Abstract
The ease of sequencing the cancer genome, identifying all somatic mutations and grafting mutation-specific T cell receptor (TCR) genes into T cells for adoptive transfer allow, for the first time, a truly tumor-specific and effective therapy. Mutation-specific TCR gene therapy might achieve optimal efficacy with least possible toxicity. Recent clinical data confirm the long-standing evidence from experimental cancer models that antigens encoded by the tumor-specific somatic mutations are potentially the best targets for adoptive T cell therapy. Open questions are, how many somatic mutations create suitable epitopes, whether only individual-specific or also recurrent somatic mutations qualify as suitable epitopes and how neoantigen-specific TCRs are most efficiently obtained. Tumor heterogeneity needs to be considered; therefore, it will be important to identify immunogenic driver mutations that occurred early, are essential for cancer cell survival and present in all cancer cells.
Introduction
In contrast to therapeutic vaccinations, adoptive T cell therapy (ATT) can very effectively eradicate large, long-established tumors in mice [1,2,3**,4] and induce sustained regression in humans [5**,6**]. The grafting of new antigen specificity onto patients’ T cells by TCR gene transfer is widely applicable to target any antigen on the cancer cells. Current trials of ATT mainly target tumor-associated antigens, which are self-antigens also expressed on normal cells. Targeting of normal cells, simultaneously with the cancer cells, can be acceptable in selected situations where the normal cells are dispensable, e.g. B cells expressing CD19 [5**]. However, for many, if not most self-antigens, severe toxicity and/or lethality can be expected when ATT has significant cytocidal effects [7*]. Targeting self-antigens by ATT shares with many drugs the lack of specificity which led many medical doctors to the bold conclusion that there is “no efficacy without toxicity”. Compared to current immunological and non-immunological therapies, targeting somatically mutated antigens by ATT should dramatically improve the risk-benefit ratio because the targets are truly tumor-specific.
Somatic mutations in cancer
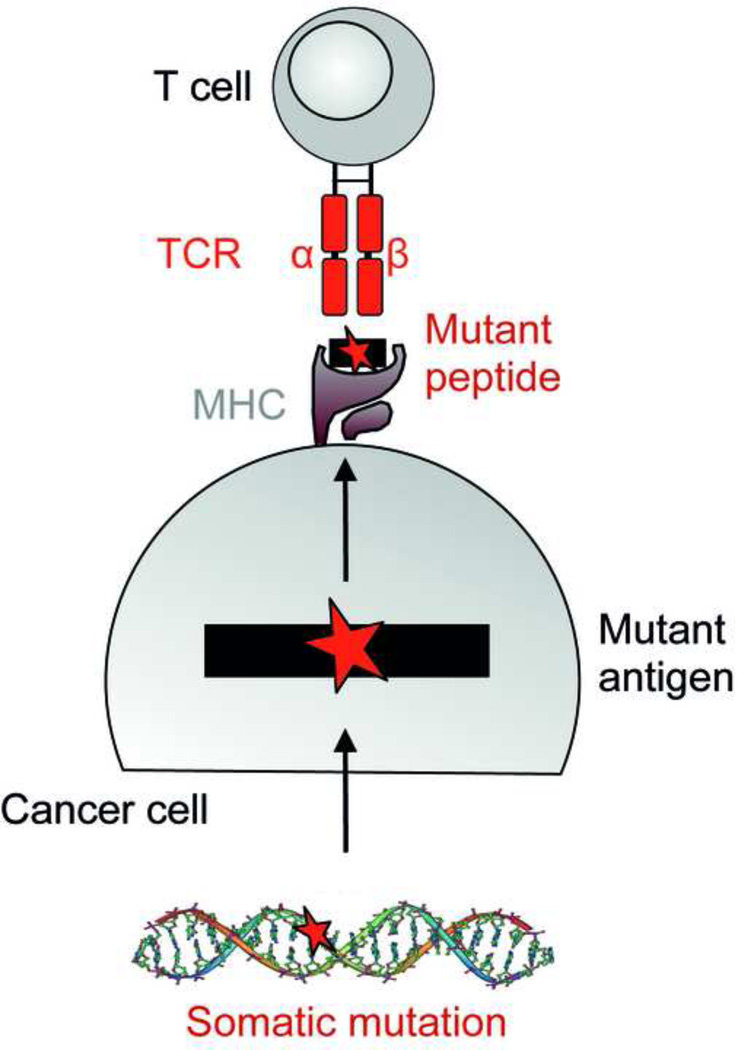
All cancers bear somatic mutations, because they are the cause of cancer. Mutations are usually random but clustering of mutations can occur. The process of Darwinian evolution selects for mutations that confer a growth advantage, e.g. increase cell proliferation, inhibit apoptosis, impair DNA repair etc. Less than 1% of the mutations occurs in coding regions and can lead to altered proteins. Only somatic mutations can encode truly tumor-specific antigens (also referred to as “neoantigens”) and are considered here. In addition, we will consider tumor virus-associated antigens that are encoded by viral genes and can also represent non-self targets on cancer cells. Most adult solid tumors have acquired around 50 mutations at the time of primary diagnosis [8*]. Pediatric cancers and some leukemias have significantly less mutations (less than 10 on average). Cancers of carcinogen-exposed tissue, ultraviolet (UV)-associated melanoma of the skin and tobacco smoke-associated lung cancer, often carry 100–200 somatic mutations. Some cancers with genomic instability, e.g. colon cancer with microsatellite instability, can carry more than 1000 mutations [8*]. Most mutations leading to an altered protein are non-synonymous point mutations causing a single amino acid substitution [9] (Figure 1). Chromosomal translocations and frameshift mutations resulting from small deletions/inversions are less frequent, and result in fusion peptides. The vast majority of all of these mutations are individually specific. Some occur repeatedly in different cancers (recurrent mutation), and the likelihood that they are involved in malignant transformation is high (so-called cancer-driving mutation). Even when mutations affect the same codon of an oncogene, e.g. RAS Gly12, different amino acids may replace the wild type amino acid and therefore lead to different mutant peptides. Cancer-driving mutations are probably present in all cancers, difficult to select against and can be individual-specific [10]. The number of cancer-driving mutations has been suggested to be ~140 [8*], but this may be an underestimate [11].
Figure 1. Mutation-specific T cells.
Somatic mutations in cancer may create a neoepitope that can be recognized by T cells.
Likelihood of creating a mutant epitope
Mutant peptides resulting from somatic tumor-specific mutation may or may not create a target on the cancer cell and/or induce an immune response in the host. Obviously, the probability of creating a targetable and/or immunogenic epitope increases with the number of mutations. Many factors like antigen expression level and processing, peptide trimming and transport, and binding of the peptide to MHC with sufficient affinity determine whether a mutant peptide is a suitable epitope. Because all these criteria have to be fulfilled, the probability for a non-synonymous point mutation or fusion peptide to create a tumor-specific antigen is low. A low probability appears to be true even when estimating the frequency of immunogenic epitopes encoded by vaccinia virus [12]. Of the ~100 vaccinia-encoded peptides that bound with an affinity IC50 of 100 nM or lower to HLA-A2, 50% elicited CD8 T cell responses in HLA-A2 transgenic mice. Of these, only 15% were naturally processed, and of these, 11% elicited CD8 T cell responses upon vaccinia virus infection; this means, only 1/14 of the predicted binders created an epitope. There is no reason to assume that point-mutant cancer peptides that differ less from self than vaccinia peptides would generate more frequently immunogenic epitopes. Based on these theoretical considerations, the number of mutant epitopes binding to a single MHC I allele with a IC50 of 100 nM or less and eliciting a CD8 T cell response would be about four to five mutant epitopes for an average human cancer harboring around 50 mutations. This number increases to 20–25 because the outbred patient population has typically six different MHC I alleles. However, more epitopes may serve as a target than induce an immune response because many or most targetable epitopes are recessive, i.e. the host may not respond to such an epitope until the dominant epitope is lost [13,14]. On the other hand, some HLA alleles appear to be able to bind more epitopes than other HLA alleles, which makes calculation difficult [15]. Furthermore, frameshift mutations may increase the number of potential epitopes by causing long novel reading frames [16].
Evidence for immunogenic mutant epitopes
From methylcholanthrene (MCA)- and UV-induced experimental cancer models, the existence of transplantation rejection antigens is long known [17,18]. These antigens were specific for each individual tumor and truly tumor-specific since they were caused by somatic mutations, first shown for UV-induced [9] and then for human [19,20] and finally MCA-induced cancers [21]. Despite this decade-long knowledge, immunotherapy remained focused on non-mutated self-antigens shared between cancers. However, with the already mentioned exception of CD19, targeting self-antigens (also called tumor-associated antigens) by immunotherapy, e.g. vaccines, has shown little if any success, because the power of self-tolerance mechanisms and the dangers of self-destruction have been underestimated [22*]. By contrast, the advent of fast and affordable cancer genome sequencing, the wealth of somatic mutations resulting from the cancer genome project and reliable algorithms to predict the binding affinity of mutant peptides to MHC I molecules has recently vitalized the interest in the potential immunogenicity of somatic mutations [6**,23–26,27*,28,29*,30,31–33*,34]. Tumor-specific somatic mutations can only be firmly determined, if germline controls are included in exome sequencing, which is routine for human but not always the case for experimental cancer lines. For example, the Jackson Laboratory C57BL/6J mouse from which B16 arose spontaneously over half a century ago [35] will certainly differ significantly from a present-day C57BL/6J mouse. This may explain why this spontaneous cancer was reported to have almost 1000 “tumor-specific somatic“ mutations [25], which is very unusual for any spontaneous cancer. Thus, much of this “mutanome” was likely caused by genetic polymorphisms [36] or mutations acquired by the B16 cells during decades of in vitro culture or in vivo passage. Certainly, even syngeneic mice suffer from genetic drift and polymorphisms, and autologous, not just syngeneic DNA, is a sine-qua-non for a germline control.
Mutations can be located in the peptide anchor position to generate an immunogenic epitope by increasing the affinity for the MHC molecule [37]. In this case, the wild-type peptide would be presented on the cell surface less effectively. If the altered amino acid is a TCR-contact residue and wild-type and mutant peptide bind with comparable affinity to the MHC [31*], then T cells responding to the wild-type self-peptide with high affinity will likely be deleted in the thymus. It has been known for decades that certain mutant peptides are shared between different cancers, and can be immunogenic (e.g. mutant RAS, BRAF or breakpoint regions of chromosomal translocations such as BCR-ABL or TEL-AML). Unfortunately, convincing experimental evidence that these conceptually attractive epitopes are efficient targets for rejection is still lacking and clinical data remain inconclusive. At least two reasons may account for the problem of identifying shared mutant epitopes that are efficient targets: peptides encoding these epitopes often have only intermediate or low affinity for MHC molecules; in addition, there often seems to be inefficient processing and transport of these peptides to be presented by MHC molecules on the cell surface [38].
Intratumoral heterogeneity and the need for targeting ancestral or multiple mutations
The more stably and homogeneously a mutant epitope is expressed in the cancer cells, the more efficiently it is targetable. However, most cancers when clinically detected consist of several genetically diverse subpopulations of cancer cells. This diversification is the result of a clonal evolution from a single common precursor cell hit by the putative “initiating event”. It seems practical to distinguish between two types of mutations, those referred to as “ancestral” or “background” mutations, and those referred to as “foreground” or “polymorphic” mutations [39]. “Ancestral” (background) mutations are shared by all cancer cells in a given tumor. For example, p53 mutations can belong into this category [39,40]. These types of mutations often affect checkpoints of cellular communication and DNA repair [41] and “prime“ the cells to proliferate and acquire additional mutations. “Polymorphic” (foreground) mutations are shared only by subpopulations of cancer cells in a given tumor. Such mutations may be involved in the transition from the least proliferative to the more aggressively growing cells and affect gene expression for example by dysregulating miRNAs [37,39,42]. Which of these mutations should be considered driver and which passenger mutations should be immaterial to the main point: the vast majority of the cancer cells in a tumor should express the targeted antigen. At initial diagnosis, a 1 cm diameter tumor is the result of 30 doublings with only 10 more doublings remaining until death of the patient. Thus, most of the genetic diversification has already occurred when the original cancer is removed. As would then be expected, analysis in vitro of cancer cell clones derived from an autochthonous, never transplanted UV-induced cancer revealed that all shared the same ancestral mutant tumor-specific antigen defined by a T cell clone. Yet, different clones of the cancer expressed different tumor-specific antigens [37,40] that were the result of “foreground” (polymorphic) mutations. As would be expected, T cells induced in recipient mice by immunizing with the uncloned original tumor select for cancer subclones that lack the polymorphic antigens. This selection is not an indication of unstable expression of a given mutant antigen but intratumoral antigenic diversity. Targeting of multiple neoepitopes by ATT using different tumor-specific T cells at the same time should reduce the risk of selecting escape variants [14].[0] Together, “ancestral” and if possible multiple mutations should be targeted whether encoded by a passenger or driver mutation. Obviously, it would be ideal if these antigens were essential and could not be lost [10,43].
Mutant epitopes can be retained in patients despite specific T cell responses
Non-destructive adaptive immune reactions, e.g. tumor-reactive IgG antibodies that imply CD4 T cell help, occur frequently in cancer patients [44]. Whether autochthonous non-virus-associated cancers spontaneously induce a destructive T cell response that can select for antigen-loss variants is still being debated [45,46]; since direct evidence from either mouse or human studies is lacking. It is long known that many UV-induced tumors that arose in immune-suppressed mice are rejected in naïve syngeneic mice and thus have a “regressor” phenotype [18]. However, regressor tumors can also develop in immunocompetent mice [47] with tumor-reactive CD8 T cells that can be induced to prevent tumor development [48]. Regressor tumors can even occur in mice with systemic tumor immunity in which SV40 large T antigen was induced by viral infection as oncogene/tumor antigen in the liver through Cre recombinase technology [49*]. The mice rejected subcutaneously injected SV40 large T-expressing cancer cells (concomitant immunity), while the autochthonous tumor progressed without losing its immunogenicity and antigenicity. Obviously, the process of cancer cell inoculation creates an artifact not reflecting autochthonous tumor growth. Thus, the T cells response to autochthonous tumors is fundamentally different from that of mice inoculated with cancer cells. The same case can be made for the Darwinian selection of cancer cells lacking the rejection antigen by a regressor tumor transplanted into normal immunocompetent mice. Such a selection occurs in many different transplant settings [2,26,37,50]. Similar to the above-mentioned sporadic cancer model, T cells did not impair tumor development in a transposon-based autochthonous cancer model [51]. Recent clinical case studies support the pre-clinical data that endogenous concomitant T cell immunity fails to select for cancer variants lacking the mutant epitope. In three individuals with ovarian cancer, the cancer genome was sequenced and 79 mutations were identified. Only a single one elicited a measurable CD8 T cell response at the time of first relapse (but not at the time of primary tumor diagnosis), but the response had disappeared at the time of second relapse even though the cancer cells had retained expression of the neoantigen [32*]. The data support the above consideration that measurable spontaneous T cell responses in the autochthonous cancer-bearing host are inefficient in selecting variants that lost immunogenic antigens. Similarly, tumor-infiltrating lymphocyte (TIL) therapy requires surgery which is performed with progressing, not regressing tumors. In some long-term responders following TIL therapy, the in vitro-expanded T cells contained a dominant population of mutation-specific T cells. Apparently, the TIL exerted their function only upon re-infusion into lymphopenic hosts, but did not spontaneously select against expression of the mutant antigen before surgery [6**]. In another melanoma patient experiencing a regression following anti-CTLA4 antibody treatment, mutation-specific CD8 T cells expanded upon therapy. Assuming that this T cell population had caused the tumor regression, the mutant antigen was not spontaneously lost before therapy [29*]. Loss of MHC I expression has been reported, rendering tumors resistant to ATT. However, in most cases MHC I expression can be upregulated by IFN-γ [52]. It is not clear whether rare tumors that have irreversibly lost MHC I expression are the result of escape from T cell pressure during tumor development. Irreversible loss of MHC I expression is often due to loss of the β2-microglobulin gene. This gene is located in proximity to a tumor suppressor gene repeatedly deleted in cancer which may explain why MHC I-negative cancer cells have a proliferative advantage [52]. Together, there is good indication that at least before therapeutic intervention, cancers retain expression of immunogenic antigens. Partial responses after immunotherapy seem to be more associated with antigen-retention, whereas relapse after complete remissions are more associated with antigen loss as observed experimentally [53**] as well as clinically [33].
High affinity of mutant peptides to MHC I required for cancer eradication
The retrospective analysis of melanoma patients responding to immunotherapy supports the data from experimental cancer models [2,53**]. A patient with a partial regression following anti-CTLA4 monoclonal antibody (mAb) treatment revealed a dominant CD8 T cell response against a mutant epitope [29*]. Similarly, the in vitro-expanded TILs of three patients that were successfully treated by the TILs contained mutation-specific CD8 T cells [6**,30]. In all cases, the mutant peptides bound with high affinity (low nM) to the respective MHC I molecules. Affinities in the 0.5 nM to 5 nM range have been shown to be critical to prevent tumor recurrence in experimental cancer models when targeting a single peptide [53**]. However, targeting a peptide that had a measured pMHCI IC50 of 186 nM also showed efficacy and caused almost complete destruction of large long-established cancers even though the cancer eventually relapsed [53**]. We propose that a combination of TCRs targeting three or more mutant peptides with <50 nM pMHC affinity (or even <200 nM) may be needed but may also be sufficient to eradicate established cancers. These epitopes should be independent of each other, i.e. encoded by different chromosomes so they cannot easily be lost simultaneously. Such an approach would be analogous to that used for combination chemotherapy of childhood leukemia. By targeting the cancers with multiple independent drugs, relapse could be prevented and cancers eradicated [54].
Source of T cell receptor
Accepting that neoepitopes are the best targets and in most cases retained on the cancer cells, the question is how to obtain TCRs for gene therapy. In principle, they can be isolated from humans or HLA-transgenic mice. In humans, TCRs can be isolated from TILs or PBLs of cancer patients (Figure 2). It may also be possible to isolate TCRs from healthy HLA-matched individuals by in vitro priming or after immunization with the mutant peptide, although these will not have been selected by the patient’s thymus for lack of self-reactivity. Based on prioritized neoepitopes with predicted high affinity for a given MHC allele, pMHC tetramers may facilitate enrichment of specific T cells. Single cell PCR [55] appears to be a reliable method to find the correct TCRαβ combination but alternative strategies have also been employed [56]. The advantage of obtaining the TCRs from the autologous host is that these TCRs have been screened for not binding self-antigens with a high affinity. However, an unresolved question is what determines the fate of neoepitope-specific T cells. In mouse models with surrogate antigens, T cell precursor frequency, TCR signaling strength, and the inflammatory milieu at the time of antigen encounter decided the T cell fate [57,58]. For example, depending on the amount of peripherally expressed antigen, the T cells were either deleted or became anergic [59]. It is not known under which immunological conditions T cells encounter mutant antigens in an evolving cancer. TCRs can be rescued from anergic T cells if they persist in the individual with progressing tumor. TCRs could also be isolated from patients responding to checkpoint inhibitor (anti-PD-1 and/or anti-CTLA4 mAbs) therapy [60–63*]. Response rates are relatively high in melanoma and lung cancer patients, who have a high mutation load. The T cell repertoire appears to broaden following anti-CTLA4 mAb but not anti-PD-1 mAb therapy, compatible with the supposed mode of action of facilitating T cell priming versus unleashing existing T cell responses, respectively [64–66]. It will be important to elucidate how frequently neoepitope-specific T cells are induced/expanded following therapy against the background of self-reactivity (and associated autoimmunity). Many patients do not respond to checkpoint inhibitors, and we do not know whether the cancers of theses patients lack suitable neoepitopes or whether the patient fails to mount a T cell response. Consistent with the clinical experience, anti-PD-1 mAb treatment as monotherapy delayed tumor growth but could not achieve complete tumor rejection in models of large established tumors or autochthonous cancers with systemic immunity but local tolerance [49*,67*]. We also do not know whether vaccination with mutated peptides with or without checkpoint inhibitors [68–70] induce mutant-specific TCRs in individuals with advanced autochthonous cancer.
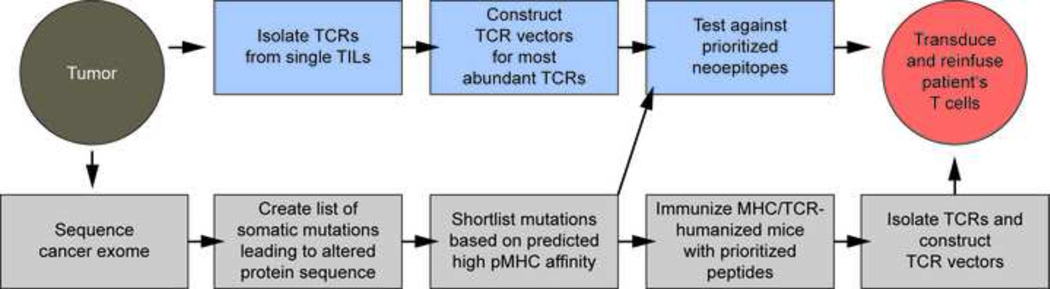
Figure 2. Strategies to isolate mutation-specific TCRs.
TCRs can either be isolated from T cells of cancer-bearing humans (top) or cancer-free HLA-transgenic mice (bottom). The advantage of obtaining TILs from humans is that their TCRs are truly specific for the neoepitope. In contrast, TCRs from HLA-transgenic mice are derived from a tumor-free environment. Note that epitope/TCR analysis in an experimental cancer model is not included but in our opinion is important to exclude therapeutically inefficient epitopes and/or TCRs.
Neoepitope-specific TCRs can be isolated from HLA-transgenic mice, preferably those additionally expressing a human TCR repertoire because of issues related to immunogenicity of non-human TCRs [71]. These mice would be immunized with the mutant peptides. An advantage is that TCRs would come from a tumor-free host, thereby excluding the possibility of neoepitope-specific T cell deletion which could occur in tumor-bearing individuals. An uncertainty is whether some neoepitope-specific TCRs obtained in the mouse may cross-react with normal human proteins, because the TCRs generated by the mouse have not been negatively selected in a human thymus. There is also the bottleneck that transgenic mice are not available for all HLA alleles. Thus, it needs to be seen whether potent neoantigen-specific TCRs are easier obtained from humans or mice.
How should TCRs be tested for efficacy in vivo?
Mutant epitopes as well as TCRs directed against them certainly differ in their power to support tumor rejection, therefore we consider pre-clinical testing as important to minimize the risk of clinical failure. In vitro assays do not necessarily predict efficacy in vivo [72]. Xenograft models with human cancer and T cells are afflicted with confounding factors (host species-specific factors necessary for T cell function), rendering human T cells notoriously ineffective in immune-deficient mice, as indicated by the absence of an acute lethal graft-versus-host disease. Furthermore, autologous T cells from the cancer patient are rarely available. Therefore, we suggest to establish syngeneic HLA-transgenic mouse cancer models for testing suitability of mutant epitopes and respective TCRs. The (mini)gene encoding the mutant epitope can be expressed in the murine HLA-expressing mouse cancer cells and T cells derived from the HLA-transgenic mice can serve as recipient for the TCR. In the clinic, TCR gene therapy is performed in individuals with established tumors, detectable at a minimal size of ~1 cm in diameter. Therefore, experimental cancers at the time of T cell transfer should be “established”. The term “established” should be clearly defined, because it is frequently used for a cancer cell inoculum few days after injection (e.g. [73–75]). In our opinion, an “established” tumor should have reached a clinically relevant diameter of 1 cm and lack the acute inflammatory reaction which is usually the case ~14 days after the inoculation [76,77]. Such tumors are much more difficult to treat than early inflammatory lesions in mice, but come closer to the clinical situation. Mutant peptide vaccines with or without checkpoint inhibitors have been tested against early lesions in mice [68–70]. It will be interesting to analyze whether such vaccinations can be used for isolating mutant peptide-specific TCRs [70], once the tumor is “established”, and whether vaccinations are therapeutically effective at that stage.
Conclusion
Grafting mutation-specific TCR genes into T cells for adoptive therapy will be possible and holds the promise of a truly tumor-specific and effective therapy. For shared (cancer-driving) mutations, TCRs would exist “off-the-shelf”, if such mutations create efficient targets for ATT. For targeting mutant peptides encoded by individual-specific somatic mutations, technological advances could significantly shorten the time interval currently needed to identify TCRs suitable for ATT.
Highlights.
T cell therapy can eradicate large tumors
Cancer-specific mutations creating neoepitopes are potentially the best target
Mutant epitopes can be retained in patients despite specific T cell responses
TCR gene therapy of cancer is feasible
Acknowledgement
Work described here was supported by a Collaborative Research Grant of the Berlin Institute of Health, the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich TR36, the Deutsche Krebshilfe, the Einstein-Stiftung Berlin, the Wilhelm Sander-Stiftung, NIH grants P01-CA97296, R01-CA22677, R01-CA37156 and the Cancer Center at the University of Chicago.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koeppen H, Singh S, Schreiber H. Genetically engineered vaccines. Comparison of active versus passive immunotherapy against solid tumors. Ann N Y Acad Sci. 1993;690:244–255. doi: 10.1111/j.1749-6632.1993.tb44013.x. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber K, Arina A, Engels B, Spiotto MT, Sidney J, Sette A, Karrison TG, Weichselbaum RR, Rowley DA, Schreiber H. Spleen cells from young but not old immunized mice eradicate large established cancers. Clin Cancer Res. 2012;18:2526–2533. doi: 10.1158/1078-0432.CCR-12-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schietinger A, Arina A, Liu RB, Wells S, Huang J, Engels B, Bindokas V, Bartkowiak T, Lee D, Herrmann A, et al. Longitudinal confocal microscopy imaging of solid tumor destruction following adoptive T cell transfer. Oncoimmunology. 2013;2:26677. doi: 10.4161/onci.26677. ** The sequence of elimination of antigen-positive cancer cells, stroma cells and bystander elimination of antigen-negative cancer cells is visualized by multi-color intravital ‘window chamber’ imaging.
- 4.Listopad JJ, Kammertoens T, Anders K, Silkenstedt B, Willimsky G, Schmidt K, Kuehl AA, Loddenkemper C, Blankenstein T. Fas expression by tumor stroma is required for cancer eradication. Proc Natl Acad Sci USA. 2013;110:2276–2281. doi: 10.1073/pnas.1218295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. ** Complete remission in 90% of patients with refractory acute lymphoblastic leukemia by CD19-redirected T cells is reported.
- 6. Robbins PF, Lu Y-C, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. ** Long-term responses in melanoma patients correlated with neoantigen-specific T cells in the in vitro expanded and infused tumor-infiltrating lymphocytes.
- 7. Hinrichs CS, Restifo NP. Reassessing target antigens for adoptive T-cell therapy. Nat Biotechnol. 2013;31:999–1008. doi: 10.1038/nbt.2725. * Comprehensive review summarizing on-target toxicities when targeting self-antigens by redirected T cells.
- 8. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. * Comprehensive review summarizing the average numbers of somatic mutations in different cancer entities.
- 9.Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity. 1995;2:45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 10.Beck-Engeser GB, Monach PA, Mumberg D, Yang F, Wanderling S, Schreiber K, Espinosa R, Le Beau MM, Meredith SC, Schreiber H. Point mutation in essential genes with loss or mutation of the second allele: relevance to the retention of tumor-specific antigens. J Exp Med. 2001;194:285–300. doi: 10.1084/jem.194.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Touw IP, Erkeland SJ. Retroviral insertion mutagenesis in mice as a comparative oncogenomics tool to identify disease genes in human leukemia. Mol Ther. 2007;15:13–19. doi: 10.1038/sj.mt.6300040. [DOI] [PubMed] [Google Scholar]
- 12.Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui H-H, Frahm N, Brander C, Peters B, Grey H, Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 13.Wettstein PJ, Bailey DW. Immunodominance in the immune response to “multiple” histocompatibility antigens. Immunogenetics. 1982;16:47–58. doi: 10.1007/BF00364441. [DOI] [PubMed] [Google Scholar]
- 14.Wortzel RD, Philipps C, Schreiber H. Multiple tumour-specific antigens expressed on a single tumour cell. Nature. 1983;304:165–167. doi: 10.1038/304165a0. [DOI] [PubMed] [Google Scholar]
- 15.Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, Sette A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol. 2013;191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer K, Nelius N, Reuschenbach M, Koch M, Weitz J, Steinert G, Kopitz J, Beckhove P, Tariverdian M, Knebel Doeberitz von M, et al. T cell responses against microsatellite instability-induced frameshift peptides and influence of regulatory T cells in colorectal cancer. Cancer Immunol Immunother. 2013;62:27–37. doi: 10.1007/s00262-012-1303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–778. [PubMed] [Google Scholar]
- 18.Kripke ML. Antigenicity of murine skin tumors induced by ultraviolet light. J Natl Cancer Inst. 1974;53:1333–1336. doi: 10.1093/jnci/53.5.1333. [DOI] [PubMed] [Google Scholar]
- 19.Wölfel T, Hauer M, Schneider J, Serrano M, Wölfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Büschenfelde KH, Beach D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 20.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wölfel C, Huber C, Wölfel T. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsutake T, Srivastava PK. The immunoprotective MHC II epitope of a chemically induced tumor harbors a unique mutation in a ribosomal protein. Proc Natl Acad Sci USA. 2001;98:3992–3997. doi: 10.1073/pnas.071523398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schietinger A, Delrow JJ, Basom RS, Blattman JN, Greenberg PD. Rescued tolerant CD8 T cells are preprogrammed to reestablish the tolerant state. Science. 2012;335:723–727. doi: 10.1126/science.1214277. * Self-reactive T cells are epigenetically imprinted for an anergic phenotype. The study illustrates the difficulty when targeting self-antigens by cancer vaccines.
- 23.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, Allison JP. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 24.Khalili JS, Hanson RW, Szallasi Z. In silico prediction of tumor antigens derived from functional missense mutations of the cancer gene census. Oncoimmunology. 2012;1:1281–1289. doi: 10.4161/onci.21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 26.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen Y-S, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anders K, Blankenstein T. Molecular pathways: comparing the effects of drugs and T cells to effectively target oncogenes. Clin Cancer Res. 2013;19:320–326. doi: 10.1158/1078-0432.CCR-12-3017. * First report suggesting mutation-specific TCR gene therapy of cancer.
- 28.Heemskerk B, Kvistborg P, Schumacher TNM. The cancer antigenome. EMBO J. 2013;32:194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJA, Behjati S, Hilkmann H, Atmioui ElD, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. * Case study of a melanoma patient with partial tumor regression following anti-CTLA4 antibody treatment, which correlated with expansion of neoantigen-specific T cells upon treatment.
- 30.Lu Y-C, Yao X, Li YF, El-Gamil M, Dudley ME, Yang JC, Almeida JR, Douek DC, Samuels Y, Rosenberg SA, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190:6034–6042. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2:522–529. doi: 10.1158/2326-6066.CIR-13-0227. * Comprehensive review of around 30 somatic mutations in cancer and how the mutation affects immunogenicity of the epitope.
- 32. Wick DA, Webb JR, Nielsen JS, Martin SD, Kroeger DR, Milne K, Castellarin M, Twumasi-Boateng K, Watson PH, Holt RA, et al. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res. 2014;20:1125–1134. doi: 10.1158/1078-0432.CCR-13-2147. * Only one out of 79 somatic mutations in 3 ovarian carcinoma patients elicited a measurable T cell response, which had disappeared at the time of relapse of the neoantigen-positive tumor. It remained unknown whether the T cells were deleted or had become anergic.
- 33. Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. * Tumor regression and recurrence after transfer of neoantigen-specific CD4 T cells isolated and in vitro expanded from tumor-infiltrating lymphocytes. Recurrent tumor had retained expression of the neoantigen.
- 34.Lu Y-C, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20:3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green E. Handbook on Genetically Standardized Jax Mice. 2nd ed. Bar Harbor, Maine: Bar Harbor Times Publishing Company; 1968. Biological Materials; pp. 1–76. [Google Scholar]
- 36.Bailey DW. How pure are inbred strains of mice? Immunol Today. 1982;3:210–214. doi: 10.1016/0167-5699(82)90093-7. [DOI] [PubMed] [Google Scholar]
- 37.Dubey P, Hendrickson RC, Meredith SC, Siegel CT, Shabanowitz J, Skipper JC, Engelhard VH, Hunt DF, Schreiber H. The immunodominant antigen of an ultraviolet-induced regressor tumor is generated by a somatic point mutation in the DEAD box helicase p68. J Exp Med. 1997;185:695–705. doi: 10.1084/jem.185.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popovic J, Li L-P, Kloetzel PM, Leisegang M, Uckert W, Blankenstein T. The only proposed T-cell epitope derived from the TEL-AML1 translocation is not naturally processed. Blood. 2011;118:946–954. doi: 10.1182/blood-2010-12-325035. [DOI] [PubMed] [Google Scholar]
- 39.Tao Y, Ruan J, Yeh S-H, Lu X, Wang Y, Zhai W, Cai J, Ling S, Gong Q, Chong Z, et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc Natl Acad Sci USA. 2011;108:12042–12047. doi: 10.1073/pnas.1108715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber K, Wu TH, Kast WM, Schreiber H. Tracking the common ancestry of antigenically distinct cancer variants. Clin Cancer Res. 2001;7:871s–875s. [PubMed] [Google Scholar]
- 41.Klein G, Imreh S, Zabarovsky ER. Why do we not all die of cancer at an early age? Adv Cancer Res. 2007;98:1–16. doi: 10.1016/S0065-230X(06)98001-4. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 43.Karanikas V, Colau D, Baurain JF, Chiari R, Thonnard J, Gutierrez-Roelens I, Goffinet C, Van Schaftingen EV, Weynants P, Boon T, et al. High frequency of cytolytic T lymphocytes directed against a tumor-specific mutated antigen detectable with HLA tetramers in the blood of a lung carcinoma patient with long survival. Cancer Res. 2001;61:3718–3724. [PubMed] [Google Scholar]
- 44.Blankenstein T, Coulie PG, Gilboa E, Jaffee EM. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12:307–313. doi: 10.1038/nrc3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 46.Qin Z, Blankenstein T. A cancer immunosurveillance controversy. Nat Immunol. 2004;5 doi: 10.1038/ni0104-3. 3–4– author reply 4–5. [DOI] [PubMed] [Google Scholar]
- 47.Willimsky G, Blankenstein T. Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance. Nature. 2005;437:141–146. doi: 10.1038/nature03954. [DOI] [PubMed] [Google Scholar]
- 48.Willimsky G, Czéh M, Loddenkemper C, Gellermann J, Schmidt K, Wust P, Stein H, Blankenstein T. Immunogenicity of premalignant lesions is the primary cause of general cytotoxic T lymphocyte unresponsiveness. J Exp Med. 2008;205:1687–1700. doi: 10.1084/jem.20072016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Willimsky G, Schmidt K, Loddenkemper C, Gellermann J, Blankenstein T. Virus-induced hepatocellular carcinomas cause antigen-specific local tolerance. J Clin Invest. 2013;123:1032–1043. doi: 10.1172/JCI64742. * Autochthonous virus-induced liver cancer in mice with tumor-destructive systemic immunity develops antigen-specific local tolerance through PD-1-dependent and PD-1-independent mechanisms. Tumor-reactive CD8 T cells from mice autochthonous liver cancer can delay autochthonous liver tumor progression in mice rendered lymphopenic through irradiation.
- 50.Ward PL, Koeppen HK, Hurteau T, Rowley DA, Schreiber H. Major histocompatibility complex class I and unique antigen expression by murine tumors that escaped from CD8+ T-cell-dependent surveillance. Cancer Res. 1990;50:3851–3858. [PubMed] [Google Scholar]
- 51.Rogers LM, Olivier AK, Meyerholz DK, Dupuy AJ. Adaptive immunity does not strongly suppress spontaneous tumors in a Sleeping Beauty model of cancer. J Immunol. 2013;190:4393–4399. doi: 10.4049/jimmunol.1203227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garrido C, Paco L, Romero I, Berruguilla E, Stefansky J, Collado A, Algarra I, Garrido F, Garcia-Lora AM. MHC class I molecules act as tumor suppressor genes regulating the cell cycle gene expression, invasion and intrinsic tumorigenicity of melanoma cells. Carcinogenesis. 2012;33:687–693. doi: 10.1093/carcin/bgr318. [DOI] [PubMed] [Google Scholar]
- 53. Engels B, Engelhard VH, Sidney J, Sette A, Binder DC, Liu RB, Kranz DM, Meredith SC, Rowley DA, Schreiber H. Relapse or Eradication of Cancer Is Predicted by Peptide-Major Histocompatibility Complex Affinity. Cancer Cell. 2013;23:516–526. doi: 10.1016/j.ccr.2013.03.018. ** High peptide-MHC I affinity prevents recurrence of antigen loss variants, as it occurs when targeting peptides with low affinity for MHC I by adoptively transferred T cells. Only high affinity peptides are cross-presented by CD11b-positive cells, which increases IFN-γ production and stroma destruction.
- 54.Skipper HE. The forty-year-old mutation theory of Luria and Delbrück and its pertinence to cancer chemotherapy. Adv Cancer Res. 1983;40:331–363. doi: 10.1016/s0065-230x(08)60683-1. [DOI] [PubMed] [Google Scholar]
- 55.Dössinger G, Bunse M, Bet J, Albrecht J, Paszkiewicz PJ, Weiβbrich B, Schiedewitz I, Henkel L, Schiemann M, Neuenhahn M, et al. MHC multimer-guided and cell culture-independent isolation of functional T cell receptors from single cells facilitates TCR identification for immunotherapy. PLoS ONE. 2013;8:e61384. doi: 10.1371/journal.pone.0061384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linnemann C, Heemskerk B, Kvistborg P, Kluin RJC, Bolotin DA, Chen X, Bresser K, Nieuwland M, Schotte R, Michels S, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat Med. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 57.Smith TRF, Verdeil G, Marquardt K, Sherman LA. Contribution of TCR signaling strength to CD8+ T cell peripheral tolerance mechanisms. J Immunol. 2014;193:3409–3416. doi: 10.4049/jimmunol.1401194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morgan DJ, Kreuwel HT, Sherman LA. Antigen concentration and precursor frequency determine the rate of CD8+ T cell tolerance to peripherally expressed antigens. J Immunol. 1999;163:723–727. [PubMed] [Google Scholar]
- 59.Redmond WL, Marincek BC, Sherman LA. Distinct requirements for deletion versus anergy during CD8 T cell peripheral tolerance in vivo. J Immunol. 2005;174:2046–2053. doi: 10.4049/jimmunol.174.4.2046. [DOI] [PubMed] [Google Scholar]
- 60. Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. * Together with [61*], [62*] and [63*], these studies show significant efficacy of T cell checkpoint inhibitors, mainly in patients with melanoma or lung cancer.
- 61. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. * See annotation to [60*].
- 62. Hamid O, Robert C, Daud A, Hodi FS, Hwu W-J, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. * See annotation to [60*].
- 63. Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon R-A, Reed K, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. * See annotation to [60*].
- 64.Robert L, Tsoi J, Wang X, Emerson R, Homet B, Chodon T, Mok S, Huang RR, Cochran AJ, Comin-Anduix B, et al. CTLA4 blockade broadens the peripheral T-cell receptor repertoire. Clin Cancer Res. 2014;20:2424–2432. doi: 10.1158/1078-0432.CCR-13-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kvistborg P, Philips D, Kelderman S, Hageman L, Ottensmeier C, Joseph-Pietras D, Welters MJP, van der Burg S, Kapiteijn E, Michielin O, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6:254ra128. doi: 10.1126/scitranslmed.3008918. [DOI] [PubMed] [Google Scholar]
- 66.Robert L, Harview C, Emerson R, Wang X, Mok S, Homet B, Comin-Anduix B, Koya RC, Robins H, Tumeh PC, et al. Distinct immunological mechanisms of CTLA-4 and PD-1 blockade revealed by analyzing TCR usage in blood lymphocytes. Oncoimmunology. 2014;3:e29244. doi: 10.4161/onci.29244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Binder DC, Engels B, Arina A, Yu P, Slauch JM, Fu Y-X, Karrison T, Burnette B, Idel C, Zhao M, et al. Antigen-specific bacterial vaccine combined with anti-PD-L1 rescues dysfunctional endogenous T cells to reject long-established cancer. Cancer Immunol Res. 2013;1:123–133. doi: 10.1158/2326-6066.CIR-13-0058. * First study to show rejection of large tumors by peptide specific vaccination and anti-PD1 mAb.
- 68.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber W-J, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J, Weinschenk T, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 71.Li L, Blankenstein T. Generation of transgenic mice with megabase-sized human yeast artificial chromosomes by yeast spheroplast-embryonic stem cell fusion. Nat Protoc. 2013;8:1567–1582. doi: 10.1038/nprot.2013.093. [DOI] [PubMed] [Google Scholar]
- 72.Spiotto MT, Rowley DA, Schreiber H. Bystander elimination of antigen loss variants in established tumors. Nat Med. 2004;10:294–298. doi: 10.1038/nm999. [DOI] [PubMed] [Google Scholar]
- 73.Mazumder A, Rosenberg SA. Successful immunotherapy of natural killer-resistant established pulmonary melanoma metastases by the intravenous adoptive transfer of syngeneic lymphocytes activated in vitro by interleukin 2. J Exp Med. 1984;159:495–507. doi: 10.1084/jem.159.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanson HL, Donermeyer DL, Ikeda H, White JM, Shankaran V, Old LJ, Shiku H, Schreiber RD, Allen PM. Eradication of established tumors by CD8+ T cell adoptive immunotherapy. Immunity. 2000;13:265–276. doi: 10.1016/s1074-7613(00)00026-1. [DOI] [PubMed] [Google Scholar]
- 75.Golumbek PT, Lazenby AJ, Levitsky HI, Jaffee LM, Karasuyama H, Baker M, Pardoll DM. Treatment of established renal cancer by tumor cells engineered to secrete interleukin-4. Science. 1991;254:713–716. doi: 10.1126/science.1948050. [DOI] [PubMed] [Google Scholar]
- 76.Schreiber K, Rowley DA, Riethmüller G, Schreiber H. Cancer Immunotherapy and Preclinical Studies: Why We Are Not Wasting Our Time with Animal Experiments. Hematol Oncol Clin N Am. 2006;20:567–584. doi: 10.1016/j.hoc.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 77.Wen FT, Thisted RA, Rowley DA, Schreiber H. A systematic analysis of experimental immunotherapies on tumors differing in size and duration of growth. Oncoimmunology. 2012;1:172–178. doi: 10.4161/onci.1.2.18311. [DOI] [PMC free article] [PubMed] [Google Scholar]