Abstract
Micromonospora is a Gram positive bacterium that can be isolated from nitrogen fixing nodules from healthy leguminous plants, where they could be beneficial to the plant. Their plant growth promoting activity in legume and non-legume plants has been previously demonstrated. The present study explores the ability of Micromonospora strains to control fungal pathogens and to stimulate plant immunity. Micromonospora strains isolated from surface sterilized nodules of alfalfa showed in vitro antifungal activity against several pathogenic fungi. Moreover, root inoculation of tomato plants with these Micromonospora strains effectively reduced leaf infection by the fungal pathogen Botrytis cinerea, despite spatial separation between both microorganisms. This induced systemic resistance, confirmed in different tomato cultivars, is long lasting. Gene expression analyses evidenced that Micromonospora stimulates the plant capacity to activate defense mechanisms upon pathogen attack. The defensive response of tomato plants inoculated with Micromonospora spp. differs from that of non-inoculated plants, showing a stronger induction of jasmonate-regulated defenses when the plant is challenged with a pathogen. The hypothesis of jasmonates playing a key role in this defense priming effect was confirmed using defense-impaired tomato mutants, since the JA-deficient line def1 was unable to display a long term induced resistance upon Micromonospora spp. inoculation. In conclusion, nodule isolated Micromonospora strains should be considered excellent candidates as biocontrol agents as they combine both direct antifungal activity against plant pathogens and the ability to prime plant immunity.
Keywords: biocontrol, defense priming, induced systemic resistance, jasmonic acid, Micromonospora, PGPR, tomato
Introduction
Most actinobacteria were considered to inhabit environments such as soil, rhizosphere, or lake sediments. However, it was later discovered that actinobacteria are closely associated with plants and they have been isolated from a great number of different plant genera, colonizing different parts of the plant (Coombs and Franco, 2003; Hasegawa et al., 2006; Qin et al., 2010; Velázquez et al., 2013). Actinobacteria have been described to promote plant growth and their beneficial effect has been reported previously in various plant species (El-Tarabily and Sivasithamparam, 2006; Franco et al., 2007).
Most legumes are engaged in a symbiotic relationship with nitrogen-fixing rhizobia hosted in root nodules. A common misconception was that nodules from leguminous plants were thought to be inhabited by only one type of microorganism, but it is now clear that they conform an ecosystem of their own: besides nodule forming bacteria, several other endophytes with PGP activities are found (Velázquez et al., 2013). Among other actinobacteria, Micromonospora strains have been found in a wide selection of leguminous plants, including Medicago sativa, the plant of choice for the isolation of Micromonospora spp. in our study. Previous studies (Martínez-Hidalgo et al., 2014b) showed that nodule isolated Micromonospora strains excel at plant growth promotion when inoculated in alfalfa. This effect was not due to biological nitrogen fixation although the nitrogen content was significantly higher than in control plants (Martínez-Hidalgo et al., 2014a).
A number of actinobacteria have been also described to reduce the negative effects of pathogens in plants, inhibiting pathogen growth via production of antibiotics, lytic enzymes or siderophores or inducing the plant defense mechanisms (Conn et al., 2008; Hirsch and Valdés, 2010; Verma et al., 2011) but very few have proven ability to promote plant growth, inhibit the growth of pathogens and also boost plant defensive capacity in plants of agronomic relevance.
Plants have developed mechanisms to detect potential aggressors and to coordinate the appropriate defense responses, including the production of toxic substances and lytic enzymes. Regulation of these responses is under phytohormone control, being salicylic acid (SA), jasmonic acid (JA) and ethylene among the major regulators (Pieterse et al., 2009). In general terms, JA coordinates responses effective against necrotrophic pathogens and chewing insects, while SA targets mainly biotrophic pathogens such as viruses, but intensive cross-talk among both pathways (generally antagonistic) allows the plant to shape the final immune response triggered against specific invaders (Pieterse et al., 2009). Upon appropriate stimulation plants can develop a state of enhanced defensive capacity, known as induced systemic resistance (ISR). Different soil beneficial microorganisms have been shown to trigger ISR in plants, usually relying on JA signaling (van Loon et al., 2006; Pozo and Azcón-Aguilar, 2007; Van Wees et al., 2008; Pieterse et al., 2014).
Induced resistance may result in direct activation of defense mechanisms – including increased basal levels of defense-related compounds, or the priming of the plant defensive capacity. In the latter, no major changes appear upon induction in the absence of a challenge, but a more efficient activation of defense mechanisms occurs upon attack. Thus, priming is a cost-effective way of increasing plant resistance (Conrath et al., 2006; Pastor et al., 2013).
The fungus Botrytis cinerea is a plant necrotrophic pathogen that colonizes senescent or dead plant tissues and causes gray mold in vegetables and softening in fruits. Its hyphae can penetrate plant tissues through wounds or natural openings and spread from previously colonized dead tissues into healthy ones. It has a broad host range of food crops, including tomato (Solanum lycopersicum), with the gray mold disease being responsible for substantial economical crop losses every year (Dean et al., 2012). Tomato is one of the most important crops in the world and it is considered an important model plant because, besides its economic importance, it display interesting features, a known genome and a considerable number of mutants and genomic tools available (Kimura and Sinha, 2008). Remarkably, B. cinerea is able to manipulate tomato defense regulatory pathways to promote fungal colonization and disease progression (El Oirdi et al., 2011; Rahman et al., 2012). B. cinerea control is usually achieved by cultural measures and application of broad spectrum fungicides.
The increasingly strict regulation on chemical pesticides and environmental and safety concerns, have evidenced the need of sustainable and safe solutions for crop protection. Thus, formulating bioinoculants with both growth and defense promotion for plants is a major goal in modern agriculture. Micromonospora strains have been isolated from leguminous root nodules and found to improve crop performance in alfalfa (Martínez-Hidalgo et al., 2014b). They are sporulating bacteria, a highly valued trait relevant for its use as bioinoculants, as it allows cultures to be stored for long periods of time without a significant loss in survival.
The aim of this study was to test the potential of Micromonospora strains isolated from alfalfa nodules as biocontrol agents exploring their antifungal properties and their ability to boost plant defense mechanisms using the agronomically relevant pathosystem tomato-B. cinerea. The potential of these strains for improving disease control in horticulture is discussed.
Materials and Methods
Bacterial and Fungal Cultures
All endophytic Micromonospora cultures were maintained in SA1 medium (Trujillo et al., 2005), while all pathogenic fungi were cultured in PDA at 24°C. B. cinerea to be used for plant bioassays was cultured similarly but on PDA supplemented with tomato leaves at 40 mg ml-1 (Vicedo et al., 2009). Micromonospora strains used in this study were isolated from M. sativa root nodules (Table 1).
Table 1.
Strains of Micromonospora used in this study and comparison of gene rrs with already described species.
| Strain (accession) | rrs identification | % | Source |
|---|---|---|---|
| AL4 (KF876221) | Micromonospora viridifaciens DSM 43909T (X92623) | 99,52 | Martínez-Hidalgo et al. (2014b) |
| AL16 (KF876222) | Micromonospora saelicesensis Lupac 09 (AJ783993) | 99,65 | Martínez-Hidalgo et al. (2014b) |
| AL20 (KF876223) | Micromonospora chokoriensis 2-19/6 (AB241454) | 99,79 | Martínez-Hidalgo et al. (2014b) |
| ALF1 (KF876224) | Micromonospora humi P0402 (GU459068) | 99,51 | Martínez-Hidalgo et al. (2014b) |
| ALF2 (KJ187181) | Micromonospora narathiwatensis BTG4-1 (AB193559) | 98,76 | This work |
| ALF4 (KF876225) | Micromonospora coxensis 2-30-b/28 (AB241455) | 99,31 | Martínez-Hidalgo et al. (2014b) |
| ALF7 (KF876233) | Micromonospora saelicesensis Lupac 09 (AJ783993) | 99,86 | Martínez-Hidalgo et al. (2014b) |
| ALFb5 (KF876226) | Micromonospora saelicesensis Lupac 09 (AJ783993) | 99,86 | Martínez-Hidalgo et al. (2014a) |
| ALFb7 (KF876227) | Micromonospora saelicesensis Lupac 09 (AJ783993) | 99,58 | Martínez-Hidalgo et al. (2014b) |
| ALFb1 (KF876228) | Micromonospora echinospora ATCC 15837 (U58532) | 97,78 | Martínez-Hidalgo et al. (2014b) |
| ALFpr18c (KF876230) | Micromonospora tulbaghiae TVU1 (EU196562) | 99,93 | Martínez-Hidalgo et al. (2014a) |
| ALFpr19a (KF876231) | Micromonospora lupini Lupac 14N (AJ783996) | 99,31 | Martínez-Hidalgo et al. (2014b) |
| ALFr4 (KF876234) | Micromonospora cremea CR30 (FN658654) | 98,62 | Martínez-Hidalgo et al. (2014b) |
In vitro Antagonistic Bioassays for Inhibition of Fungal Growth
Pathogens were selected based on their importance as plant pathogens in Spain. The species chosen were Fusarium circinatum, Sclerotinia sclerotiorum, Rhizoctonia solani, and B. cinerea.
Pathogens routinely grown in PDA cultures, were grown in petri dishes with SA1 medium (Trujillo et al., 2005) in order to confirm they could grow normally under these conditions. For the trial, the 13 selected Micromonospora strains (Table 1) were streaked in a thick line in the center of the plate and let grow for 7 days at 28°C. After this time, 1 cm plugs of PDA media containing actively growing fungi of two different species (F. circinatum and R. solani or S. sclerotiorum and B. cinerea) were located equidistant at both sides of the streak of Micromonospora sp. (Supplementary Figure S1) and incubated for 2 days at 24°C. Three replicates were performed for each fungus.
Plant Material and Micromonospora Inoculation Procedures
Five tomato (S. lycopersicum L.) genotypes were used in our studies including the three cultivars ‘Roma,’ ‘Castlemart’ and ‘Moneymaker,’ and the following lines altered in defense signaling: the JA-deficient mutant def1 (Howe et al., 1996; in background ‘Castlemart,’ gently provided by G. Howe, Michigan State University) and SA-impaired transgenic line nahG (Brading et al., 2000; in background ‘Moneymaker’ gently provided by J. Jones, John Innes Centre).
To test the capacity of Micromonospora spp. to ISR in plants against B. cinerea, tomato seeds were sterilized with sodium hypochlorite for 20 min and rinsed three times in sterile distilled water. Seeds were placed on sterile vermiculite, grown until the first true leaf appeared and then transplanted to pots with commercial substrate (Projar Seed Pro 5050, Spain). Plants were randomly distributed and grown in a greenhouse at 24/16°C with a 16/8 h photoperiod and 60% humidity, and watered three times a week with Long Ashton nutrient solution (Hewitt, 1966).
After transplantation, the tomato plants were used in two sets of experiments: to analyze long-term effect of Micromonospora spp. inoculation the plants were inoculated immediately after transplantation with each microbial strain. To analyze short-term effect of Micromonospora spp. inoculation, the plants were inoculated 24 h before the infection with the pathogen. The experiments were repeated at least twice, and for each experiment, five replicates per treatment were used.
Inoculation of tomato plants with the selected Micromonospora strains was performed with 1 ml of bacterial suspensions (109 cells per ml) of each microbial strain grown on solid medium. The bacterial suspensions were carefully strewn in the soil near the roots of the seedling using a micropipette.
Pathogen Bioassays
One month after transplantation, B. cinerea was inoculated either by adding plugs of the fungal culture to each leaflet or by spray on the leaflets of tomato plants with 106 B. cinerea conidia (in whole plants or in detached leaves; Zhai et al., 2013). The extension of the disease was measured 48 and 72 h after the challenge.
B. cinerea Bioassays in Intact Plants
Solanum lycopersicum L. ‘Roma’ were grown in the greenhouse in pots of 1 L capacity, filled with commercial substrate (Compo Sana® Universal potting soil, Compo Iberia S.L.) and inoculated with Micromonospora strains ALFb5 and ALFpr18c a month before inoculation with B. cinerea. Plugs of agar containing B. cinerea micellium were attached to seven leaflets in each plant and located in a humidity chamber kept at 20–23°C. 48 h after challenge with B. cinerea, the diameter of the necrotic lesions formed by the fungal hyphae in the leaflets was measured with the aid of a Vernier caliper (Martínez-Medina et al., 2013). Two measurements were taken for each lesion considering the biggest and the smallest diameter and the average between the two was calculated.
B. cinerea Bioassays on Detached Leaves
Tomato plants were grown as described above, and the inoculation with Micromonospora spp. was performed 30 days or 24 h before pathogen challenge.
In these experiments B. cinerea was applied to detached leaves. Two leaves of each plant were detached with a sharp blade, placed in plastic trays on wet paper and challenged with B. cinerea by applying 1 cm diameter plugs of PDA media containing actively growing mycelia of B. cinerea obtained from 3 weeks-old culture plates. The leaves were then placed in a humidity chamber and kept at 20–23°C with constant light. Disease damage level was scored after 72 h and three levels of damage were established, according to the severity of the symptoms: mild for necrosis extending 1–2 mm from the plug, moderate for necrosis ranging from 3 to 5 mm from the plug and severe for lesions that extended for more than one third of the leaflet (Supplementary Figure S2).
Gene Expression Analysis
Quantitative analysis of defense related gene expression. To evaluate the effects of Micromonospora spp. inoculation on defense gene expression upon pathogen attack, B. cinerea was applied to detached leaves of tomato plants that had been inoculated with Micromonospora spp. at transplanting, 30 days before the challenge with the fungus. Two leaves of each plant were detached, placed in a humidity chamber as described above and challenged by spraying a spore suspension of B. cinerea on the leaf surface. This inoculation method ensures homogeneous and simultaneous contact of the leaf tissue with the pathogen, allowing a more precise quantification of changes in gene expression levels. Spores collected from 15 days-old cultures were incubated in Gambor’s B5 medium (Duchefa, Haarlem, The Netherlands) supplemented with 10 mM sucrose and 10 mM KH2PO4 for 2 h in the dark without shaking (Vicedo et al., 2009). The suspension was adjusted to 5 × 106 spores ml-1. Controls were mock inoculated in a similar way. All leaves were kept under high humidity and harvested at 72 h.
Total RNA was extracted from tomato leaves taken from four different treatments: uninoculated and unchallenged control plants (Control), plants treated only with Micromonospora sp. (pr18c), plants challenged only with B. cinerea (Botrytis) and plants inoculated with Micromonospora sp. and then challenged with B. cinerea (pr18c+Botrytis).
The RNA was extracted following the Tri-Reagent (Sigma) protocol following the manufacturer’s procedure. Contaminating DNA was removed with RQ1 DNase (Promega), purified through a silica column using the NucleoSpin RNA Clean-up kit (Macherey- Nagel). cDNA was synthesized with 3 μg of purified total RNA using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer’s instructions. The conditions of RT-qPCR experiments and the relative quantification of specific mRNA levels was performed according to López-Ráez et al. (2010), using tomato gene specific primers previously described, coding for Pathogenesis related protein PR1a (ID M69247, PR1a; Martínez-Medina et al., 2013), Proteinase inhibitor II (ID K03291, Pin II; Uppalapati et al., 2005), Lipoxygenase A (ID U09026, LoxA; López-Ráez et al., 2010) and elongation factor 1α (ID X14449, SlEF; Rotenberg et al., 2006).
Expression values were normalized using the housekeeping gene SlEF, which encodes for the tomato elongation factor-1α. At least three independent biological replicates were analyzed per treatment, and each qPCR reaction was performed in duplicate.
Detection of Micromonospora sp. in Plant Tissues
Presence of Micromonospora in plant roots and shoots was assessed by amplification of Micromonospora DNA in the samples by PCR. Primers to amplify the gyrB gene, which encodes for the subunit B of DNA gyrase, were designed in our laboratory on the basis of available sequences of this gene in the Micromonospora genus. The primer sequences are: F: TCGACGGCAAGGCGTACG and R: CGCAGCTTCTCSATGTCG. Genomic DNA of root or leaf samples was extracted using NucleoSpin® Plant II columns (Macherey Nagel, Duren, Germany). DNA quality and PCR performance were confirmed in all samples by amplification of the tomato SlEF gene. Bacterial DNA obtained from a pure Micromonospora culture was used as a positive control for gyrB amplification. gyrB amplification conditions were as follows: 9 min at 95°C, 35 cycles of 1 min at 94°C, 1 min at 62°C and 2 min at 72°C, followed by 7 min final extension at 72°C. PCR products were electrophoresed in 1% agarose gels containing ethidium bromide (0.5 μg/ml) using modified Tris-Acetate EDTA buffer (Millipore, Cork, Ireland).
Statistical Analysis
The statistical analysis of data was performed using SPSS software, version 21 (SPSS, Inc., Chicago, IL, USA). The data on lesion diameter in tomato leaves was processed with one-way analysis of variance (ANOVA). The statistical significance of the results was determined using Fisher’s LSD test (P < 0.05).
Association between severity of leaf fungal damage and inoculation treatments was examined using Chi-square tests, followed by z-tests to compare the inoculation groups at each damage level. A criterion of P < 0.05 for statistical significance was used for all analyses and P-values were corrected for multiple tests using the Bonferroni correction.
Results
Micromonospora Strains Inhibit the Growth of Fungal Plant Pathogens In Vitro
The strains under study were isolated from alfalfa root nodules and have been found to have great genetic diversity according to the rrs gene sequences (Table 1).
Selected Micromonospora isolates were tested for their ability to inhibit the growth of the damaging fungal pathogens F. circinatum, S. sclerotiorum, R. solani, and B. cinerea (Supplementary Figure S1).
All of the 13 isolates assayed inhibited the growth of one or more of the selected pathogenic fungi (Table 2). Only two strains of Micromonospora were capable of inhibiting growth of F. circinatum (ALF4 and ALFb7), five strains (AL4, AL16, AL20, ALFpr18c y ALFr4) inhibited S. sclerotinum, seven strains were inhibitory to R. solani (AL20, ALF1, ALF2, ALFb5, ALFb7, ALFpr18c y ALFpr19a) and 10 strains inhibited B. cinerea (AL4, AL20, ALF1, ALF2, ALF7, ALFb1, ALFb5, ALFpr18c, ALFpr19a y ALFpr4). The results suggest the potential of these strains to control fungal diseases through direct effects on the pathogen.
Table 2.
Antimicrobial activity of selected strains of Micromonospora against four selected plant pathogenic fungi.
| Strains | AL4 | AL16 | AL20 | ALF1 | ALF2 | ALF4 | ALF7 | ALFb1 | ALFb5 | ALFb7 | ALFpr18c | ALFpr19a | ALFr4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fusarium circinatum | - | - | - | - | - | + | - | - | - | + | - | - | - |
| Sclerotinia sclerotiorum | + | + | + | - | - | - | - | - | - | - | + | - | + |
| Rhizoctonia solani | - | - | + | + | + | - | - | - | + | + | + | + | - |
| Botrytis cinerea | + | - | + | + | + | - | + | + | + | - | + | + | + |
Micromonospora strains ALFb5 and ALFpr18c, were selected for in planta studies as they inhibited a large number of fungal pathogens (three for ALFpr18c and two for ALFb5) and were previously shown to efficiently promote plant growth (Martínez-Hidalgo et al., 2014b).
Micromonospora sp. Induce Systemic Resistance against B. cinerea in Tomato
The two selected Micromonospora strains (ALFpr18c and ALFb5) were tested for its efficiency to increase tomato resistance against B. cinerea.
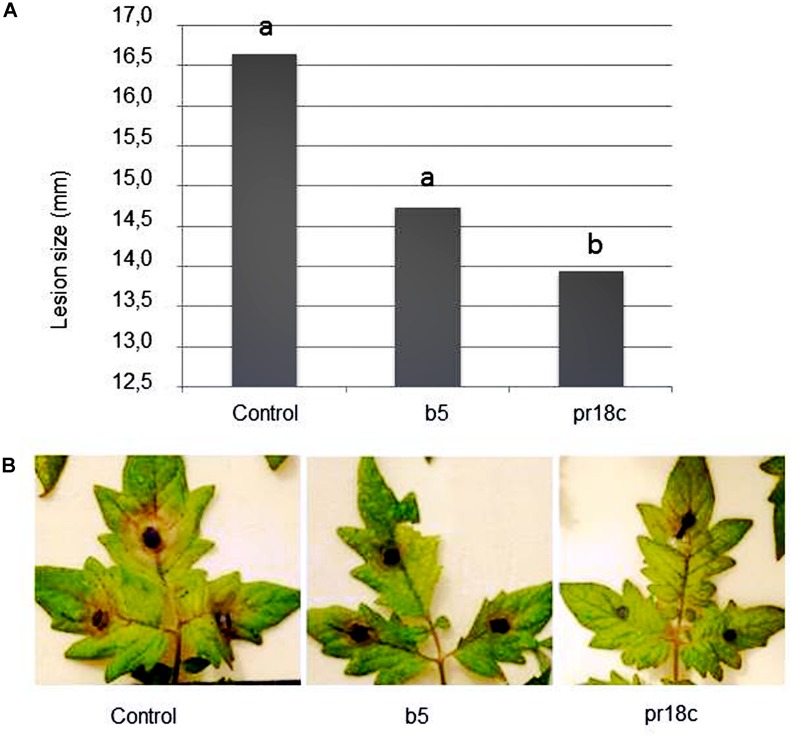
S. lycopersicum L. ‘Roma’ plants were root inoculated with Micromonospora strains at transplanting or 24 h prior challenge with B. cinerea, as described in materials and methods. Plants treated with any of the two Micromonospora strains looked healthier than control plants, even though only plants treated with Micromonospora sp. ALFpr18c showed less disease symptoms at the long term (Figure 1).
FIGURE 1.
Effect of root inoculation with selected Micromonospora strains (ALFb5 and ALFpr18c) on tomato resistance against Botrytis cinerea. (A) Symptom severity in leaves of tomato plants upon challenge with B. cinerea was determined by measuring the diameter of the necrotic lesions 42 h post-pathogen inoculation. Plants were grown in the absence of Micromonospora (Control) or inoculated at transplanting with Micromonospora strain ALFpr18c (pr18c) or strain ALF b5 (b5). Data not sharing a common letter are significantly different (Fisher’s protected LSD test at P ≤ 0.05). (B) Examples of B. cinerea symptom development in leaves of Micromonospora spp. inoculated (b5 or PR18c) or non-inoculated (control) plants.
Remarkably, this significant reduction on the severity of symptoms caused by the pathogen was also evidenced even when pathogen inoculation was performed on detached leaves in a different experiment. As Micromonospora sp. ALFpr18c was consistently the most efficient strain in reducing disease severity, it was selected for the follow up studies.
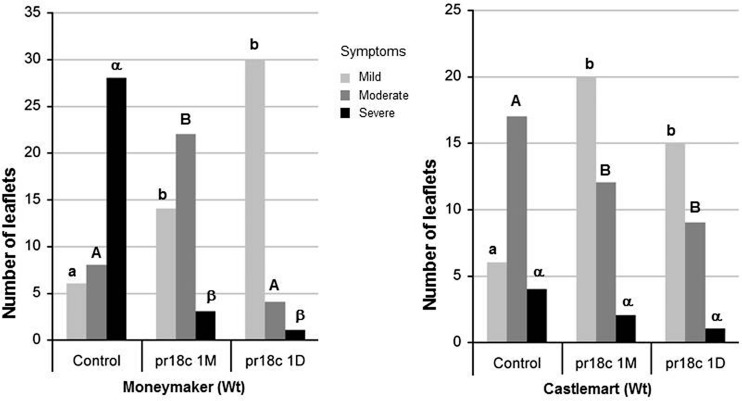
We extended our analysis to other tomato cultivars to find out if the protection by Micromonospora sp. ALFpr18c was a consistent effect for tomato and not a cultivar-specific response, choosing two well-characterized cultivars with defense impaired mutants available: ‘Castlemart’ and ‘Moneymaker.’ The sensitivity to B. cinerea of the two tomato cultivars differ significantly, being ‘Castlemart’ less severely affected than ‘Moneymaker’ (X2 = 18,871, P = 0.001; Figure 2). Remarkably, inoculation with Micromonospora spp. resulted in a significant reduction of the disease symptoms in both cultivars. The ISR by ALFpr18c against B. cinerea was effective regardless the timing of the inoculation of the bacteria, 30 days or 24 h before the challenge with the pathogen (Figure 2). Micromonospora sp. protected plants challenged with B. cinerea by reducing the severity of damage caused by the pathogen, as it was shown by a significant decrease in the number of leaflets with the highest level of damage (Figure 2). Indeed, there was a statistically significant association between levels of leaf fungal damage and inoculation treatments for ‘Castlemart’ (X2 = 8,374, P = 0.015) and for ‘Moneymaker’ (X2 = 29,824, P = 0.001).
FIGURE 2.
Effect of Micromonospora ALFpr18c root inoculation on B. cinerea disease development in different tomato cultivars (Solanum lycopersicom L. ‘Moneymaker’ and ‘Castlemart’). Necrosis severity caused by Botrytis was scored using a three levels disease scale: mild, moderate and severe, as shown in Supplementary Figure S2. The number of leaflets within each category is shown. For each tomato cultivar, disease damage level was compared between the control and the bacterial treatments. Control, plants not inoculated with Micromonospora; pr18c 1M and pr18c 1D, plants inoculated with Micromonospora sp. pr18c 1 month or 1 day before challenge with B. cinerea, respectively. Bars not sharing a common letter (lowercase for mild, uppercase for moderate and greek for severe symptoms) are significantly different using Bonferroni corrected Chi-square tests, followed by z-tests (P ≤ 0.05).
The spatial separation between both microorganisms (Micromonospora was inoculated in the soil, close to the roots, and the pathogen in the leaves) suggest that the protection is related to the activation of plant defense mechanisms. However, to rule out a possible direct antifungal activity of the bacteria through an eventual colonization of the leaf tissue in the long term treatments, we evaluated the presence of the bacteria in roots and leaves of inoculated and non-inoculated tomato plants using Micromonospora gene gyrB specific primers. GyrB amplification, and therefore, the presence of Micromonospora sp. ALFpr18c was evidenced in the roots of the plants inoculated at transplanting, 30 days before harvesting, confirming effective root colonization by Micromonospora. However, no bacterial gene amplification was detected in the leaves of the same plants, confirming the bacteria localization in the roots and the physical separation of the resistance inducer and the pathogen (Supplementary Figure S3).
Micromonospora sp. Induced Resistance is Related to Priming of Jasmonate-Regulated Responses
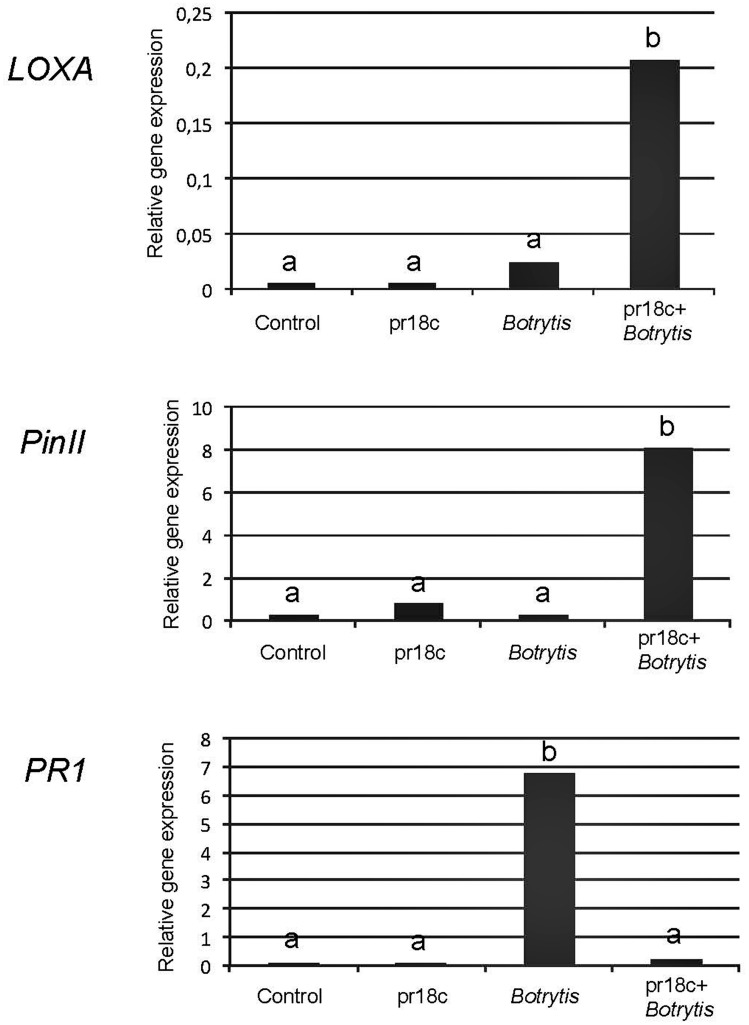
To understand the mechanism behind this long term induced resistance, and to determine what defense signaling pathways were involved, we compared the response against B. cinerea in plants inoculated or not with the strain Micromonospora sp. ALFpr18c 30 days before the challenge with the pathogen. Plant defense responses to Botrytis are known to depend on the interplay between SA and JA dependent responses, and those regulated by JA have been proposed as the major players in resistance (El Oirdi et al., 2011). Accordingly, we monitored the expression levels of well-characterized marker genes for both signaling pathways in leaves from the different treatments. Non-challenged plants showed no differences in the expression levels of any of the marker genes analyzed regardless of the presence of Micromonospora sp. (Figure 3). In contrast, important differences were found in the transcription levels of marker genes from both pathways between non-inoculated plants and plants preinoculated with Micromonospora spp. upon challenge with B. cinerea (Figure 3).
FIGURE 3.
Expression levels of the defense related genes PR1, LoxA, and PinII in leaves of tomato plants. Relative expression levels for PR1, LoxA, and PinII were determined by qPCR and normalized against the constitutive SlEF (tomato elongation factor α). Control: uninoculated, unchallenged plants; pr18c: plants inoculated with Micromonospora strain ALFpr18c; Botrytis: uninoculated plants challenged with B. cinerea; pr18c+Botrytis: plants previously inoculated with Micromonospora strain ALFpr18c and challenged with B. cinerea. Micromonospora inoculation was performed at transplanting, a month before infection with Botrytis. Data not sharing a common letter are significantly different according to Fisher’s protected LSD test at P ≥ 0.05.
The PR1 gene, encoding for an acidic form of the Pathogenesis Related Protein 1, is well-recognized as a marker of SA dependent responses. The expression of PR1 was strongly induced in control plants upon challenge with the pathogen (sevenfold), but the increase upon B. cinerea challenge was much lower in plants previously inoculated with Micromonospora sp.
Regarding the JA regulated signaling pathway, the expression of the marker genes PinII, encoding for Proteinase inhibitor II, known to be involved in the plant resistance to B. cinerea, and LoxA, coding for a JA-inducible lipoxygenase involved in the biosynthesis of oxylipins was monitored. Transcript levels of both PinII and LoxA genes were significantly higher in plants inoculated with Micromonospora sp. and then challenged with B. cinerea than in the rest of treatments (Figure 3). Remarkably, control plants without Micromonospora sp. but challenged with B. cinerea showed very low levels of expression of both JA-marker genes, indicating a poor activation of the efficient (JA regulated) defense mechanisms in the absence of Micromonospora ALFpr18c.
Micromonospora sp. Induced Resistance against B. cinerea is Compromised in Defense-Deficient Tomato Mutants
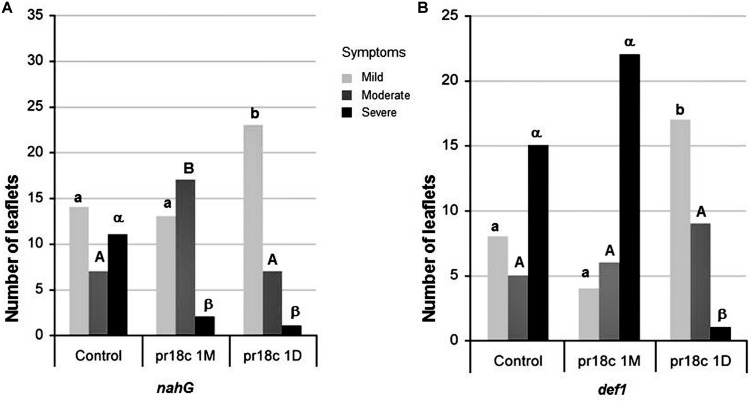
The ability of Micromonospora sp. ALFpr18c to induce resistance against B. cinerea was analyzed in tomato plants impaired in JA or SA signaling (Figure 4). Tomato plants were challenged with B. cinerea 30 days after root inoculation with Micromonospora sp., as described in Section “Materials and Methods.”
FIGURE 4.
Effect of Micromonospora ALFpr18c root inoculation on disease development caused by B. cinerea in different defense deficient tomato lines. (A) SA-deficient nahG and (B) JA-deficient def 1 plants. Necrosis severity caused by B. cinerea was scored using a three levels disease scale: mild, moderate and severe, as shown in Supplementary Figure S2. Control, plants not inoculated with Micromonospora; pr18c 1M and pr18c 1D, plants inoculated with Micromonospora sp. ALFpr18c 1 month or 1 day before challenge with B. cinerea, respectively. Within each genotype, bars not sharing a common letter (lowercase for mild, uppercase for moderate and greek for severe symptoms) are significantly different using Bonferroni corrected Chi-square tests, followed by z-tests (P ≤ 0.05).
To address the possible role of the SA regulated defense pathway in the Micromonospora ISR, nahG plants and their corresponding wild type (‘Moneymaker’) were used. nahG plants, unable to accumulate SA, were significantly less susceptible to B. cinerea than their corresponding wild type (X2 = 9,499, P = 0.009). Thus, SA appears to have a negative role in disease resistance, in agreement with recent reports showing that B. cinerea manipulate SA signaling to promote infection and disease progression (El Oirdi et al., 2011).
When studying the effect of ALFpr18c on nahG mutant, there is a statistically significant association between levels of leaf fungal damage and inoculation treatments: (X2 = 10,434, P = 0.005); Micromonospora sp. inoculated nahG plants showed a significant reduction on the severity of symptoms caused by the pathogen, as evidenced by the decrease in the percentage of leaves with higher severity symptoms (Figure 4). No significant differences were found (X2 = 0,190, P = 0.909) when comparing ALFpr18c-inoculated nahG or wild type (Moneymaker) plants, confirming that Micromonospora induced resistance was not impaired in the SA deficient mutant.
To analize the relevance of JA signaling on Micromonospora ISR, def1 mutants, deficient in JA regulated responses and their corresponding wild type cultivar (‘Castlemart’) were used. Differences in the basal resistance of def1 and its parental line (‘Castlemart’) were clear, being def1 plants significantly more susceptible to B. cinerea than the corresponding wild type cultivar (X2 = 13,186, P = 0.001; Figure 4), supporting the key role of JA in resistance to B. cinerea. When analyzing the effect of ALFpr18c on def1 mutant, there is not a statistically significant association between levels of leaf fungal damage and inoculation treatments: (X2 = 2,493, P = 0.288). Accordingly, plant protection against the pathogen by the strain ALFpr18c is lost in the JA deficient mutant.
Taken all together, our results show that JA has a positive role in tomato resistance against B. cinerea, and these responses are effectively primed by Micromonospora sp.
We also tested the efficacy of the short term induction of resistance, comparing disease severity when Micromonospora ALFpr18c inoculation was performed 24 h before the challenge. Plants treated with ALFpr18c showed less severe lesions than control plants regardless the tomato line tested, even in the case of the JA compromised mutant def1, unable to maintain long term ISR (Figure 4).
Discussion
In the context of the new agricultural sustainability directives that have been outlined by the European Union, the search for effective bioinoculants is a major goal in agronomic research. The Micromonospora strains presented here, isolated from alfalfa root nodules showed very good plant growth promoting activity (Martínez-Hidalgo et al., 2014a). Plant growth promotion is often achieved by both improved plant nutrition (Vessey, 2003; Velázquez et al., 2013) and enhanced stress tolerance, for example, reducing disease damages. In this study we evaluated the potential of Micromonospora strains for biological control of pathogens. We evaluate the two different ways by which Micromonospora spp. could act as biocontrol agents, by testing their direct inhibitory activity against plant pathogens, and their ability to boost plant defense mechanisms.
Most of the Micromonospora strains tested showed a clear inhibitory effect on multiple plant pathogenic fungi in in vitro antagonistic assays (Table 2). Production of secondary metabolites is common in Actinobacteria, many of them with antibiotic effect (El-Tarabily et al., 1997; Gadelhak et al., 2005; Hirsch and Valdés, 2010). Production of antimicrobial substances by biocontrol agents is associated to plant protection by selective inhibition of fitopathogenic fungi, thus avoiding crop losses linked to diseases. Root diseases caused by soil pathogens are a major problem in agriculture and antagonism in the rhizosphere is an effective tool to decrease their incidence and damage. Inoculation with Micromonospora spp. may contribute to this antagonistic effect. The metabolic versatility of Micromonospora spp. is very high, they are able to produce multiple metabolites, such as antibiotics (Weinstein et al., 1966), antitumorals (Igarashi et al., 2007) or lytic enzymes, like chitinases, or proteases that could inhibit germination of B. cinerea spores or suppress fungal growth (Frankowski et al., 2001; Kamensky et al., 2003). Further studies will be needed to discover the metabolites or enzymes responsible for the observed inhibitory effects of our selected strains.
Besides a direct antimicrobial effect, that can contribute to reduce pathogens propagation in the soil, Micromonospora sp. ALFpr18c is able to stimulate plant defenses and ISR against foliar pathogens. Here we show that root inoculation with Micromonospora sp. enhanced disease resistance to the foliar pathogen B. cinerea. For every tomato cultivar tested in the different trials (‘Roma,’ ‘Moneymaker’ and ‘Castlemart’), the extent and severity of the symptoms caused by B. cinerea in plants pretreated with Micromonospora sp. was significantly lower than in untreated controls.
Micromonospora sp. inoculation was performed in the soil, near the root system, while B. cinerea was directly applied to leaves, so that there was no contact between both microorganisms. Absence of bacteria on the leaves was confirmed by DNA amplification analysis. Thus, the direct antagonism of the bacteria on the fungus in our experimental conditions is very unlikely. The protection observed was, therefore, likely related to an effect on the plant defense mechanisms, further confirmed by the lack of Micromonospora triggered ISR in the def1 defense deficient mutant lines.
The two Micromonospora strains tested in plant bioassays, ALFpr18c and ALFb5, reduced disease progression when inoculated 24 h before challenging the plants with B. cinerea. However, only ALFpr18c was able to induce long term systemic resistance, since inoculation at transplanting -30 days before challenge- with these bacteria, but not with ALFb5, did reduce disease severity significantly. For this reason, ALFpr18c was selected as an effective inducer of durable resistance and used to uncover the mechanisms underlying such effect.
Plant defense responses to B. cinerea are coordinated by the interplay of the JA and SA regulated signaling pathways (El Oirdi et al., 2011), the two major branches of defense related signaling. Quantitative analysis of transcript levels of marker genes for both pathways and the use of tomato lines defective in the signaling pathways allow to determine which pathways are activated in the different treatments, as previously described for other microbial inducers of plant resistance (Martínez-Medina et al., 2013). In the absence of pathogen attack, the bacteria do not have a clear impact on these major defense signaling pathways in the leaves (Figure 3). In contrast, the pathogen B. cinerea triggers a strong induction of the SA pathway, as previously described (El Oirdi et al., 2011). It has been proven that B. cinerea is sensitive to JA regulated responses, but not to those regulated by SA. Since SA and JA pathways are mutually antagonistic, the pathogen manipulate SA signaling to downregulate JA dependent responses to promote disease and necrosis spreading (El Oirdi et al., 2011; Rahman et al., 2012). The negative role of SA in tomato resistance against B. cinerea is here confirmed by the fact that the SA impaired line nahG was significantly less susceptible to B. cinerea than its corresponding wild type background (Figure 4), so SA pathway impairment renders the plant less susceptible to the fungus. Remarkably, the plants previously treated with the bacteria showed a minimal induction of the SA pathway upon challenge, thus the bacteria prevented the SA response triggered by the pathogen. In contrast, JA regulated responses were strongly upregulated in the preinoculated plants (Figure 3), particularly, Pin II that encodes for a proteinase inhibitor with strong inhibitory effects on B. cinerea (El Oirdi et al., 2011). As Micromonospora sp. itself does not induce a response from the plant, but this increase in defense was only evident upon challenge, the results support that Micromonospora sp. ISR against B. cinerea through priming of JA regulated defense responses. Priming is a cost efficient way of inducing resistance since defenses are only boosted upon pathogen attack, but they remain in basal levels in the absence of pathogen pressure (Conrath et al., 2006). Other beneficial microorganisms prime JA dependent responses, appearing as a common mechanism for efficient resource management in beneficial plant-microbe interactions (Van Wees et al., 2008; Selosse et al., 2014). It has been postulated that during initial stages of the interaction, beneficial soil microorganisms are perceived as potential aggressors by the plants, thus triggering some general defense responses. Later on, the beneficial bacteria deal with the plant immune system modulating plant defenses to achieve successful colonization, and as result, defenses may remain under “alert,” or primed (Zamioudis and Pieterse, 2012).
The analysis of the induction of resistance in tomato lines altered in the JA and SA signaling pathways further support this notion. The different lines showed a decrease in the severity of B. cinerea symptoms when inoculated with Micromonospora sp. Plants that were inoculated 24 h before the challenge with the pathogen show a reduction in disease severity in all tomato lines, including the JA and SA signaling deficient lines nahG and def1. Thus, this protection is probably due to an early general defense response to the bacteria that affect B. cinerea, but differs from the long term protective effects. Differences in the mechanisms underlying early and late responses associated to priming have been described, with early responses associated to the accumulation of reactive oxygen species and cell wall reinforcements, and late responses being under phytohormone control (Pastor et al., 2013), and a similar distinction may be applying here. In fact, the early unspecific defense response from the plant upon colonization by beneficial bacteria seems frequent. Even Rhizobium, known for establishing a very well-coordinated symbiosis with legumes, triggers a peak of defense pathway activation during the early stages of the interaction (Santos et al., 2001; Soto et al., 2006).
When Micromonospora sp. was inoculated at transplanting, 30 days before the challenge, disease severity was significantly reduced in the different tomato cultivars tested lines. However, the induction of resistance was completely lost in the JA deficient mutant def1, that was even more severely affected when inoculated with the bacteria.
In summary, the transcriptional analysis and the genetic approach with tomato deficient lines evidenced that the durable systemic resistance induced by Micromonospora sp. ALFpr18c is based on priming of JA regulated defense responses. JA dependent defenses, although mainly effective against necrotrophs, may also affect hemibiotrophs and even biotrophs (Pozo et al., 2005), so the spectrum of efficiency of Micromonospora sp. induced resistance deserves further exploration.
In this article, we provided evidences of the potential of Micromonospora strains as biocontrol agents for long lasting crop protection against phytopathogenic fungi. The priming activity of Micromonospora spp. is sustained in time -more than a month in our experiments- without significantly reducing its effectiveness, therefore inoculation with these bacteria could be performed at the time of sowing without a reduction in the effectiveness of the protection over time. These results, together with the direct antifungal potential evidenced for these strains, their proven role as plant probiotic bacteria and their sporulation capacity makes bacteria from the genus Micromonospora a very promising source of multifunctional bioinoculants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by MICINN Grant AGL2010-17380 and AGL-2012-39923. We thank Estefania Berrio for excellent technical support. PM-H was supported by a fellowship from CSIC JAE-PRE.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00922
References
- Brading P. A., Hammond-Kosack K. E., Parr A., Jones J. D. (2000). Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J. 23 305–318. 10.1046/j.1365-313x.2000.00778.x [DOI] [PubMed] [Google Scholar]
- Conn V. M., Walker A. R., Franco C. M. M. (2008). Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol. Plant Microbe 21 208–218. 10.1094/MPMI-21-2-0208 [DOI] [PubMed] [Google Scholar]
- Conrath U. U., Beckers G. J. M. G., Flors V. V., García-Agustín P. P., Jakab G. G., Mauch F. F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe 19 1062–1071. 10.1094/MPMI-19-1062 [DOI] [PubMed] [Google Scholar]
- Coombs J. T., Franco C. M. (2003). Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69 5603–5608. 10.1128/AEM.69.9.5603-5608.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R., Van Kan J. A. L., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13 414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T. A., Rigano L., El Hadrami A., Rodriguez M. C., Daayf F., et al. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23 2405–2421. 10.1105/tpc.111.083394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tarabily K. A., Hardy G. E. S. J., Sivasithamparam K., Hussein A., Kurtböke I. (1997). The potential for the biological control of cavity spot disease of carrots caused by Pythium coloratum, by streptomycete and non-streptomycete Actinomycetes in Western Australia. New Phytol. 137 495–507. 10.1046/j.1469-8137.1997.00856.x [DOI] [PubMed] [Google Scholar]
- El-Tarabily K. A., Sivasithamparam K. (2006). Non-streptomycete actinomycetes as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Soil Biol. Biochem. 38 1505–1520. 10.1016/j.soilbio.2005.12.017 [DOI] [Google Scholar]
- Franco C., Michelsen P., Percy N., Conn V., Listiana E., Moll S., et al. (2007). Actinobacterial endophytes for improved crop performance. Aust. Plant Pathol. 36 524–531. 10.1071/AP07067 [DOI] [Google Scholar]
- Frankowski J. J., Lorito M. M., Scala F. F., Schmid R. R., Berg G. G., Bahl H. H. (2001). Purification and properties of two chitinolytic enzymes of Serratia plymuthica HRO-C48. Arch. Microbiol. 176 421–426. 10.1007/s002030100347 [DOI] [PubMed] [Google Scholar]
- Gadelhak G. G., El-Tarabily K. A., Al-Kaabi F. K. (2005). Insect control using chitinolytic soil actinomycetes as biocontrol agents. Int. J. Agr. Biol. 7 627–633. [Google Scholar]
- Hasegawa S., Meguro A., Shimizu M., Nishimura T., Kunoh H. (2006). Endophytic actinomycetes and their interactions with host plants. Actinomycetologica 20 72–81. 10.3209/saj.20.72 [DOI] [Google Scholar]
- Hewitt E. J. (1966). Sand and water culture methods used in the study of plant nutrition. Commonwealth Agric. Bureaux 3 104 10.1017/S0014479700021852 [DOI] [Google Scholar]
- Hirsch A. M., Valdés M. (2010). Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol. Biochem. 42 536–542. 10.1016/j.soilbio.2009.11.023 [DOI] [Google Scholar]
- Howe G. A., Lightner J., Browse J., Ryan C. A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8 2067–2077. 10.1105/tpc.8.11.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y., Trujillo M. E., Martínez-Molina E., Yanase S., Miyanaga S., Obata T., et al. (2007). Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg. Med. Chem. Lett. 17 3702–3705. 10.1016/j.bmcl.2007.04.039 [DOI] [PubMed] [Google Scholar]
- Kamensky M., Ovadis M., Chet I., Chernin L. (2003). Soil-borne strain IC14 of Serratia plymuthica with multiple mechanisms of antifungal activity provides biocontrol of Botrytis cinerea and Sclerotinia sclerotiorum diseases. Soil Biol. Biochem. 35 323–331. 10.1016/S0038-0717(02)00283-283 [DOI] [Google Scholar]
- Kimura S., Sinha N. (2008). Tomato (Solanum lycopersicum): a model fruit-bearing crop. CSH Protoc. 3 1–9. 10.1101/pdb.emo105 [DOI] [PubMed] [Google Scholar]
- López-Ráez J. A., Verhage A., Fernández I., García J. M., Azcón-Aguilar C., Flors V., et al. (2010). Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J. Exp. Bot. 61 2589–2601. 10.1093/jxb/erq089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hidalgo P., Galindo-Villardón P., Igual J. M., Martínez-Molina E. (2014a). Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). A new promising plant probiotic bacteria. Sci. Rep. 4:6389 10.1038/srep06389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hidalgo P., Olivares J., Delgado A., Bedmar E., Martínez-Molina E. (2014b). Endophytic Micromonospora from Medicago sativa are apparently not able to fix atmospheric nitrogen. Soil Biol. Biochem. 74 201–203. 10.1016/j.soilbio.2014.03.011 [DOI] [Google Scholar]
- Martínez-Medina A., Fernández I., Sánchez-Guzmán M. J., Jung S. C., Pascual J. A., Pozo M. J. (2013). Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 4:206 10.3389/fpls.2013.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor V., Luna E., Mauch-Mani B., Ton J., Flors V. (2013). Primed plants do not forget. Environ. Exp. Bot. 94 46–56. 10.1016/j.envexpbot.2012.02.013 [DOI] [Google Scholar]
- Pieterse C. M. J., Leon-Reyes A., Van der Ent S., Van Wees S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 308–316. 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Zamioudis C., Berendsen R. L., Weller D. M., Van Wees S. C. M., Bakker P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52 347–375. 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed] [Google Scholar]
- Pozo M. J., Azcón-Aguilar C. (2007). Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant. Biol. 10 393–398. 10.1016/j.pbi.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Pozo M. J., Loon L. C., Pieterse C. M. J. (2005). Jasmonates – Signals in plant-microbe interactions. J. Plant Growth Regul. 23 211–222. [Google Scholar]
- Qin S., Xing K., Jiang J.-H., Xu L.-H., Li W.-J. (2010). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic Actinobacteria. Appl. Microbiol. Biotechnol. 89 457–473. 10.1007/s00253-010-2923-2926 [DOI] [PubMed] [Google Scholar]
- Rahman T. A. E., Oirdi M. E., Gonzalez-Lamothe R., Bouarab K. (2012). Necrotrophic pathogens use the salicylic acid signaling pathway to promote disease development in tomato. Mol. Plant Microbe Interact. 25 1584–1593. 10.1094/MPMI-07-12-0187-R [DOI] [PubMed] [Google Scholar]
- Rotenberg D., Thompson T. S., German T. L., Willis D. K. (2006). Methods for effective real-time RT-PCR analysis of virus- induced gene silencing. J. Virol. Methods 138 49–59. 10.1016/j.jviromet.2006.07.017 [DOI] [PubMed] [Google Scholar]
- Santos R., Hérouart D., Sigaud S., Touati D., Puppo A. (2001). Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant Microbe 14 86–89. 10.1094/MPMI.2001.14.1.86 [DOI] [PubMed] [Google Scholar]
- Selosse M.-A., Bessis A., Pozo M. J. (2014). Microbial priming of plant and animal immunity: symbionts as developmental signals. Trends Microbiol. 22 607–613. 10.1016/j.tim.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Soto M. J., Sanjuán J., Olivares J. (2006). Rhizobia and plant-pathogenic bacteria: common infection weapons. Microbiology 152 3167–3174. 10.1099/mic.0.29112-0 [DOI] [PubMed] [Google Scholar]
- Trujillo M. E., Fernández-Molinero C., Velázquez E., Kroppenstedt R. M., Schumann P., Mateos P. F., et al. (2005). Micromonospora mirobrigensis sp. nov. Int. J. Syst. Evol. Microbiol. 55 877–880. 10.1099/ijs.0.63361-63360 [DOI] [PubMed] [Google Scholar]
- Uppalapati S. R., Ayoubi P., Weng H., Palmer D. A., Mitchell R. E., Jones W., et al. (2005). The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 42 201–217. 10.1111/j.1365-313X.2005.02366.x [DOI] [PubMed] [Google Scholar]
- van Loon L. C., Geraats B. P. J., Linthorst H. J. M. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11 184–191. 10.1016/j.tplants.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Van Wees S. C. M., Van der Ent S., Pieterse C. M. J. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11 443–448. 10.1016/j.pbi.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Velázquez E., Martínez-Hidalgo P., Carro García L., Alonso-Vega P., Peix A., Trujillo M. E., et al. (2013). “Nodular endophytes: an untapped diversity,” in Beneficial Plant-Microbial Interactions (Boca Raton, FL: CRC Press LLC.), 214–236. 10.1007/s00203-011-0733-736 [DOI] [Google Scholar]
- Verma V. C., Singh S. K., Prakash S. (2011). Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss. J. Basic Microbiol. 51 550–556. 10.1002/jobm.201000155 [DOI] [PubMed] [Google Scholar]
- Vessey J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255 571–586. 10.1023/A:1026037216893 [DOI] [Google Scholar]
- Vicedo B., Flors V., de la O Leyva M., Finiti I., Kravchuk Z., Real M. D., et al. (2009). Hexanoic acid-induced resistance against Botrytis cinerea in tomato plants. Mol. Plant Microbe 22 1455–1465. 10.1094/MPMI-22-11-1455 [DOI] [PubMed] [Google Scholar]
- Weinstein M. J., Luedemann G. M., Oden E. M., Wagman G. H. (1966). Halomicin, a new Micromonospora-produced antibiotic. Antimicrob. Agents Chemother. 7 435–441. [DOI] [PubMed] [Google Scholar]
- Zamioudis C., Pieterse C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe 25 139–150. 10.1094/MPMI-06-11-0179 [DOI] [PubMed] [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., et al. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9:e1003422 10.1371/journal.pgen.1003422 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.