Abstract
Background
We sought to characterize the expression of CD200, a membrane protein that functions in immune evasion, to examine its correlations with cancer stem cell-like features and analyze its response to chemotherapy and radiation, in human papillomavirus (HPV) positive and negative head and neck squamous cell carcinomas (HNSCCs).
Methods
CD200 expression was analyzed in several HNSCC cell lines. CD200 was over-expressed in HPV(+) murine tonsil epithelial cells, its effects on Shh and Bmi-1 examined in vitro, and tumor growth and response to chemoradiation analyzed in vitro and in vivo.
Results
CD200 was diversely expressed, consistently associated with expression of Bmi-1 and Shh. Overexpression of CD200 induced Bmi-1 and Shh. Tumor grew similarly between C57BL/6 and Rag1−/− C57BL6 mice. ,. CD200 expression enhanced the resistance to chemoradiation only in vivo.
Conclusions
CD200 was related to cancer-stem-cell features and modulates response to chemoradiation in vivo. Attenuating this might be a potential therapeutic strategy.
Keywords: oropharyngeal cancer, human papillomavirus, CD200, immune tolerance, neoplastic stem cells
Head and neck squamous cell carcinoma (HNSCC) is the eighth most common cause of cancer death worldwide.(1) This cancer is caused by both human papilloma viral and non-viral changes each with their own epidemiologic risks. Recent studies indicate that the incidence of HPV positive (+) HNSCCs is on the rise, increasing 60-85% in recent years.(2, 3) HPV(+) HNSCCs respond better to ionizing radiation, with and without the addition of chemotherapy, and are thus more curable than their HPV negative (−) counterparts.(4) However, treatment failures still occur in this group and the failure rate is increasing due to increased incidence. 20-40% of HPV(+) and 40-60% of HPV(−) HNSCCs exhibit poor response to treatment and patients suffer with disease recurrence. Thus, a greater understanding of the mechanisms driving these disease processes are necessary to provide better treatment options and improve patient outcomes for this expanding disease population.(4)
Cancer stem cell (CSC) or cells possessing CSC properties may explain some aspects of treatment resistance phenotypes encountered during therapy. Recent evidence indicates intratumoral CSC subsets are preferentially resistant to the effects of both radiation(5) and chemotherapy.(6) Moreover, CSC's may aid in immune evasion during immune-related tumor clearance.(7) It is possible that similar CSC subsets in HPV(+) or HPV(−) HNSCCs may drive poor prognosis and therapeutic response. However, the expression and distribution of this subset of CSCs within HNSCC tumors and their relationship to HPV infection are yet to be fully elucidated.
One marker associated with stem cell properties is, CD200. Formerly known as OX-2, CD200 is a highly conserved type I transmembrane glycoprotein with two extracellular immunoglobulin superfamily (IgSF) domains, a single transmembrane domain and a short cytoplasmic tail with no defined function.(8) CD200 is normally expressed by neurons, hair follicle epithelial cells, kidney glomeruli, syncitiotrophoblast, endothelial cells, some T and B lymphocytes, and thymocytes.(9) CD200 is a ligand that binds to its receptor (CD200R1), which is expressed on cells of the monocyte/macrophage lineage, on T lymphocytes, and on monocyte-derived dendritic cells.(9) The interaction of CD200 with CD200R1 triggers an immunosuppressive signal leading to inhibition of macrophages(10, 11), induction of regulatory T cells(12), switching of cytokine profiles from Th1 to Th2(13) and inhibition of tumor-specific T cell immunity. Treatment with a blocking monoclonal antibody to CD200 reverses this immunosuppressive phenotype.(14)
Several cancer cell lines and/or tissues express CD200 including ovarian, testicular, renal cell carcinoma, melanoma, malignant mesothelioma, neuroblastoma, chronic lymphocytic leukemia, prostate, breast, and colon cancers.(15) A growing body of evidence indicates that the CD200/CD200R1 pathway is exploited by the cancer cells to initiate immune evasion and tumor progression in several human solid tumors(15), CLL (16), AML(17), and multiple myeloma (MM).(18) Consistent with this, CD200 appears to be a prognostic factor in multiple myeloma(18) and in acute myeloid leukemia.(17)
In addition, CD200 is co-expressed with surface markers of CSCs found on prostate (CD44+), breast (CD44+CD24−), colon cancer (CD133+), and glioblastoma (CD133). These studies suggest that CD200 serves as a putative marker of CSC populations.(19) Expression of CD200 on CSCs together with its known capability to activate signaling pathways that ultimately result in immune evasion would significantly enhance tumor cell proliferation. Activation of sonic hedgehog (Shh)(20) and B-cell-specific Moloney murine leukemia virus integration site 1 (Bmi-1)(21) signaling cascades is typically associated with a proliferative response, increased DNA repair mechanism, and self-renewal characteristics of CSCs. Little is known, however, regarding the relationship between CD200 expression, these markers and a proliferative phenotype. Moreover, while CD200-mediated signaling and its downstream effects on immune evading mechanisms have been studied, little is known regarding CD200's activation of proliferative signals. Although CD200 was originally known as ‘pro-tumor’ protein, this ‘pro-tumor’ effect of CD200/CD200R1 interaction was occasionally challenged. CD200 induction reduced tumor formation and lung metastasis in a murine model of B16 melanoma cells.(22) The role of CD200 especially in wild-type tumor situation might vary depending on tumor type and its microenvironment, and is thus occasionally debated.
Herein, using in vitro models and an in vivo preclinical mouse model of HPV(+) tonsil cancers(23), we define the native expression patter of CD200 and explore its biologic functions in terms of tumor growth and treatment response in HNSCC.
MATERIALS AND METHODS
Cell Culture
Wild type mouse tonsil epithelial cells (MTECs) were isolated from C57BL/6 male mice as previously described.(23) MTECs were immortalized by stable expression of HPV16 E6, E7 together with Ras (MEER), generating a murine model for HPV (+) OSCCs as previously described.(23) MTEC and MEER cells were maintained with E-media (DMEM/Hams F12, 10% fetal calf serum, 1% penicillin/streptomycin, 0.5 μg/ml hydrocortisone, 8.4 ng/ml cholera toxin, 5 μg/ml transferrin, 5 μg/ml insulin, 1.36 ng/ml tri-iodo-thyonine, and 5 ng/ml EGF). The HNSCC tumor cell lines, UMSCC-1, -19, and -84 (HPV negative), UMSCC-47, and UPCI-SCC90 (HPV positive), as well as HEK293T cells were maintained with Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum and 1% penicillin/streptomycin.
Antibodies
Anti-CD200 (R & D Systems, Minneapolis, MN, AF2724), anti-Shh (Santa Cruz Biotechnology, Santa Cruz, CA, sc-9024), anti-Bmi-1 (clone F6, Millipore Corp., Billerica, MA, 05-637), anti-β actin (Sigma-Aldrich, St. Louis, MO, A2228), and anti-GAPDH (Ambion, Austin, TX, AM4300) were used for immunoblotting, immunocytochmistry, and immunohistochemistry. As secondary antibodies, HRP-conjugated anti-rabbit (GE Healthcare, Piscataway, NJ, NA934), anti-mouse (R & D Systems, HAF007), and anti-goat (Santa Cruz Biotechnology, sc-2020) were used for western blot analysis. Secondary antibodies for immunofluorescent localization were as follows: Alexa conjugates [anti-mouse (A11001); anti-goat (A11055)], and Alexa568 conjugates [anti-rabbit (A11011); anti-mouse (A11019)] were from Invitrogen Co (Carlsbad, CA).
Cell lysis immunoblot analysis
Cells were harvested at a confluency of 70%, proteins extracted at 4°C with lysis buffer, and soluble protein assayed by BCA protein assay (Pierce Biotechnology, Rockford, IL) as previously described(23). 50 μg of total cellular protein was loaded on SDS-PAGE gels, proteins transferred to PVDF membranes, blocked with either 5% milk or bovine serum albumin and probed for 2 hours at room temperature or overnight at 4°C with anti-CD200 at 1:1,000; anti-shh at 1:200; anti-Bmi-1 at 1:500; anti-β actin at 1:5000 and anti-GAPDH 1:2000 dilution. Bound antibody was detected with appropriate HRP-conjugated secondary antibody and visualized by standard luminescence reaction and exposure to Kodak film (Pierce Biotechnology). Protein bands were quantified and analyzed using quantitative densitometry, with ImageJ version 1.47u (rsbweb.nih.gov/ij).
Flow Cytometric Analyses
For BMI-1 and shh protein staining, cells were fixed with 4% paraformaldehyde and permeabilzed with Cytofix/Cytoperm kit (BD Biosciences, San Jose, CA). Cells were prepared in PBS plus 0.2% Tween 20, were blocked with PBS supplemented with 5% BSA, and incubated at 4°C for 45 min with PE-labeled anti-shh antibody (cat# IC4641P, BD Bioscience) or PE-labeled anti-BMI-1 antibody (cat# 562528, BD Bioscience). Then, cells were analyzed by FACS Calibur (BD Bioscience). For CD200 protein staining, cells, which were prepared by washing and subsequently being blocked with PBS plus 5% BSA, were reacted with anti-CD200 primary antibody (cat# 552512, BD Bioscience). After being washed with PBS plus 0.2% Tween 20, cells were labeled with FITC-conjugated secondary antibody (cat# 553896, BD Bioscience) for flow cytometric analyses.
Transfection
HEK293T cells were transfected with pUNO1-hCD200 (human) (Invivogen, San Diego, CA) or pUNO1-mCD200 (mouse) (Invivogen), using Polyfect transfection reagent as manufacturer's instructions. Twenty-four to 48 hours post-transfection, cells were lysed as described above. These cells were utilized as positive controls for human and mouse CD200 respectively. K562 nuclear lysate (ProSci Inc., Poway, CA, XBL-10435) served as positive control for Bmi-1 while IMR-32 cell lysate (Santa Cruz Biotechnology, sc-2409) served as a positive control for Shh.
Immunocytochemistry
Cells were seeded on eight-well glass-bottom slides (Lab-Tek II Chamber Slide System; Nalge Nunc International, Tokyo, Japan). The next day, cells were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 (Thermo Scientific Inc., Waltham, MA, #28314), blocked with Superblock (Thermo Scientific Inc., #37515), and incubated with primary antibodies, generally at a 1:100 dilution, for 2 hr at 37 °C. Secondary antibody, either Alexa Fluor 488- or 546-conjugated IgG was then incubated at a 1:100. Coverslips were mounted using VectaShield-4',6-diamidino-2-phenylindole mounting medium (VectaShield-DAPI; Vector Laboratories Inc., Burlingame, CA). Negative controls included omission of the primary antibody. Cells were analyzed by confocal microscopy (Olympus IX81 system equipped with argon-Kr/HeNe lasers, Olympus, Center Valley, PA).
Transfection and generation of stable CD200 overexpressing cell lines
HEK293T and MEER cells were transfected with 2 μg of DNA at 60% to 80% confluence using Polyfect reagent (Qiagen, Hilden, Germany, #301105). The expression plasmids, pUNO1-hCD200 (Invivogen) for human, and pUNO1-mCD200 (Invivogen) for mouse experiments were used. Clones were placed under selection with 1-20 μg/ml of blasticidin (MP Biomedicals, Solon, OH, #150477) and cells maintained at the highest concentration of blasticidin until 100% of uninfected control cells had died, about 2 weeks. Surviving colonies were ring cloned and tested. Over 20 clones were isolated and tested.
Proliferation assay
Cell lines were plated in 35 mm dish at a density of 2×104 cells in medium without selection agent. 24, 48, 72, 96, 120, and 144 hours following initial plating, cells were trypsinized, resuspended, and counted with Coulter counter (Beckman Coulter, Inc., Brea, CA). All samples were assayed in triplicate.(24)
Clonogenic assay
In vitro resistance to cisplatin or radiation was evaluated as previously described.(25) Briefly, 200 cells of each cell line were seeded per 60 mm plate. Cisplatin (0.025, 0.05, 0.1, 0.25, 0.5, and 1.0 μg/mL in dimethyl sulfoxide) was added 5 hours later and maintained for 24 hours. For radiation experiments, cells were irradiated once (0, 2, 4, 6, and 8 Gy). Cells were allowed to grow until colonies of untreated controls reached more than 50 cells (10-14 days). After staining with Comassie blue, colonies with more than 15 cells were counted. Each experiment was performed in triplicate. Surviving fraction was calculated using the following formula: (colonies counted)/ (cells plated× plating efficiency /100). Plating efficiency was defined as the number of cells present 24 hours after plating divided by the original number of cells plated.
In vivo assay
All experiments were performed in accordance with institutional and national guidelines and regulations; the protocol was approved by the Institutional Animal Care and Use Committee at Sanford Research. Using an 18-gauge needle, C57BL/6 mice (immune competent) and Rag1−/−C57BL6 mice (immune incompetent, lacking B and T lymphocytes) were injected 1×106 cells in the subcutaneous tissue of the right hind limb (10 mice per treatment condition; the experiment was repeated 3 times averaging the results). Volume of tumor was estimated by weekly caliper measurements and using the equation: Volume=width2×depth. To the treated group, 8 Gy of weekly external beam radiation and concomitant intraperitoneal injection of cisplatin (10 mg/m2, dissolved in 500 μl of normal saline) were given 3 times. Treatment began 2 weeks after tumor cell injection. Mice were euthanized when the tumor size was greater than 20 mm in greatest dimension or when emaciated. Mice were considered tumor free when they showed no evidence of tumor for 3 months.
Immunohistochemistry
Staining was mainly done using MACH4 Universal HRP-Polymer Kit (Biocare Medical, Concord, CA, M4U534). All reagents and instruments used were purchased from Biocare Medical, unless otherwise stated. Formalin-fixed, paraffin-embedded murine tissues were deparaffinized in xylene and dehydrated. Antigen was then unmasked using Rodent Block M (RBM961) in Decloaking Chamber. Endogenous peroxidases were quenched using Peroxidazed 1 (PX968). Tissues were then blocked using Background Sniper (BS966). Incubation with primary antibody (1:50) was performed for 30 minutes at room temperature. Secondary antibody (1:50) incubation and detection were carried out with a Betazoid DAB Chromogen Kit (BDB2004). As a negative control, staining with secondary antibody alone was used. Tissue was counterstained with hematoxylin. After dehydration, slides were mounted with Echomount (EM897L) and photographed with a microscope. Histologic findings were analyzed by a pathology specialist in head and neck cancer.
Statistical analysis
Comparisons between groups were made using the 2-tailed, paired Student's t test or the Mann Whitney U test. Comparisons of serially changing values over time or dose between groups were done using general linear model (univariate or multivariate). Mouse survival curves were constructed using Kaplan–Meier method with a 60-day cutoff. The Log-rank or Breslow's test was used to compare the homogeneity of survival rates between categories. Two-tailed p values <.05 were considered significant. STATA/SE version 10.1 (StataCorp LP, College Station, TX) were used.
RESULTS
CD200 is diversely expressed in wild type HNSCC cell lines, correlated with the expression of Shh and Bmi-1
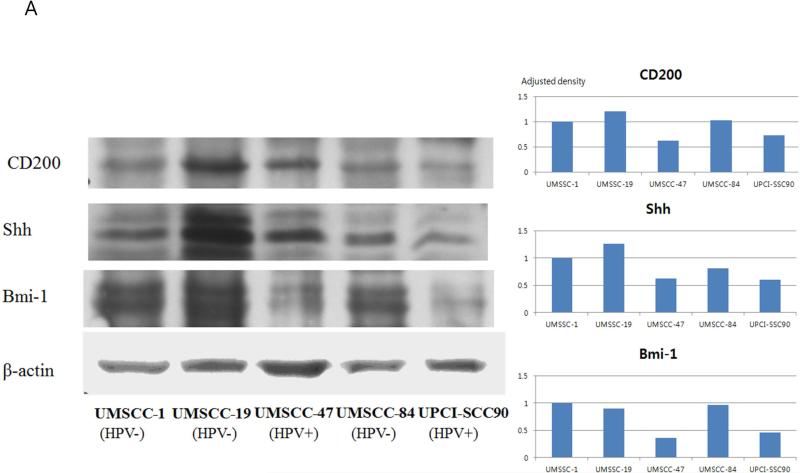
To initially assess the expression of CD200, several HNSCC cell lines were analyzed by western blot, as well as quantitative densitometry. CD200 was ubiquitously expressed across all HNSCC cell lines analyzed, running at the predicted molecular weight of 46 kDa (Figure 1A). Interestingly, however, the level of CD200 expression varied among the cell lines. UMSCC-19 (HPV(−)) demonstrated the strongest signal followed by −47 (HPV(+)); −1 (HPV(−)); −84 (HPV(−)), while UPCI-SCC90 (HPV(+)), showed the weakest expression; these data suggest that CD200 expression is not altered by HPV status.
FIGURE 1.
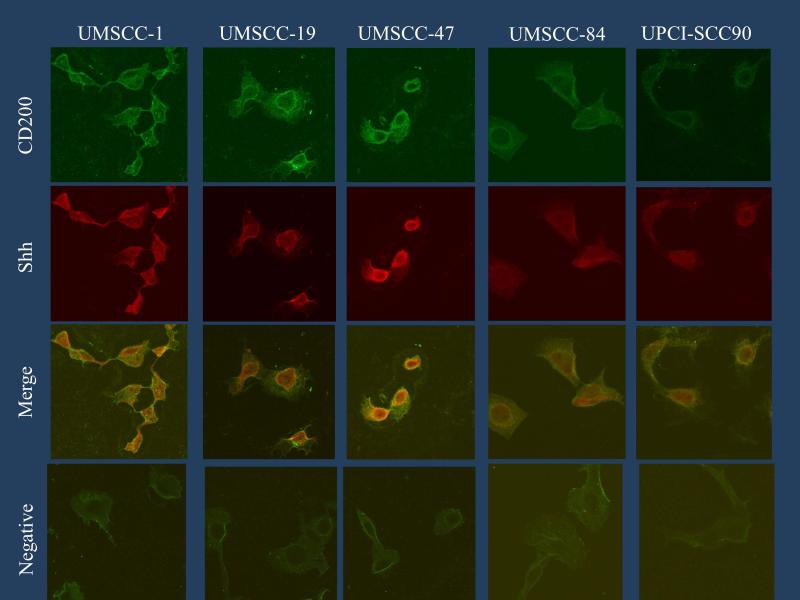
CD200 expression in HNSCC cell lines and association with the expressions of Shh and Bmi-1. (A) Western blot analyses of CD200, Shh, and Bmi-1 and actin controls are shown for multiple HNSCC cell lines with various HPV status. Adjusted densities of each blots were graphed, being those of UMSCC-1 as a reference. (B) Immunofluorescence of the same cancer cells.
While CD200 is co-expressed with CSC surface markers in various cancers(7, 19), we wondered if the differential expression of CD200 evident across the HNSCC cell lines analyzed correlated with expressions of the proliferative markers typically associated with CSC subsets. These markers include sonic hedgehog (Shh) and B-cell-specific Moloney murine leukemia virus integration site 1 (Bmi-1).(20) (21) Western blot analysis for Bmi-1 and Shh demonstrate that these markers consistently correlated with the differential expression pattern of CD200 in five different HNSCC cell lines in western blot, and this pattern was also consistently confirmed on the quantitative densitometry analysis (Figure 1A).
HNSCC cell lines were processed for immunofluorescence (IF) for CD200 and Shh as an additional validation of their co-expression patterns. The IF data strongly supports the western blot analysis suggesting that the differential expression pattern of CD200 in HNSCC cell lines consistently correlated with the expression of Shh (Figure 1B). As expected, CD200 localized to the cell membrane and cytoplasm while Shh localization was predominantly nuclear. The expression of CD200 was the strongest in UMSCC-19 and UMSCC-47, followed by UMSCC-1, and UMSCC-84 and UPCI-SCC90 showed weaker signal. The signal intensity of Shh was concordant with the signal intensity of CD200, similar to the correlation evident by western blot (Figure 1A, 1B). Flow cytometric analysis was also performed on these HNSCC cell lines. Cells were fixed and incubated with anti-mouse CD200 and subsequent FITC-conjugated anti-Rat IgG2a. CD200 was diversely expressed, also on FACS analysis, similar to western blot. For Bmi-1 and Shh, cells were fixed, permeabilized, and permeabilized and stained with either PE-labeled anti–human Bmi-1 or PE-labeled anti-Human Shh. The association between CD200, Bmi-1, and Shh, on FACS, seen both on cell count or mean fluorescence, was not as definite as that on western blot or IF (Supplementary Figure 1).
Over-expression of CD200 induces over-expression of Shh and Bmi-1
To better define whether CD200 regulates expression of Bmi-1 and Shh, we evaluated their expression following transfection of HEK293T cells with a CD200 expression plasmid. HEK293T cells were transfected with control plasmid (GFP) or the human CD200 cDNA (PUNO1-hCD200). Over-expression of CD200 induced an increase in Shh and Bmi-1 expression (Figure 2).
FIGURE 2.
Ectopic expression of CD200 correlates with increased expression of Shh and Bmi-1. After transfection of 293T cells expression of CD200, Shh and Bmi-1 were compared using western blot.
Over-expression of CD200 in a murine model of HPV+ HNSCC induces significant phenotypic changes in vitro
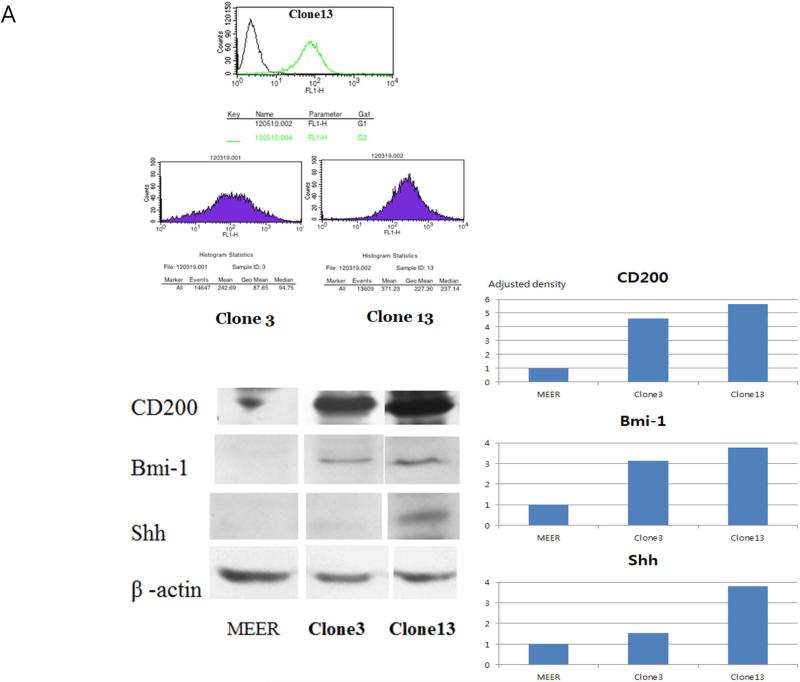
To investigate whether CD200 expression and its associated induction of Shh and Bmi-1 expression result in phenotypic changes associated with oncogenesis, we stably expressed CD200 in a previously described murine model of HPV+ HNSCC. Two CD200 clones were selected for analysis, one with weaker expression (clone 3), the other with significantly stronger expression of CD200 (clone 13). This pattern was also evident on western blot and its quantitative densitometry analysis. This expression pattern was also validated by flow cytometry (Figure 3A).
FIGURE 3.
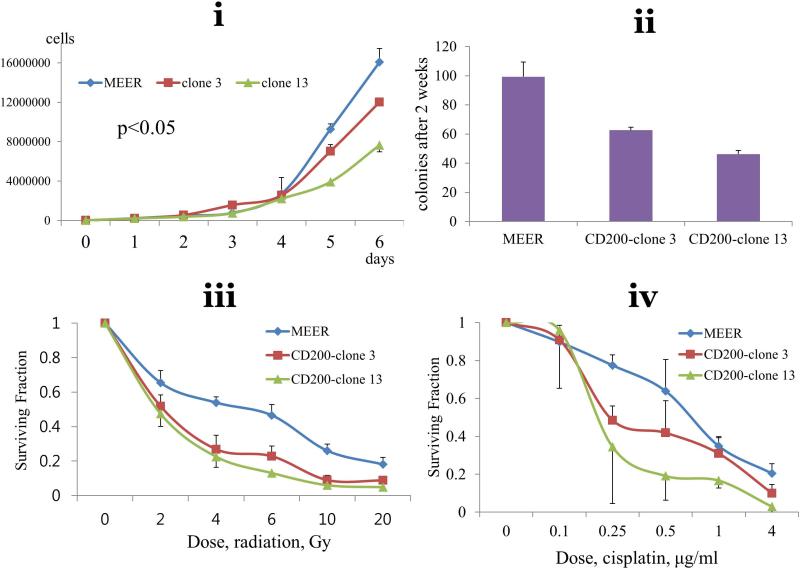
Transfection of CD200 to mouse HPV(+) cell lines. (A) Surface and total CD200 was quantified in two stable clones (3,13) using flow cytometry and western blot. Western blots of these clones were examined to correlate CD200 expression to expression of Shh and Bmi-1. Adjusted densities of each blots were graphed, being those of UMSCC-1 as a reference. (B) i. Cell proliferation, ii. colony formation, and clonogenic assay at varying doses of iii. radiation and iv. cisplatin compare growth characteristics of these cells.
To define the role of CD200 in cellular proliferation, clone 13 (high CD200 expression) and clone 3 (low CD200 expression) were compared to control parental (MEER) cells using a standard proliferation assay. Figure 3B illustrates in vitro effect of CD200 was not significant alone enough to drive phenotypic change. Clone 13 (high CD200) demonstrated the slowest proliferation, followed by clone 3 (low CD200), and MEER cells (parental) (p<0.05) (Figure 3B) in in vitro setting without treatment. Colony forming assay showed a similar non-significant trend. We next asked whether CD200 expression altered sensitivity to cisplatin or radiation. Colony forming assays following exposure to elevating doses of cisplatin or radiation demonstrated that CD200 didn't induce resistance to chemotherapy or radiation in vitro (Figure 3B).
CD200 transfection induces in vivo phenotypic changes
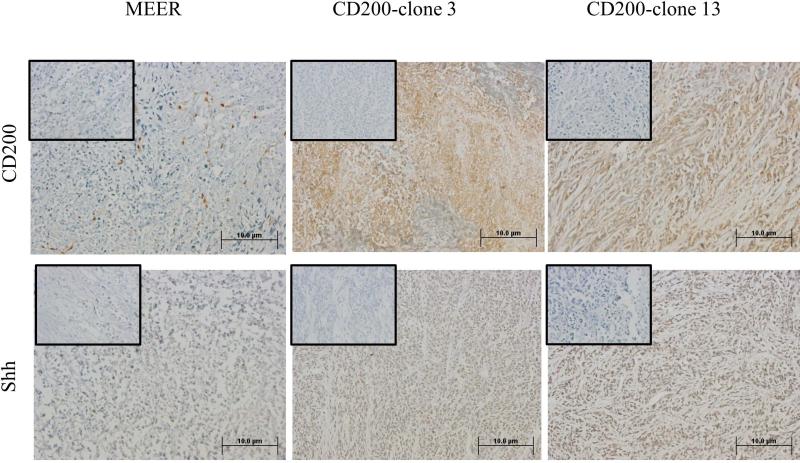
To investigate the consequences of CD200 on growth in vivo, we injected MEER (control), clone 3 (low CD200 expressor), and clone 13 cells (high CD200 expressor) into C57BL/6 mice (N=8 per group). All mice developed tumors. The tumor growth between 3 groups; MEER, clone 3, and clone 13, were not significantly different (p>0.05) (Supplementary Figure 2A). Similarly, survival between these three groups was not significantly different (p>0.05). Tissue samples was analyzed by immunohistochemistry and demonstrated that CD200 expression was similar in vivo and in vitro. Immunohistochemical staining of CD200 showed similar expression pattern to initial cell lines. Of note, CD200 staining closely correlated with that of Shh in this in vivo setting, as shown in the immunohistochemical staining (20X) of anti-Shh in the same tissue (Figure 4A). High power view (40X) showed CD200 membrane/cytoplasmic staining, whereas Shh staining is nuclear (Figure 4B).
FIGURE 4.
Immunohistochemical of tumor sections, after tumor growth study with no treatment of indicated cells in wild type C57BL6 mice for CD200 and Shh at two magnifications. [(A) 20X; (B) 40X)]. Negative control with secondary only is shown in the small inset box.
To test the function of the immune response in CD200 mediated tumor growth, the above in vivo experiments were repeated in Rag1−/−C57BL/6 mice which are unable to mount an adaptive immune response. Similar to wild-type C57Bl/6 mice, neither tumor growth nor survival were significantly different between MEER, clone 3, and clone 13 (p>0.05) (Supplementary Figure 2B). These data show that the host immune response does not cause the change of the tumor growth and survival pattern of these cells with different CD200 expression.
Expression of CD200 impairs the response for chemoradiation
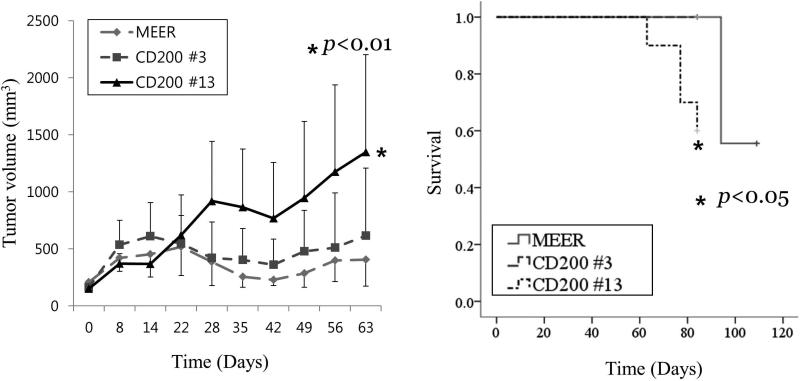
To evaluate if expression of CD200 modulates the response to conventional chemoradiation, we treated C57BL/6 mice 2 weeks after tumor injection with 8 Gy of weekly external beam radiation and concomitant cisplatin 10 mg/m2. Different from the results seen in the in vivo proliferation assay without treatment, clone 13, strongly expressing CD200, grew significantly faster than MEER (control) or clone 3 (weak expresser) after chemoradiation (p<0.01) (Figure 5). Survival of clone 13 was significantly worse than MEER and clone 3 (p<0.01). Taken together, these data suggest that CD200 expression modulates the response to chemoradiation in vivo.
FIGURE 5.
Tumor growth and survival in C57BL6 mice of CD200 clones, and MEER cell after treatment with chemoradiation (3 weekly treatments with radiation (8 Gy) and cisplatin (10mg/m2)).
DISCUSSION
Although the expression of CD200 in a variety of human cancer cells has been previously appreciated, its expression in HNSCCs and correlation with HPV status remained undefined.(15, 19) To our knowledge, this is the first study focused on the extent of CD200 expression in HPV(+) and (−) HNSCC. Our data show that CD200 protein is expressed to varying degrees by a variety of HPV(+) and (−) HNSCC cell lines. The data suggest that HPV status does not directly affect CD200 expression.
Surprisingly, while the literature suggests that CD200 expression correlates with some CSC markers, studies focused on the role of CD200 and cellular proliferation are lacking. We show that the extent of CD200 expression correlates with that of Bmi-1 and Shh, two markers of cellular proliferation. In addition, Bmi-1 and Shh are regarded as key molecules for CSCs. Thus, our data suggest that CD200 may impart a CSC phenotype via its modulation of Bmi-1 and Shh. This association was confirmed on IF, western blot, as well as quantitative densitometry analysis. FACS analysis showed similarly diverse expression of CD200 in HNSCC cell lines. On flow cytometric analysis, Bmi-1 and Shh were also expressed, although the expression seeemed not as significant. As Bmi-1 and Shh is not membrance proteins, permeabilization was needed for proper flow cytometirc analysis, which might be the reason of inconsistent result. Absolute cell count on FACS is not as consistent parameter as blot density on western blot shown here. Further studies on the association between these molecules, mechanisms, and its oncologic implications would be warranted.
Although our initial hypothesis was CD200 would enhance cell growth, and resistance against cisplatin and radiation even in in vitro setting, our data illustrated in vitro effect of CD200 was not significantly enough to drive phenotypic change. Moreover, initial in vivo study without treatment demonstrated no difference in tumor growth or survival of CD200 clones when injected into wild type or immune compromised mice (Supplementary Figure 2), similar to in vitro growth study. Both in immune competent and deficient mice, CD200 alone was not enough to strongly drive phenotypic and survival changes. Considering the pattern of tumor growth and survival without treatment are similar between immune competent and deficient mice, we interpret that CD200 may modulate tumor growth by direct activation of cellular proliferation (possibly via activation of downstream signaling), not through modulation of the adaptive immune response, which has been proposed as the predominant function of CD200, at least in HPV(+) HNSCC model used in this study.
However, importantly, we show that CD200 modulates the response to chemoradiation. High CD200 expression (clone 13) significantly impaired survival compared to the control, weak CD200 clone (clone 3), transfected with the same plasmid (Figure 5). Following chemoradiation, mice bearing tumors with high CD200 expression grew significantly faster than the control, MEER. These data suggest that CD200 mediates resistance to chemoradiation also in this murine model of HPV(+) HNSCC. These data also suggest that attenuating CD200 expression together with conventional treatment might be a potential strategy to improve treatment outcomes. The mechanism of this resistance is not clear. Whether immune evasion mechanism of CD200 or direct cell proliferation modulation might affect the chemoradioresistance, should be further investigated.
To conclude, we demonstrated CD200 protein was diversely expressed in various wild type HNSCC, with its intrinsic role for inducing Bmi-1 and Shh, known marker related to cellular proliferation. Furthermore, CD200 expression significantly induced resistance to chemoradiation. Attenuating the action of CD200 might be a potent strategy to enhance the treatment outcome of HNSCC.
Supplementary Material
Acknowledgements
This work was supported by NIH/NIDCR Grant (7R01DEO18386-03, to Dr. Lee) and by National Cancer Center Grant, Korea (NCC-1210220, 1310381, to Dr. Jung).
Footnotes
This manuscript was presented in 8th International Conference on Head and Neck Cancer, Toronto, ON, Canada, July 23, 2012.
REFERENCES
- 1.Shibuya K, Mathers CD, Boschi-Pinto C, Lopez AD, Murray CJ. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000. BMC Cancer. 2002;2:37. doi: 10.1186/1471-2407-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Termine N, Panzarella V, Falaschini S, et al. HPV in oral squamous cell carcinoma vs head and neck squamous cell carcinoma biopsies: a meta-analysis (1988-2007). Ann Oncol. 2008;19(10):1681–90. doi: 10.1093/annonc/mdn372. [DOI] [PubMed] [Google Scholar]
- 3.Nasman A, Attner P, Hammarstedt L, et al. Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: an epidemic of viral-induced carcinoma? Int J Cancer. 2009;125(2):362–6. doi: 10.1002/ijc.24339. [DOI] [PubMed] [Google Scholar]
- 4.Licitra L, Perrone F, Bossi P, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24(36):5630–6. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 5.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 6.Todaro M, Perez Alea M, Scopelliti A, Medema JP, Stassi G. IL-4-mediated drug resistance in colon cancer stem cells. Cell Cycle. 2008;7(3):309–13. doi: 10.4161/cc.7.3.5389. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki BT, Farrar WL. Cancer stem cells, CD200 and immunoevasion. Trends Immunol. 2008;29(10):464–8. doi: 10.1016/j.it.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Barclay AN, Clark MJ, McCaughan GW. Neuronal/lymphoid membrane glycoprotein MRC OX-2 is a member of the immunoglobulin superfamily with a light-chain-like structure. Biochem Soc Symp. 1986;51:149–57. [PubMed] [Google Scholar]
- 9.Wright GJ, Jones M, Puklavec MJ, Brown MH, Barclay AN. The unusual distribution of the neuronal/lymphoid cell surface CD200 (OX2) glycoprotein is conserved in humans. Immunology. 2001;102(2):173–9. doi: 10.1046/j.1365-2567.2001.01163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoek RM, Ruuls SR, Murphy CA, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science. 2000;290(5497):1768–71. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 11.Jenmalm MC, Cherwinski H, Bowman EP, Phillips JH, Sedgwick JD. Regulation of myeloid cell function through the CD200 receptor. J Immunol. 2006;176(1):191–9. doi: 10.4049/jimmunol.176.1.191. [DOI] [PubMed] [Google Scholar]
- 12.Gorczynski RM. CD200 and its receptors as targets for immunoregulation. Curr Opin Investig Drugs. 2005;6(5):483–8. [PubMed] [Google Scholar]
- 13.Gorczynski L, Chen Z, Hu J, et al. Evidence that an OX-2-positive cell can inhibit the stimulation of type 1 cytokine production by bone marrow-derived B7-1 (and B7-2)-positive dendritic cells. J Immunol. 1999;162(2):774–81. [PubMed] [Google Scholar]
- 14.Gorczynski RM, Cohen Z, Fu XM, Lei J. Anti-rat OX-2 blocks increased small intestinal transplant survival after portal vein immunization. Transplant Proc. 1999;31(1-2):577–8. doi: 10.1016/s0041-1345(98)01563-2. [DOI] [PubMed] [Google Scholar]
- 15.Moreaux J, Veyrune JL, Reme T, De Vos J, Klein B. CD200: a putative therapeutic target in cancer. Biochem Biophys Res Commun. 2008;366(1):117–22. doi: 10.1016/j.bbrc.2007.11.103. [DOI] [PubMed] [Google Scholar]
- 16.McWhirter JR, Kretz-Rommel A, Saven A, et al. Antibodies selected from combinatorial libraries block a tumor antigen that plays a key role in immunomodulation. Proc Natl Acad Sci U S A. 2006;103(4):1041–6. doi: 10.1073/pnas.0510081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonks A, Hills R, White P, et al. CD200 as a prognostic factor in acute myeloid leukaemia. Leukemia. 2007;21(3):566–8. doi: 10.1038/sj.leu.2404559. [DOI] [PubMed] [Google Scholar]
- 18.Moreaux J, Hose D, Reme T, et al. CD200 is a new prognostic factor in multiple myeloma. Blood. 2006;108(13):4194–7. doi: 10.1182/blood-2006-06-029355. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki BT, Mistree T, Hurt EM, Kalathur M, Farrar WL. Co-expression of the toleragenic glycoprotein, CD200, with markers for cancer stem cells. Biochem Biophys Res Commun. 2007;364(4):778–82. doi: 10.1016/j.bbrc.2007.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ischenko I, Seeliger H, Schaffer M, Jauch KW, Bruns CJ. Cancer stem cells: how can we target them? Curr Med Chem. 2008;15(30):3171–84. doi: 10.2174/092986708786848541. [DOI] [PubMed] [Google Scholar]
- 21.Jiang L, Li J, Song L. Bmi-1, stem cells and cancer. Acta Biochim Biophys Sin (Shanghai) 2009;41(7):527–34. doi: 10.1093/abbs/gmp040. [DOI] [PubMed] [Google Scholar]
- 22.Talebian F, Liu JQ, Liu Z, et al. Melanoma cell expression of CD200 inhibits tumor formation and lung metastasis via inhibition of myeloid cell functions. PloS one. 2012;7(2):e31442. doi: 10.1371/journal.pone.0031442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoover AC, Spanos WC, Harris GF, Anderson ME, Klingelhutz AJ, Lee JH. The role of human papillomavirus 16 E6 in anchorage-independent and invasive growth of mouse tonsil epithelium. Arch Otolaryngol Head Neck Surg. 2007;133(5):495–502. doi: 10.1001/archotol.133.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson JD, Chen YN, Cornell-Kennon SA, et al. Validating cancer drug targets. Nature. 2006;441(7092):451–6. doi: 10.1038/nature04873. [DOI] [PubMed] [Google Scholar]
- 25.Spanos WC, Nowicki P, Lee DW, et al. Immune response during therapy with cisplatin or radiation for human papillomavirus-related head and neck cancer. Arch Otolaryngol Head Neck Surg. 2009;135(11):1137–46. doi: 10.1001/archoto.2009.159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.