Abstract
Neutrophils are an emerging cellular source of IFN-γ, a key cytokine that mediates host defense to intracellular pathogens. Production of IFN-γ by neutrophils, in contrast to lymphoid cells, is Toll-like receptor (TLR) and IL-12-independent and the events associated with IFN-γ production by neutrophils are not understood. In this report, we show that mouse neutrophils express IFN-γ during their lineage development in the bone marrow niche at the promyelocyte stage independently of microbes. IFN-γ accumulates in primary neutrophilic granules and is released upon induction of degranulation. The developmental mechanism of IFN-γ production in neutrophils arms the innate immune cells prior to infection and assures the potential for rapid release of IFN-γ upon neutrophil activation, the first step during responses to many microbial infections.
Introduction
Induction of immune responses to microbial pathogens is generally divided into distinct stages, in which the recognition of microbial molecules mediated by innate immune receptors results in maturation of dendritic cells and macrophages, followed by secretion of activating cytokines that regulate the effector phase of host defense (1, 2). This model is particularly well studied in the context of TLR-mediated immunity, in which DC-specific TLR activation results in production of IL-12 that subsequently regulates IFN-γ production by NK and T cells (3). In vivo experiments with the protozoan parasite Toxoplasma gondii formally tested this model and revealed that while NK and CD4+ T cell IFN-γ production was regulated by TLR11-dependent activation of MyD88 (4–8), there is an unforeseen component of IFN-γ-dependent host response mediated by neutrophils (9). In contrast to NK (10, 11), T cells (5, 12, 13), and innate lymphoid cells (14), the ability of neutrophils to produce IFN-γ does not depend on IL-12 or TLR-dependent parasite recognition (9), and therefore how neutrophils produce IFN-γ is incompletely understood. This knowledge is crucial, since while neutrophils have long been viewed as effector cells that mediate their protective effects via the release of lytic granules, recent evidence suggests that neutrophils secrete cytokines which have pleotropic effects on host defense to microbial infections (15, 16).
Here we investigated how neutrophils produce IFN-γ. Our data revealed that neutrophil IFN-γ production is regulated during hematopoietic development of neutrophil precursor cells and that IFN-γ accumulates in primary granules produced in the promyelocyte stage of neutrophil development. The accumulation of IFN-γ in primary granules is independent of microbial infection or the endogenous microbiota. Neutrophil IFN-γ-effector mechanisms are achieved by activation induced degranulation. This represents a broad innate immune mechanism of IFN-γ-mediated host defense required for host protection from intracellular pathogens.
Materials and Methods
Mice
C57BL/6, germ-free C57BL/6, and TLR11−/− mice have been previously described in experimental toxoplasmosis (5, 6, 17). All experiments were performed with protocols approved by the Institutional Animal Care and Use Committees at UT Southwestern.
Ex Vivo analysis of IFN-γ production by neutrophils
C57BL/6 and TLR11−/− mice were infected intraperitoneally with 20 cysts of T. gondii (ME49 strain) as previously described (6, 9). Cells were not given any stimulation or golgi inhibitors unless otherwise indicated. For indicated experiments, peritoneal cells were incubated for 5 hours at 37C in media with or without GolgiPlug 1μg/ml. To block protein translation, peritoneal cells were incubated in media with GolgiPlug and increasing concentrations of Cycloheximide, with or without PMA (50ng/ml) and ionomycin (750ng/ml). To induce neutrophil degranulation, cells were activated with 10 nM fMLP, 100ng/ml LPS, 50ng/ml PMA, and/or 750 ng/ml ionomycin, incubated for 1 hour at 37C, supernatants collected for IFN-γ ELISA (eBioscience) and cells stained for flow cytometry.
Flow Cytometry
For bone marrow (BM) and peritoneal cells, single cell suspensions were stained in PBS plus 1% FBS and 0.5mM EDTA. Blood samples were directly stained and erythrocytes lysed with ACK lysing buffer. After extracellular staining, samples were fixed and permeabilized for intracellular staining using the FoxP3 staining buffer kit by eBioscience, per the manufactures instructions. The following antibodies were used for staining and were from eBioscience unless otherwise indicated: αNK1.1, αCD34, αIFN-γ (XMG1.2; BD Bioscience), αLy-6G (BD Bioscience), αCD34, αCD117, αCD16/32, αSca-1, αCD19, αF4/80, αTer119, αCD11b, αGr-1, and isotype IgG1κ. These data were acquired on FACSCalibur and FACSCanto cytometers or on the MoFlo cytometer and analyzed with FlowJo.
Quantitative Real Time-PCR
RNA from defined immature neutrophils was isolated using the PureLink RNA Mini kit. cDNA synthesis was done using SuperScript III RT. Samples were analyzed on MyiQ Real-Time PCR System and data were processed for relative expression to control gene HPRT.
Microscopy
Magnetic bead sort-purified Ly-6G+ BM cells were fixed in 4% PFA, blocked in PBS with 2% BSA and 0.2% TritonX-100, and stained with antibodies of interest. The following antibodies were used for microscopy analysis: αIFN-γ Alexa488 (XMG1.2; eBioscience), αMyeloperoxidase (ab9535; Abcam), αLactoferrin (ab135710; Abcam), αGelatinase (ab38898; Abcam), and donkey anti-rabbit IgG Alexa568 (A10042;Life Technologies). Images were acquired with Leica TCS SPE or SP5 laser scanning confocal microscopes. Downstream images were examined with ImageJ, Z stacks underwent 3D blind deconvolution with AutoQuant X software, colocalization analysis (Manders coefficient (18)) and 3D modeling done with Imaris. Statistical Analysis.
All statistical analysis on bar graphs is done using the standard unpaired t-test and error bars shown are mean ± SD.
Results
Constitutive production of IFN-γ by neutrophils in the absence of microbial stimulation
T. gondii elicits potent innate and adaptive IFN-γ responses from multiple cell types including neutrophils, innate lymphoid cells, NK, and T cells (19). While TLR activation and IL-12 production by DCs is required for IFN-γ production by lymphoid cells, how neutrophils mediate TLR and IL-12 independent IFN-γ production is largely unknown (20). Therefore, we began to address this question by examining the appearance of IFN-γ-producing neutrophils triggered by T. gondii infection.
Neutrophils are present in large numbers in the peritoneal exudate cells (PEC) isolated from the site of T. gondii infection in WT mice and they stain positive for IFN-γ (Fig. 1A). Yet, the amount of IFN-γ detected in neutrophils by intracellular staining was not increased by in vitro incubation with GolgiPlug, a protein transport inhibitor that leads to intracellular accumulation of IFN-γ seen in NK and T cells (Supplemental Fig. 1A). To boost IFN-γ expression, neutrophils and T cells isolated from T. gondii infected mice were stimulated with PMA and ionomycin. As expected, stimulated T cells produced large amounts of IFN-γ that was inhibited in a dose dependent manner by the de novo protein synthesis inhibitor cycloheximide (CHX) (Supplemental Fig. 1B). In contrast, stimulation of neutrophils failed to increase their IFN-γ positivity and CHX treatment had no dose-dependent effects on the levels of IFN-γ detected in neutrophils (Supplemental Fig. 1B). Taken together these data revealed that neutrophils expressed IFN-γ prior to isolation from T. gondii infected mice.
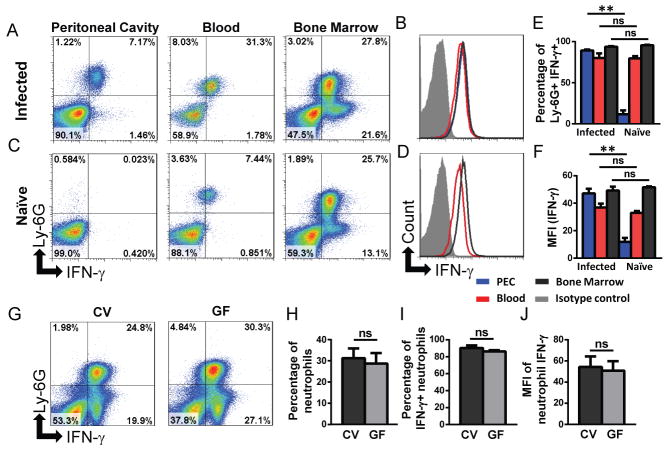
Fig. 1. Neutrophils express IFN-γ independently of pathogens or microbes.
(A–B) C57BL/6 mice (n=5) were infected i.p. with T. gondii or (C–D) left untreated and peritoneal cavity cells, blood, and BM were examined for the presence of Ly6G+ IFN-γ+ neutrophils day 5 post-infection. Histograms in (B, D) show MFI of neutrophil IFN-γ in the peritoneal cavity (blue), blood (red), and BM (black) in infected and naïve mice respectively. Isotype control stain (filled histogram) is shown for BM neutrophils. (E) IFN-γ+ neutrophils and (F) MFI of neutrophil IFN-γ was quantified from (A–B). (G) BM from naïve conventional (CV) or germ-free (GF) C57BL/6 mice (n=3) was examined for the presence of IFN-γ+ neutrophils. Quantification of data seen in (G), the percentage of total BM that are Ly6G+ neutrophils (H), the percentage of IFN-γ+ neutrophils (I), and MFI of neutrophil IFN-γ (J). The data shown are representative of >3 independent experiments, each involving 2–4 mice per group and error bars shown are mean ± SD. *p<0.05, **p<0.001, ns - not significant.
To address the question of whether neutrophil IFN-γ production is limited to the site of infection in vivo, WT mice were infected with T. gondii and several tissues were examined for neutrophil IFN-γ-positivity including the peritoneal cavity, peripheral blood, and bone marrow (BM). Neutrophils stained positive for IFN-γ at a similar level in all locations examined (Fig. 1A, B, E). T. gondii parasites are largely confined to the site of experimental infection during the first 5 days, thus these data suggest that an indirect sensing of infection may be responsible for IFN-γ production by neutrophils. However T. gondii infected TLR11−/− mice also have IFN-γ+ neutrophils in all tissues examined (Supplemental Fig. 1C). Additionally, the analysis of neutrophils in naïve mice revealed IFN-γ+ neutrophils in the blood and BM but not in the peritoneal cavity (Fig. 1C–1E, and Supplemental Fig. 1C). The amount of IFN-γ-positivity by neutrophils in the BM and blood was comparable in naïve and T. gondii-infected mice (Fig. 1E–1F). Lack of IFN-γ+ neutrophils in the peritoneal cavity of naïve mice is not surprising, given that neutrophils are not present in the peritoneal cavity of naïve mice. Nevertheless, when thioglycollate was used to elicit neutrophils into the peritoneum of otherwise naïve mice, neutrophils stained uniformly positive for IFN-γ (Supplemental Fig 1D.) When combined, these experiments suggested that, neutrophils constitutively produce IFN-γ independent of infection.
One possible explanation for the appearance of IFN-γ producing neutrophils in naïve mice is that microbial products derived from intestinal microbiota could be stimulating neutrophils in peripheral tissues to produce IFN-γ, since commensal bacteria are involved in the activation of mature neutrophils (21, 22). To examine this possibility we quantified the presence of IFN-γ+ neutrophils in the BM of germ-free C57BL/6 mice. We observed that the colonization status of mice had no effect on the appearance of IFN-γ+ neutrophils (Fig. 1G–J). Thus, IFN-γ production by neutrophils is regulated differently from lymphoid cells and does not require the presence of T. gondii or other microbial stimuli. Instead, T. gondii infection triggers neutrophil recruitment to the site of infection, explaining the selective appearance of IFN-γ-positive neutrophils in peritoneal cavity of the infected but not naïve mice (Fig. 1A, C).
Developmental regulation of IFN-γ production by neutrophils
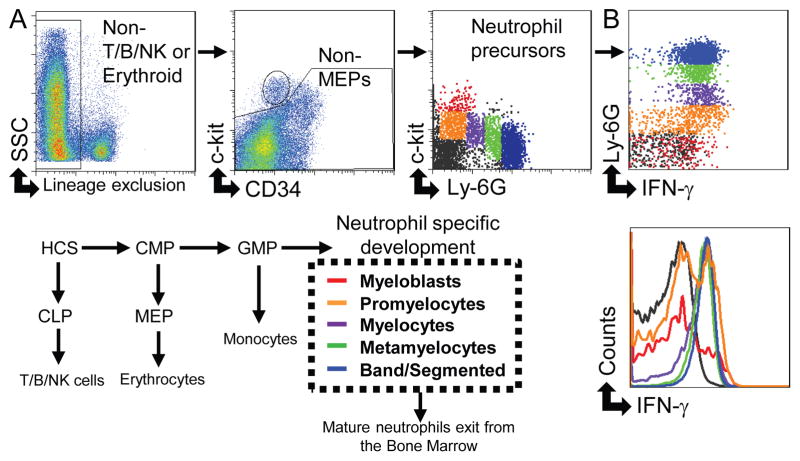
Analysis of BM neutrophils revealed that in addition to IFN-γ+Ly6Ghigh cells, there is an appearance of Ly-6Gnegative and Ly-6Glow cells that produce IFN-γ in germ-free, naïve, and T. gondii-infected mice (Fig. 1). The presence of IFN-γ+ in the BM suggests a model of developmental regulation for IFN-γ production by neutrophils. To obtain insight into this hypothesis, we investigated the stages of neutrophil development and their association with IFN-γ production. Flow cytometric analysis of neutrophil precursors in BM excluded T, B, and NK cells, followed by the analysis of c-Kit, CD34, and Ly-6G-expressing cells (Fig. 2A). The cells were divided into 5 populations; myeloblasts (red), promyelocytes (orange), myelocytes (purple), metamyelocytes (green), and mature neutrophils (blue) (Fig. 2). To confirm the successful identification of neutrophil precursors, the immature neutrophil populations were sort purified and analyzed for stage-specific proteins by qRT-PCR, including proteinase 3, a primary granule protein expressed mostly in early promyelocytes, lactoferrin, a secondary granule protein expressed mostly in metamyelocytes, and gelatinase a tertiary granule protein expressed mostly in late band cells (Supplemental Fig. 2A) (23). Examination of immature neutrophils for IFN-γpositivity by flow cytometry revealed that neutrophils acquire IFN-γ-positivity at the promyelocyte stage, exhibiting a nearly equal split of IFN-γ-positive and IFN-γ-negative cells at this stage (Fig. 2B). Subsequent neutrophil stages were uniformly IFN-γ-positive (Fig. 2B).
Fig. 2. Developing neutrophils express IFN-γ at the promyelocyte stage.
(A) The gating strategy to identify neutrophil precursor cells excludes other potentially IFN-γ+ cells by negative selection (CD3e, CD19, NK1.1, and Ter119), with the gating of the neutrophil precursor populations shown in the third box from the left. Neutrophils develop from the granulocyte-monocyte progenitor (GMP) through a series of stages shown in red (myeloblasts), orange (promyelocytes), purple (myelocytes), green (metamyelocytes), and blue (band/segmented neutrophils). (B) Neutrophil precursor populations were examined for expression of IFN-γ by flow cytometry. These data are representative of at least 3 independent experiments each involving 2–4 mice per group.
We also examined if earlier hematopoietic cells were capable of producing IFN-γ. Lineage-negative sca-1+ c-kit+ (LSKs), which included the long-term HSC, short-term HSC, and multi-potent progenitors, did not stain positive for IFN-γ (Supplemental Fig. 2B). Additionally, the immediate precursor to all granulocytes, the GMPs, also did not stain positive for IFN-γ (Supplemental Fig. 2B). These data revealed that IFN-γ expression in the BM is restricted to the neutrophil-specific lineage development (Fig. 2).
Neutrophil IFN-γ accumulates in primary granules
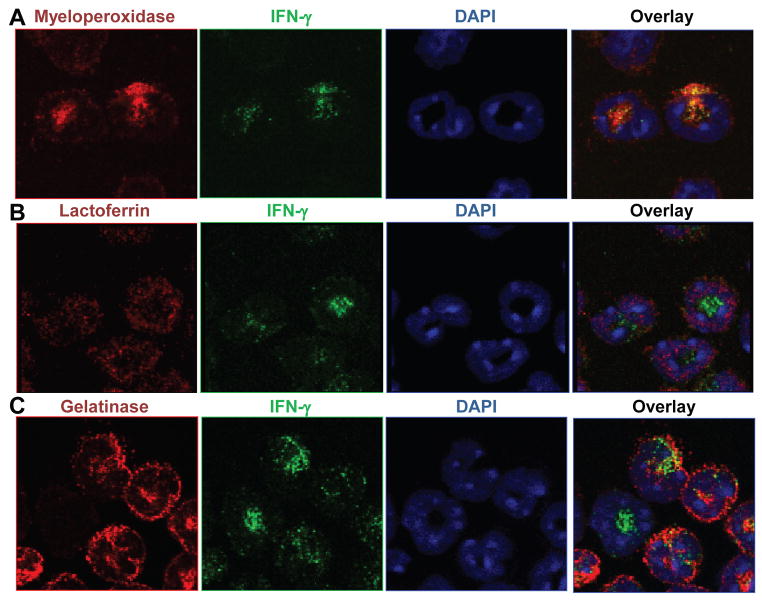
Given that promyelocyte stage neutrophils produce IFN-γ (Fig. 2B) and that immature neutrophils begin packaging granules as they transition to the promyelocyte stage (24), we hypothesized that neutrophil IFN-γ could be stored in granules. To examine the localization of IFN-γ, we utilized immunofluorescence detection of IFN-γ together with known granular proteins. Simultaneous visualization of IFN-γ with myeloperoxidase, lactoferrin, or gelatinase which localize in primary, secondary, and tertiary granules respectively, revealed colocalization between IFN-γ and myeloperoxidase (Fig. 3A). Minimal to no colocalization was observed between IFN-γ and lactoferrin or gelatinase (Fig. 3B, C). Quantitative colocalization analysis, using Manders coefficient, confirmed that IFN-γ is localized in the same granules that contain myeloperoxidase (Fig. 3). These results formally established that IFN-γ is largely contained within the primary granules of neutrophils.
Fig. 3. Neutrophil IFN-γ colocalizes with granule protein myeloperoxidase.
Ly6G+ neutrophils were purified from the BM of naïve C57BL/6 mice, fixed to slides, and stained with αIFN-γ-Alexa488 (green) and DAPI (blue). The samples were additionally stained with (A) αMyeloperoxidase, (B) αLactoferrin, or (C) αGelatinase and a secondary antibody conjugated to Alexa568 (red). These data are representative of >3 independent experiments. Colocalization analysis produced a Manders coefficient average of 0.60 for IFN-γ and Myeloperoxidase and 0.17 for IFN-γ and Gelatinase; a Manders coefficient value >0.5 is considered colocalization.
Neutrophil degranulation releases IFN-γ
To further examine the physiological significance of IFN-γ distribution in primary granules, we tested if induction of neutrophil degranulation would result in IFN-γ release. We first induced neutrophil degranulation with fMLP and LPS and observed that this stimulation resulted in tertiary granule, but little primary granule, degranulation and minor loss of IFN-γpositivity (Fig. 4A, B, C). However, the combined treatment of neutrophils with fMLP, LPS, PMA, and ionomycin resulted in primary granule degranulation as seen by the loss of myeloperoxidase staining in the stimulated neutrophils (Fig. 4B). Importantly, primary granule degranulation also resulted in a nearly complete loss of intracellular IFN-γ staining and simultaneous release of IFN-γ into the cell culture supernatant (Fig. 4C, D). Taken together, our data revealed a developmental mechanism for IFN-γ expression in immature neutrophils and that the release of IFN-γ is regulated by activation-induced primary granule degranulation.
Fig. 4. Degranulation stimuli induce release of neutrophil-derived IFN-γ.
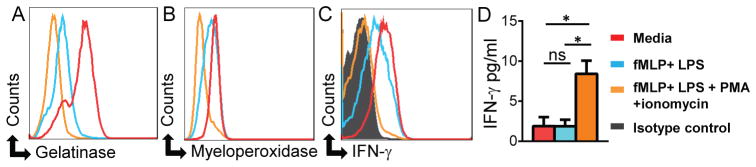
Neutrophils from BM of C57BL/6 mice (n=3) were incubated for one hour with: media alone (red), fMLP + LPS (blue), or fMLP + LPS + PMA + ionomycin (orange) and expression of (A) gelatinase, (B) myeloperoxidase, and (C) IFN-γ was analyzed by intracellular staining. (D) IFN-γ secretion by activated neutrophils in the same experiments was analyzed by ELISA. These data are representative of >3 independent experiments and error bars shown are mean ± SD.*p<0.0001, ns - not significant.
Discussion
In this report we have identified developmental regulation of IFN-γ expression in neutrophil precursor cells which represents a novel mechanism for IFN-γ-mediated host defense. While IFN-γ production by innate lymphoid cells, NK, and T cells is largely regulated by IL-12-dependent induction of IFN-γ expression after infection, neutrophil IFN-γ is produced prior to infection in immature neutrophils at the promyelocyte stage. This mechanism pre-arms neutrophils ensuring a rapid response to pathogens. Regulated secretion of IFN-γ from neutrophil granules prevents spontaneous effects of IFN-γ in the absence of microbial infections. While IFN-γ expressing neutrophils are in blood and bone marrow, the effector mechanisms of neutrophil-derived IFN-γ are restricted to the sites of infection since their IFN-γ secretion requires microbial or inflammatory environment driven degranulation of primary granules. Granular localization of IFN-γ may explain the confusion in the literature regarding the ability of neutrophils to produce IFN-γ, since both isolation-induced degranulation or insufficient granular protein fixation will result in an inability to detect neutrophil IFN-γ. In addition to the priming effects of IFN-γ on other cells types, IFN-γ can prime neutrophils themselves for increased migration, pathogen clearance, antigen presentation abilities, and cytokines production (25), the complexities of which will need to be revisited in light of neutrophil derived IFN-γ.
The release of IFN-γ by neutrophils represents a bona fide type I innate immune response that can potentially be elicited by a variety of pathogens and host inflammatory responses sufficient to trigger neutrophil primary granule degranulation. In this regard it is of interest that neutrophil IFN-γ expression is developmentally regulated which is distinct from their cytokine and stimuli-induced production of IL-22 and IL-17, two additional cytokines that were also, until recently, considered to be lymphoid cell-specific effector molecules (26, 27). Both IL-22 and IL-17 production by neutrophils is restricted to mucosal tissues and in the case of IL-17, the transcription factor RORγt is a master regulator of IL-17 expression by neutrophils (27) and lymphoid cells (28). The localization of IFN-γ within primary granules of neutrophils provides an activation-dependent regulatory mechanism, since stronger inflammatory signals at the site of infection or inflammation are involved in primary granule degranulation. Overall, in this report we unveiled an arm of IFN-γ-mediated innate immunity that is prepared for immediate effector responses and is regulated by neutrophil recruitment and activation.
Supplementary Material
Acknowledgments
This work was supported by NIH A1085263 and the Burroughs Wellcome Fund. to F.Y. and DK070855 to L.V.H. C.R.S. and E.B. were supported in part by NIH A1005284.
References
- 1.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 4.Scanga CA, Aliberti J, Jankovic D, Tilloy F, Bennouna S, Denkers EY, Medzhitov R, Sher A. Cutting edge: MyD88 is required for resistance to Toxoplasma gondii infection and regulates parasite-induced IL-12 production by dendritic cells. J Immunol. 2002;168:5997–6001. doi: 10.4049/jimmunol.168.12.5997. [DOI] [PubMed] [Google Scholar]
- 5.Raetz M, Hwang SH, Wilhelm CL, Kirkland D, Benson A, Sturge CR, Mirpuri J, Vaishnava S, Hou B, Defranco AL, Gilpin CJ, Hooper LV, Yarovinsky F. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-gamma-dependent elimination of Paneth cells. Nat Immunol. 2013;14:136–142. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, Sher A. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- 7.Jankovic D, Kullberg MC, Hieny S, Caspar P, Collazo CM, Sher A. In the absence of IL-12, CD4(+) T cell responses to intracellular pathogens fail to default to a Th2 pattern and are host protective in an IL-10(−/−) setting. Immunity. 2002;16:429–439. doi: 10.1016/s1074-7613(02)00278-9. [DOI] [PubMed] [Google Scholar]
- 8.LaRosa DF, Stumhofer JS, Gelman AE, Rahman AH, Taylor DK, Hunter CA, Turka LA. T cell expression of MyD88 is required for resistance to Toxoplasma gondii. Proc Natl Acad Sci U S A. 2008;105:3855–3860. doi: 10.1073/pnas.0706663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturge CR, Benson A, Raetz M, Wilhelm CL, Mirpuri J, Vitetta ES, Yarovinsky F. TLR-independent neutrophil-derived IFN-gamma is important for host resistance to intracellular pathogens. Proc Natl Acad Sci U S A. 2013;110:10711–10716. doi: 10.1073/pnas.1307868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldszmid RS, Caspar P, Rivollier A, White S, Dzutsev A, Hieny S, Kelsall B, Trinchieri G, Sher A. NK cell-derived interferon-gamma orchestrates cellular dynamics and the differentiation of monocytes into dendritic cells at the site of infection. Immunity. 2012;36:1047–1059. doi: 10.1016/j.immuni.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou B, Benson A, Kuzmich L, DeFranco AL, Yarovinsky F. Critical coordination of innate immune defense against Toxoplasma gondii by dendritic cells responding via their Toll-like receptors. Proc Natl Acad Sci U S A. 2011;108:278–283. doi: 10.1073/pnas.1011549108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson DC, Matthews S, Yap GS. IL-12 signaling drives CD8+ T cell IFN-gamma production and differentiation of KLRG1+ effector subpopulations during Toxoplasma gondii Infection. J Immunol. 2008;180:5935–5945. doi: 10.4049/jimmunol.180.9.5935. [DOI] [PubMed] [Google Scholar]
- 13.Yap G, Pesin M, Sher A. Cutting edge: IL-12 is required for the maintenance of IFN-gamma production in T cells mediating chronic resistance to the intracellular pathogen, Toxoplasma gondii. J Immunol. 2000;165:628–631. doi: 10.4049/jimmunol.165.2.628. [DOI] [PubMed] [Google Scholar]
- 14.Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, Domingues RG, Veiga-Fernandes H, Arnold S, Busslinger M, Dunay IR, Tanriver Y, Diefenbach A. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 17.Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe. 2009;6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturge CR, Yarovinsky F. Complex immune cell interplay in the gamma interferon response during Toxoplasma gondii infection. Infect Immun. 2014;82:3090–3097. doi: 10.1128/IAI.01722-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarovinsky F. Innate immunity to Toxoplasma gondii infection. Nat Rev Immunol. 2014;14:109–121. doi: 10.1038/nri3598. [DOI] [PubMed] [Google Scholar]
- 21.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karmarkar D, Rock KL. Microbiota signalling through MyD88 is necessary for a systemic neutrophilic inflammatory response. Immunology. 2013;140:483–492. doi: 10.1111/imm.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lominadze G, Powell DW, Luerman GC, Link AJ, Ward RA, McLeish KR. Proteomic analysis of human neutrophil granules. Mol Cell Proteomics. 2005;4:1503–1521. doi: 10.1074/mcp.M500143-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Ellis TN, Beaman BL. Interferon-gamma activation of polymorphonuclear neutrophil function. Immunology. 2004;112:2–12. doi: 10.1111/j.1365-2567.2004.01849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, Chaplin DD, Weaver CT. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci U S A. 2013;110:12768–12773. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor PR, Roy S, Leal SM, Jr, Sun Y, Howell SJ, Cobb BA, Li X, Pearlman E. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORgammat and dectin-2. Nat Immunol. 2014;15:143–151. doi: 10.1038/ni.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.