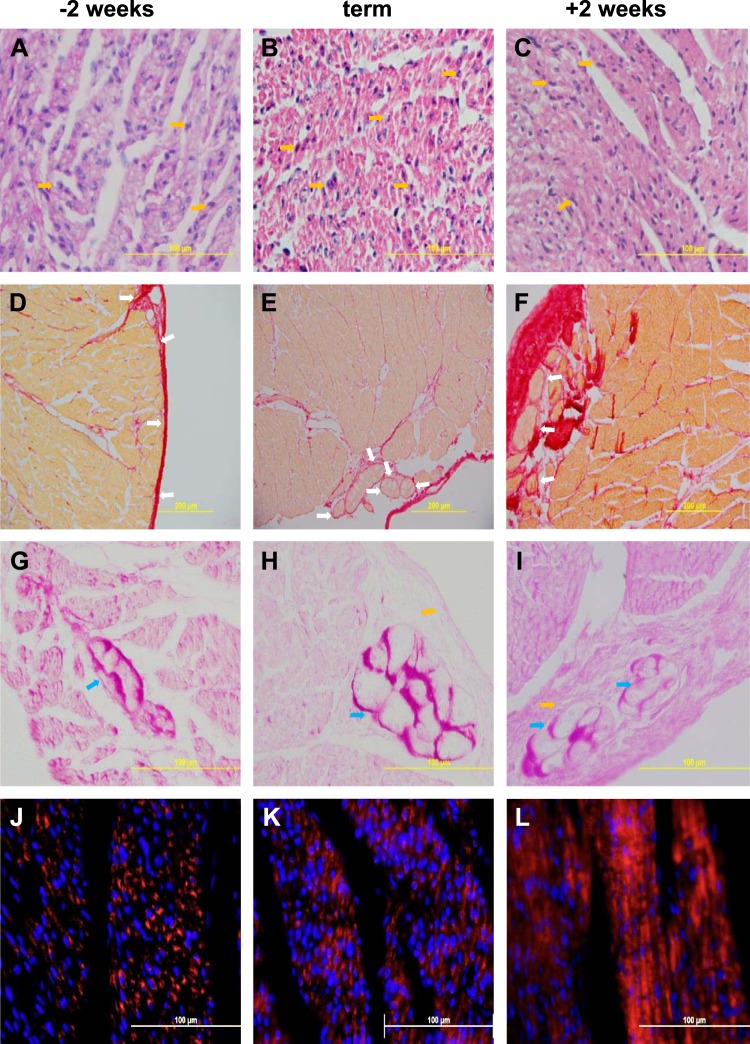
Histology was performed to confirm some of the processes modeled in our transcriptomic analyses. Cross sections of hearts (∼6 mm thick) were cut ∼10–15 mm from the apex of the heart. Sections were fixed in 4% phosphate-buffered paraformaldehyde for 24 h before long-term storage in 70% reagent alcohol. The tissue was embedded in paraffin wax and sectioned at 5 μm. Picrosirius red staining for collagen was performed as previously described (55), as was hematoxylin-and-eosin staining (21). Cytochrome c oxidase IV (COX IV) staining was performed with rabbit monoclonal anti-COX IV antibody (Cell Signaling #4850) at 1:150 dilution for 2 h at room temperature following antigen retrieval in a pressure cooker in citrate buffer and Bloxall and 5% normal horse serum treatment to prevent nonspecific staining. The secondary antibody was Cy5-labeled goat anti-rabbit IgG (Molecular Probes AlexaFluor 647 #A21245) used at 10 μg/ml for 2 h at room temperature. Hoechst 33342 at 1 μg/ml was added to the first wash step (10 min) after the secondary antibody step. The slides were mounted in Fluoromount G, and the septa imaged on an Olympus BX 41 microscope with a DP 71 camera with appropriate filters. The merged blue and red images were adjusted to decrease the blue intensities to 50% to correct for the difference in intensities of the two stains.
RESULTS
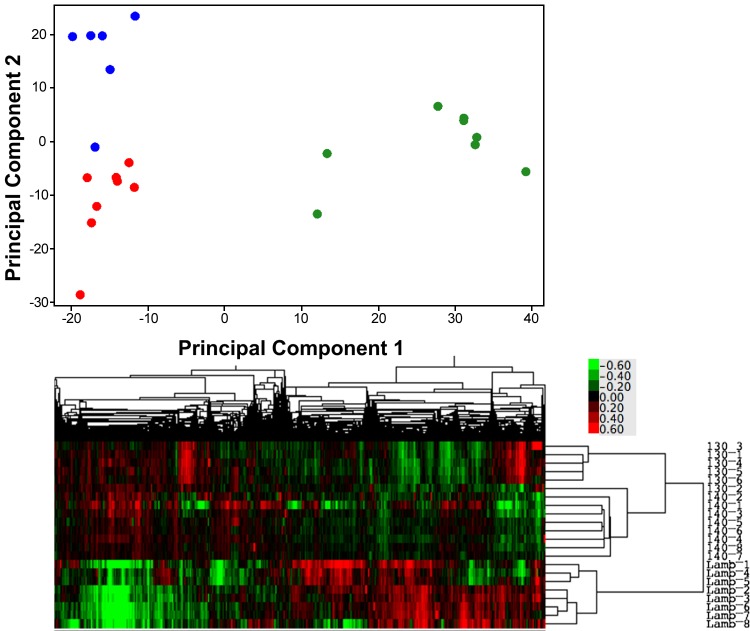
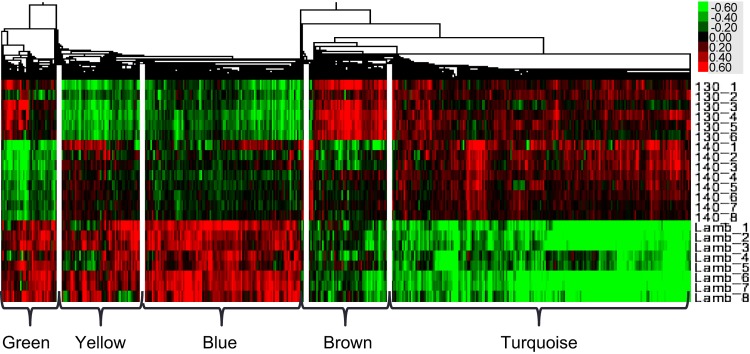
Unsupervised PCA of the normalized gene expression distribution showed that the principal component of variability separated two groups within the samples, the lambs, and the fetal ages; the second principal component of the variability separated the two gestational ages (130-days gestation and term, Fig. 1). The separation of the groups within the data can also be visualized in the heat map in Fig. 1. Of 7,345 unique genes, 1,763 genes were identified as DR by BETR in the septum between fetuses at 130-days gestation, fetuses at term, and lambs at 14 days of life. The 1,763 DR genes could be broadly grouped into five categories based on the pattern of gene coexpression changes over the perinatal period. For simplicity of discussion, these patterns of gene expression have been color-coded (as shown in Fig. 2). Four genes (ADCY9, COQ3, PDE3B, and WBSCR16) were differentially expressed but were not coexpressed with other genes and so were not analyzed any further. The patterns of gene expression will be described in a quasi-chronological pattern, beginning with patterns with the highest expression in gestation, followed by those that were highest in extrauterine life, and finally finishing with a pattern that dipped at term but was higher at the other times. The top 10 transcription factors modeled for each pattern of gene expression, using the human genome as the reference set, are seen in Table 1, along with the findings, if any, for the transcription factors GR, thyroid hormone receptor (T3R), and ERR1. See Supplemental Table S2 for a complete list of transcription factors significantly (P < 0.05) associated with each coexpression pattern in Webgestalt analyses.1
Fig. 1.
Top: principal component plot showing the distribution of gene expression in cardiac septum along the first 2 principal components of the variance in the data identified in the unsupervised analysis of the normalized, background corrected gene expression levels detected on the microarrays. 130-days gestation = blue, term = red, and 2 wk old = green circles. Bottom: heat map representation of the unsupervised gene expression data for each animal, the closer their data are on the y-axis, the more closely related their gene expression levels. 130_1 through 130_6 are the 6 130-day gestation samples, 140_1 through 140_8 the 8 term samples, and Lamb_1 through Lamb_8 are the 2-wk-old samples. The colored scale for the heat maps, bottom right, shows the fold change in expression levels of the gene from the normalization point, where green is decreased, and red increased, expression.
Fig. 2.
Heat map showing the clusters of covarying gene expression detected in the expression data by WGCNA analysis, labeled with colors to represent the different patterns of gene coexpression over time. 130_1 through 130_6 are the 6 130-day gestation samples, 140_1 through 140_8 the 8 term samples, and Lamb_1 through Lamb_8 are the 2 wk old samples. The colored scale for the heat maps, top right, shows the fold change in expression levels of the gene from the normalization point, where green is decreased, and red increased, expression.
Table 1.
Top 10 transcription factors and GR, T3R, and ERR1 identified by Webgestalt analysis for each gene expression pattern
| Name | Sequence | #Genes/Total | Adj. P Value | |
|---|---|---|---|---|
| Brown | ||||
| 1 | NFAT | TGGAAA | 27/1,871 | 4.5e−10 |
| 2 | SPI | GGGCGGR | 31/2,891 | 7.3e−9 |
| 3 | LEF1 | CTTTGT | 24/1,939 | 7.3e−8 |
| 4 | unknown | AACTTT | 23/1,859 | 1.3e−7 |
| 5 | unknown | RNGTGGGC | 15/758 | 2.4e−7 |
| 6 | MAZ | GGGAGGRR | 24/2,250 | 6.7e−7 |
| 7 | CEBPGAMMA | 9/252 | 2.1e−6 | |
| 8 | FOXO4 | TTGTTT | 21/2,037 | 8.4e−6 |
| 9 | AREB6 | CAGGTA | 13/780 | 1.2e−5 |
| 10 | EGR1 | 8/266 | 3.14e−5 | |
| GR | 6/276 | 0.001 | ||
| T3R | 4/248 | 0.014 | ||
| ERR1 | TGACCTY | 8/1,023 | 0.017 | |
| Turquoise | ||||
| 1 | SPI | GGGCGGR | 182/2,891 | 3e−44 |
| 2 | MAZ | GGGAGGRR | 133/2,250 | 1.3e−28 |
| 3 | LEF1 | CTTTGT | 112/1,939 | 5.5e−23 |
| 4 | EI2 | CAGGTC | 124/2,450 | 8.6e−21 |
| 5 | ELK1 | SCGGAAGY | 80/1,176 | 2.2e−20 |
| 6 | FOXO4 | TTGTTT | 102/2,037 | 1.3e−16 |
| 7 | unknown | AACTTT | 92/1,859 | 1.3e−14 |
| 8 | MYC | CACGTG | 61/1,015 | 5.2e−13 |
| 9 | NFY | GATTGGY | 65/1,141 | 7.1e−13 |
| 10 | unknown | GGGYGTGNY | 46/655 | 4.2e−12 |
| GR | 12/276 | 0.014 | ||
| T3R | 18/248 | 1.5e-5 | ||
| ERR1 | TGACCTY | 49/1,023 | 1.4e-7 | |
| Yellow | ||||
| 1 | SPI | GGGCGGR | 22/2,891 | 1.2e−7 |
| 2 | ELK1 | SCGGAAGY | 12/1,176 | 3.4e−5 |
| 3 | ERR1 | TGACCTY | 11/1,023 | 4.2e−5 |
| 4 | STAT1 | 4/248 | 0.014 | |
| 5 | SF1 | TGACCTTG | 4/243 | 0.014 |
| 6 | STAT5B | 4/239 | 0.014 | |
| 7 | USF | 4/237 | 0.014 | |
| 8 | SP1 | 4/252 | 0.014 | |
| 9 | MEF2 | 4/268 | 0.015 | |
| 10 | MYC | CACGTG | 7/1,015 | 0.019 |
| GR | not detected | |||
| T3R | not detected | |||
| ERR1 | TGACCTY | see 3 above | ||
| Blue | ||||
| 1 | SPI | GGGCGGR | 127/2,891 | 1.38e−30 |
| 2 | ERR1 | TGACCTY | 61/103 | 6.1e−20 |
| 3 | ELK1 | SCGGAAGY | 62/1,176 | 8.9e−18 |
| 4 | NRF1 | RCGCANGCGY | 53/894 | 2.6e−17 |
| 5 | EI2 | GAGGTG | 88/2,450 | 3.1e−15 |
| 6 | MAZ | GGGAGGRR | 83/2,250 | 5.1e−15 |
| 7 | NFAT | TGGAAA | 73/1,871 | 2.4e−14 |
| 8 | MYC | CACGTG | 51/1,015 | 5.8e−14 |
| 9 | NFY | GATTGGY | 52/1,141 | 1.3e−12 |
| 10 | GABP-B | MGGAAGTG | 39/744 | 3.6e−11 |
| GR | 9/202 | 0.0051 | ||
| T3R | not detected | |||
| ERR1-Q2 | TGACCTY | 14/257 | 9.6e-5 | |
| Green | ||||
| 1 | LEF1 | CTTTGT | 27/1,939 | 5.6e−11 |
| 2 | NFAT | TGGAAA | 26/1,871 | 9.1e−11 |
| 3 | FOXO4 | TTGTTT | 22/2,037 | 5.4e−7 |
| 4 | unknown | AACTTT | 19/1,859 | 1e−5 |
| 5 | SOX9 | 8/236 | 1e−5 | |
| 6 | STAT | 8/248 | 1e−5 | |
| 7 | ISRE | 8/246 | 1e−5 | |
| 8 | ETS2 | RYTTCGTG | 14/1,074 | 1.9e−5 |
| 9 | MAZ | GGGAGGRR | 20/2,250 | 2.3e−5 |
| 10 | E2A | 7/241 | 7.4e−5 | |
| GR | 6/276 | 0.0007 | ||
| T3R | not detected | |||
| ERR1 | TGACCTY | not detected | ||
Transcription factors associated with each pattern of gene coexpression were identified by Webgestalt and the human genome as the reference set. The top 10 most significant transcription factors are shown with their DNA binding sequence, the number of genes regulated by the transcription factor suggested in our data set against the total number of possible genes, and the P value adjusted for multiple comparisons. The potential involvement of glucocorticoid receptor (GR), thyroid hormone receptor (T3R), and estrogen-related receptor (ERR1) in the regulation of the coexpressed genes is also shown.
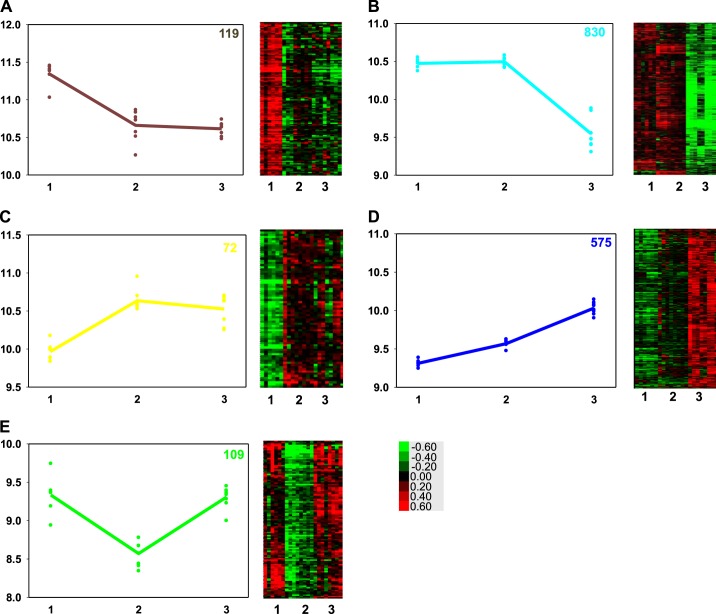
The set of DR genes with expression highest at 130-days gestation, declining by term, and staying unchanged relative to term or showing a slight further decrease by 2 wk of age contained 119 uniquely identified genes by Webgestalt from 125 DR genes (brown in Figs. 2 and 3A). The BPs associated with this gene set are broadly defined as developmental process (55 genes, adj. P value = 2.8 × 10−3) and include heart development (12 genes, adj. P value = 8.1 × 10−3) including cardiac septum development (5 genes, adj. P value = 1.4 × 10−2) and nervous system development (30 genes, adj. P value = 8 × 10−4). Cell adhesion (17 genes, adj. P value = 3.06 × 10−2) was also related to this gene set. See Supplemental Table S1 for the complete list of biological processes modeled for the five temporal patterns of coexpressed DR genes with Webgestalt. ECM-receptor interaction was identified as an enriched KEGG pathway in this temporal pattern of coexpressed genes (5 genes, adj. P value = 0.02). This induction by 130 days is reflected in the established pattern of collagen deposition as observed histologically (Fig. 4, D–I). The transcription factors most associated with this pattern of decreasing gene expression are nuclear factor of activated T-cells (NFAT), SP1, and LEF1, although GR, T3R, and ERR1 were also implicated, but with lower significance (Table 1). Protein interaction analysis of this gene coexpression pattern revealed genes related to central nervous system development dependent on calcium ion binding in the biogenesis of lysosome-related organelle complex 1 (14 genes in H. sapiens module 199, adj. P value = 0.026), and two genes involved in trans-membranous receptor protein tyrosine phosphatase activity in the synapse (H. sapiens module 291, adj. P value = 0.033).
Fig. 3.
The 5 panels of this figure show on the left a graph of the average gene expression of all the genes identified in a pattern (colored line) of gene expression for each animal in the time point sampled (colored symbols), and on the right, the heat map showing the relative expression level of each gene in each animal. 1 = 130-days gestation (n = 6); 2 = term (n = 8); and 3 = 2-wk-old (n = 8). Expression pattern A = brown, B = turquoise, C = yellow, D = blue, E = green. The number in the top right of each graph is the number of genes identified in each expression pattern. The colored scale for the heat maps, bottom right, shows the fold change in expression levels of the gene from the normalization point, where green is decreased and red increased expression.
Fig. 4.
Representative illustrations of the histological changes in the heart, occurring from 2 wk before term (A, D, G, J), to term (B, E, H, K) and to 2 wk postnatally (C, F, I, L). Top row, A–C: hematoxylin and eosin staining illustrating increased density of the myocardium; dividing nuclei are seen at all ages (examples shown yellow arrows). 2nd row, D–F: picrosirius red staining for collagen (red), showing increased density of the myocardium and increased collagen (red staining); Purkinje fibers in the collagen can be seen at all ages (examples shown with white arrows), although Purkinje fibers are more difficult to identify in the younger fetus because of the relatively low collagen deposition around them. 3rd row, G–I: periodic acid-Schiff staining illustrating Purkinje fibers (blue arrows) containing plentiful glycogen (purple); collagen surrounds them, orange arrows (H, I). 4th row, J–L: COX IV (red) staining in mitochondria and nuclear DNA stained with Hoechst 33342 (blue) show increased mitochondrial density and organization with maturation.
The set of DR genes that were highly expressed in the fetus at 130 days and term but decreased dramatically by 2 wk of postnatal age, contained 830 recognized genes from 863 DR genes (turquoise in Figs. 2 and 3B). The BP assigned to the gene set could be broadly associated with three major processes: cellular component organization or biogenesis, localization, and metabolic process. Cellular component organization or biogenesis included cellular macromolecular complex disassembly (37 genes, adj. P value = 7.6 × 10−8); most of these 37 genes were ribosomal genes, and this was a subset of cellular component disassembly at the cellular level (42 genes, adj. P value = 1 × 10−4), and this was in turn a subset of macromolecular complex subunit organization (101 genes, adj. P value = 5.5 × 10−3). An enriched KEGG pathway modeled from the genes in this expression pattern was ribosome (32 genes, adj. P value = 1.25 × 10−12). Protein-protein interaction modeling in Webgestalt suggests that the turquoise pattern of ribosomal biogenesis was of cytosolic ribosomes (ribosomal small subunit biogenesis in H. sapiens module 230, with 32 genes, adj. P value = 1.3 × 10−8); in contrast, genes related to mitochondrial ribosome biogenesis were increased at term and remained stable postnatally (yellow pattern in Fig. 2C, see below). Included in the genes in the BP, metabolism, in the pattern with high fetal gene expression were two major processes, protein metabolism and RNA metabolism (translation (84 genes, adj. P value = 5 × 10−4), RNA metabolic process (72 genes, adj. P value = 2.7 × 10−3) and RNA catabolic process (42 genes, adj. P value = 1.05 × 10−7). Protein metabolism included translational initiation (39 genes, adj. P value 3.08 × 10−9), translational elongation (34 genes, adj. P value = 1.91 × 10−10), and translation termination (31 genes, adj. P value = 1.91 × 10−10); translation termination was a subset of both cellular component organization or biogenesis and metabolism. Another significant BP was de novo posttranslational protein folding (13 genes adj. P value 4.9 × 10−3). The BP localization contained subgroups involved with intracellular transport (162 genes, adj. P value = 3 × 10−4) and targeting of proteins to membranes, particularly to ribosomal membranes (33 genes, adj. P value = 1.01 × 10−9). Two of the significant molecular functions suggested for the genes in this pattern of expression were neurotransmitter receptor activity (9 genes, adj. P value = 3.4 × 10−3) and G protein-coupled peptide receptor activity (14 genes, adj. P value = 4.1 × 10−3). Neuroactive ligand-receptor interaction was an enriched KEGG pathway for this gene expression pattern (24 genes, adj. P value = 1.7 × 10−3). Thus, this pattern of gene expression appears associated with the synthesis of machinery necessary for cytoplasmic protein synthesis, and protein synthesis, folding and transport within the cell, and possibly involves assembling and correctly positioning signaling molecules. Consistent with this, there is an increasing organization and density of the cardiac myocytes (Fig. 4, A–F) and a relative increase in multinucleate cells between preterm and postnatal hearts (Fig. 4, A–C). The transcription factors associated with decreased gene expression in the 2 wk old lambs include SP1, MAZ, LEF1. There is also involvement of ERR1, T3R, and NFKB at lower significance levels (i.e., fewer genes) and GR and PPAR at even lower, but still significant, levels (Table 1).
The smallest set of genes had increased expression at term and remained at the same level of expression at 2 wk of age (72 genes uniquely recognized by Webgestalt from 73 DR genes; yellow, Figs. 2 and 3C). The biological processes modeled from these genes fell into six broad categories: small molecule metabolic process (27 genes, adj. P value = 3.4 × 10−3), catabolic process (23 genes, adj. P value = 4.7 × 10−3), generation of precursor metabolites and energy (10 genes, adj. P value = 3.4 × 10−3), coenzyme metabolic process (6 genes, adj. P value = 1.46 × 10−2), mitochondrion organization (8 genes, adj. P value = 3.4 × 10−3), and transport, a nonsignificant broad category that included mitochondrial ATP synthesis-coupled proton transport (3 genes, adj. P value = 3.4 × 10−3). The BP catabolic process included carboxylic acid catabolic process (7 genes, adj. P value = 3.4 × 10−3), while the BPs oxidation-reduction process and generation of precursor metabolites and energy encompassed the categories respiratory electron transport chain (5 genes, adj. P value = 3.4 × 10−3) and oxidative phosphorylation (3 genes, adj. P value 8.5 × 10−3). ATP biosynthetic process (4 genes, adj. P value = 4.7 × 10−3), ADP biosynthesis process (2 genes, adj. P value = 3.4 × 10−3), and ubiquinone biosynthetic process (2 genes, adj. P value = 2.17 × 10−2) were also significant for this gene expression pattern. Protein interaction analysis largely supported the above findings (e.g., acetyl-coA biosynthetic process from pyruvate; H. sapiens module 46, adj. P value = 9.13 × 10−5), TCA cycle (H. sapiens module 234, adj. P value = 0.022) but suggested that both large and small subunits of the mitochondrial ribosome were being synthesized and participating in mitochondrial protein synthesis (H. sapiens modules 568 and 235, both adj. P value = 0.032) and that mitochondrial carbohydrate derivative transport (H. sapiens module 551, adj. P value = 0.046) genes were coexpressed with this pattern. The mitochondrial ribosomes' involvement with this increasing pattern of expression in late gestation contrasts with the cytosolic ribosomal involvement in the decreasing pattern of expression. Transcription factors important for controlling the increases in gene expression were SP1, ELK1, and ERR1 (Table 1). There was no direct effect of GR or T3R noted for this pattern.
The second largest set of differentially expressed genes progressively increased in expression from the 130-day fetus through term and to 14 days postnatal life. This set contained 589 genes, 575 of which could be assigned to unique gene names (blue, Figs. 2 and 3D). The principal BP associated with this set of genes was broadly related to metabolism (396 genes, adj. P value = 4.15 × 10−2), but this encompassed subsets such as mitochondrion organization (23 genes, adj. P value = 1.18 × 10−2), lipid oxidation (17 genes, adj. P value = 2.9 × 10−3), carboxylic acid metabolic process (62 genes, adj. P value = 1.18 × 10−2), mitochondrial transport (18 genes, adj. P value = 4.7 × 10−3), autophagic vacuole assembly (7 genes, adj. P value = 1.8 × 10−2), and cofactor biosynthetic process (16 genes, adj. P value = 1.81 × 10−2). The cellular components involved in the DR genes in this set include mitochondrion (104 genes, adj. P value = 1.85 × 10−5 and sarcoplasmic reticulum (8 genes, adj. P value = 1.67 × 10−2), as might be expected given the BPs, but also autophagic vacuole (6 genes, adj. P value = 4.18 × 10−2) and peroxisome (15 genes, adj. P value = 1.25 × 10−2). The relative change in organization of mitochondria is also evident in histologic sections (Fig. 4, J–L). The primary transcription factors associated with this pattern of gene expression are SP1, ERR1, and ELK1 (Table 1). GR and PPAR were also involved but are less significant; there was no significant contribution of T3R to this pattern of gene expression. Protein interaction modeling suggested regulation of fatty acid oxidation associated with AMP activated protein kinase complex activity (H. sapiens module 741, 7 genes, adj. P value = 0.0067). Activated AMPK stimulating fatty-acid oxidation in muscle (8 genes, adj. P value = 3 × 10−4) was one of two enriched pathway commons (PC) pathways for this gene expression pattern, suggesting genes with this expression pattern support fatty acid metabolism. This is confirmed by the other enriched PC pathway of this expression pattern, import of palmitoyl-coA into the mitochondrial matrix (6 genes, adj. P value = 8 × 10−4).
The fifth pattern of DR genes had decreased expression at term that increased again by 2 wk of extrauterine life (green, Figs. 2 and 3E). This gene set consisted of 109 DR genes yielding 107 genes that could be uniquely identified and modeled in Webgestalt. These genes modeled six basic processes: developmental process (51 genes, adj. P value = 4 × 10−4), single multicellular organism process (59 genes, adj. P value = 4.81 × 10−5), response to stimulus (64 genes, adj. P value = 1.86 × 10−5), regulation of biological quality (37 genes, adj. P value = 5 × 10−4), immune system processes (33 genes, adj. P value = 8.09 × 10−5), and establishment or maintenance of cell polarity (7 genes, adj. P value = 1.5 × 10−3). The major transcription factors associated with this pattern of gene expression are LEF1, NFAT, and FOXO4 (Table 1), but it is interesting that GR was of higher importance as a transcription factor regulating this gene expression pattern than any of the other patterns (6 genes, adj. P value = 0.0007, Table 1). Protein interaction analysis suggested many interactions typically related to immune system function, for example, actin polymerization or depolymerization in lamellipodium (H. sapiens module 111, 13 genes, adj. P value = 0.02), but genes involved with JAK-STAT cascade signaling involved in growth hormone signaling pathways (H. sapiens module 25, 21 genes, adj. P value = 0.038) were also significant.
DISCUSSION
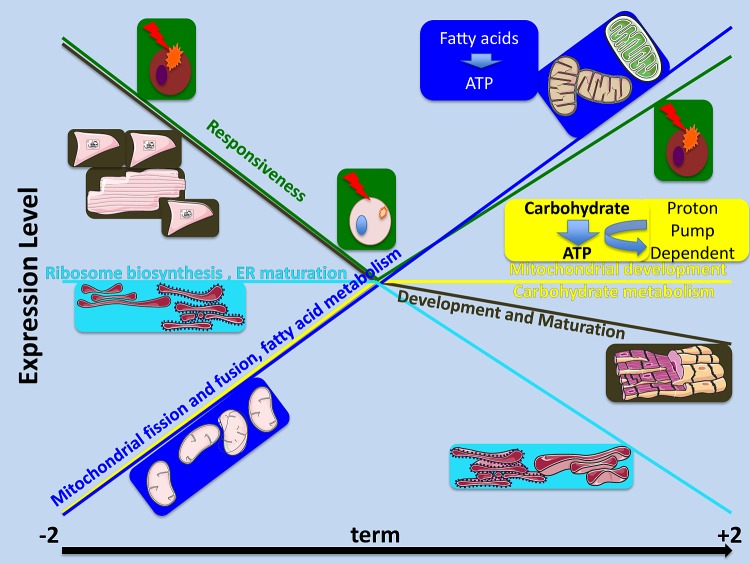
Transcriptomic analyses have revealed five patterns of changing gene expression over the last 2 wk of gestation and the first 2 wk of extrauterine life. These patterns are consistent with anatomical and cellular differentiation of the heart followed by growth through protein synthesis and reorganization, and metabolic processes that increase the potential to use lipids rising throughout the period, whereas those allowing use of carbohydrates peaking before birth. There also is a pattern suggesting that immune activation in the heart is suppressed at term. These patterns have been summarized in Fig. 5.
Fig. 5.
A summary illustration of the most significant patterns of gene expression discovered over time from 2 wk before term (−2), to term, and to 2 wk postnatally (+2). The color-coded gene expression patterns are shown as colored lines; images representing the major processes modeled for the expression pattern are shown in boxes with background colors matching the line drawn for their expression pattern.
There are some limitations to the analyses as performed. Discrete sampling times do not enable determination of precise timing of the changes in gene expression, and there are a relatively small number of genes in some of the gene coexpression pattern groups (e.g., yellow). Reliance on gene ontology terms for the analyses necessarily biases results toward well-studied areas and their processes, such as the brain and immune system. However, there are clear trends in the gene expression pattern analyses that complement previous knowledge of heart development in the perinatal sheep and indicate a major role of the transcription factors in the ERR family and MYC in the process. Involvement of these transcription factors has been well characterized in the rodent (1, 20, 26, 33, 36) but is less well appreciated in the perinatal sheep heart.
The temporal pattern of gene expression in which expression is highest at ∼15 days before birth and then declines through term and 2 wk of life (brown) includes genes in BPs associated with differentiation of the heart, including regulation of cell differentiation and neuronal differentiation (Fig. 5). Purkinje fibers arise as specialized cardiomyocytes and acquire some neuronal characteristics, for example, ubiquitin carboxyl-terminal hydrolase-1 (also known as PGP9.5) and contactin 2 (53). Development of the septum would be expected to reflect these BPs as it includes Purkinje and myocytes as well as neuronal components around blood vessels and myocytes. Thus genes in this pattern of expression were anatomically associated with development of all aspects of the cardiovascular system including the heart, the atria, ventricles, and reassuringly, the septa. Cellular adhesion and ECM genes were revealed in this pattern of expression, implying that closer associations between cells to augment impulse conduction and generation of greater force of contraction are also largely complete by 130 days of gestation. This also agrees with the observations that the last gross anatomical feature of the heart to develop was the collagen sheath around individual cardiomyocytes, and the bundles of two to eight, Purkinje fibers, seen in the sheep (9, 46, 51, 74). Histology of the septum shows Purkinje fibers are evident at all ages examined (Fig. 4, D–I), although collagen deposition and organization of Purkinje fibers in bundles appear to be greater after 130 days.
The transcription factors associated with this pattern include NFATs, a family of calcium/calcineurin-regulated transcription factors that often act in concert with AP1 to effect gene transcriptional regulation. In nonimmune cells they can also act with ANP and BNP and in skeletal muscle with slow-twitch genes and the IP3 receptor. NFAT has been shown to be important for heart valve formation and differentiation of muscle fiber types, as well as via interactions with CRELD1 and calcineurin, necessary for VEGF-dependent proliferation of endocardial cells (43). C/EBPgamma, a transcription factor important for epicardial activation (27), and EGR1, involved in excitation-contraction coupling (52) and cardiac fibroblast apoptosis (76), were also implicated in controlling the genes in this coexpression pattern. Thus BPs of the set of genes following this trajectory of expression suggest that the anatomical development of the heart is largely complete by 130 days of gestation; except for collagen deposition, most cells have differentiated to their final phenotype and adhered to one another.
The second pattern of gene expression reflecting the next step in development is that of constant gene expression throughout late gestation, but declining expression after birth. This pattern is shown by the largest set of genes (turquoise, Fig. 5). The BPs inferred for the group suggest that the growth by protein synthesis and transport of nascent proteins to their proper localization within the cells of the heart slows after birth. The protein complexes in this expression pattern are predominantly ribosomal, but also cytoskeletal, suggesting that rearrangement of ribosomes and the cytoskeleton is complete soon after birth. These genes were associated with the transcription factors SP1, MAZ, LEF1, MYC, and NFY but also ERR1, T3R and NFKB, and GR and PPAR. NFY interacts with calcineurin and is important for mediating the actions of verapamil to block L-type calcium channel activity and decrease the proapoptotic TXNIP in cardiomyocytes (10). It is also increased in NKx2-5 knockout mice that display complex cardiac phenotypes (38).
Despite this pattern of gene expression associated with protein synthesis, the heart weight-to-body weight ratios of 130 day-gestation fetuses and newborn lambs are similar (∼0.7%) (7), although there is an increase in the fraction of myocardium that is cardiac muscle (∼76% to 85%) (66) (Fig. 4, A–F). Since fetuses and lambs are growing rapidly throughout this time period, the pattern of increased expression of genes associated with protein synthesis likely reflects the switch from hyperplastic to hypertrophic growth (15, 37), beginning at about 110 days gestation through early extrauterine life (29) in the sheep, but at about postnatal day 3–4 of the rat (37). This switch can be driven by a fall in IGF2 (seen in the brown expression group) acting at the IGF1 receptor that drives proliferation (68) and a rise in thyroid hormone activity. MYC, a transcription factor associated with this coexpression pattern, decreases at the switch from proliferation to differentiation and prior to cell hypertrophy in the developing heart. MYC is important for controlling proliferation, because its overexpression causes an increase in cardiomyocyte number with normal maturation (28). Thyroid hormone precursor (T4) levels, tissue deiodinases, and T3R rise beginning at about 135 days gestation, slowing the proliferation driven by IGF as gestation advances (12, 23, 68). However, the binucleation that follows cessation of proliferation begins at about 110 days of gestation, and so it is likely that this is transition is not driven solely by thyroid hormone-bound T3R, as suggested by the relatively small number of genes driven by TRE. Indeed, this process is complex as it was recently shown that at 100 days of gestation, IGF1 combined with T3 decreased the phosphorylation of ERK and AKT (growth and differentiation effectors respectively downstream of T3R and IGF1R) but increased their phosphorylation at 135 days gestation; these hormones both increase the phosphorylation of ERK and AKT when administered alone at either age (12). MYC overexpression enhances the ability of T3R activity to increase hypertrophic growth in the differentiated cardiomyocyte (57). This coexpression pattern (high throughout gestation and decreasing after birth) is the only one of the three patterns in which MYC is modeled as a transcription factor targeting genes that also has significant T3R gene targets, suggesting that they may interact to decrease proliferation and increase hypertrophic growth toward the end of gestation. It could also be driven by LEF1 that interacts with ß-catenin to cause hypertrophy in both adult and neonatal mouse hearts (13).
A third pattern was displayed by the set of genes whose expression was decreased at term relative to either 2 wk before term or 2 wk after birth (green, Fig. 5). The BPs associated with these genes were developmental, and responses to many kinds of stimuli, including immune, temperature, stress, virus, wounding, and chemical stimuli. This suggests the blunting of the responses to these stimuli at term. In the cow both innate and acquired immune responses are lowest between 3 wk prenatally and 3 wk postnatally (40). Considering that glucocorticoids in the sheep fetus peak at term (60), the pattern of blunted responses may result from transcriptional repression due to the high steroid levels at this time, and indeed the GR was implicated as a fairly important transcription factor of this gene expression pattern. Other studies, in this case in models of caloric restriction to increase longevity, have suggested that a decrease in genes regulating immune function pathways, including inflammation (2), occurs in concert with activation of beneficial pathways associated with metabolic health (e.g., the mitochondrial electron transport system, cytosolic ribosomes, and regulation of mitochondrial redox potential). This suggests that immune function is related to metabolic health. Thus, the dip in immune responsiveness modeled at term in this study may be a mechanism to promote metabolic changes during the challenging switch to extrauterine life (42).
Mitochondrial organization, structure, and metabolism figure prominently in two gene expression patterns (yellow and blue). Genes with an increasing expression from day 135 to term but no further increase after term (yellow) include genes controlling mitochondrial organization, the respiratory electron chain, and ATP, ADP, ubiquinone biosynthesis, and carboxylic acid catabolism. The transcription factor MYC, an important regulator of metabolic processes, is known to increase glycolysis (as well as mitochondrial mass, OXPHOS, and electron chain transport function) (26). MYC is a transcription factor identified as a regulator of genes following this pattern (yellow) of protein expression (adj. P value = 0.019), although it is also implicated in controlling genes with the turquoise and blue patterns of expression, as befits its major role in metabolic regulation (as indicated by staining for COX IV, a mitochondrial enzyme; Fig. 4, J–L). This suggests that as the cells of the heart become terminally differentiated in late gestation but are still actively growing and organizing, they are also increasing their ability to synthesize ATP from sugars. The capacity to generate ATP through these pathways then remains constant from term through the first 2 wk of life (Fig. 5). In contrast, expression of genes associated with the ability to use fatty acids continuously rises throughout the time period of our study (blue pattern). Indeed, an increasing ability to use fatty acids is indicated by increased expression of genes associated with lipid oxidation, the carnitine shuttle, and the organization of mitochondria and mitochondrial transport. This is consistent with the ability of the postnatal heart to utilize free fatty acids, which are a more efficient fuel than glucose (i.e., more ATP per molecule metabolized) but require more oxygen per unit of energy. The fetal heart predominantly uses glucose and lactate as energy sources, although the fetal sheep heart can use lipids if they are supplied before birth (4). Postnatally the supply of fatty acids increases, and the heart switches away from carbohydrates as the main energy source (22) to preferential use of fats during the first few weeks of life, so that by 15 days of age about 80% of the oxygen used by the LV is used for long-chain fatty acid oxidation (4, 49). Thus, this pattern of gene expression clearly reflects this ongoing process in the fetal and neonatal heart.
The metabolic switch has been associated with the ERRs, especially ESRRG, in mouse knockout studies (1). ESRRG does not have a specific gene set in the Broad Institute's database, presumably because it binds to the same motif as ESRRA; thus we cannot identify which ERR is important in our gene expression patterns. This is unfortunate because ESRRA knockout mice are relatively normal because of compensation by cofactor upregulation, whereas ESRRG knockout mice die immediately after birth after failing to complete the metabolic shift to fatty acid metabolism among a myriad of other problems (1). Nevertheless, genes containing the binding motif in their promotor region for this transcription factor family were overrepresented in all but one of our expression patterns, but most significantly in the metabolically associated expression patterns, highlighting their importance in perinatal heart maturation. Various cofactors of ERRs are found in different expression patterns for example RARA and CREB1 (20) in the pattern with highest gestational expression (brown) and STAT3 (20) in the continuously rising pattern of gene expression (blue). NRF1, another cofactor in the continuously rising (blue) pattern, has been suggested to maintain the expression pattern set up by a more short-lived interaction of ERR and PPARGC1A gene products on transcription (20). This suggests that the pattern of coactivator expression with ERRs may define when and how the ERRs affect heart maturation.
These two patterns of gene expression also include genes that alter mitochondrial structure and function. The mitochondrial organization regulated in the pattern with high prenatal gene expression is concerned with mitochondrial respiratory chain component assembly (2 genes, adj. P value 10−3), and regulation of the mitochondrial membrane potential in an antiapoptotic fashion (2 genes, adj. P value = 1.4 × 10−2), a different role in mitochondrial organization from that seen with the progressively increasing pattern of gene expression. The mitochondrial organization that occurs with this pattern of expression is suggestive of remodeling of the mitochondrial system as it involved fusion, fission, morphogenesis, apoptosis, organization of the mitochondrial membranes, and targeting of proteins to mitochondria (Fig. 4, J–L). The mitochondrial genome, the mitochondrial nucleoid, a membrane-bound complex, was also involved as would be expected if mitochondrial fission and fusion occurred. Mitochondrial fission requires contact with the endoplasmic reticulum (24), and it has been estimated that 10–20% of the mitochondrial surface participates in endoplasmic reticulum (ER) contacts in the rat liver, an organ with a highly dynamic mitochondrial network (50). Increases in expression of genes associated with mitochondrial organization followed the increase in expression of genes involved with the organization of the ER, thereby setting up the potential for mitochondrion-ER interaction. For example, the protein product of the MFF gene, within the continuously rising (blue) pattern of gene expression, localizes to the site of fission and recruits other proteins to produce fission (24).
At term, the mitochondria of the cardiomyocytes are organized into two pools, one around the poles of the nucleus and the other associated in a very orderly fashion with the sarcomeres (7). The mitochondrial network of a cell is in constant flux, being a dynamic, energy-dependent structure that responds quickly to the state of the cell and is very dependent on the cell type. The groundwork for the cardiomyocyte mitochondrial reticulum is laid by the genes with the yellow expression pattern and built upon with the genes in the blue pattern of gene expression, both in terms of the “anatomy” of the structure and the energy sources that it can use to fuel the cell and maintain its own structure.
Since the energetic state of the cell and the biophysical forces on the cardiomyocyte regulate its mitochondrial network, it is likely that a bidirectional flow of signals from mitochondrion to cell and cell to mitochondrion shapes the network and determines the functionality of the cardiomyocyte. One of the most highly significant transcription factors associated with the progressively increasing (blue) pattern of gene expression, NRF1, has been described as one of the key regulators of nuclear-mitochondrial communication. It is transcriptionally activated downstream of PPARGC1A, the so-called master regulator of mitochondrial biogenesis, that acts through ESRRA (64) and GABPB (aka NRF2) to induce NRF1 expression (45), but NRF1 is also a coactivator partner with PPARGC1A, revealing their complex interaction in mitochondrial biogenesis (reviewed in Ref. 63). Another transcription factor known to interact with PPARGC1A, PPAR, was also important in this pattern of gene expression. Transcription of PPARGC1A is strongly induced in the postnatal mouse, but nearly absent before birth (35). Primary cultures of embryonic mouse cardiomyocytes treated with a GR agonist show myofibril maturation and increased mitochondrial oxygen consumption that is blocked by PPARGC1A knockdown, suggesting that glucocorticoids act through induction of PPARGC1A (58). The pattern of increasing expression of genes regulated by factors either downstream of, or coactivators with, PPARGC1A throughout the study period suggest it may be important in maturation of cardiac function in the perinatal sheep. However, there was no change in expression of PPARGC1A over the time course we studied, although it is possible that PPARGC1A is induced earlier than 130 days in the sheep or that these time points miss short-lived changes in PPARGC1A expression.
MEF2, a family of transcription factors with gene targets in the expression pattern that rises to term then remains constant (yellow), is reported to control expression of PPARGC1A and to be involved with cardiac differentiation, postnatal growth (32), cardiac muscle development in response to stress, mitochondrial biogenesis (48), extracellular matrix protein expression (39), and vascular development of the heart (6). The transcription factor MYC regulates mitochondrial positioning within the cell (26) and was significantly associated with this pattern of gene expression, but it also has other major effects on mitochondria that are also consistent with this rising pattern of gene expression, such as a role in normal cardiac development (28) and increased expression following T3R stimulation prior to hypertrophic growth that is magnified by overexpression in transgenic mice (57).
The rising pattern of expression suggests that there is large-scale mitochondrial remodeling in the septum of the perinatal sheep fetus that may occur earlier in ontogeny than in the rat, but this would need to be confirmed by further studies. The different developmental stages of the newborn sheep and rodent may contribute to these differences, as may the relative size, shape, and contraction rate of the cardiomyocyte in large animals compared with small and relatively immature rodents, which is suggested to affect the EC coupling within the cell (65) and, in turn, affect their metabolic and mitochondrial systems.
Both GREs and T3RE are found in the mitochondrial genome and have been shown to affect mitochondrial DNA transcription when bound by their respective hormone receptors; GR and TR are also localized in mitochondria but may have a slightly different structure from the cytosolic receptor (5, 17, 19, 47). The genes in the continuously rising pattern of gene expression (blue) includes a significant number associated with GR as a transcription factor, but the pattern that rises in the perinatal period and then remains constant (yellow pattern) does not, and neither seems to depend on T3R. This implies that the increase in mitochondrial remodeling could be driven by GRE both on the nuclear and on the mitochondrial DNA, but that thyroid hormone, well known to effect mitochondrial biogenesis postnatally (for example Ref. 70) may do so by interactions with other factors rather than the T3R for example PPARs (44) (PPAR, 11 genes, adj. P = 0.0025, blue pattern).
In summary, transcriptomic analysis of gene coexpression patterns illustrates the sequential development of cardiomyocyte structure and metabolism that is necessary for maturation of cardiomyocytes and for the transition in function occurring in the postnatal period. In particular, temporal patterns of the maturation were suggested by the modeling. During this period of development the anatomical differentiation of the septum slows, and then the ER matures and becomes closely associated with the evolving mitochondrial network. The ability of the cells to metabolize carbohydrates matures prenatally, but the ability to efficiently use fatty acids rises throughout this time, in contrast to the rodent where this occurs postnatally. Also, the timing of the coexpression of genes regulated by the transcription factors MYC and ESRRs was different in the sheep compared with rodents. In addition, there is a period of decreased immune and cellular responsiveness, likely controlled by high glucocorticoid levels perinatally, that may influence metabolic health in the transition to extrauterine life.
GRANTS
This work was supported by National Institutes of Health Grants DK-60820 and HD-57871.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: E.M.R., M.B.R., C.E.W., and M.K.-W. conception and design of research; E.M.R., A.A., C.E.W., and M.K.-W. performed experiments; E.M.R., M.B.R., and M.K.-W. analyzed data; E.M.R., M.B.R., C.E.W., and M.K.-W. interpreted results of experiments; E.M.R. and M.B.R. prepared figures; E.M.R. and M.K.-W. drafted manuscript; E.M.R., M.B.R., A.A., C.E.W., and M.K.-W. edited and revised manuscript; E.M.R., M.B.R., A.A., C.E.W., and M.K.-W. approved final version of manuscript.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, Evans RM. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab 6: 13–24, 2007. [DOI] [PubMed] [Google Scholar]
- 2.Anderson RM, Weindruch R. Metabolic reprogramming, caloric restriction and aging. Trends Endocrinol Metab 21: 134–141, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aryee MJ, Gutierrez-Pabello JA, Kramnik I, Maiti T, Quackenbush J. An improved empirical Bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation). BMC Bioinformatics 10: 409, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartelds B, Knoester H, Smid GB, Takens J, Visser GH, Penninga L, van der Leij FR, Beaufort-Krol GC, Zijlstra WG, Heymans HS, Kuipers JR. Perinatal changes in myocardial metabolism in lambs. Circulation 102: 926–931, 2000. [DOI] [PubMed] [Google Scholar]
- 5.Bassett JH, Harvey CB, Williams GR. Mechanisms of thyroid hormone receptor-specific nuclear and extra nuclear actions. Mol Cell Endocrinol 213: 1–11, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Bi W, Drake CJ, Schwarz JJ. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol 211: 255–267, 1999. [DOI] [PubMed] [Google Scholar]
- 7.Brook WH, Connell S, Cannata J, Maloney JE, Walker AM. Ultrastructure of the myocardium during development from early fetal life to adult life in sheep. J Anat 137: 729–741, 1983. [PMC free article] [PubMed] [Google Scholar]
- 8.Burrell JH, Boyn AM, Kumarasamy V, Hsieh A, Head SI, Lumbers ER. Growth and maturation of cardiac myocytes in fetal sheep in the second half of gestation. Anat Rec A Discov Mol Cell Evol Biol 274: 952–961, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Canale E, Smolich JJ, Campbell GR. Differentiation and innervation of the atrioventricular bundle and ventricular Purkinje system in sheep heart. Development 100: 641–651, 1987. [DOI] [PubMed] [Google Scholar]
- 10.Cha-Molstad H, Xu G, Chen J, Jing G, Young ME, Chatham JC, Shalev A. Calcium channel blockers act through nuclear factor Y to control transcription of key cardiac genes. Mol Pharmacol 82: 541–549, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattergoon NN, Giraud GD, Louey S, Stork P, Fowden AL, Thornburg KL. Thyroid hormone drives fetal cardiomyocyte maturation. FASEB J 26: 397–408, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattergoon NN, Louey S, Stork PJ, Giraud GD, Thornburg KL. Unexpected maturation of PI3K and MAPK-ERK signaling in fetal ovine cardiomyocytes. Am J Physiol Heart Circ Physiol 307: H1216–H1225, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Shevtsov SP, Hsich E, Cui L, Haq S, Aronovitz M, Kerkela R, Molkentin JD, Liao R, Salomon RN, Patten R, Force T. The beta-catenin/T-cell factor/lymphocyte enhancer factor signaling pathway is required for normal and stress-induced cardiac hypertrophy. Mol Cell Biol 26: 4462–4473, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J, Suzuki KT, Sakuma T, Shewade L, Yamamoto T, Buchholz DR. Unliganded thyroid hormone receptor alpha regulates developmental timing via gene repression in Xenopus tropicalis. Endocrinology 156: 735–744, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clubb FJ Jr, Bishop SP. Formation of binucleated myocardial cells in the neonatal rat: an index for growth hypertrophy. Lab Invest 50: 571–577, 1984. [PubMed] [Google Scholar]
- 16.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics 20: 1453–1454, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Demonacos C, Djordjevic-Markovic R, Tsawdaroglou N, Sekeris CE. The mitochondrion as a primary site of action of glucocorticoids: the interaction of the glucocorticoid receptor with mitochondrial DNA sequences showing partial similarity to the nuclear glucocorticoid responsive elements. J Steroid Biochem Mol Biol 55: 43–55, 1995. [DOI] [PubMed] [Google Scholar]
- 18.Dostert A, Heinzel T. Negative glucocorticoid receptor response elements and their role in glucocorticoid action. Curr Pharm Des 10: 2807–2816, 2004. [DOI] [PubMed] [Google Scholar]
- 19.Du J, Wang Y, Hunter R, Wei Y, Blumenthal R, Falke C, Khairova R, Zhou R, Yuan P, Machado-Vieira R, McEwen BS, Manji HK. Dynamic regulation of mitochondrial function by glucocorticoids. Proc Natl Acad Sci USA 106: 3543–3548, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab 5: 345–356, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Feng X, Reini S, Richards E, Wood CE, Keller-Wood M. Cortisol stimulates proliferation and apoptosis in the late gestation fetal heart: differential effects of mineralocorticoid and glucocorticoid receptors. Am J Physiol Regul Integr Comp Physiol 305: R343–R350, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher DJ, Heymann MA, Rudolph AM. Myocardial consumption of oxygen and carbohydrates in newborn sheep. Pediatr Res 15: 843–846, 1981. [DOI] [PubMed] [Google Scholar]
- 23.Forhead AJ, Fowden AL. Thyroid hormones in fetal growth and prepartum maturation. J Endocrinol 221: R87–R103, 2014. [DOI] [PubMed] [Google Scholar]
- 24.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science 334: 358–362, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giraud GD, Louey S, Jonker S, Schultz J, Thornburg KL. Cortisol Stimulates Cell Cycle Activity in the Cardiomyocyte of the Sheep Fetus. Endocrinology 147: 3643–3649, 2006. [DOI] [PubMed] [Google Scholar]
- 26.Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, Prochownik EV. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS One 7: e37699, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, Bassel-Duby R, Olson EN. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science 338: 1599–1603, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol 10: 3709–3716, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonker SS, Zhang L, Louey S, Giraud GD, Thornburg KL, Faber JJ. Myocyte enlargement, differentiation, and proliferation kinetics in the fetal sheep heart. J Appl Physiol 102: 1130–1142, 2007. [DOI] [PubMed] [Google Scholar]
- 30.Kassel O, Herrlich P. Crosstalk between the glucocorticoid receptor and other transcription factors: molecular aspects. Mol Cell Endocrinol 275: 13–29, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Keller-Wood M, Feng X, Wood CE, Richards E, Anthony RV, Dahl GE, Tao S. Elevated maternal cortisol leads to relative maternal hyperglycemia and increased stillbirth in ovine pregnancy. Am J Physiol Regul Integr Comp Physiol 307: R405–R413, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolodziejczyk SM, Wang L, Balazsi K, DeRepentigny Y, Kothary R, Megeney LA. MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Curr Biol 9: 1203–1206, 1999. [DOI] [PubMed] [Google Scholar]
- 33.Lai L, Wang M, Martin OJ, Leone TC, Vega RB, Han X, Kelly DP. A role for peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1) in the regulation of cardiac mitochondrial phospholipid biosynthesis. J Biol Chem 289: 2250–2259, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest 106: 847–856, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol 29: 339–345, 2002. [DOI] [PubMed] [Google Scholar]
- 37.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 28: 1737–1746, 1996. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Cao Y, Wu Y, Chen W, Yuan Y, Ma X, Huang G. The expression profile analysis of NKX2-5 knock-out embryonic mice to explore the pathogenesis of congenital heart disease. J Cardiol [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 39.Lockhart MM, Wirrig EE, Phelps AL, Ghatnekar AV, Barth JL, Norris RA, Wessels A. Mef2c regulates transcription of the extracellular matrix protein cartilage link protein 1 in the developing murine heart. PLoS One 8: e57073, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallard BA, Dekkers JC, Ireland MJ, Leslie KE, Sharif S, Vankampen CL, Wagter L, Wilkie BN. Alteration in immune responsiveness during the peripartum period and its ramification on dairy cow and calf health. J Dairy Sci 81: 585–595, 1998. [DOI] [PubMed] [Google Scholar]
- 41.Martin NP, Marron Fernandez de Velasco E, Mizuno F, Scappini EL, Gloss B, Erxleben C, Williams JG, Stapleton HM, Gentile S, Armstrong DL. A rapid cytoplasmic mechanism for PI3 kinase regulation by the nuclear thyroid hormone receptor, TRbeta, and genetic evidence for its role in the maturation of mouse hippocampal synapses in vivo. Endocrinology 155: 3713–3724, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin OJ, Lai L, Soundarapandian MM, Leone TC, Zorzano A, Keller MP, Attie AD, Muoio DM, Kelly DP. A role for peroxisome proliferator-activated receptor gamma coactivator-1 in the control of mitochondrial dynamics during postnatal cardiac growth. Circ Res 114: 626–636, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mass E, Wachten D, Aschenbrenner AC, Voelzmann A, Hoch M. Murine Creld1 controls cardiac development through activation of calcineurin/NFATc1 signaling. Dev Cell 28: 711–726, 2014. [DOI] [PubMed] [Google Scholar]
- 44.McClure TD, Young ME, Taegtmeyer H, Ning XH, Buroker NE, Lopez-Guisa J, Portman MA. Thyroid hormone interacts with PPARalpha and PGC-1 during mitochondrial maturation in sheep heart. Am J Physiol Heart Circ Physiol 289: H2258–H2264, 2005. [DOI] [PubMed] [Google Scholar]
- 45.Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA 101: 6570–6575, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morita T, Shimada T, Kitamura H, Nakamura M. Demonstration of connective tissue sheaths surrounding working myocardial cells and Purkinje cells of the sheep moderator band. Arch Histol Cytol 54: 539–550, 1991. [DOI] [PubMed] [Google Scholar]
- 47.Morrish F, Buroker NE, Ge M, Ning XH, Lopez-Guisa J, Hockenbery D, Portman MA. Thyroid hormone receptor isoforms localize to cardiac mitochondrial matrix with potential for binding to receptor elements on mtDNA. Mitochondrion 6: 143–148, 2006. [DOI] [PubMed] [Google Scholar]
- 48.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med 8: 1303–1309, 2002. [DOI] [PubMed] [Google Scholar]
- 49.Neely JR, Morgan HE. Relationship between carbohydrate and lipid metabolism and the energy balance of heart muscle. Annu Rev Physiol 36: 413–459, 1974. [DOI] [PubMed] [Google Scholar]
- 50.Ohta K, Okayama S, Togo A, Nakamura K. Three-dimensional organization of the endoplasmic reticulum membrane around the mitochondrial constriction site in mammalian cells revealed by using focused-ion beam tomography. Microscopy (Oxf) 63, Suppl 1: i34, 2014. [DOI] [PubMed] [Google Scholar]
- 51.Ono N, Yamaguchi T, Ishikawa H, Arakawa M, Takahashi N, Saikawa T, Shimada T. Morphological varieties of the Purkinje fiber network in mammalian hearts, as revealed by light and electron microscopy. Arch Histol Cytol 72: 139–149, 2009. [DOI] [PubMed] [Google Scholar]
- 52.Pacini L, Suffredini S, Ponti D, Coppini R, Frati G, Ragona G, Cerbai E, Calogero A. Altered calcium regulation in isolated cardiomyocytes from Egr-1 knock-out mice. Can J Physiol Pharmacol 91: 1135–1142, 2013. [DOI] [PubMed] [Google Scholar]
- 53.Pallante BA, Giovannone S, Fang-Yu L, Zhang J, Liu N, Kang G, Dun W, Boyden PA, Fishman GI. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol 3: 186–194, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabaglino MB, Richards E, Denslow N, Keller-Wood M, Wood CE. Genomics of estradiol-3-sulfate action in the ovine fetal hypothalamus. Physiol Genomics 44: 669–677, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reini SA, Dutta G, Wood CE, Keller-Wood M. Cardiac corticosteroid receptors mediate the enlargement of the ovine fetal heart induced by chronic increases in maternal cortisol. J Endocrinol 198: 419–427, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richards EM, Wood CE, Rabaglino MB, Antolic A, Keller-Wood M. Mechanisms for the adverse effects of late gestational increases in maternal cortisol on the heart revealed by transcriptomic analyses of the fetal septum. Physiol Genomics 46: 547–559, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robbins RJ, Swain JL. C-myc protooncogene modulates cardiac hypertrophic growth in transgenic mice. Am J Physiol Heart Circ Physiol 262: H590–H597, 1992. [DOI] [PubMed] [Google Scholar]
- 58.Rog-Zielinska EA, Craig MA, Manning JR, Richardson RV, Gowans GJ, Dunbar DR, Gharbi K, Kenyon CJ, Holmes MC, Hardie DG, Smith GL, Chapman KE. Glucocorticoids promote structural and functional maturation of foetal cardiomyocytes: a role for PGC-1alpha. Cell Death Differ 22: 1106–1116, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rog-Zielinska EA, Thomson A, Kenyon CJ, Brownstein DG, Moran CM, Szumska D, Michailidou Z, Richardson J, Owen E, Watt A, Morrison H, Forrester LM, Bhattacharya S, Holmes MC, Chapman KE. Glucocorticoid receptor is required for foetal heart maturation. Hum Mol Genet 22: 3269–3282, 2013. [DOI] [PubMed] [Google Scholar]
- 60.Rose JC, Meis PJ, Urban RB, Greiss FC. In vivo evidence for increased adrenal sensitivity to adrenocorticotropin-(1–24) in the lamb fetus late in gestation. Endocrinology 111: 80–85, 1982. [DOI] [PubMed] [Google Scholar]
- 61.Rudolph AM. Congenital Diseases of the Heart: Clinical-physiologic Considerations. Wiley-Blackwell, 2009. [Google Scholar]
- 62.Saatcioglu F, Lopez G, West BL, Zandi E, Feng W, Lu H, Esmaili A, Apriletti JW, Kushner PJ, Baxter JD, Karin M. Mutations in the conserved C-terminal sequence in thyroid hormone receptor dissociate hormone-dependent activation from interference with AP-1 activity. Mol Cell Biol 17: 4687–4695, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638, 2008. [DOI] [PubMed] [Google Scholar]
- 64.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA 101: 6472–6477, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiels HA, Galli GL. The sarcoplasmic reticulum and the evolution of the vertebrate heart. Physiology (Bethesda) 29: 456–469, 2014. [DOI] [PubMed] [Google Scholar]
- 66.Smolich JJ. Ultrastructural and functional features of the developing mammalian heart: a brief overview. Reprod Fertil Dev 7: 451–461, 1995. [DOI] [PubMed] [Google Scholar]
- 67.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004. [DOI] [PubMed] [Google Scholar]
- 68.Sundgren NC, Giraud GD, Schultz JM, Lasarev MR, Stork PJ, Thornburg KL. Extracellular signal-regulated kinase and phosphoinositol-3 kinase mediate IGF-1 induced proliferation of fetal sheep cardiomyocytes. Am J Physiol Regul Integr Comp Physiol 285: R1481–R1489, 2003. [DOI] [PubMed] [Google Scholar]
- 69.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145: 224–241, 2011. [DOI] [PubMed] [Google Scholar]
- 70.Walrand S, Short KR, Heemstra LA, Novak CM, Levine JA, Coenen-Schimke JM, Nair KS. Altered regulation of energy homeostasis in older rats in response to thyroid hormone administration. FASEB J 28: 1499–1510, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41: W77–W83, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang KC, Brooks DA, Thornburg KL, Morrison JL. Activation of IGF-2R stimulates cardiomyocyte hypertrophy in the late gestation sheep fetus. J Physiol 590: 5425–5437, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen L, Shi YB. Unliganded Thyroid Hormone Receptor alpha Controls Developmental Timing in Xenopus tropicalis. Endocrinology 156: 721–734, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Young MJ, Moussa L, Dilley R, Funder JW. Early inflammatory responses in experimental cardiac hypertrophy and fibrosis: effects of 11 beta-hydroxysteroid dehydrogenase inactivation. Endocrinology 144: 1121–1125, 2003. [DOI] [PubMed] [Google Scholar]
- 75.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33: W741–W748, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zins K, Pomyje J, Hofer E, Abraham D, Lucas T, Aharinejad S. Egr-1 upregulates Siva-1 expression and induces cardiac fibroblast apoptosis. Int J Mol Sci 15: 1538–1553, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.