Abstract
Background
Chromosomal instability is a hallmark of cancer. Chromosomal imbalances, like amplifications and deletions, influence the transcriptional activity of genes. These imbalances affect not only the expression of genes in the aberrant chromosomal regions, but also that of related genes, and may be relevant to the cancer status.
Material/Methods
Here, we used the 7 publicly available microarray studies in breast cancer tissues and propose a general and unsupervised method by using the gene expression data and related gene information to systematically identify aberrant chromosomal regions. This method aimed to identify the chromosomal regions where the genes and their related genes both show consistent changes in the expression levels. Such patterns have been reported to be associated with the chromosomal aberrations and may be used in cancer diagnosis.
Results
We compared 488 tumor and 222 normal samples from 7 microarray-based human breast cancer studies and detected the amplifications of 8q11.21, 14q32.11, 4q21.23, 18q11.2, Xq28, and the deletions of 3p24.1, 10q23.2 (BSCG1), 20p11.21, 9q21.13, and 1q41, which may be involved in the novel mechanisms of tumorigenesis. In addition, several known pathogenic genes, transcription factors (TFs), and microRNAs (miRNAs) associated with breast cancer were found.
Conclusions
This approach can be applied to other microarray studies, which provide a new and useful method for exploring chromosome structural variations in different types of diseases.
MeSH Keywords: Breast Neoplasms, Male; Chromosomal Instability; Gene Expression
Background
DNA microarray technology provides a powerful tool for characterizing genes expression on a genome-wide scale. It has been widely used for the analysis of genomic changes detection and clinical diagnosis [1]. Many efforts have been made to analyze and interpret the public accessibility of data, such as the identification of differentially expressed genes and the development of data mining methods [2–4]. However, it is difficult to identify the real pathogenesis of cancer just from the perspective of alterations in the expression levels of individual genes. Using integrated analysis of the differentially expressed genes, we need to identify the real pathogenesis of cancer from a functional pathway perspective. Chromosome aberrations are associated with various cancers and it is very important to identify aberrant chromosomal regions from the whole-genome perspective in cancer. Now, chromosomal localization has been integrated in several methods that aim to detect differential gene expression of adjacent genes. PGE [5], MACAT [6], and ChroCoLoc [7], have been instrumental for integrating chromosomal location information into the gene expression data analysis to detect differential gene expression of adjacent genes. These methods mapped the differentially expressed or expressed genes in the array to the chromosome and explored the consistent expression levels of those genes by using different data mining methods. These methods may ignore genes that are expressed at low levels or unexpressed genes in the chromosome, which may be caused many of false-positive results. These methods identified the adjacent genes on the chromosome with consistent expression levels. We know that if the cancer is related with some aberrant chromosomal regions, the changes of gene expression levels may be performed by genes in the aberrant chromosomal regions and their related genes. So it is appropriate to simultaneously explore the changes of gene expression levels of gene in the aberrant chromosomal regions and their related genes. Therefore, with just genomic array data of genes within the aberrant regions, it is difficult to identify all genes whose expression may be associated with the disease.
To identify aberrant chromosomal regions, we present a general method that integrates genome-wide gene expression data and contextual information about genes in a chromosomal region and their related genes. Firstly, this method considers the information of detected genes in the array and also considered the information of undetected genes in the array for the chromosomal regions. Secondly, this method not only uses the information of gene expression profiles, but also incorporates other relevant information, such as gene ontology (GO), KEGG, and protein-protein interaction (PIP), to mine aberrant chromosome regions. Additionally, the method also takes account of miRNAs and TFs regulatory effects on gene expression changes in the regions. We used this method to analyze the 7 independent human breast cancer microarrays and identified several frequent aberrant regions, including those with putative tumorigenic effects. This approach can be applied to any microarray study. It provides a general approach for exploring genome-wide expression in many disease types and could identify groups of patients with distinct regional patterns of gene and interrelated-gene expression. It also allows the identification of associations between the clinical breast cancer parameters and helps improve patient satisfaction in relation to disease diagnosis and therapy.
Material and Methods
Material
Chromosomal locations of genes were obtained from NCBI and GeneCards databases [8,9]. We downloaded 42 564 human gene symbols from the NCBI database (Build 35.1) and 34 463 gene locations were extracted from the GeneCards database online by use of a Java program. The chromosomal locations of 1240 miRNAs and 939 pre-miRNAs were downloaded from the PicTar database. The GeneOntolgy data were downloaded from http://www.geneontology.org/GO.downloads.database.shtml#dl, and contained 9633 distinct Biological Processes, 1570 distinct Cellular Components and 7063 distinct Molecular Functions [10]. High-throughput protein-protein interaction data were downloaded from the BIND, DIP, IntAct, MINT, and HPRD databases [11–15].
Protein-protein interaction data from these databases were integrated to reduce the number of false positives due to the use of different prediction algorithms for different databases. Consequently, only interaction relationships identified in at least 2 databases were used. We obtained 73 976 protein-protein interaction relationships for 11 435 genes obtained after integration. Pathway information for 5379 genes was obtained from the KEGG database (ftp://ftp.genome.jp/pub/kegg/pathway_gif/organisms/hsa/) [16]. All human miRNAs and target genes data were downloaded from the miRBase, mirGen, miRDB, and miRANDA databases [10,17,18]. To reduce false positives, only targets that appeared in at least 2 databases were analyzed. After integration, 9393 regulatory relationships were obtained from 296 miRNAs and 3772 target genes.
Data sets
The 7 publicly available microarray studies in breast cancer tissues were compiled and prepared for analysis. These microarray studies were performed using different platforms and different populations, as shown in Table 1. To determine the chromosomal locations of the corresponding probes on the microarrays, we mapped the probes to the gene symbol identifier via the GeneCards database. To avoid ambiguities, when multiple probes matched the same gene symbol, only the average expression value was used.
Table 1.
Seven gene expression profiles for breast cancer tissues.
| Study (reference) | Number of samples | Number of gene symbols used | Platform | Main focus | |
|---|---|---|---|---|---|
| 1 | Hawthorn et al., 2010 | 20 | 19,803 | U133_Plus_2 | IDC (expression analysis) |
| 2 | Chen et al., 2008 | 185 | 19,803 | U133_Plus_2 | Malignancy-risk gene signature |
| 3 | Pedraza et al., 2008 | 58 | 19,803 | U133_Plus_2 | Gene expression signatures |
| 4 | Yu et al., 2008 | 341 | 12,632 | U133A | Modulation metastasis and survival |
| 5 | Alimonti et al., 2010 | 47 | 19,803 | U133_Plus_2 | Breast tumor expression |
| 6 | Yao et al., 2011 | 142 | 16,546 | cDNA | Recovery of biological information |
| 7 | Richardson et al., 2007 | 62 | 19,803 | U133_Plus_2 | Expression data |
Definition of the indexes
The genes distance index (GDI) is defined as the distance measure and it means the genetic distance which any 2 differentially expressed genes should be smaller than. Its value is a positive integer which can be set freely. The genes’ expression coverage rate (GECR) is defined as coverage rate measure, which is equal to the ratio of the number of expressed genes of the microarray to the total number of genes in the candidate chromosomal regions. The candidate chromosomal regions whose measured value of GECR is greater than the set value are retained. The value can be set freely and the range of GECR is from 0 to 1.The rank index (RI) is defined as the measure of global gene expression level; it is a ratio value which can be set freely. The set value means the ranking percentage of genes expression in microarray.
Algorithm flow
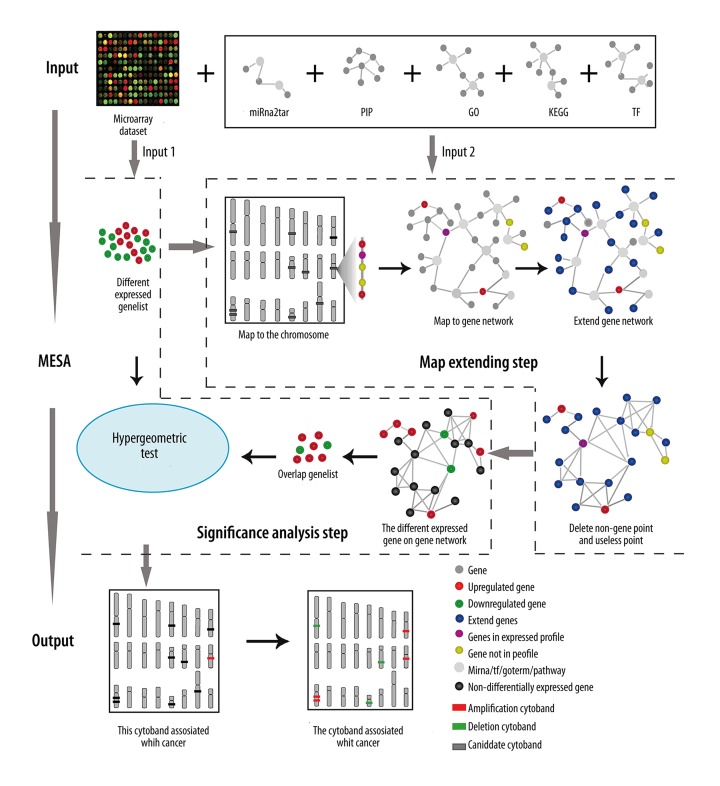
We developed an intuitive method to detect aberrant chromosomal regions based on gene expression and their related gene’s information. Firstly, this method orientated those chromosomal regions where genes displayed significant and consistent changes in expression levels. Secondly, it examined whether the expression levels of genes in those chromosomal regions and their related genes are significantly related to all differently expressed genes detected in microarray. The purpose of this method was to identify those chromosomal regions where the changes in gene expression levels caused by chromosomal aberrations were consistent with the changes measured by microarrays. As shown in Figure 1, a detailed flow was as follows: (1) differentially expressed genes were extracted from microarray data; (2) candidate aberrant chromosomal regions were identified based on the chromosomal location of differentially expressed genes. If the interval between any two differentially expressed genes was smaller than the value of GDI in a chromosomal region, it was identified as a candidate aberrant chromosomal region; and (3) the candidate chromosomal region was retained if the genes met the following conditions: the value of the number of expressed genes of the microarray in the candidate chromosomal regions divided by the total number of genes in the candidate chromosomal region was greater than GECR. The rank of the expression level of differentially expressed and expressed genes was smaller than RI in the expression array. Next, the genes in the retained candidate chromosomal region and their relative genes in the same GO term, same KEGG pathway, regulated by the same transcription factor and by the same miRNA with the same protein interactions, were put into a new gene set called an expanded gene set.
Figure 1.
Algorithm flow of methods used to identify aberrant chromosomal regions. See Material and Methods for details.
Hypergeometric tests were used to determine whether the expanded genes set were significantly related to the differentially expressed genes set. The expanded gene set was considered to be significantly related to the differentially expressed gene set, and the corresponding chromosomal region was considered as a potentially cancer-related chromosomal region, if the p value was <0.01.
Relationships between differentially expressed gene sets and expanded gene sets
Hypergeometric tests were used to determine if the expanded genes set were significantly related to the differentially expressed genes set, which were detected in microarray. The p value for the test was defined as follows:
Where N was the number of genes in the microarray; K was the number of genes in the expanded gene set, M was the number of all differentially expressed genes set, which were detected in microarray; and X was the number of differentially expressed genes in the expanded gene set. If p<0.01, the expanded genes set would be considered to be significantly related to the differentially expressed genes set, which were detected in microarray and the corresponding chromosomal region was considered to be a potentially cancer-related chromosomal region.
Definition of the intersection ratio index (IRI)
The intersection ratio index (IRI) was defined to assess the similarity degree between the identified chromosomal regions for any 2 result sets. One of the result sets represents that when we set the fixed values for the values of GDI, GECR, RI, the cancer-related chromosomal regions set we get by our method for the same microarray. The value of IRI was between 0 and 1 and was equal to the ratio of the number of intersections between 2 results sets to the number of chromosome regions of the result whose number of chromosome regions was smaller. The higher IRI value is, the more similar the 2 result sets are.
Meta-analysis
We analyzed the microarray data from different platforms with different values which combined GDI, GECR, and RI. All chromosomal regions identified in each study were then merged to produce a list of non-overlapping continuous chromosomal regions, referred to as meta-regions, which begin from the start base position to the end base position of overlapping chromosomal segments. Finally, the number of times each meta-region was found in each study with different values combination of GDI, GECR, and RI was calculated.
The more times a meta-region was found, the more likely it was to be a breast cancer-related abnormal chromosomal region.
Results
Chromosomal regions with distinct gene and related-gene expression patterns
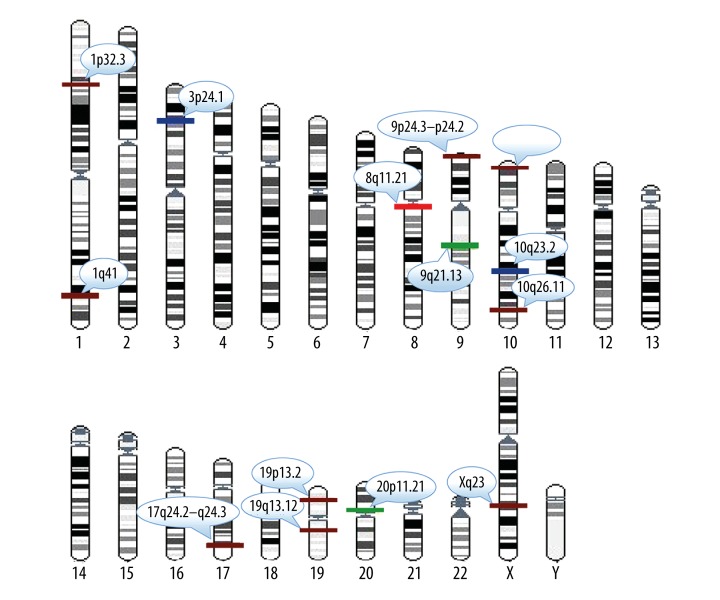
We systematically compared the genes and their related genes’ expression levels in the chromosomal regions and identified the aberrant chromosomal regions from 7 independent breast cancer studies comprising 710 samples for their relative expression patterns (Table 1). Here, differentially expressed genes of any 2 data set from the 7 independent breast cancer studies were identified by the significance analysis of microarray (SAM) method with false discovery rate (FDR) <0.05(Chumbley and Friston 2009). Here, the values of GDI, GECR, and RI were set at 2, 0.8, and 0.3, respectively, and we then identified chromosomal regions for the 7 breast cancer studies separately. The meta-analysis enabled us to prioritize the predominant chromosomal regions found across a collection of 710 samples. We chose those meta-regions in which the number of times the meta-region was found in the 7 studies was greater than or equal to 4 (Figure 2). The results showed that the identified chromosomal regions included well known aberrant regions in breast cancer, such as 8q11.21, 20p11.21, 3p24.1, 10q23.2, 1p32.3, 9q21.13, and 17q24.2-q24.3. In addition, known pathogenic genes, TFs, and miRNAs associated with breast cancer were found, including MCM4 and CEBPD in 8q11.21 [19–21], THBD and CD93 in 20p11.21 [22,23], ANXA1 and ALDH1A1 in 9q21.13 [24–26], SNCG in 10q23.2 [27], KCNJ16, ABCA10 and ABCA6 in 17q24.2-q24.3 [28], transforming growth factor β receptor 2 (TGFBR2) in 3p24.1 [29], and hsa-miR-761 in 1p32.3 [30]. For example, the region of 8q11.21, found in all but 1 study, showed abnormal amplification in breast cancer. MCM4 in 8q11.21 was differentially expressed in breast cancers. MCM4 is a subunit of the MCM replication helicase complex (MCM2-7), which is essential for DNA replication, genome stability, and DNA damage response. An MCM4 mutation subtype, Phe345Iso, has been shown to be involved in the development of breast cancer in female mice. CEBPD is a tumor suppressor gene and was found to be up-regulated in the region. The chromosomal region 3p24.1 is very prominent, and is a region frequently associated with chromosomal abnormalities in breast cancer. It was deleted in at least 6 data sets in the current study. Transforming growth factor β receptor 2 (TGFBR2) in 3p24.1 has also been shown to be absent in breast cancer. TGF-β family members are potent inhibitors of the growth of many epithelial cells. Transmembrane signaling by TGF-β occurs via a complex of serine/threonine (Ser/Thr) kinases. As a transcription factor, TGFBR2 regulates many target genes associated with breast cancer and is involved in breast cancer development and cancer cell growth [29,31]. The 1p32.3 region contains the miRNA hsa-miR-761, which has been shown to be associated with breast cancer by next-generation sequencing of miRNAs. These results demonstrated that this method was able to identify not only pathogenic genes in breast cancer, but also cancer-related TFs and miRNAs.
Figure 2.
A karyotype to show the identified aberrant chromosome regions. Red rectangles in regions represent the amplified regions of chromosomes detected in a total of 6 data sets. Green rectangles in regions represent deleted regions of chromosomes detected in a total of 6 data sets. Dark blue rectangles in regions represent deleted regions of chromosomes detected in a total of 5 data sets. Crimson rectangles in regions represent deleted regions of chromosomes detected in a total of 4 data sets.
When we expanded the related genes in these GO terms to the expanded gene set, for expanding the most related genes, we chose that the distances of the paths from these terms to the root node of the GO tree were greater than 7 and the number of genes in these terms was greater than 100. When we expanded the genes that were in the same pathway as the genes in the candidate chromosomal regions to form an expanded gene set, and we found that there were different numbers of genes in different pathways. It is unreasonable to expand all the genes in a pathway into an expanded gene set, because there may be little relation between genes that are far apart in the same pathway. To extract genes that were more highly correlated with the genes in the candidate chromosomal regions to include in an expanded gene set, each pathway was converted to an undirected graph with enzymes as nodes. Depth-first traversal and breadth-first traversal methods [1] were then used, in which the genes in the candidate chromosomal regions were taken as root nodes and all genes in the same pathway were listed according to the order of traversal in the metabolic pathway. A total of 30% of the listed genes, which are near to the root-node gene in the metabolic pathway, were extended into the expanded gene set.
Although the chromosomal region location was based on gene mapping, with the start position of the first gene used as the start location and the end position of the final gene as the termination location, this could lead to some errors. First, many genes have different chromosomal locations according to different databases. We used the GeneCards V3 chromosome location by default. Second, we compared expression abundance of probes on different platforms, not gene expression levels. Because of alternative splicing, probe abundance does not necessarily represent gene abundance. Finally, increased or decreased gene expression may result from factors other than chromosome amplification or deletion. Gene’s regulation by TFs and miRNAs is usually very complex, and these factors may regulate mRNA expression in synergy. Abnormal expression of TFs and/or miRNAs can therefore lead gene expression to be changed in the target region.
Tumor-associated gene expression
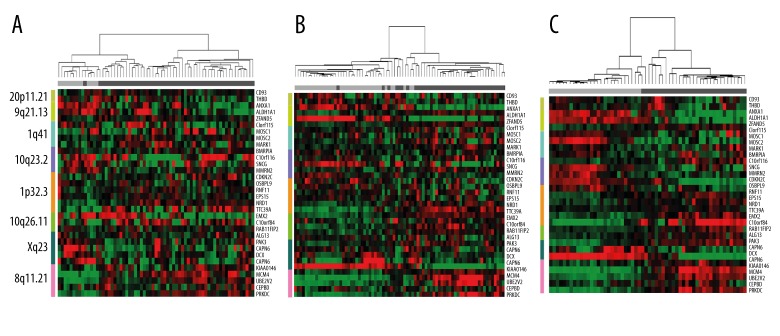
Because disease progression may be correlated with the accumulation of genetic alterations, such as DNA amplification, we assumed that groups of samples with increased gene expression in the identified chromosomal regions might represent tumor-related groups. To validate this assumption, we clustered the samples using the expression profiles of the genes in the identified chromosome regions by the hierarchical clustering method. Variations in chromosomal regions 20p11.21, 9q21.13, 1q41, 10q23.2 1p32.3, 10q26.11, Xq23, and 8q11.21 were found in at least 4 sets of expression profiles. Clustering analysis of gene expression levels in these regions was performed in 3 data sets, which contained the tumor status of the samples (for data sets 2, 3, 4, shown in Table 1). We found that expression profiles within these chromosomal regions could be used to distinguish between normal and breast-cancer samples. The clustering results shown in Figure 3 suggest an association between tumor status and gene expression profile. Normal samples appeared to align with lower gene expression (Figure 3, shown in green), whereas tumor samples correlated with higher expression (Figure 3, shown in red) in those identified chromosome regions. We also found that common genes in those chromosome regions seemed to be associated with breast cancer. For example, SNCG in 10q23.2 encodes a specific protein in breast cancer and expression of the gene is highly correlated with breast cancer occurrence. In our algorithm, SNCG expression was reduced in all expression profiles. BCSG1 is barely expressed in normal breast tissue and benign lesions, but is highly expressed in most tumor tissues, including invasive breast cancer and ovarian cancer. Abnormal expression of BCSG1 is related to tumor cell growth, invasion and metastasis, and deletion of SNCG has been reported to be an independent indicator of good prognosis in breast cancer [27]. CDKN2C, OSBPL9, RNF11, EPS15, NRD1, and TTC39A were found on chromosome 1p32.3. These genes covered a compact region of 1 Mb and were found to be down-regulated. CDKN2C encodes cyclin-dependent kinase inhibitor 2C, also known as p18, which mainly inhibits the activity of cyclin-dependent kinase 4 (CDK4). CDK4, in turn, is mainly involved in the cell cycle and cell proliferation. CDKN2C thus plays a major role in cell cycle arrest, as a negative regulator of cell proliferation in tumorigenesis. CDKN2C has been shown to be expressed at a low level in breast cancer and prompts unlimited tumor proliferation [32,33].
Figure 3.
Three studies (data sets 2, 3, and 4, shown in Table 1) demonstrate the hierarchical clustering of the sample expression profiles (A, B, and C, respectively). Columns represent the samples, and rows represent the genes. The colored bar at the left represents the regions: 20p11.21 (yellow-green), 9q21.13 (light green), 1q41 (light blue), 10q23.2 (purple), 1p32.3 (orange), 10q26.11 (emerald), Xq23 (dark green) and 8q11.21 (pink). Standardized gene expression values are displayed as colored boxes; red=high; green=low. The horizontal bar at the top indicates the classification of the samples: gray=normal; black=cancer.
The chromosomal regions 1q41 and 9q21.13 were also identified using our algorithm. Although there are no previous reports linking deletion of this chromosome region to breast cancer, we have shown close relationships between ANXA1 [34] and ALDH1A1 [35] and breast cancer, as key genes in breast cancer diagnosis.
Gene function terms and novel genes which are potentially tumor-related are identified
The method has not only identified lots of genes that have been well known to correlate with breast cancer, but also identified some gene function terms and novel genes that could potentially be correlated with breast cancer. We got 57 non-redundant genes from the aberrant chromosomal regions from 7 gene expression data set and mapped those genes to the Gene Ontology to identify the potentially correlated function terms. The results showed that there were 5 enrichment GO terms and 16 non-redundant genes in those GO terms. The genes symbols showed as bold in Table 2, meaning that these genes were reported in the literature and altered in breast cancer cells obtained from the Genes-to-Systems Breast Cancer Database. Hypergeometric tests were used to determine whether 16 non-redundant genes in Go terms were significantly related to the breast cancer. Here, N was the number of genes in the genome, K was the number of all genes obtained from the Genes-to-Systems Breast Cancer Database, M was the number of 16 non-redundant genes in Go terms, and X was the number of genes obtained from the Genes-to-Systems Breast Cancer Database in 16 genes. The p value was <0.00001 and the result showed that 16 genes were significantly related to breast cancer. Genes for AKR1C3, ABCA9, and MAP2K6 were not reported in the Genes-to-Systems Breast Cancer Database and they were identified as the novel genes that were potentially tumor-related. Aldo-keto reductase 1C3 (AKR1C3), which was known as a biomarker and therapeutic target for Castration-Resistant Prostate Cancer, had high expression in breast cancer and recently was regarded as a potential anti-cancer drug target in both CRPC and ER-positive breast cancer [36,37]. The combined expression pattern of ABCA8 or ABCA9 was associated with particularly poor outcome in Epithelial Ovarian Cancer and the expression of ABCA9 showed the differently expression in breast cancer [38]. MAP2K6 gene showed low expression in breast cancer. Those novel genes will test in the future works and be applied to the breast cancer.
Table 2.
Gene ontology annotation results for genes in the aberrant chromosomal regions.
| GO terms | GO category | p-values | Gene symbol |
|---|---|---|---|
| GO: 0047115 | Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity | 0.009 | AKR1C2; AKR1C3 |
| GO: 0042626 | ATPase activity, coupled to transmembrane movement of substances | 0.003 | ABCA5; ABCA6; ABCA8; ABCA9 |
| GO: 0016818 | Hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides | 0.0005 | ABCA5; ABCA6; ABCA8; ABCA9; MCM4; SMARCA2 |
| GO: 0005524 | Adenyl nucleotide binding | 0.004 | ABCA5; ABCA6; ABCA8; ABCA9; MAP2K6; MCM4; PAK3; SMARCA2; TGFBR2 |
| GO: 0005215 | Transporter activity | 0.002 | ABCA5; ABCA6; ABCA8; ABCA9; AKR1C2; AKR1C3; ALDH1A1; FXYD1; FXYD3; KCNJ16; MCM4; VLDLR |
The identification of aberrant chromosomal regions from training and testing data sets were consistent
To test the performance of this algorithm, we divided the 7 sets of gene expression data into training data set and test data set. Six data sets were extracted from 7 data sets as the training set, and the remaining data set was retained as the test set. The cross-validation by leaving-one method was used for the consistency verification of the algorithm and the 7 analysis results were obtained by using the algorithm. The percentage of overlapping regions on chromosomal regions in any 2 results was at least 76%. The analysis results showed that the chromosome regions identified by the algorithm in the training set and the test set had strong consistency and the algorithm had a strong applicability for the different gene expression data sets.
Robustness of the method
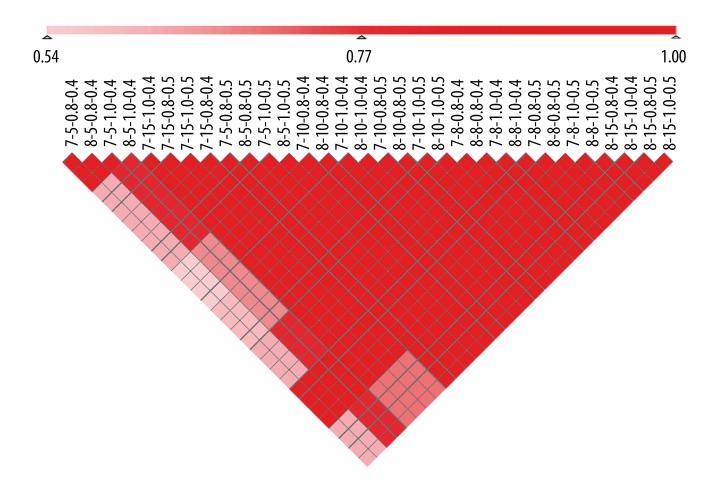
To analyze the robustness of the results from 7 independent breast cancer studies, we examined the intersections between the identified chromosomal regions for any 2 result sets with the different values combination of GDI, GECR, and RI. We focused on RI (0.4, 0.5), GDI (5, 8, 10, and 15) (see Material and Methods), GO terms, which the distances of the paths from these terms to the root node of the GO tree were equal to 7 or 8, KEGG pathways, with a total of 80% of listed genes close to the root-node gene in the pathway. A total of 32 identified chromosome regions result sets were obtained with the different values’ combination of GDI, GECR, and RI. Then, we calculated IRI values (see Material and Methods) for any 2 of the result sets, and found that the intersection ratios between any 2 result sets were all greater than 0.8 (Figure 4). The analysis of the results showed that this method could identify chromosomal aberrant regions robustly.
Figure 4.
Robustness analysis results. Color depth represents the intersection ratio, which ranged from 0.54 to 1. Parameters: The distances of the paths from these terms to the root node of the GO tree; the number of consecutive non-differentially expressed genes; extended percentage of KEGG; p-values for genes in the expression profile.
Discussion
We established a general and unsupervised method for identifying aberrant chromosomal regions in cancers by using the information from the genes expression, genes function, and protein-protein interaction. The analysis of the results showed that those chromosomal regions known for frequent chromosome aberrance regions were identified. When the values of GDI, GECR, and RI were set at 2, 0.8, and 0.3, the chromosome arrant regions for 8q11.21, 20p11.21, 3p24.1, 10q23.2, 1p32.3, 9q21.13, and 17q24.2-q24.3 were identified through the meta-analysis and those chromosome regions were proved to correlate with the breast cancer. In addition, some known and novel pathogenic genes, TFs, and miRNAs associated with breast cancer were also found. The genes of MCM4 and CEBPD in 8q11.21, THBD and CD93 in 20p11.21, ANXA1 and ALDH1A1, SNCG in 10q23.2, KCNJ16, ABCA10 and ABCA6 in 17q24.2-q24.3, TGFBR2 in 3p24.1, and hsa-miR-761 in 1p32.3 were proved to correlate with the breast cancer. The genes for r AKR1C3, ABCA9, and MAP2K6 were identified as the novel genes that were potentially tumor-related and those genes may provide a new target for breast cancer therapy.
This method is robust and can get strong consistent results among different datasets. We identified 32 aberrant chromosome regions sets under the different values combination of GDI (5, 8, 10, and 15), GECR (0.8), RI (0.4, 0.5) and examined the intersections between any 2 result sets. As the intersection ratios between any 2 results is more than 0.8, the result showed that this method could identify chromosomal aberrant regions robustly. Although we verified the stability and robustness of the algorithm, it still requires further testing and adjustment of the parameters for the identification of chromosomal regions affected by multiple parameters. We also divided the gene expression data into training data and test data sets, and used the cross-validation by leaving-one method to verify the consistency. At least 76% chromosome overlap regions between any 2 results were obtained which showed that the method could be used for the different gene expression data platform.
We clustered the samples using the expression profiles of the genes in the identified chromosome regions. The normal samples and tumor samples were clustered into different clusters and they had different gene expression profiles, so the identified chromosomal regions could be used as the biomarkers of breast cancer. These chromosome regions may provide an effective approach to identify breast cancer with fluorescence in situ hybridization.
Conclusions
Here, we provide a new applicable method for identifying chromosome structural variations in different types of diseases and gene expression data platforms. This method can identify aberrant chromosomal regions and will thus provide the basis for further experimental studies, as well as helping in clinical diagnosis. Bioinformatics data analysis using our method will allow the identification of potential aberrant chromosomal regions based on gene expression profiles. Further studies using this approach will aid clinical diagnosis and improve our understanding of complex diseases.
Footnotes
Source of support: Grant No. 61300116 from the National Science Foundation of China and Grant No. QC2013C019, No. QC2013C063 from the Natural Science Foundation of Heilongjiang Province, China and Grant NO.12541319, No. 12541475 from the Scientific Research Fund of Heilongjiang Provincial Education Department and No. 2014M551268 from the China Postdoctoral Science Foundation
Statement
No additional external funding was received for this study. No competing financial interests exist.
References
- 1.Govindarajan R, Duraiyan J, Kaliyappan K, et al. Microarray and its applications. J Pharm Bioallied Sci. 2012;4:S310–12. doi: 10.4103/0975-7406.100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carulli JP, Artinger M, Swain PM, et al. High throughput analysis of differential gene expression. J Cell Biochem Suppl. 1998;30–31:286–96. [PubMed] [Google Scholar]
- 3.To KY. Identification of differential gene expression by high throughput analysis. Comb Chem High Throughput Screen. 2000;3:235–41. doi: 10.2174/1386207003331616. [DOI] [PubMed] [Google Scholar]
- 4.Segata N, Blanzieri E, Priami C. Towards the integration of computational systems biology and high-throughput data: supporting differential analysis of microarray gene expression data. J Integr Bioinform. 2008;5(1) doi: 10.2390/biecoll-jib-2008-87. [DOI] [PubMed] [Google Scholar]
- 5.De Preter K, Barriot R, Speleman F, et al. Positional gene enrichment analysis of gene sets for high-resolution identification of overrepresented chromosomal regions. Nucleic Acids Res. 2008;36:e43. doi: 10.1093/nar/gkn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toedling J, Schmeier S, Heinig M, et al. MACAT – microarray chromosome analysis tool. Bioinformatics. 2005;21:2112–13. doi: 10.1093/bioinformatics/bti183. [DOI] [PubMed] [Google Scholar]
- 7.Blake J, Schwager C, Kapushesky M, et al. ChroCoLoc: an application for calculating the probability of co-localization of microarray gene expression. Bioinformatics. 2006;22:765–67. doi: 10.1093/bioinformatics/btk022. [DOI] [PubMed] [Google Scholar]
- 8.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris MA, Clark J, Ireland A, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xenarios I, Salwinski L, Duan XJ, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–5. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis RC, Hogue CW. Searching, viewing, and visualizing data in the Biomolecular Interaction Network Database (BIND) Curr Protoc Bioinformatics. 2006;Chapter 8(Unit 8.9) doi: 10.1002/0471250953.bi0809s12. [DOI] [PubMed] [Google Scholar]
- 13.Chatr-aryamontri A, Ceol A, Palazzi LM, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–74. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerrien S, Alam-Faruque Y, Aranda B, et al. IntAct – open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–65. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human Protein Reference Database – 2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion 101–3, 119–28, 244–52. [PubMed] [Google Scholar]
- 17.Megraw M, Sethupathy P, Corda B, et al. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–55. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–17. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connelly MA, Zhang H, Kieleczawa J, et al. The promoters for human DNA-PKcs (PRKDC) and MCM4: divergently transcribed genes located at chromosome 8 band q11. Genomics. 1998;47:71–83. doi: 10.1006/geno.1997.5076. [DOI] [PubMed] [Google Scholar]
- 20.Mosse YP, Greshock J, Margolin A, et al. High-resolution detection and mapping of genomic DNA alterations in neuroblastoma. Genes Chromosomes Cancer. 2005;43:390–403. doi: 10.1002/gcc.20198. [DOI] [PubMed] [Google Scholar]
- 21.Tang D, Sivko GS, DeWille JW. Promoter methylation reduces C/EBPdelta (CEBPD) gene expression in the SUM-52PE human breast cancer cell line and in primary breast tumors. Breast Cancer Res Treat. 2006;95:161–70. doi: 10.1007/s10549-005-9061-3. [DOI] [PubMed] [Google Scholar]
- 22.Maglott DR, Feldblyum TV, Durkin AS, et al. Radiation hybrid mapping of SNAP, PCSK2, and THBD (human chromosome 20p) Mamm Genome. 1996;7:400–1. doi: 10.1007/s003359900120. [DOI] [PubMed] [Google Scholar]
- 23.Steinberger P, Szekeres A, Wille S, et al. Identification of human CD93 as the phagocytic C1q receptor (C1qRp) by expression cloning. J Leukoc Biol. 2002;71:133–40. [PubMed] [Google Scholar]
- 24.Wang LP, Bi J, Yao C, et al. Annexin A1 expression and its prognostic significance in human breast cancer. Neoplasma. 2010;57:253–59. doi: 10.4149/neo_2010_03_253. [DOI] [PubMed] [Google Scholar]
- 25.Ali HR, Dawson SJ, Blows FM, et al. Cancer stem cell markers in breast cancer: pathological, clinical and prognostic significance. Breast Cancer Res. 2011;13:R118. doi: 10.1186/bcr3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khoury T, Ademuyiwa FO, Chandrasekhar R, et al. Aldehyde dehydrogenase 1A1 expression in breast cancer is associated with stage, triple negativity, and outcome to neoadjuvant chemotherapy. Mod Pathol. 2012;25:388–97. doi: 10.1038/modpathol.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Spence MJ, Zhang YL, et al. Transcriptional suppression of synuclein gamma (SNCG) expression in human breast cancer cells by the growth inhibitory cytokine oncostatin M. Breast Cancer Res Treat. 2000;62:99–107. doi: 10.1023/a:1006418219012. [DOI] [PubMed] [Google Scholar]
- 28.Bertucci F, Orsetti B, Negre V, et al. Lobular and ductal carcinomas of the breast have distinct genomic and expression profiles. Oncogene. 2008;27:5359–72. doi: 10.1038/onc.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan AR, Alexe G, Reiss M. Transforming growth factor-beta signaling: emerging stem cell target in metastatic breast cancer? Breast Cancer Res Treat. 2009;115:453–95. doi: 10.1007/s10549-008-0184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Q, Lu Z, Li H, et al. Next-generation sequencing of microRNAs for breast cancer detection. J Biomed Biotechnol. 2011;2011:597145. doi: 10.1155/2011/597145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucke CD, Philpott A, Metcalfe JC, et al. Inhibiting mutations in the transforming growth factor beta type 2 receptor in recurrent human breast cancer. Cancer Res. 2001;61:482–85. [PubMed] [Google Scholar]
- 32.Lapointe J, Lachance Y, Labrie Y, et al. A p18 mutant defective in CDK6 binding in human breast cancer cells. Cancer Res. 1996;56:4586–89. [PubMed] [Google Scholar]
- 33.Bostrom J, Meyer-Puttlitz B, Wolter M, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159:661–69. doi: 10.1016/S0002-9440(10)61737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Graauw M, van Miltenburg MH, Schmidt MK, et al. Annexin A1 regulates TGF-beta signaling and promotes metastasis formation of basal-like breast cancer cells. Proc Natl Acad Sci USA. 2010;107:6340–45. doi: 10.1073/pnas.0913360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahlert C, Bergmann F, Beck J, et al. Low expression of aldehyde dehydrogenase 1A1 (ALDH1A1) is a prognostic marker for poor survival in pancreatic cancer. BMC Cancer. 2011;11:275. doi: 10.1186/1471-2407-11-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamid AR, Pfeiffer MJ, Verhaegh GW, et al. Aldo-keto reductase family 1 member C3 (AKR1C3) is a biomarker and therapeutic target for castration-resistant prostate cancer. Mol Med. 2013;18:1449–55. doi: 10.2119/molmed.2012.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou W, Limonta P. AKR1C3 inhibition therapy in castration-resistant prostate cancer and breast cancer: Lessons from responses to SN33638. Front Oncol. 2014;4:162. doi: 10.3389/fonc.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedditch EL, Gao B, Russell AJ, et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Natl Cancer Inst. 2014;106(7):766–76. doi: 10.1093/jnci/dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]