Abstract
α-glucans such as starch and glycogen are abundant in the human oropharynx, the main site of group A Streptococcus (GAS) infection. However, the role in pathogenesis of GAS extracellular α-glucan binding and degrading enzymes is unknown. The serotype M1 GAS genome encodes two extracellular proteins putatively involved in α-glucan binding and degradation; pulA encodes a cell-wall anchored pullulanase and amyA encodes a freely secreted putative cyclomaltodextrin α-glucanotransferase. Genetic inactivation of amyA, but not pulA, abolished GAS α-glucan degradation. The ΔamyA strain had a slower rate of translocation across human pharyngeal epithelial cells. Consistent with this finding, the ΔamyA strain was less virulent following mouse mucosal challenge. Recombinant AmyA degraded α-glucans into β-cyclomaltodextrins that reduced pharyngeal cell transepithelial resistance, providing a physiologic explanation for the observed transepithelial migration phenotype. Higher amyA transcript levels were present in serotype M1 GAS strains causing invasive infection compared to strains causing pharyngitis. GAS proliferation in a defined α-glucan-containing medium was dependent on the presence of human salivary α-amylase. These data delineate the molecular mechanisms by which α-glucan degradation contributes to GAS host-pathogen interaction including how GAS employs human salivary α-amylase for its own metabolic benefit.
Keywords: Streptococcus, salivary amylase, pharyngitis, translocation
Introduction
To successfully colonize or infect humans, bacteria must obtain nutrients from their host environment but understanding of the molecular mechanisms utilized by microbes to proliferate in vivo is limited (Xu et al., 2003, Sonnenburg et al., 2006, Shelburne et al., 2008b). Given their presence at high concentrations in diverse human environments, complex carbohydrates may serve as key carbon sources for infecting microbes (Iyer & Camilli, 2007, Moyrand et al., 2007, Shelburne et al., 2006). Genome-wide investigations have demonstrated that genes encoding proteins involved in complex carbohydrate acquisition and utilization are highly expressed in vivo by diverse bacterial pathogens (Roos & Klemm, 2006, Rollenhagen & Bumann, 2006, Son et al., 2007, Cho & Caparon, 2005). Further, for multiple human pathogens, genes encoding proteins involved in complex carbohydrate binding, transport, and metabolism contribute to microbial pathogenesis (Iyer & Camilli, 2007, Moyrand et al., 2007, Munoz-Elias & McKinney, 2005). Therefore, investigation of microbial complex carbohydrate utilization pathways may generate novel insights into host-pathogen interaction.
Group A Streptococcus (GAS) causes infections in humans ranging from uncomplicated pharyngeal or skin infections to life-threatening bacteremia, pneumonia, and necrotizing fasciitis (Cunningham, 2000). The major site of GAS infection and colonization in humans is the oropharynx (Peter & Smith, 1977). α-glucans such as starch and glycogen are polysaccharides comprised of repeating D-glucose monomers linked by α-bonds and are present at high concentrations in the human oropharynx (Mormann & Muhlemann, 1981). As their molecular weight is typically > 100,000, to serve as an energy source α-glucans must be digested by extracellular enzymes to form smaller molecules that can be transported into the bacterial cell and enter into energy production pathways. Investigations in the 1950’s determined that some GAS strains are capable of starch degradation, but the enzyme(s) responsible for GAS α-glucan degradation and subsequent transport are unknown (Crowley, 1950).
α-glucans may be degraded into glucoses linked in a linear fashion (i.e. maltodextrins) by enzymes termed amylases or pullulanases (Bertoldo & Antranikian, 2002) (Fig. S1). Alternatively, cyclomaltodextrin α-glucanotransferase (CGTase) enzymes degrade α-glucans into cyclic chains composed of glucose, i.e. cyclomaltodextrins (Qi & Zimmermann, 2005). The GAS serotype M1 strain MGAS5005 encodes at least two extracellular proteins putatively capable of α-glucan digestion (Ferretti et al., 2001, Hytonen et al., 2003, Lei et al., 2000, Sumby et al., 2005). PulA (M5005_SPy1682) is a cell-wall anchored pullulanase that in its purified form binds to and degrades glycogen and starch to linear maltodextrins (Hytonen et al., 2003, van Bueren et al., 2007), and AmyA (M5005_SPy1065) is a freely-secreted putative CGTase (Lei et al., 2000). It is known that strain MGAS5005 transports and catabolizes linear maltodextrins up to 7 glucoses in length suggesting that a PulA initiated pathway for α-glucan catabolism could function in GAS (Shelburne et al., 2007a). Alternatively, the genome of strain MGAS5005 also encodes proteins with similarity to enzymes in other bacterial species that are putatively involved in the transport of cyclomaltodextrins and their subsequent linearization to maltodextrins (Fiedler et al., 1996). However, no physiological study of cyclomaltodextrin production and utilization in GAS or any other Gram-positive pathogen has been performed. Thus, analysis of the GAS genome suggests that either AmyA or PulA or both could be critical to GAS α-glucan degradation.
Because of their predicted function and extracellular location, AmyA and PulA have been suggested to be GAS virulence factors (Ferretti et al., 2001, Hytonen et al., 2003, van Bueren et al., 2007, Hytonen et al., 2006). Moreover the ability of GAS to degrade α-glucans has been postulated to be contribute to GAS pathogenesis (Crowley, 1959, van Bueren et al., 2007). Therefore, we sought to define the molecular mechanisms by which GAS degrades α-glucans and to determine if α-glucan degradation contributes to GAS host-pathogen interaction. Our results demonstrate that AmyA-mediated α-glucan digestion influences GAS transepithelial migration and that GAS has adapted to use a host enzyme to initiate α-glucan catabolism.
Results
Analysis of GAS genome regions encoding genes putatively involved in extracellular α-glucan degradation
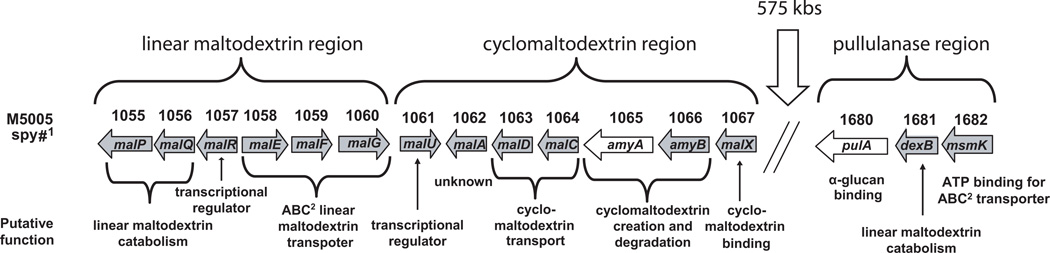
In the sequenced genome of the serotype M1 strain MGAS5005, there are two genes encoding putative extracellular α-glucan degrading enzymes, AmyA, a freely secreted, putative CGTase, and PulA, a cell-surface pullulanase (Sumby et al., 2005). amyA is one of 7 contiguous genes that encode proteins putatively involved in cyclomaltodextrin formation, transport, catabolism and gene regulation (Fig. 1). The pulA gene is about 575 kb away from amyA and is located next to genes putatively involved in linear maltodextrin degradation and transport. For the purposes of this manuscript, we will refer to the region comprising the open reading frames M5005_spy1061 to M5005_spy1067 as the cyclomaltodextrin (CMD) gene region and the region comprising M5005_spy1680 to M5005_spy1682 as the pullulanase gene region. The CMD gene region is immediately downstream of six contiguous open reading frames, M5005_spy1055 to M5005_spy1060, shown to be involved in linear maltodextrin transport and metabolism, which will be referred to herein as the linear maltodextrin (LMD) gene region (Shelburne et al., 2007a).
Fig. 1.
Schematic of the linear maltodextrin, cyclomaltodextrin and pullulanase gene regions in GAS serotype M1 strain MGAS5005 (Sumby et al., 2005). amyA and pulA genes are indicated in white. M5005_spy numbers refer to open reading frame in the serotype M1 strain MGAS5005. ABC = ATP-binding cassette.
Only GAS strains encoding AmyA degrade starch
Previous investigators have associated the pathogenesis of GAS infection with starch degrading activity, but there is no information regarding the mechanism by which GAS breaks down starch or other α-glucans (Crowley, 1959). To begin to investigate the molecular basis of GAS α-glucan metabolism, we determined the starch degradation capacity for 72 GAS strains comprising 28 of the most common M serotypes isolated in a recent survey of GAS North American pharyngeal isolates (Shulman et al., 2004). Strains from 10 of the 28 tested M serotypes, including the serotype M1 strain MGAS5005, degraded starch (Table S1) (Fig. 2). Next we used PCR to test for the presence of the amyA and pulA genes in all 72 strains. The pulA gene was amplified from all strains studied (data not shown). In contrast, amyA was only amplified from strains of the M serotypes that hydrolyzed starch (Table S1). Therefore, the presence of amyA, but not pulA, correlated perfectly with the ability of GAS to degrade starch.
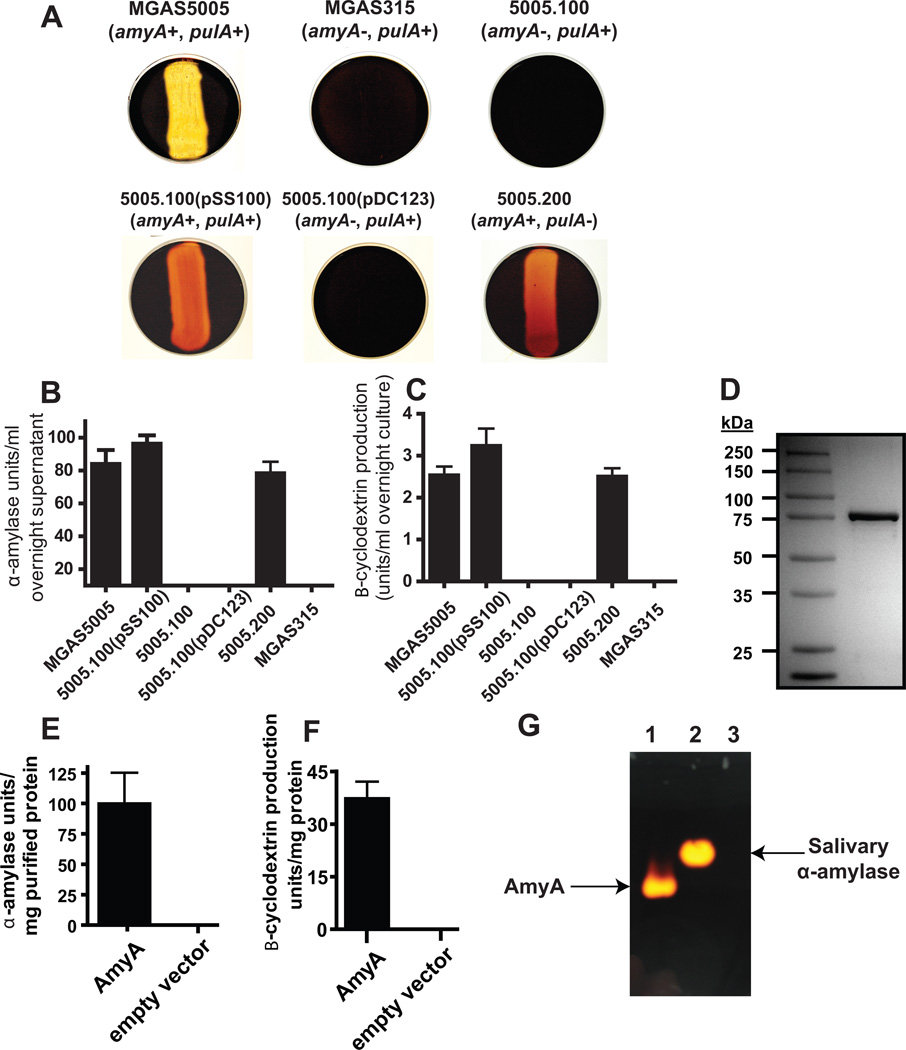
Fig. 2.
AmyA α-glucan digestion results in β-cyclomaltodextrin formation. (A) GAS strains of indicated amyA and pulA genotype were grown overnight on THY agar plates supplemented with 0.5% starch. Iodine was added to plates and the presence of clearing was assessed for evidence of starch hydrolysis. (B) Colorimetric analysis of iodine staining following growth in THY supplemented with 1% starch was used to assess starch degrading activity in indicated GAS strains as described in Experimental procedures. (C) β-cyclomaltodextrin production from starch by various strains was assessed using colorimetric analysis as described in Experimental procedures. (D) GAS AmyA was cloned and overexpressed in E. coli and then purified to apparent homogeneity as described in Experimental procedures. Proteins were separated on a 12% polyacrylamide gel. (E) Degradation of starch by purified AmyA with empty pBAD-hisA vector as negative control. (F) Production of B-cyclomaltodextrin by purified AmyA with empty pBAD-hisA vector as negative control. (G) Purified AmyA was run under non-denaturing conditions as described in Experimental procedures and subsequently allowed to diffuse into a 1% starch agar plate. Iodine staining confirmed that starch degradation (bright areas) was limited to the gel locations corresponding to AmyA. Lanes are as follows; 1 = 5 µl purified AmyA, 2 = 5 μl human salivary α-amylase, 3 = 5 µl empty vector. All starch degrading experiments were performed in triplicate on four separate occasions with data graphed being mean and error bars representing standard deviation.
AmyA is the major GAS extracellular α-glucan degrading enzyme and degrades α-glucans to cyclic maltodextrins
To assess whether AmyA is required for GAS α-glucan degradation, we created a ΔamyA isogenic mutant strain (strain 5005.100, Table 1) from the parental serotype M1 strain MGAS5005 via non-polar insertional mutagenesis (Lukomski et al., 2000). Additionally, we used a plasmid capable of replicating in GAS to create the genetically complemented strain 5005.100(pSS100) (Chaffin & Rubens, 1998). The doubling time during exponential growth and final growth densities of MGAS5005 in THY medium, glucose-medium, maltodextrin-medium, or in human saliva were not statistically different from strain 5005.100 (Table S2). Insertional mutagenesis of amyA abolished the ability of strain MGAS5005 to hydrolyze starch using starch/THY plates whereas providing amyA in trans to strain 5005.100 restored starch hydrolysis activity (Fig. 2). No significant difference in starch degradation was observed between parental strain MGAS5005 and its ΔpulA isogenic mutant derivative (strain 5005.200) (Fig. 2). We have previously shown that, in strain MGAS5005, pulA transcript level is increased approximately 75-fold during growth in a maltodextrin-medium compared to a standard laboratory medium, which raises the possibility that the lack of observed PulA contribution to α-glucan degradation might have resulted from low PulA expression (Shelburne et al., 2007b). However, we still could detect no difference in starch degradation between strain MGAS5005 and strain 5005.200 when starch/maltodextrin-medium plates were assayed demonstrating that increasing PulA expression did not reveal a role for PulA in GAS α-glucan digestion under the conditions tested (Fig. S2A).
Table 1.
Strains and plasmids.
| Strain or plasmid | Description | Genotype | Reference |
|---|---|---|---|
| Strains | |||
| MGAS5005 | Invasive isolate, serotype M1 | ΔcovS nt80 | (Sumby et al., 2005) |
| MGAS5005(pDC123) | MGAS5005 with empty vector | MGAS5005, Cmr | (Shelburne et al., 2006) |
| 5005.100 | ΔamyA strain | MGAS5005 amyA::spc | This study |
| 5005.100(pDC123) | ΔamyA strain with empty vector | ΔamyA, Spcr Cmr | This study |
| 5005.100(pSS100) | ΔamyA strain with complementation vector | ΔamyA/amyA+, Spcr Cmr | This study |
| 5005.200 | ΔpulA strain | MGAS5005 pulA::spc | This study |
| MGAS294 | Invasive isolate, serotype M1 | V286F in covS | (Sumby et al., 2006) |
| MGAS315 | Invasive isolates, serotype M3 | Wild-type | (Beres et al., 2002) |
| MGAS2217 | Invasive isolate, serotype M1 | 1 bp insert in covS | (Sumby et al., 2006) |
| MGAS2221 | Pharyngeal isolate, serotype M1 | Wild-type | (Sumby et al., 2006) |
| MGAS5322 | Pharyngeal isolate, serotype M1 | Wild-type | (Sumby et al., 2006) |
| MGAS5392 | Pharyngeal isolated, serotype M1 | Wild-type | (Sumby et al., 2006) |
| MGAS6184 | Invasive isolate, serotype M1 | A397V in covS | (Sumby et al., 2006) |
| MGAS9506 | Pharyngeal isolate, serotype M | Wild-type | (Sumby et al., 2006) |
| Klebsiella oxytoca M5a1 | Wild-type | (Fiedler et al., 1996) | |
| Plasmids | |||
| pSL60-1 | Vector containing aad9 gene | Spcr | (Lukomski et al., 2000) |
| pDC123 | Chloramphenicol resistance vector | Cmr | (Chaffin & Rubens, 1998) |
| pSS100 | amyA complementation vector | Cmr | This study |
| pBAD_amyA | pBAD vector with mature amyA sequence | Ampr | This study |
We next sought to quantitate the contribution of AmyA to GAS α-glucan degradation and to determine whether the resultant digestion products were linear or cyclomaltodextrins. Similar to our findings using agar plates, the amyA-positive strains MGAS5005 and 5005.100(pSS100) degraded starch to a significantly greater extent than the amyA-negative strains 5005.100 and 5005.100(pDC123) when grown in either THY or a maltodextrin-medium (Fig. 2B, Fig. S2B). Incubation of strain MGAS5005 with starch or glycogen did not result in an appreciable increase in reducing (i.e. linear) sugar formation (data not shown). Rather, degradation of starch by strain MGAS5005 or 5005.100(pSS100) resulted in the production of β-cyclomaltodextrins (Fig. 2C, Fig. S2C). To definitively establish the role of AmyA in the production of β-cyclomaltodextrins, we overexpressed and purified mature GAS AmyA (Fig. 2D). Consistent with our strain data, purified AmyA degraded starch to β-cyclomaltodextrins (Fig. 2E, 2F). Next, purified AmyA and human salivary α-amylase, the major human α-glucan degrading enzyme in the oropharynx, were run in a non-denaturing gel and then allowed to diffuse overnight into a starch agar plate. Staining of the plate showed that starch degradation occurred only at the site of the band representing purified AmyA and human salivary α-amylase (Fig. 2G). Taken together, we conclude that under the tested conditions AmyA is the major GAS α-glucan degrading enzyme and that the digestion of α-glucans by AmyA results in the formation of cyclic maltodextrins.
Loss of genes in the cyclomaltodextrin region likely occurred because of transposon duplication resulting in two GAS lineages
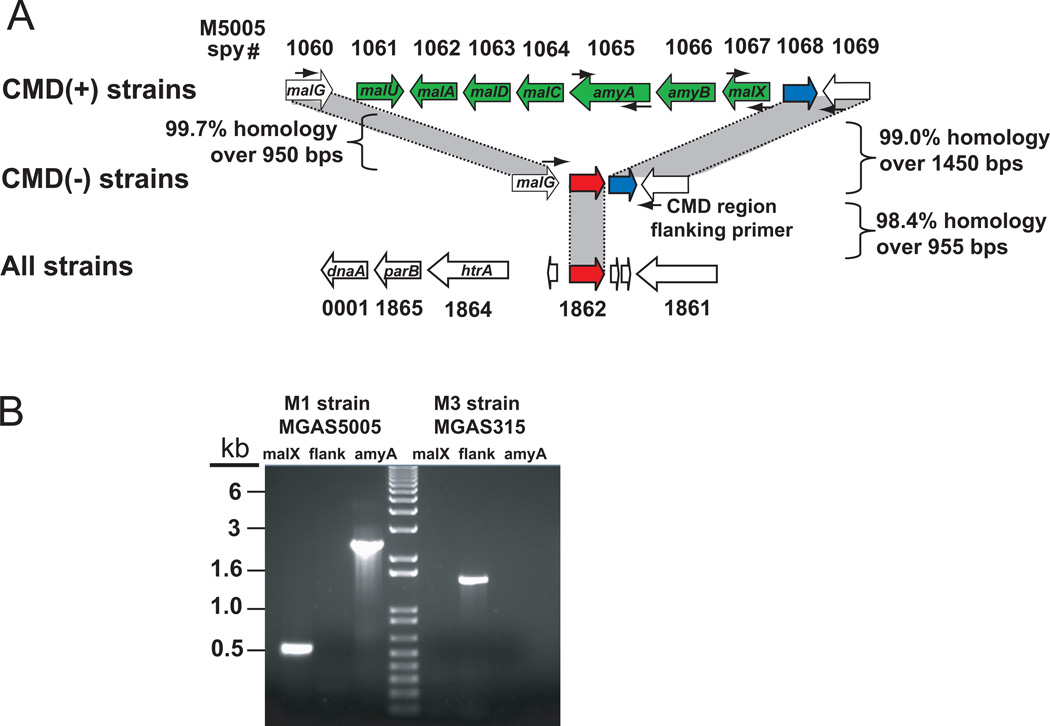
Our finding that amyA is present only in a limited number of M serotypes raised the question of how such a genetic arrangement came to occur. To address this we used PCR amplification spanning the CMD region to determine if all seven CMD genes are absent from the 37 strains of 16 M serotypes for which amyA amplification failed. The resultant amplicons indicated that the CMD region genes (i.e. M5005_spy1061 to M5005_spy1067) are absent en bloc from all 37 strains (Fig. 3, data not shown). The G+C content of the CMD region (9.3 kbp) is 38.08% whereas the G+C content of the core GAS genome is 38.59%. This difference is not statistically significant by either χ2 (P = 0.60) or Student’s t-test (P = 0.11) analyses. Moreover the dinucleotide composition (i.e. “genomic signature”) of the CMD region does not differ significantly from the core genome (Table S3). These findings are consistent with the CMD genes being either the ancestral GAS condition or acquired from an organism with a GAS-like nucleotide composition. BLAST analysis found that among fully sequenced bacteria, the GAS CMD region genes are conserved in content and genome context only in a strain of the closely related group C Streptococcus zooepidemicus (Beres et al., 2008).
Fig. 3.
Schematic of cyclomaltodextrin (CMD) region genes in various GAS strains. (A) Overview of CMD region in sequenced GAS strains. CMD(+) sequenced strains include serotype M1, M2, M4, and M28 strains (Ferretti et al., 2001, Sumby et al., 2005, Beres et al., 2006, Green et al., 2005). CMD(-) sequenced strains include serotype M3, M5, M6, M12, M18, and M49 strains (Beres et al., 2002, McShan et al., 2008, Nakagawa et al., 2003, Holden et al., 2007, Banks et al., 2004, Smoot et al., 2002, Beres et al., 2006). CMD(+) and CMD(-) strains share CMD flanking region genes and a transposase gene (M5005_spy1068) shown in blue. All GAS strains sequenced to date have a second transposase gene (M5005_spy1862, shown in red) found near the origin of replication that is duplicated in place of the CMD region genes in CMD (-) strains. (B) PCR reactions to determine presence or absence of CMD genes. Lane designations: Serotype M1 strain MGAS5005 with malX primers (1), with CMD region flanking primers (2), with amyA primers (3); serotype M3 strain MGAS315 with malX primers (4), with CMD region flanking primers (5), with amyA primers (6). Small black arrows showing primer locations appear in (A).
To gain further insight into the molecular basis of CMD region presence or absence, we compared the sequence of gene regions flanking the CMD genes in CMD-negative and CMD-positive strains (Fig. 3A). The nucleotide position of discontinuity between the CMD positive and negative genomes is the same for both the 5’- (100 bps downstream of malG) and 3’-end breakpoints (413 bps upstream of M5005_spy1068). That is, the indel site is in the exact same nucleotide position in all fully sequenced GAS strains. This finding is consistent with a single ancestral genetic event as repeated acquisition at the exact same site in all strains would be extremely unlikely. Analysis of the residual genetic material between the cognates of M5005_spy1060 (malG) and M5005_spy1069 in the sequenced CMD-negative strains revealed that all CMD-negative strains have two transposase-like elements in this region. Cognates of the transposase M5005_spy1068 (shown in blue in Fig. 3A) are present in the same location in all GAS strains, regardless of CMD gene status and have a high degree of sequence identity (shown in Fig. 3A). In contrast, the transposase gene immediately downstream of malG is present at this loci only in CMD-negative strains. This transposase gene (shown in red in Fig. 3A) shares ∼98% nucleotide identity over 955 bps with a transposase gene present near the origin of replication (M5005_spy1862, parB/dnaA gene region). The transposase near the origin of replication is present in all sequenced GAS strains. Thus, the CMD-negative strains have two copies of this transposase gene (one near the origin of replication and one in lieu of the CMD genes) whereas the CMD-positive strains only have the copy near the origin of replication. Therefore the genetic content of the CMD-negative strains suggests that the putative ancestral event leading to the loss of the CMD genes likely involved homologous recombination among transposases resulting in en bloc deletion of M5005_spy1061 to M5005_spy1067 leading to two branches of GAS (e.g. CMD-positive and CMD-negative strains). Of note, the gene encoding the fibronectin binding protein Fba is present in the same M protein serotype distribution as amyA, further supporting the concept of two distinct GAS lineages (Terao et al., 2001).
amyA transcript levels are high during the stationary phase of growth, and amyA is expressed in humans with pharyngitis
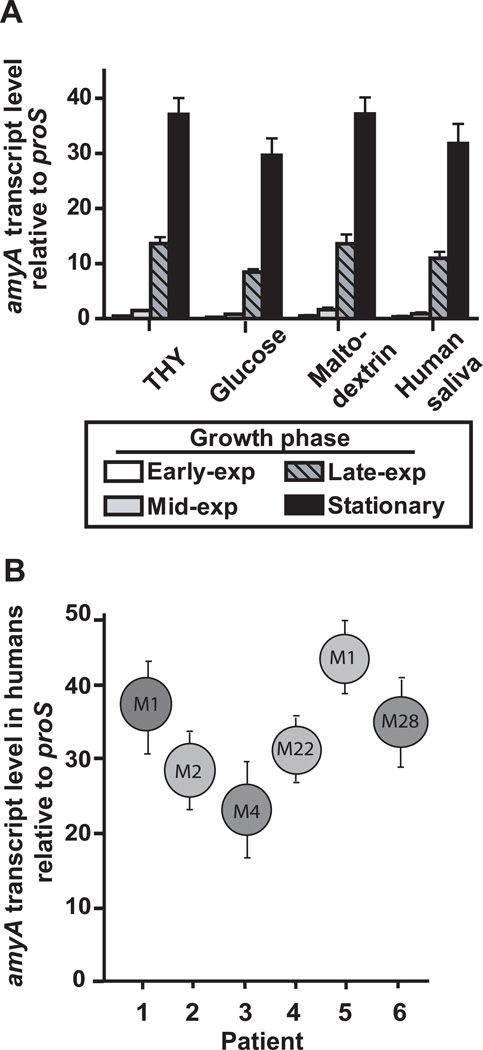
To begin to study the role of AmyA in host-pathogen interaction, we determined amyA transcript levels under a variety of conditions including from patients with GAS pharyngitis. In contrast to previous investigations of the LMD region genes and pulA, we found negligible medium-dependent differences in amyA transcript levels for strain MGAS5005 (Fig. 4) (Shelburne et al., 2007b). Under all conditions tested, maximal amyA transcript levels were noted to be present during the stationary phase of growth (Fig. 4A). Similarly, high amyA transcript levels were found in patients with GAS pharyngitis caused by strains of M serotypes that contain the CMD genes (Fig. 4B). Thus, the high amyA transcript levels observed in patients with pharyngitis are likely due to constitutive expression in stationary phase rather than in vivo induction as has been observed for other carbohydrate utilization genes (Shelburne et al., 2008a).
Fig. 4.
Analysis of amyA transcript levels. (A) Serotype M1 strain MGAS5005 was grown to early-exponential (exp), mid-exponential, late-exponential, and stationary growth phase in indicated media in quadruplicate biologic replicates on three separate occasions (total of 12 samples at each time point). RNA was isolated, converted to cDNA, and transcript levels for amyA and the endogenous control gene proS were determined using TaqMan real-time PCR. Data graphed are mean +/− standard deviation. (B) amyA transcript levels for 6 patients with GAS pharyngitis were determined by TaqMan real-time PCR. The M serotype of the infecting GAS strain is shown in the circle. Data graphed are mean +/− standard deviation with each experiment done with quadruplicate technical replicates on three separate occasions (total of 12 data points per sample).
Insertional inactivation of amyA does not affect the ability of GAS to colonize the mouse oropharynx but does affect the development of bacteremia following mucosal challenge
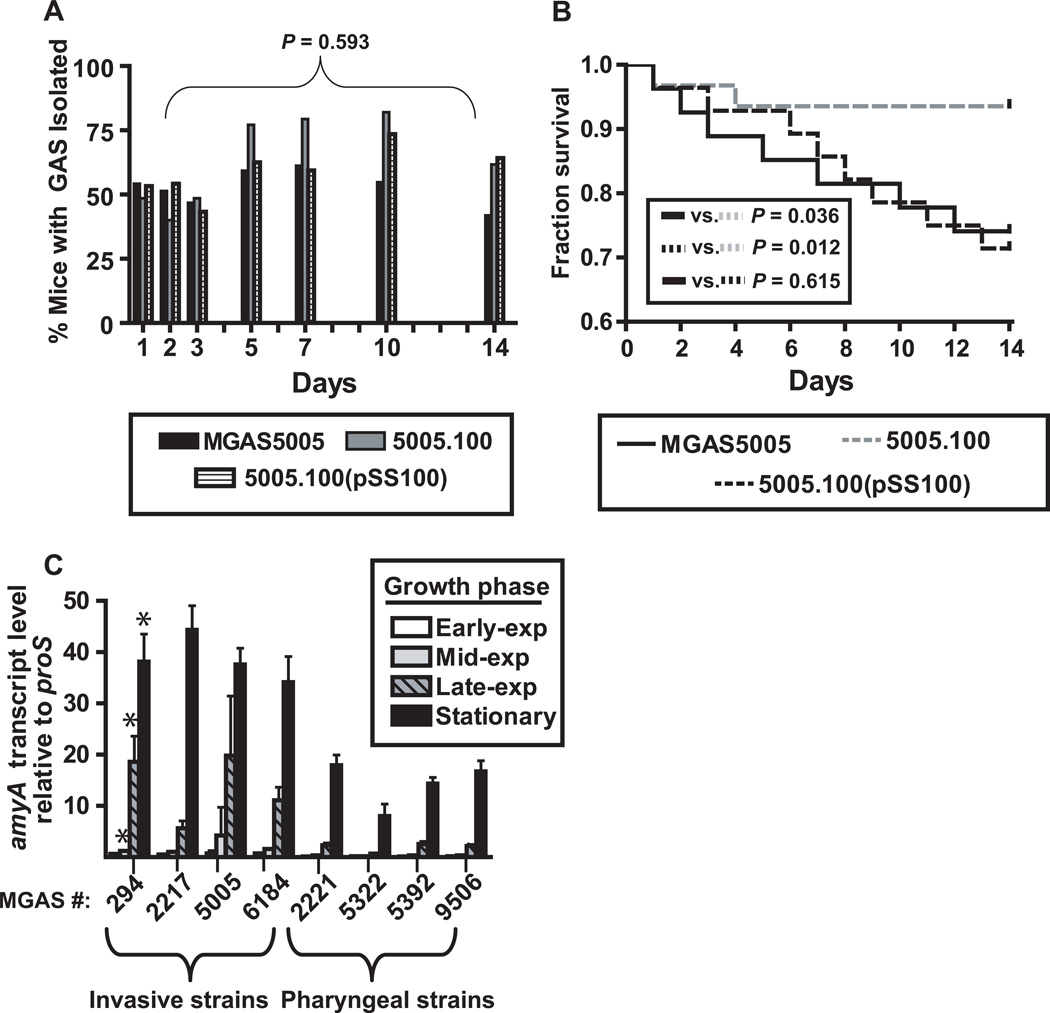
To this point, we had established that as the leading cause of pharyngitis, GAS is among a select few Gram-positive bacterium that encode AmyA and that amyA is expressed at high levels during GAS pharyngitis. Therefore, we next tested the hypothesis that AmyA contributes to GAS murine oropharyngeal colonization. To this end, we inoculated the nasopharynx of 35 adult outbred CD-1 mice each with either strain MGAS5005, 5005.100, or 5005.100(pSS100) and assayed for colonization by performing serial swabs of the oropharynx over a period of 14 days to ascertain the number of GAS present. Surprisingly, there was no statistically significant difference in either the proportion of mice testing positive for GAS colonization or in the average number of CFUs recovered per mouse between the three strains (Fig. 5A, data not shown). However, the overall mortality rate for mice infected with strains MGAS5005 and 5005.100(pSS100) was higher compared to strain 5005.100 (Fig. 5B). Analysis of the blood of dead or dying mice invariably revealed high numbers of GAS whereas healthy animals did not have GAS in their bloodstream. Taken together, these data do not support the hypothesis that AmyA contributes to GAS oropharyngeal colonization in the mouse. Rather, the data suggest that AmyA facilitates entry of GAS into the bloodstream.
Fig. 5.
AmyA does not contribute to GAS colonization of the mouse oropharynx but does increase murine mortality following mucosal challenge. Adult outbred CD-1 mice (n = 35 per group) were intransally inoculated with ∼1.0x107 CFU with indicated GAS strains. Mice oropharynges were swabbed daily and plated onto BSA. Plates were incubated for 24 hr, and β-hemolytic colonies were counted and tested for GAS carbohydrate antigen using latex agglutination. (A) Percentage of mice with GAS isolated by day. P value refers to repeated measures analysis. (B) Percent mice surviving graphed by data with P value referring to χ2 test done on day 14 of experiment. (C) Analysis of amyA transcript levels during growth in THY at indicated growth phase in invasive vs. pharyngeal GAS strains (for strain description see Table 1). * = P < 0.05 at the mid-logarithmic, late-logarithmic, and stationary growth phases when comparing invasive vs. pharyngeal strains. Transcript level analysis was done using quadruplicate biologic replicates on three separate occasions with data graphed being mean +/− standard deviation.
amyA transcript levels are increased in invasive serotype M1 GAS strains, and AmyA contributes to epithelial translocation of strain MGAS5005
Serotype M1 GAS strains can be broadly divided into pharyngeal and invasive isolates, with many invasive isolates having mutations in the control of virulence (CovR/S) two-component gene regulatory system resulting in derepression of genes encoding factors critical to the pathogenesis of invasive infection (Sumby et al., 2006). For example, strain MGAS5005 has a nucleotide insertion mutation in the covS gene that results in a truncated, non-functional CovS protein (Sumby et al., 2006). Given that amyA contributed to strain MGAS5005 bloodstream invasion, we next used real-time QRT-PCR to test the hypothesis that M1 strains with an “invasive” CovR/S system phenotype have higher transcript levels of amyA compared to strains with a “pharyngeal” phenotype (see Table 1 for strain description). For the mid-exponential, late-exponential, and stationary growth phases, we found that the “invasive” M1 strains had significantly higher levels of amyA transcript compared to the “pharyngeal” strains (Fig. 5C). These data are consistent with a previous investigation that found higher amyA transcript levels in a ΔcovR isogenic mutant strain compared to the parental strain MGAS5005 (Graham et al., 2002).
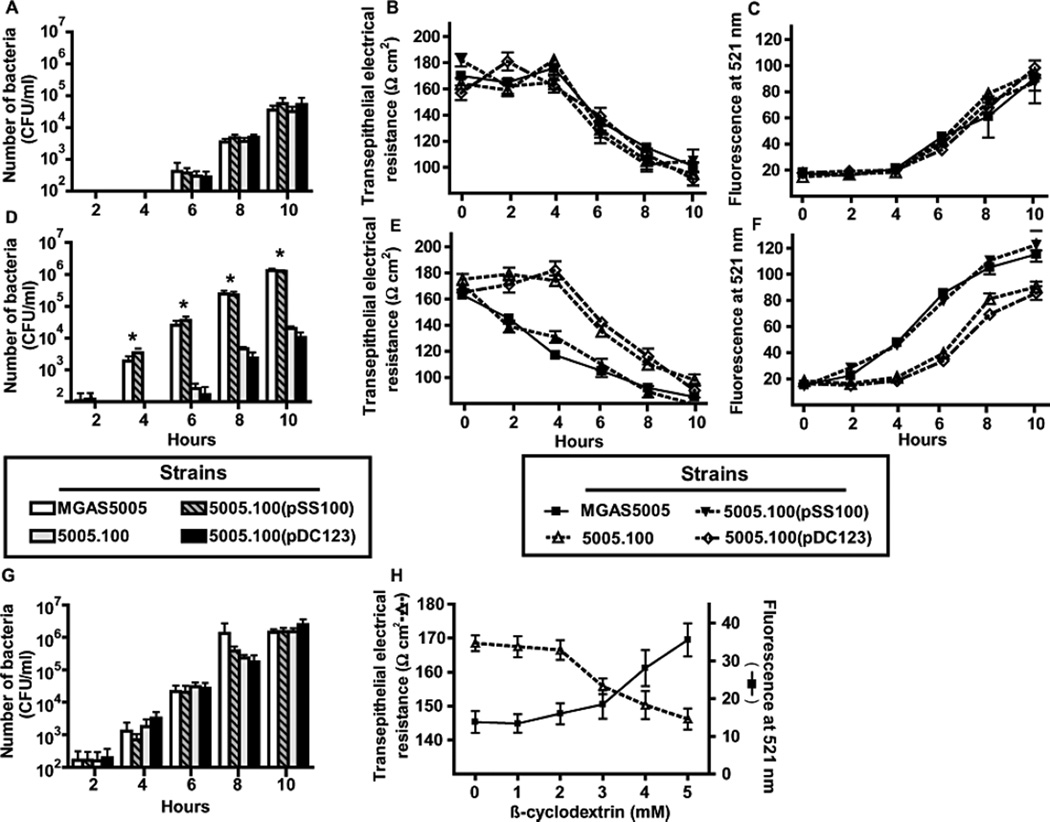
The presence of GAS in the bloodstream of the experimentally infected mice indicated that GAS had breached the airway epithelial cell barrier. Therefore, we next tested the hypothesis that strains MGAS5005, 5005.100, and 5005.100(pSS100), and 5005.100(pDC123) differ in epithelial translocation capacity. There was no significant difference in the ability of the four strains to cross a pharyngeal epithelial cell monolayer when the experiment was performed in a standard minimal medium (Fig. 6A). For each of the strains, the translocation of GAS across the pharyngeal cell layer was associated with an increase in epithelial cell permeability as determined by transepithelial electrical resistance (TEER) and fluorescein-dextran assays (Fig. 6B, 6C) which is consistent with a previous investigation of GAS epithelial cell translocation (Cywes & Wessels, 2001). When starch was included in the medium, strain MGAS5005 and 5005.100(pSS100) translocated earlier and at higher numbers compared to the strains lacking amyA (Fig. 6D). The earlier appearance of the amyA-positive strains was temporally related to a decrease in TEER and increase in fluorescein-dextran measurements indicating an accelerated loss of integrity of the epithelial cell monolayer (Fig. 6E, 6F). The addition of starch to the minimal medium did not increase GAS apical cell density for any of the strains compared to minimal medium alone (data not shown). Moreover, there were no significant CFU differences between the four strains in the apical compartment at any time point tested, indicating that increased bacterial burden did not account for the observed differences in cells in the basolateral compartment (data not shown). Therefore, we conclude that, in the presence of an α-glucan, AmyA augments the translocation capacity of an invasive M1 serotype GAS strain by a growth-independent mechanism.
Fig. 6.
AmyA increases GAS transepithelial invasion in the presence of starch. GAS translocation across a D562 epithelial cell monolayer were performed as described in Experimental procedures using indicated strains. For (A), (B), and (C) experiments were performed in standard minimal medium. (A) Number of translocated GAS by indicated strain. (B) and (C) are indicators of epithelial integrity using transepithelial resistance (TEER) and fluorescent-dextran. For (D), (E), and (F) experiments were performed in minimal medium with 0.5% starch added. (G) 5 mM β-cyclomaltodextrins were added to the starch-medium. (H) TEER and fluorescent measurements in the presence indicated concentrations of β-cyclomaltodextrin without GAS. Transepithelial invasion assays were performed in six replicates per strain on four separate occasions. Data graphed are mean with error bars representing standard deviation. Asterisks indicate a P value of < 0.05 using a repeated measures analysis.
The contribution to transepithelial invasion by AmyA is likely due to the production of cyclomaltodextrins
Cyclomaltodextrins interact with eukaryotic cell membranes leading to altered permeability and even toxicity (Matilainen et al., 2008). We hypothesized that cyclomaltodextrin formation by the AmyA(+) strains contributed to their accelerated translocation rate compared to AmyA(-) strains. Indeed, the addition to the medium of β-cyclomaltodextrins at a concentration consistent with the β-cyclomaltodextrin production capacity of strain MGAS5005 restored the translocation rate of the AmyA(-) strains to wild-type levels (Fig. 6G). To determine if cyclomaltodextrins directly affected epithelial cell permeability, we incubated D562 pharyngeal epithelial cells with β-cyclomaltodextrins. The presence of β-cyclomaltodextrins, but not linear maltodextrins, led to increased permeability as assessed by accumulation of fluorescein-dextran in the basolateral compartment of the transwell and by a decrease in TEER (Fig. 6H). Therefore, we conclude that AmyA mediated α-glucan digestion results predominantly if not exclusively in the formation of cyclic rather than linear maltodextrins (see Fig. 2). Moreover, cyclomaltodextrins produced by AmyA α-glucan digestion increase pharyngeal epithelial cell barrier permeability resulting in enhanced GAS translocation capacity.
GAS utilizes human salivary α-amylase to initiate catabolism of α-glucans
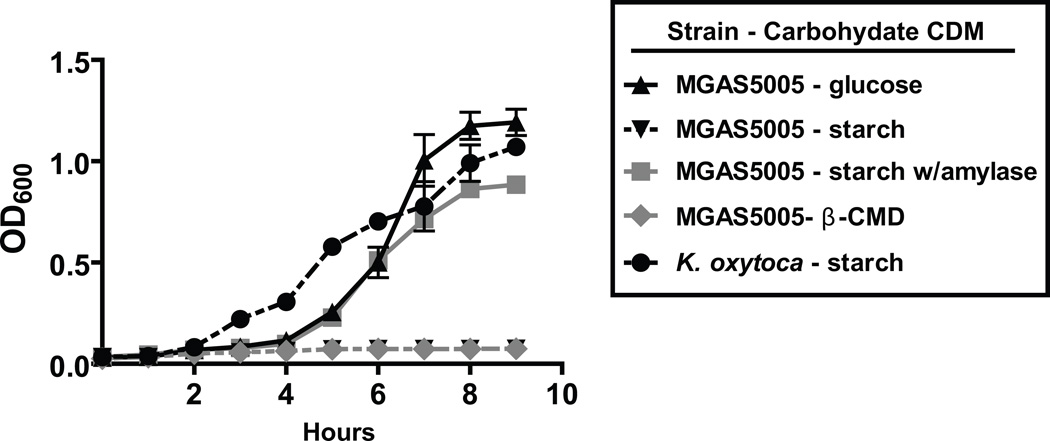
During the translocation experiments, we noted that the addition of starch to minimal medium did not increase GAS cell density, suggesting that GAS was not catabolizing the cyclomaltodextrins produced by AmyA. We tested this hypothesis by growing strains MGAS5005 and 5005.100 in a carbohydrate free chemically defined medium supplemented with various α-glucans. Strains MGAS5005 and 5005.100 were unable to grow in a medium in which soluble starch, glycogen, or pullulan was the sole carbon source (Fig. 7, data not shown). In contrast, the three α-glucan-media were capable of supporting bacterial growth as demonstrated by the growth of Klebsiella oxytoca strain M5a1, a bacterium that catabolizes starch via an actively secreted CGTase (Fig. 7) (Fiedler et al., 1996). We next tested the hypothesis that the lack of α-glucan catabolism was due to the inability of GAS to catabolize cyclomaltodextrins. Indeed, we found that strain MGAS5005 was unable to grow when cyclomaltodextrins (either α-, β-, γ-types) were the sole carbon source, with K. oxytoca again serving as the positive control (Fig. 7, data not shown) (Pajatsch et al., 1998). To ensure that the results were not unique to strain MGAS5005, we tested strains from the 10 distinct amyA+ M serotypes for growth in starch- and cyclomaltodextrin-media. None of the strains was able to catabolize starch or cyclomaltodextrins (data not shown). These data demonstrate that whereas amyA containing GAS strains can degrade α-glucans via AmyA, under the conditions tested GAS is unable to catabolize the cyclomaltodextrins generated from α-glucan digestion.
Fig. 7.
GAS uses human salivary α-amylase to initiate α-glucan catabolism. Data graphed are growth curves of GAS in various carbohydrate supplemented chemically defined media. Serotype M1 strain MGAS5005 was grown overnight in THY and then added at a 1:100 dilution into designated medium with growth monitored via OD600 readings. Human salivary α-amylase was added at physiologic concentrations when indicated. β-CMD = cyclomaltodextrins. Klebsiella oxytoca, which is capable of growth in starch, is included as a positive control. All growth experiments were performed in triplicate on four separate occasions with mean ± standard deviation graphed.
The inability of GAS to catabolize α-glucans suggested that it might rely on a host enzyme to initiate α-glucan degradation. Human salivary α-amylase degrades α-glucans to linear maltodextrins and has the highest concentration of enzymes present in human saliva (approximately 200 U/mL) (Humphrey & Williamson, 2001). In light of the known ability of GAS to proliferate in human saliva, we hypothesized that GAS would grow in an α-glucan-medium supplemented with human salivary α-amylase (Shelburne et al., 2005a). As hypothesized, the addition of physiologic doses of purified salivary α-amylase to either a starch- or glycogen-medium resulted in growth of strain MGAS5005 (Fig. 7). Neither the addition of α-glucan without α-amylase nor α-amylase without α-glucan resulted in detectable growth of strain MGAS5005 (data not shown). Thus, the data indicate that strain MGAS5005 can use α-glucans for growth when the host provides an enzyme that initiates degradation of α-glucans to linear maltodextrins, which have been shown previously to serve as a GAS carbon source (Kaczmarek & Rosenmund, 1977, Shelburne et al., 2007a).
Discussion
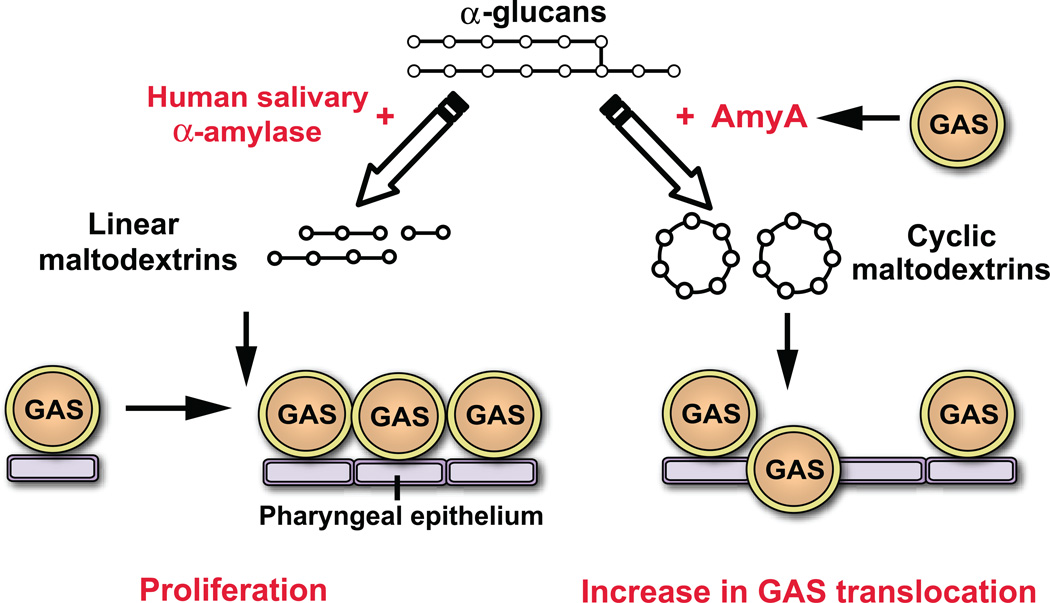
As numerous genome-wide analyses have suggested that complex carbohydrate utilization proteins contribute to infectivity, there has been significant investigation recently into the relationship between complex carbohydrate utilization and virulence in an array of human microbial pathogens (Iyer & Camilli, 2007, Moyrand et al., 2007, Munoz-Elias & McKinney, 2005). Genome-wide studies have identified streptococcal α-glucan degrading enzymes as putative virulence factors, but detailed analysis of the role of such enzymes in microbial pathogenesis have been lacking (Hava & Camilli, 2002). Therefore, we sought to understand the molecular mechanism of GAS extracellular α-glucan utilization and whether α-glucan degradation contributes to GAS host-pathogen interaction. Our data demonstrate that the freely secreted, α-glucan degrading CGTase AmyA influences GAS pathogenesis through a catabolism independent mechanism (Fig. 8).
Fig. 8.
Schematic of hypothesized role for α-glucan degrading enzymes in the pathogenesis of GAS pharyngitis. Degradation of α-glucans (such as starch or glycogen) by human salivary α-amylase results in the production of linear maltodextrins which can be used as an energy source for GAS proliferation. Alternatively, GAS AmyA α-glucan digestions leads to cyclic maltodextrin formation resulting in a decrease in epithelial transepithelial resistance and subsequent GAS translocation.
One of our key discoveries was that AmyA is the primary GAS α-glucan degrading enzyme and that cyclomaltodextrins produced by AmyA α-glucan degradation disrupt human pharyngeal epithelial cell integrity resulting in enhanced bacterial translocation. The development of invasive disease following GAS pharyngitis is a rare, but potentially lethal complication (Carapetis et al., 2005). It was recently discovered that, at least for serotype M1 and M3 strains, some GAS strains causing invasive disease have mutations in the CovR/S system which result in derepression of numerous virulence factors such as amyA (Sumby et al., 2006, Miyoshi-Akiyama et al., 2006). In addition to amyA, expression of genes encoding the hyaluronic acid capsule is increased in invasive GAS strains (Sumby et al., 2006). A previous investigation identified the hyaluronic acid capsule as being a critical contributor to GAS translocation capacity (Cywes & Wessels, 2001). Similar to that study, we found that GAS epithelial cell translocation was associated with a loss of integrity of the epithelial cell layer suggesting that entry of GAS into deeper host tissues results from epithelial cell disruption rather than passage through intact host cells, such as occurs for Streptococcus pneumoniae (Beisswenger et al., 2007). If alterations in the CovR/S system occur prior to epithelial translocation, then increased expression of the hyaluronic acid capsule along with increased cyclomaltodextrin production by AmyA may contribute to the subsequent development of invasive disease for AmyA-containing strains.
We determined that an entire genetic lineage of GAS strains lack the amyA gene and other CMD region genes and that gene loss was likely due to a duplication of a transposon located near the origin of replication resulting in en bloc deletion of the CMD region. The fact that all GAS strains sequenced to date that lack the CMD region genes differ from CMD-positive strains starting at the exact same site strongly suggests that a one-time genetic event occurred during GAS evolution leading to two separate GAS branches. Despite the fact that AmyA-positive GAS stains produce cyclomaltodextrins from starch, GAS appears unable to catabolize the cyclomaltodextrins, suggesting a loss of transport ability or subsequent degradative processes. Examination of the GAS genome (Fig. 1) reveals that all genes necessary for CMD transport and utilization seem to be present and intact. Indeed, we have recently found that some closely related group C and G streptococcal strains are able catabolize starch and cyclomaltodextrins and have a similar CMD gene region to GAS (Shelburne, unpublished data). Thus, the reason for the inability of AmyA-encoding GAS strains to catabolize cyclomaltodextrins remains unclear and is an area of active investigation. The high level of salivary α-amylase in the human oropharynx means that GAS strains infecting or colonizing the oropharynx will be exposed to an α-glucan degrading enzyme thereby obviating the need for GAS to initiate α-glucan digestion. We have previously demonstrated that GAS had adapted its maltodextrin transport mechanism to optimally bind and transport linear maltodextrins resulting from α-glucan digestion by human salivary α-amylase (Fig. 8) (21). Therefore, the CMD genes may have been critical when GAS existed under different metabolic conditions, but GAS now appears to rely on its human host and the linear maltodextrin gene region for α-glucan catabolism, obviating the need for the cyclomaltodextrin pathway (Fig. S1). These findings suggest that the loss of AmyA and other CMD region genes may be part of the evolutionary process of GAS adaptation to its human host.
In summary, we have discovered the molecular basis for GAS α-glucan degradation and identified how GAS α-glucan degradation contributes to host-pathogen interaction. These data extend the growing understanding of the close links between nutrient catabolism and microbial pathogenesis in humans. Further investigation of these links may provide the basis for preventive or therapeutic strategies.
Experimental procedures
Bacterial strains and culture media
Bacterial strains and plasmid used in this study are listed in Table 1 and Table S1. The genomes of the invasive serotype M1 strain MGAS5005 and pharyngeal serotype M1 strain MGAS2221 have been sequenced (Sumby et al., 2005, Sumby et al., 2006). GAS M serotype was determined by sequencing of the emm gene. Klebsiella oxytoca strain M5a1 was obtained from ATCC (Fiedler et al., 1996). GAS was grown on Trypticase soy agar containing 5% sheep blood (BSA; Becton Dickinson) or in Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY; Difco) whereas K. oxytoca was grown in Luria-Bertani broth (LB; Difco). Spectinomycin or chloramphenicol (Sigma) antibiotic was added when appropriate to THY agar or broth at a concentration of 150µg/mL and 4 µg/mL, respectively. Ultra-pure carbohydrates (Sigma) were added at a concentration of 0.5% (wt/vol) to a carbohydrate-free preparation of a commercially available chemically-defined medium (CDM; JR Biosciences) (Vise et al., 2003). For the purposes of this manuscript, we will use the terms glucose-medium, maltotriose-medium, starch-medium etc., to refer to the carbohydrate-free CDM supplemented with the specified carbohydrate. We have previously demonstrated that GAS cannot grow in the CDM without the addition of exogenous carbohydrates (Shelburne et al., 2007a). When indicated, purified human salivary α-amylase (Sigma) was added to CDM at physiologic concentration (200 U/mL) (Takai et al., 2004, Soderling et al., 1993).
Analysis of α-glucan degradation and cyclomaltodextrin production by GAS strains
GAS starch degrading activity on solid medium was assessed by plating 10 µl of overnight THY culture onto THY or CDM-maltodextrin agar plates containing 0.5% soluble starch (Sigma). The plates were incubated overnight at 37°C, flooded with iodine, and examined for a zone of clearing (Glor et al., 1988). Starch and glycogen degradation by various GAS strains were assessed in a liquid medium consisting of THY or CDM supplemented with 0.5% wt/vol soluble starch via iodine staining with absorbance measured at 660 nm using α-glucan digestion by human salivary α-amylase as the standard (Thiemann et al., 2004). The production of linear maltodextrins from the digestion of starch or glycogen was assessed by measuring reducing sugar formation using the dinitrosalicyclic acid assay with α-glucan digestion by human salivary α-amylase as the standard (Miller, 1959). β-cyclomaltodextrin spectrophotometric assays with purified cyclomaltodextrins as the standard were performed as described with 1 unit defined as the activity needed to produce 1 mg of β-cyclomaltodextrin from 1 mL of 0.5% wt/vol starch solution in 15 min (Makela et al., 1987). All α-glucan degradation experiments were done at least in triplicate on three separate occasions.
Creation of isogenic mutant and complemented mutant strains
Strain 5005.100 (ΔamyA isogenic mutant) was created by non-polar insertional mutagenesis from strain MGAS5005 using the PCR splicing overlap extension method described by Kuwayama et al. with the spectinomycin resistance cassette being amplified from plasmid pSL60 (Kuwayama et al., 2002, Lukomski et al., 2000). A similar strategy was used to create strain 5005.200 (ΔpulA isogenic mutant). The isogenic mutant strains were analyzed by Southern hybridization and DNA sequencing to confirm that the proper genetic construct was obtained (Fig. S3). For complementation purposes, we used plasmid pDC123 which encodes the chloramphenicol acetyl transferase (cat) gene for antibiotic selection and the phoZ gene for blue-white screening via alkaline phosphatase activity (Chaffin & Rubens, 1998). The complete amyA gene along with ∼ 250 bps upstream and ∼15 bps downstream were amplified from strain MGAS5005 chromosomal DNA using primers that introduced EcoR1 and BamH1 cut sites at the 5’ and 3’ ends respectively. The resulting PCR product was digested with EcoR1 and BamH1 (Invitrogen) and directionally cloned into the EcoR1 and BamH1 sites of plasmid pDC123. The resulting plasmid, named pSS100, was used to transform competent ΔamyA cells to create strain 5005.100(pSS100). pDC123 lacking the amyA gene was used to transform strain MGAS5005 and the strain 5005.100 as controls.
Overexpression and purification of AmyA and assessment of α-glucan degrading activity
PCR primers that introduced XhoI and EcoR1 restriction endonuclease sites were used to amplify the mature (i.e. lacking the secretion signal sequence) amyA gene from strain MGAS5005. The resultant PCR product was digested with XhoI and EcoR1 and ligated into the pBAD-hisA plasmid (Invitrogen) that had also been digested with the same enzymes. The ligation product, called pBAD-amyA, was cloned into Novablue (Novagen) and then BL21 (Novagen) competent Escherichia coli cells with selection via ampicillin resistance. pBAD-amyA was analyzed via sequencing to ensure that the correct construct had been obtained. To isolated purified protein, a Ni-NTA Fast Kit (Qiagen) was employed per the manufacturer’s instructions with the AmyA protein being present in the soluble fraction. Size exclusion chromatography was used to complete purification (Fig. 2D). The amount of AmyA present was determined using a Bradford assay (Bio-Rad). The same procedure was performed using the empty vector pBAD-hisA to ensure that any observed activity was not due to carry over from the plasmid. α-glucan degrading activity was analyzed by incubating varying amounts of purified protein with 1 mL of a 1% starch solution for 1 hr at 37°C and then using iodine staining. Determination of α-glucan degradation products were done using the dinitosalicyclic acid method (for linear maltodextrins) and phenolpthaline method (for cyclic maltodextrins) as noted above (Miller, 1959, Makela et al., 1987). For solid-phase determination of purified AmyA activity, indicated amounts of purified protein were loaded into a 12% Tris-Glycine pH 10.4 non-denaturing gel and run for 4 h at 15 mA. The gel was then placed on top of a 1% of starch-agar plate, left for 24 h to allow for protein diffusion into the plate, and stained with iodine to assess for starch degradation.
Assessment of amyA and pulA distribution and allelic diversity
The presence of amyA (M5005_spy1065 or spy1302 in serotype M1 strain SF370) and pulA (M5005_spy1680 or spy1972 in strain SF370) in diverse GAS strains was determined by PCR amplification. Chromosomal DNA was isolated using a DNeasy kit (Qiagen). PCR primers used for amyA and pulA amplification and sequencing were designed on the basis of the serotype M1 strain MGAS5005 genome (Sumby et al., 2005). As a positive control, all strains were assessed for the presence of the malE gene, previously found to be present in all strains used in this study (Shelburne et al., 2006). The amyA nucleotide sequence was determined from data obtained from both DNA strands using an Applied Biosystems 3730XL DNA Analyzer. Sequences were assembled and edited using Sequencher v4.5, and the inferred amino acid sequences were aligned using ClustalW. All sequence polymorphisms identified relative to strain MGAS5005 were confirmed by a second independent iteration of the entire DNA sequence determination process. Strains for which amyA failed to amplify using the initial primer pair were re-tested with an alternate amyA primer pair. Strains for which amyA failed to amplify using both the initial and alternate primers pairs, were further examined using a third pair of primers located in sequence flanking the cyclomaltodextrin genes that is conserved among the genomes of 12 sequenced GAS strains. Resultant flanking primer amplicons were examined for having a size (c.a. 1 kb) consistent with the lack of the cyclomaltodextrin utilization genes such as is the case for the M3 strain MGAS315 genome (Beres et al., 2002).
Assessment of gene similarities, codon usage, and nucleotide composition
Sequence alignments for assessing allelic diversity and identification of single nucleotide polymorphisms (SNPs) were performed using SeqScape v2.5 (Applied Biosystems) and MUMmer (Kurtz et al., 2004). Local best fit alignments of the α-glucan utilization genes studied with the sequenced GAS strains and the NCBI non-redundant sequence database were performed using BLAST (Altschul et al., 1990). Determination of nucleotide composition (mono- and di- nucleotide frequencies), and codon usage was determined using MacVector and CodonW (Rastogi, 2000). Global pairwise and multi sequence alignments were performed using ClustalW (Chenna et al., 2003).
RNA isolation and transcript level analysis
For transcript level analysis under diverse growth conditions, various GAS strains were grown as indicated. RNA was isolated and purified using an RNeasy kit (Qiagen) (Shelburne et al., 2005b). The quality and the concentration of RNA were assessed with an Agilent 2100 Bioanalyzer and by analysis of the A260/A280 ratio. cDNA was reverse transcribed from RNA using Superscript III (Invitrogen) following the manufacturer’s instructions. TaqMan quantitative real-time PCR (QRT-PCR) was performed with an ABI Thermocycler 7700 (Applied Biosystems) using the ΔCT method of analysis with the proS gene used as the internal control (Chaussee et al., 2001). To determine amyA transcript levels in vivo, GAS RNA was isolated from throat swabs obtained from six patients with GAS pharyngitis and analyzed as previously described (Virtaneva et al., 2003). The throat swabs were obtained from patients who gave their informed consent prior to participation in the study according to a protocol for human subjects approved by the Baylor College of Medicine Institutional Review Board. To ensure accuracy, all QRT-PCR primers and probes were tested to confirm similar amplification efficiencies using GAS genomic DNA as template. Primers and probes used are listed in Table S4. All transcript level analysis experiments were performed in quadruplicate on three separate occasions.
Growth of GAS strains in various media
All growth experiments were done in triplicate on three separate occasions for a total of nine replicates. For studies comparing GAS growth in THY or CDM, starter cultures were grown overnight in THY. Overnight cultures were inoculated into fresh medium to achieve a uniform starting OD600 of 0.01. Growth was monitored by measuring optical density at 600 nm. GAS was grown in human saliva as previously described (Shelburne et al., 2005a). Healthy volunteers who gave their informed consent prior to participation in the study donated saliva according to a protocol for human subjects approved by the Baylor College of Medicine Institutional Review Board. Saliva pooled from at least four donors was used to minimize the effects of donor variation on study results.
Mouse pharyngeal mucosal challenge
All experiments in mice were performed according to a protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee and were conducted with adult (18- to 20-g) female outbred CD-1 Swiss mice (Harlan Sprague-Dawley Inc.) (Shelburne et al., 2006, Lukomski et al., 2000). GAS strains were grown in THY and harvested at an OD600 of approximately 0.5. The cells were washed once with sterile phosphate buffered saline (PBS) and suspended in PBS at 1 × 108 colony forming units (CFU)/ml. Both nares of each mouse were inoculated with 50 µL of the GAS suspension (total inoculation 100 µl, 1 × 107 CFU) with 35 mice in each group. The throat of each mouse was swabbed before inoculation and then at specific time periods thereafter. The throat swabs were streaked onto BSA, grown overnight, and β-hemolytic colonies were enumerated. For each plate, five of the resultant β-hemolytic colonies were then tested for the presence of GAS carbohydrate antigen via latex agglutination with all colonies testing positive indicating that all of the β-hemolytic colonies were GAS (BD Biosciences).
Transepithelial invasion assays
D562 cells (human oropharyngeal cell line, ATCC) were grown to confluence on 24 mm collagen Transwell inserts with 3.0 µm pores in 6 well cell culture plates (Corning Costar) in Eagle’s minimal essential medium (ATCC) with 10% fetal bovine serum (Lonza) supplemented with penicillin and streptomycin (100 IU/mL and 100 µg/mL respectively, Mediatech). The integrity of the D562 monolayer epithelial barrier was determined by measuring the transepithelial resistance (TEER) using the Millicell-Electrical Resistance System (Millipore). The TEER value for the confluent D562 monolayer was ∼170 Ω cm2. Monitoring of cell barrier integrity was also performed using FITC-dextran (Sigma) which was added to the apical chamber at 1 µM final concentration with measurement done in the basolateral chamber by removing 100 µl and scanning at 521 nm in a Synergy HT microplate reader (Biotek). An intact barrier was generally achieved 14 days after inoculation of the cells. GAS translocation across the epithelial cell barrier was determined by growing GAS to mid-logarithmic growth phase, washing with PBS, and adding GAS resuspended in antibiotic free minimal medium to the apical chamber at a multiplicity of infection of 1 CFU per epithelial cell. The lower chamber was monitored hourly for the presence of GAS by removal of 100 µl of media followed by direct plating onto BSA with the lower limit of detection being 10 CFU/ml. Starch was added to the apical chamber at a 0.5% wt/vol when indicated. 5 mM β-cyclomaltodextrin (Sigma) was added at indicated concentrations where noted. The choice of 5 mM β-cyclomaltodextrin was based on the capacity of strain MGAS5005 to generate β-cyclomaltodextrins from α-glucans (i.e presumed physiologic concentration). The integrity of the epithelial cell barrier was followed using TEER and FITC measurements. All experiments were performed in six replicates for each strain on four separate occasions.
Statistical analysis
Growth, amylase activity, and RNA transcript levels were compared using Student’s 2-sided t-test. For the murine mucosal challenge model, a repeated measures analysis was used to test for differences in the percent of animals colonized and in the number of CFU recovered per animal. Difference in the transepithelial invasion of various strains was determined using a repeated measured analysis. Statistical significance was assigned a 2-sided P value of 0.05 using Bonferroni’s adjustment for multiple comparisons when appropriate. Statistical calculations were performed using NCSS software version 2004.
Supplementary Material
Acknowledgments
This work was supported by American Heart Association grant 0565133Y (S.A.S) and National Institute Allergy and Infectious Diseases K08 Career Development Award AI-064564 (S.A.S.). Portions of this work were presented at the 2007 American Society of Clinical Investigation/American Academy of Physicians annual meeting in Chicago, IL and the 2008 Infectious Diseases Society of America annual meeting in Washington DC. We thank Kathryn Stockbauer for help with figure generation.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, DeLeo FR, Martin JM, Somerville GA, Musser JM. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. Journal of Infectious Diseases. 2004;190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- Beisswenger C, Coyne CB, Shchepetov M, Weiser JN. Role of p38 MAP kinase and transforming growth factor-beta signaling in transepithelial migration of invasive bacterial pathogens. J Biol Chem. 2007;282:28700–28708. doi: 10.1074/jbc.M703576200. [DOI] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus . Proc Natl Acad Sci U S A. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sesso R, Pinto SW, Hoe NP, Porcella SF, Deleo FR, Musser JM. Genome sequence of a Lancefield group C Streptococcus zooepidemicus strain causing epidemic nephritis: new information about an old disease. PLoS ONE. 2008;3:e3026. doi: 10.1371/journal.pone.0003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu MY, Smoot JC, Porcella SF, Parkins LD, Campbell DS, Smith TM, McCormick JK, Leung DY, Schlievert PM, Musser JM. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci U S A. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoldo C, Antranikian G. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr Opin Chem Biol. 2002;6:151–160. doi: 10.1016/s1367-5931(02)00311-3. [DOI] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Chaffin DO, Rubens CE. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- Chaussee MS, Watson RO, Smoot JC, Musser JM. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes . Infect Immun. 2001;69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KH, Caparon MG. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes . Mol Microbiol. 2005;57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- Crowley N. The degradation of starch by strains of group A streptococci having related antigens. J Gen Microbiol. 1950;4:156–170. doi: 10.1099/00221287-4-2-156. [DOI] [PubMed] [Google Scholar]
- Crowley N. The association of starch-accumulating strains of group A streptococci with acute nephritis and acute rheumatic fever. J Hyg (Lond) 1959;57:235–247. doi: 10.1017/s0022172400020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywes C, Wessels MR. Group A Streptococcus tissue invasion by CD44-mediated cell signalling. Nature. 2001;414:648–652. doi: 10.1038/414648a. [DOI] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes . Proc Natl Acad Sci U S A. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler G, Pajatsch M, Bock A. Genetics of a novel starch utilisation pathway present in Klebsiella oxytoca . J Mol Biol. 1996;256:279–291. doi: 10.1006/jmbi.1996.0085. [DOI] [PubMed] [Google Scholar]
- Glor EB, Miller CH, Spandau DF. Degradation of starch and its hydrolytic products by oral bacteria. J Dent Res. 1988;67:75–81. doi: 10.1177/00220345880670011501. [DOI] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, LeFebvre RB, Musser JM. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Scott A, Cherevach I, Chillingworth T, Churcher C, Cronin A, Dowd L, Feltwell T, Hamlin N, Holroyd S, Jagels K, Moule S, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Skelton J, Whitehead S, Barrell BG, Kehoe M, Parkhill J. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain Manfredo. Journal of Bacteriology. 2007;189:1473–1477. doi: 10.1128/JB.01227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SP, Williamson RT. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001;85:162–169. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- Hytonen J, Haataja S, Finne J. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect Immun. 2003;71:784–793. doi: 10.1128/IAI.71.2.784-793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytonen J, Haataja S, Finne J. Use of flow cytometry for the adhesion analysis of Streptococcus pyogenes mutant strains to epithelial cells: investigation of the possible role of surface pullulanase and cysteine protease, and the transcriptional regulator Rgg. BMC Microbiol. 2006;6:18. doi: 10.1186/1471-2180-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Camilli A. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae . Mol Microbiol. 2007;66:1–13. doi: 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek MJ, Rosenmund H. The action of human pancreatic and salivary isoamylases on starch and glycogen. Clin Chim Acta. 1977;79:69–73. doi: 10.1016/0009-8981(77)90462-4. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002;30:E2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, Mackie S, Lukomski S, Musser JM. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect Immun. 2000;68:6807–6818. doi: 10.1128/iai.68.12.6807-6818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela M, Korpela T, Laakso S. Colorimetric determination of beta-cyclodextrin: two assay modifications based on molecular complexation of phenolphtalein. J Biochem Biophys Methods. 1987;14:85–92. doi: 10.1016/0165-022x(87)90043-1. [DOI] [PubMed] [Google Scholar]
- Matilainen L, Toropainen T, Vihola H, Hirvonen J, Jarvinen T, Jarho P, Jarvinen K. In vitro toxicity and permeation of cyclodextrins in Calu-3 cells. J Control Release. 2008;126:10–16. doi: 10.1016/j.jconrel.2007.11.003. [DOI] [PubMed] [Google Scholar]
- McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, Jia H, Kenton S, Najar F, Wu H, Scott J, Roe BA, Savic DJ. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes . J Bacteriol. 2008;190:7773–7785. doi: 10.1128/JB.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426. [Google Scholar]
- Miyoshi-Akiyama T, Ikebe T, Watanabe H, Uchiyama T, Kirikae T, Kawamura Y. Use of DNA arrays to identify a mutation in the negative regulator, csrR, responsible for the high virulence of a naturally occurring type M3 group A Streptococcus clinical isolate. J Infect Dis. 2006;193:1677–1684. doi: 10.1086/504263. [DOI] [PubMed] [Google Scholar]
- Mormann JE, Muhlemann HR. Oral starch degradation and its influence on acid production in human dental plaque. Caries Res. 1981;15:166–175. doi: 10.1159/000260514. [DOI] [PubMed] [Google Scholar]
- Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol. 2007;64:771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Kurokawa K, Yamashita A, Nakata M, Tomiyasu Y, Okahashi N, Kawabata S, Yamazaki K, Shiba T, Yasunaga T, Hayashi H, Hattori M, Hamada S. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Research. 2003;13:1042–1055. doi: 10.1101/gr.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajatsch M, Gerhart M, Peist R, Horlacher R, Boos W, Bock A. The periplasmic cyclodextrin binding protein CymE from Klebsiella oxytoca and its role in maltodextrin and cyclodextrin transport. J Bacteriol. 1998;180:2630–2635. doi: 10.1128/jb.180.10.2630-2635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter G, Smith AL. Group A streptococcal infections of the skin and pharynx (second of two parts) N Engl J Med. 1977;297:365–370. doi: 10.1056/NEJM197708182970706. [DOI] [PubMed] [Google Scholar]
- Qi Q, Zimmermann W. Cyclodextrin glucanotransferase: from gene to applications. Appl Microbiol Biotechnol. 2005;66:475–485. doi: 10.1007/s00253-004-1781-5. [DOI] [PubMed] [Google Scholar]
- Rastogi PA. MacVector. Integrated sequence analysis for the Macintosh. Methods Mol Biol. 2000;132:47–69. doi: 10.1385/1-59259-192-2:47. [DOI] [PubMed] [Google Scholar]
- Rollenhagen C, Bumann D. Salmonella enterica highly expressed genes are disease specific. Infect Immun. 2006;74:1649–1660. doi: 10.1128/IAI.74.3.1649-1660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos V, Klemm P. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect Immun. 2006;74:3565–3575. doi: 10.1128/IAI.01959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Fang H, Okorafor N, Sumby P, Sitkiewicz I, Keith D, Patel P, Austin C, Graviss EA, Musser JM, Chow DC. MalE of group A Streptococcus participates in the rapid transport of maltotriose and longer maltodextrins. J Bacteriol. 2007a;189:2610–2617. doi: 10.1128/JB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Granville C, Tokuyama M, Sitkiewicz I, Patel P, Musser JM. Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun. 2005a;73:4723–4731. doi: 10.1128/IAI.73.8.4723-4731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Keith DB, Davenport MT, Horstmann N, Brennan RG, Musser JM. Molecular characterization of group A Streptococcus maltodextrin catabolism and its role in pharyngitis. Mol Microbiol. 2008a;69:436–452. doi: 10.1111/j.1365-2958.2008.06290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Okorafor N, Sitkiewicz I, Sumby P, Keith D, Patel P, Austin C, Graviss EA, Musser JM. Regulation of polysaccharide utilization contributes to the persistence of group A Streptococcus in the oropharynx. Infect Immun. 2007b;75:2981–2990. doi: 10.1128/IAI.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Sumby P, Sitkiewicz I, Granville C, DeLeo FR, Musser JM. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci U S A. 2005b;102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Sumby P, Sitkiewicz I, Okorafor N, Granville C, Patel P, Voyich J, Hull R, DeLeo FR, Musser JM. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect Immun. 2006;74:4605–4614. doi: 10.1128/IAI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, Davenport MT, Keith DB, Musser JM. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008b;16:318–325. doi: 10.1016/j.tim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman ST, Tanz RR, Kabat W, Kabat K, Cederlund E, Patel D, Li Z, Sakota V, Dale JB, Beall B. Group A streptococcal pharyngitis serotype surveillance in North America, 2000–2002. Clin Infect Dis. 2004;39:325–332. doi: 10.1086/421949. [DOI] [PubMed] [Google Scholar]
- Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, Sturdevant DE, Ricklefs SM, Porcella SF, Parkins LD, Beres SB, Campbell DS, Smith TM, Zhang Q, Kapur V, Daly JA, Veasy LG, Musser JM. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling E, Pienihakkinen K, Alanen ML, Hietaoja M, Alanen P. Salivary flow rate, buffer effect, sodium, and amylase in adolescents: a longitudinal study. Scand J Dent Res. 1993;101:98–102. doi: 10.1111/j.1600-0722.1993.tb01096.x. [DOI] [PubMed] [Google Scholar]
- Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci U S A. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Yamaguchi M, Aragaki T, Eto K, Uchihashi K, Nishikawa Y. Effect of psychological stress on the salivary cortisol and amylase levels in healthy young adults. Arch Oral Biol. 2004;49:963–968. doi: 10.1016/j.archoralbio.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Terao Y, Kawabata S, Kunitomo E, Murakami J, Nakagawa I, Hamada S. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol Microbiol. 2001;42:75–86. doi: 10.1046/j.1365-2958.2001.02579.x. [DOI] [PubMed] [Google Scholar]
- Thiemann V, Donges C, Prowe SG, Sterner R, Antranikian G. Characterisation of a thermoalkali-stable cyclodextrin glycosyltransferase from the anaerobic thermoalkaliphilic bacterium Anaerobranca gottschalkii . Arch Microbiol. 2004;182:226–235. doi: 10.1007/s00203-004-0717-x. [DOI] [PubMed] [Google Scholar]
- van Bueren AL, Higgins M, Wang D, Burke RD, Boraston AB. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat Struct Mol Biol. 2007;14:76–84. doi: 10.1038/nsmb1187. [DOI] [PubMed] [Google Scholar]
- Virtaneva K, Graham MR, Porcella SF, Hoe NP, Su H, Graviss EA, Gardner TJ, Allison JE, Lemon WJ, Bailey JR, Parnell MJ, Musser JM. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71:2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vise PD, Kodali K, Hoe N, Paszczynski A, Musser JM, Daughdrill GW. Stable isotope labeling of a group A Streptococcus virulence factor using a chemically defined growth medium. Protein Expr Purif. 2003;32:232–238. doi: 10.1016/S1046-5928(03)00235-3. [DOI] [PubMed] [Google Scholar]
- Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci U S A. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.