Abstract
Individuals diagnosed with chromosome 8p inverted duplication deletion (invdupdel(8p)) manifest a wide range of clinical features and cognitive impairment. The purpose of this study is to employ array CGH technology to define more precisely the cytogenetic breakpoints and regions of copy number variation found in several individuals with invdupdel(8p), and compare these results with their neuropsychological characteristics. We examined the cognitive-behavioral features of two male and two female children, ages 3–15 years, with invdupdel(8p). We noted cognitive deficits that ranged from mild to severe, and adaptive behavior composites that ranged from significantly to substantially lower than adequate levels. CARS scores, a measure of autistic behavior, identified three children with autism or autistic-like features. Three of the four children exhibited attention deficits and hyperactivity consistent with a DSM-IV-TR diagnosis of ADHD. One child showed extreme emotional lability. Interestingly, intellectual disability was not correlated with deletion size, nor was the deletion location associated with the autistic phenotype. On the other hand, the duplication length in 8p21.1/8p22 was associated with cognitive deficit. In addition, a small locus of over-expression in 8p21.3 was common for all three participants diagnosed as autistic. A limitation of the study is its small sample size. Further analyses of the deleted and over-expressed regions are needed to ascertain the genes involved in cognitive function and, possibly, autism.
Keywords: Subtelomeric deletion, Inverted duplication deletion 8p21–23, Intellectual disability, Autism, CGH microarray
Introduction
It has been estimated that 2.5–3% of all live births will result in some form of intellectual disability (ID). It is also estimated that 30–40% of those infants born with ID will have been produced by some genetic anomaly, the majority of which, at first glance, appear to be idiopathic in nature. Recent studies suggest that approximately 4–9% of individuals with idiopathic ID have subtelomeric rearrangements or deletions (Anderlid et al. 2002; Koolen et al. 2004; Shao et al. 2008; Wu et al. 2010), all of which suggests that approximately 1:1000–1:1500 neonates will show evidence of cognitive deficits arising from subtelomeric deletions. From the results of their analysis of subtelomeric rearrangements, Rooms et al. (2007) argued that no single mechanism seemed to account for these breakpoints, but that diverse molecular processes may produce these outcomes. Other researchers have observed that causes for the high proportion of ID in subtelomeric regions may be due to gene-rich loci residing at the telomeres, concurrent with higher than expected occurrences of chromosomal rearrangements (Bergemann et al. 2005).
Several subtelomeric rearrangements once thought to be rare now appear to be somewhat more common. One such region is associated with the short arm of chromosome 8. Floridia et al. (1996) found two types of rearrangements: Aduplication from the centromere toD8S552, with a deletion from D8S349 to the telomere; second, a duplication from 8p11.2 or 8p21 toD8S552,with a deletion fromD8S349 to the telomere. Floridia et al. categorized these two types, respectively, as dicentric inverted duplication (dicinvdup(8p)) and inverted duplication (invdup(8p)). Based on cases reported at the time, these researchers estimated the prevalence of either dicentric or monocentric invdup(8p) at 1:22,000–30,000. However, Yu et al. (2010) examined 966 individuals with developmental disorders and found 10 with copy number variants (CNVs) on the short arm of 8p. For either inverted duplication at 8p, the clinical aspects include dysmorphic craniofacial features, e.g., prominent forehead, hypotonia, congenital heart disease, agenesis of the corpus callosum, and ID (Shimokawa et al. 2004). Most often, invdup(8p) arises de novo, however, transmission to offspring from translocation carriers has been observed, often associated with a more variable cognitive phenotype that has included normal adaptive functioning (Sklower-Brooks et al. 1998) or mild developmental delay (Yenamandra et al. 1999; Moog et al. 2000). Shimokawa et al. (2004) found the inherited invdupdel(8p) to be maternal in origin in the five patients they examined.
Interestingly, Stec et al. (2001) noted that the Wolf-Hirschhorn syndrome candidate gene, WHSC1, is nearly homologous to the WHSC1L1 gene that localizes to chromosome 8p11.2. Using CGH microarray technology, Barber et al. (2008) found a 3.75 Mb duplication between 8p23.1 and the olfactory receptor/defensin repeats. Also using microarray technology, Buysse et al. (2009) examined a 13 month old infant with developmental delay and found a smaller, 3.4 Mb, duplication limited to the 8p22 region.
A similar phenotype associated with invdupdel(8p) has been observed among individuals with a partial deletion near the telomere on chromosome 8p (del8p21), and on smaller segments (8p23.1 → ter). Digilio et al. (1998) delineated the features for deletion 8p syndrome, citing growth impairment, craniofacial anomalies, congenital heart defects, genital anomalies, and varying degrees of ID, characteristics found in the invdupdel(8p) phenotype. Often, individuals diagnosed with del8p21 present with a less severe phenotype and mild ID (Fryns et al. 1989; Hutchinson et al. 1992; Wu et al. 1996; Claeys et al. 1997; Hand et al. 2010). Also published is an account of an individual with normal cognitive ability (Gilmore et al. 2001). On the other hand, researchers find ADHD and aggressive behavior among many of those examined (Hutchinson et al. 1992; Wu et al. 1996; Claeys et al. 1997; de Vries et al. 2001). Most often, the deletion arises de novo. However, parents with a balanced translocation (8; 20)(p23; p13) have been shown to transmit the 8p deletion to their offspring (de Vries et al. 2001).
In addition to its previously identified clinical features, inversion duplication on the short arm of chromosome 8 was found to be associated with autism (Papanikolaou et al. 2006). Marshall et al (2008) examined 427 individuals who had been diagnosed with autism; and, using CGH microarray technology, found 27 instances of de novo alterations, one of which involved the DLGAP2 gene at locus 8p23.3. Ozgen et al. (2009) cytogenetically examined 4 patients diagnosed with autism and found copy number variants in the invdupdel(8p) region 8p23.1 → p21.1. In addition to the DLGAP2 gene, these researchers identified two other candidate genes, MCPH1 and NEF3, which may be associated with autism. Glancy et al. (2009) examined a child with autism and detected an inversion duplication 8p23.1–p23.2. Recently, Chien et al. (2010) discovered a 2.4 Mb deletion at 8p23.2 → ter associated with autism in two males they examined.
Apart from the several reports cited, we are unaware of other studies published using CGH microarray technology to associate the molecular-genetic features invdupdel(8p) to its cognitive-behavioral phenotype, nor has there been a systematic investigation of the cognitive-behavioral features associated with invdupdel(8p). Therefore, the purpose of this study was to provide a comprehensive assessment of the cognitive-behavioral features of children with invdupdel(8p), and attempt to establish an association with its molecular-genetic features. Specifically, we employed array CGH technology to define more precisely the cytogenetic breakpoints and CNV regions found in four individuals with this cytogenetic condition, and compare these results with their phenotypic features as assessed by a comprehensive neurobehavioral battery.
Method
Subjects
Seven participants who had been previously diagnosed cytogenetically with invdup(8p23) were recruited from various locations in the US. They were enrolled as part of our longitudinal cognitive-behavioral study of children with subtelomeric deletions. Aspects of their cognitive-behavioral features were published previously (Fisch et al. 2010). Parents for two of the seven children declined to have blood drawn for DNA testing. The family of a third child had moved from their residence and could not be located. The remaining four children ranged in age from 3 to 15 years, and were assessed behaviorally and genetically. Two participants were male and two were female.
Materials
Participants were administered a comprehensive neurobehavioral battery which consisted of five standardized, valid and reliable instruments to assess behavior. To the extent that they had sufficient language to be tested, participants were administered the Stanford-Binet (4th Edition) (SBFE; Thorndike et al. 1986). In the meanwhile, their parents or caregivers completed the Child Behavior Checklist (CBCL; Achenbach 1991) and Conners Parent Rating Scale-Revised (CPRS-R; Conners 2003). Afterward, parents/caregivers were interviewed using the Vineland Adaptive Behavior Scale (VABS; Sparrow, Balla, and Cicchetti 1984) and Child Autism Rating Scale (CARS; Schopler et al. 1988).
Cognitive-Behavioral Measures
Where possible, cognitive abilities were assessed using the SBFE. The SBFE contains standard area scores (SAS) for four major areas of assessment: verbal reasoning (VR), abstract/visual reasoning (AVR), quantitative reasoning (QR), and short-term memory (STM). Adaptive behavior skills were assessed with the VABS, which contains four domains: communication, daily living skills (DLS), socialization, and motor skills for children less than 6 years of age. The VABS also contains two scales to assess Maladaptive Behavior.
Attention/Activity
Activity levels and attentiveness were assessed using the CPRS-R. The CPRS-R examines several scales associated with behavioral genotypes: oppositional, cognitive problems/inattention, hyperactivity, anxiety or shyness, perfectionism, social problems, and psychosomatic problems. More importantly, DSM-IV-TR symptoms/indices specifically associated with attention deficit disorder (ADD) and attention deficit/hyperactivity disorder (ADHD) can be obtained from the CPRS-R. ADD or ADHD scale scores at or above the 95th percentile are consistent with a DSM-IV-TR diagnosis of ADD or ADHD, respectively.
Emotionality/Temperament
To evaluate the child’s emotionality and temperament, parents completed the CBCL. The CBCL assesses emotional behavior along two major dimensions: internalizing, and externalizing behaviors. Internalizing behaviors contain scales that assess the following: withdrawn, somatic complaints, anxious/depressed, social problems, thought problems, attention. Externalizing behaviors contain two scales: delinquent, aggressive. For each of these scales, raw scores are converted into standardized T-scores. T-scores ≥ 70 indicate clinically significant aberrant behavior. T-scores between 65 and 69 denote unusually high, subclinical behavior.
Autism
To ascertain the child’s status with regard to autism, the CARS was employed. The CARS consists of 15 subscales associated with DSM-IV criteria defining autistic behavior. Each item is rated from 0 (behavior typical for a child that age) to 4 (extremely abnormal behavior for a child that age). Item scores are summed and summated scores ≥30 are considered in the autism range. In terms of diagnostic sensitivity and specificity, the CARS is comparable to the ADOS-G, and exhibits slightly higher measure of agreement with the “gold standard” of autism, clinical judgment, than the ADOS-G (Ventola et al. 2006).
Procedure
To obtain measures of their cognitive abilities, one of us (G.S.F.) administered the SBFE to the participating children. It should be noted that two children presented with extremely limited language skills, or were diagnosed as autistic without language skills. Therefore, they could not be administered the SBFE. However, all participants’ parents or caregivers were interviewed using the VABS and CARS; and, all parents and caregivers also completed the CPRS-R and CBCL. Duration for each behavioral assessment was about 2 h. All assessments were scored and compiled by one of us (G.S.F.).
DNA Analysis
Blood was obtained from all 4 participants and examined at the M.I.N.D. Institute at the University of California, Davis using CGH microarray analysis.
Microarray Analysis
Agilent’s Human Genome CGH Microarray Kit 244A
These microarrays consist of roughly 236,000 60-mer oligonucleotide probes that cover both exon and intron sequences. These high resolution arrays have an average spatial resolution of 8.9 kB. The entire content is printed on a standard 1″ × 3″ slide using Agilent’s Sure Print technology.
Target Preparation
Samples were prepared using the direct method according to the manufacturer’s Oligonucleotide Array-Based CGH for Genomic DNA Analysis protocol (Version 6.1, available at http://www.chem.agilent.com), to which minor changes were incorporated. Commercially available human male genomic DNA (Promega, Madison, WI) was used as a reference sample. Hybridization took place in a rotisserie oven for 72 h, set to 65°C and the rotation speed at 20 rpm. The washing and scanning of the slides took place in an ozone free area to prevent the degradation of the cyanine 5 dye. In turn, the slides were washed according to the wash procedure B. After washing, the slides were dried and then scanned on an Agilent High-Resolution C scanner.
Data Analysis
Image files generated from the scanner were analyzed using Agilent’s Feature Extraction Software 9.1. This software allows a standardize method of extracting data using a standard protocol (CGH_105_Jan09). The QC report generated was checked to ensure proper hybridization and placement of the grid by the software. Data generated by the feature extraction tool were loaded into Agilent Genomic Workbench 5.0 to allow visualization of the data. A smoothing average of 10 data points was used.
Results
Microarray Analysis
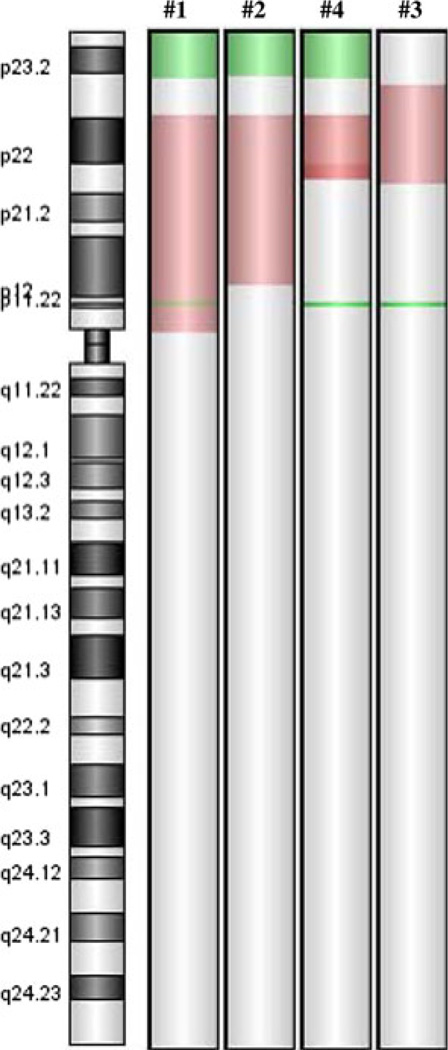
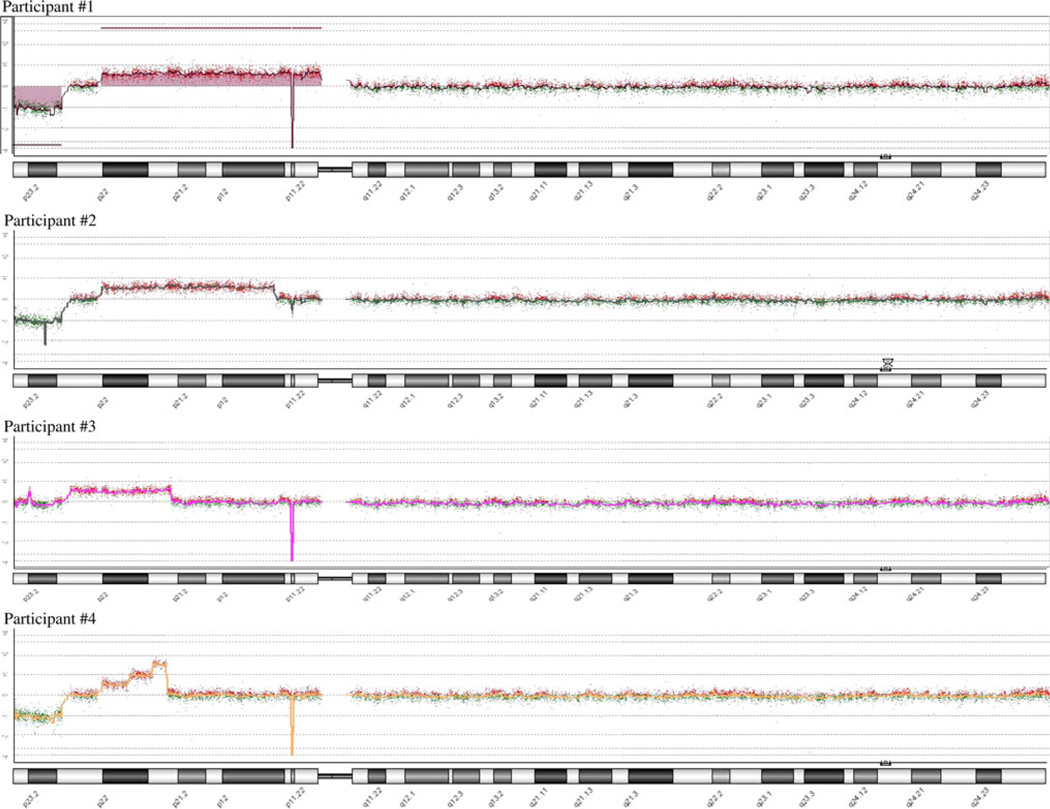
Comparison of the microarray results among the patients showed regions of consistency where chromosomal deletions and duplications occurred (Fig. 1). Three of the 4 participants have terminal deletions with the breakpoints near the boundary between 8p22 and 8p23.1. However, neither the existence of this deletion nor its size correlated with the severity of the clinical outcome. That is, participants #2 and #4 with the deletion were less severely affected than participant #3 without the deletion. Three participants: #1, #4, and #3 have an additional small deletion at 8p11.23p11.22 (39,356,395–39,505,456) that is most likely a CNP (copy number polymorphism) involving ADAM5P and ADAM3A genes and are non-pathogenic. These three participants have a homozygous deletion of the region, while participant #2 has a heterozygous deletion of the same region. The effect of this deletion is unknown, but was not associated with the level of cognitive deficit. Duplicated segments show greater variability among all participants. A common core duplicated region was found in all four participants that includes all of 8p22 and extends midway into 8p21.3. One participant showed an interesting triple stepwise increase in duplication of adjacent segments within this exact region, with no other duplicated regions involved. Two other participants have duplications that extend from this core region, albeit in opposite directions, and both are more severely affected physically and cognitively (Fig. 2). ISCN physical positions for the aberrations on 8p and approximate copy numbers for participants are presented in Table 1. We did not see other aberrations in pathogenic areas of the genome. Parents of all participants showed normal microarray patterns.
Fig. 1.
CHG microarray of chromosome 8 for participants #1, #2, #4, and #3. Green areas indicate deleted portions of the telomere. Red areas indicate over-expression (color figure online)
Fig. 2.
Individual participants CHG microarray analysis of chromosome 8
Table 1.
ISCN 2009 physical positions for the aberrations on 8p and approximate copy numbers for participants #1–#4
| Participant | ISCN |
|---|---|
| 1 | arr 8p23.3p23.1(90,616-6,913,476)X1 dn,8p23.1p11.1(12,547,803–43,647,263)X2*3 dn,8p11.23p11.22(39,356,395–39,505,456)X0 |
| 2 | arr 8p23.3p23.1(166,252–6,913,476)X1 dn,8p23.1p11.1(12,547,803–37,028,346)X2*3 dn, 8p11.23p11.22(39,356,395–39,505,456)X1 |
| 3 | arr 8p23.1p21.3(8,117,071–22,366,537)X2*3 dn,8p11 .23p11.22(39,356,395–39,505,456)X0 |
| 4 | arr 8p23.3p23.1(166,252–6,913,476)X1 dn,8p23.1p21.3(12,511,655–21,726,774)X2*5 dn, 8p11 .23p11.22(39,356,395–39,505,456)X0 |
Participant #1
#1 Is a 15-year old female who is relatively tall for her age. Her face was somewhat long and eyes unusually wide open. She often stared at people or into space for long periods. She lacked expressive speech and language; therefore, the SBFE was not administered to her. Her overall VABS score, which is strongly correlated with the SBFE, was <20, and indicated much lower than adequate levels of adaptive behavior. Although she exhibited several autistic-like behaviors, her initial CARS score, 27.5, was in the non-autistic range. However, on re-examination, her CARS score was 35.5, indicating mild-to-moderate autism. Her CPRS-R scores showed that she is in the 99th percentile for hyperactivity, and in the 98th percentile for restlessness and impulsivity. At these levels, her behavior is consistent with a DSM-IV diagnosis of ADHD. Her CBCL results show that all but one of her emotionality and temperament T-scores were within the normal range, but her T-score for attention 75, is statistically unusual and clinically significant, confirming results obtained by the CPRS-R.
Participant #2
#2 Is a 3-year old male with a large head and prominent forehead, who was also big for his age. Otherwise, no other obvious dysmorphic craniofacial features were evident. He was lethargic, did not point or gesture, and made no eye contact. He did not want his face to be touched. His speech/language was extremely limited therefore, the SBFE was not administered. The VABS, which is strongly correlated with IQ, was 51, and indicated much lower than adequate levels of adaptive behavior. CBCL scores noted subclinical thought problems (T = 67) and significant withdrawal from others (T = 74). CPRS-R scores also noted significant anxious/withdrawn behaviors (100th percentile), psychosomatic problems (94th percentile), and emotional lability (95th percentile). These behaviors on the CARS produced a score of 32 (mildly autistic).
Participant #3
#3 Is a 13-old female who was severely developmentally and intellectually disabled for her age. Although her expressive speech and language were also severely impaired, her language skills were sufficient for the examiner to administer the SBFE. Her test composite, 36, was at the floor value for the SBFE. Her adaptive behavior composite score was 20, which is also at the floor value for the VABS, and indicated much lower than adequate levels of adaptive behavior. Her CARS score was 45.5, indicating severe autism. Her T-scores on the CBCL denoted severe thought problems (T = 86), withdrawn (T = 74), and social problems (T = 75). Scores for cognitive problems, hyperactivity, anxious/shy, perfectionistic, somatic problems, ADHD symptoms, restless/impulsive, and emotionally labile scales on the CPRS-R were all above the 95th percentile.
Participant #4
#4 Is a 15-year old male and was the least impaired individual in the sample. His craniofacial features appeared normal. He displayed the clearest expressive speech and language. However, after he had been taken off Ritalin, he began to stutter. His test composite on the SBFE, 56, revealed mild intellectual deficits. His adaptive behavior composite on the VABS, 59, indicated lower than adequate levels of adaptive behavior. His CARS score, 21, demonstrated that he was not autistic. Scores on the CPRS-R showed that he was in the 95th percentile for hyperactivity and his scores were consistent with a DSM-IV diagnosis of ADHD. The CBCL noted some thought and social problems, but were not statistically significant.
Discussion and Summary
We examined the molecular-genetic and cognitive-behavioral features of 2 male and 2 female children, ages 3–15 years, initially diagnosed cytogenetically with invdupdel(8p23). Of the 4 participants we tested using CGH microarrays, 2 exhibited classic invdupdel(8p), the rear-rangement in participant #4 corresponds to the invdupdel(8p) mechanism but is more complex than a “classical” invdupdel(8p), while the fourth displayed only the duplication in the region between 8p22 and midway into 8p21.3. The break point for the deleted region from 8p22 → pter was the same for all 3 participants with the deletion, but was not associated with severity of cognitive deficit. On the other hand, the size of the duplicated region did correlate well with the degree of cognitive deficit. The complex structure found in participant #4 is similar to that observed in amplified regions found in cancers and likely results from the mechanism of breakage/fusion/bridge.1 As a result, the presence of multiple copies may complicate interpretation of the genotype-phenotype analysis.
In their review of chromosome 8p, Tabarés-Seisdedos and Rubenstein (2009) reported that the region contains 484 genes and 110 pseudogenes, and that there are 76 genes from 8p21.1 → pter known to be involved with brain function and development, brain disorders, neuropsychiatric disorders, or cancer. They also note that the 8p region is a ‘hotspot’ that contains many CNVs, some of which are associated with autism and schizophrenia. In the deleted portion, 8p22 → pter, there are 4 genes associated with brain function and development (DLGAP2, CLN8, ARHGEF10, mir-124-1), 3 associated with neuropsychiatric disorders (ARHGEF10, mir-1241-1, mir-598) and 2 associated with cerebral disorders (MCPH1, CTSB). In the duplicated region, 8p22/8p21.3, there are 15 genes involved with brain function and development (FGF20, ChGn, VMAT/SLC18A1, GFRA2, DOK2, NPM2, FGF17, PPP3CC, EGR3, ADAM28, ADAM7, NEF3, NEFL, GNRH1, DPYSL2, CLU, SCARA3, PNOC, ZNF395, FZD3, HMBOX1, KIF13B), 12 genes implicated in neuropsychiatric disorders (PCM1, NAT2, VMAT/SLC18A1, FGF17, mir-320, PPP3CC, EGR3, NEF3, DPYSL2, ADRA1A, PTK2B, CHRNA2, FZD3), and 14 genes related to cerebral disorders (FGF20, PDGFRL, NAT1, NAT2, LPL, GFRA2, PHYHIP, PPP3CC, PEBP4, NEFL, PNMA2, DPYSL2, ADRA1A, CHRNA2, CLU). Previously, researchers have cited DLGAP2, MCPH1, and NEF3 as possible candidate genes for autism (Marshall et al. 2008; Ozgen et al. 2009).
Agenesis of the corpus callosum (ACC) is a frequent finding in individuals with invdupdel(8p) and del(8p) (Hutchinson et al. 1992; Minelli et al. 1993; Yenamandra et al. 1999). In their review of social and behavioral problems tied to ACC, Badaruddin et al. (2007) note that these individuals are often developmentally delayed or effect some form of intellectual disability. In addition, the behavioral, communication, and social problems typically observed in individuals with ACC are linked with pervasive developmental disabilities and autism. Parents of the 2 female participants reported that their children had been diagnosed with ACC, both of whom were also autistic. Behavior consistent with a diagnosis of autism was identified in three of the four children examined.
Three of the four children exhibited attention deficits and hyperactivity consistent with a DSM-IV-TR diagnosis of ADHD. One child showed extreme emotional lability. ADHD and aggressive behavior is also often observed among children with del(8p) (Hutchinson et al. 1992; Wu et al. 1996; Claeys et al. 1997; de Vries et al. 2001), but not reported in studies of individuals with invdupdel(8p). Based on previous studies of invdupdel(8p) and del8p21, and the similarities noted in their respective phenotypes, one might have conjectured that the cognitive deficits observed result from the terminal deletion. To the contrary, we found that cognitive deficit was not correlated with deletion size, but to the extent of overexpression in 8p21.1/8p22. Nor was deletion location specifically associated with the autistic phenotype. However, we did note a small region between 8p21.2 and 8p22 in which NEF3 is expressed, and overexpression occurs in all three participants with autism. Ozgen et al. (2009) reported that NEF3 is a candidate gene associated with autism.
There are obvious limitations to our study. Our sample size was smaller than we originally sought to analyze using CGH arrays, especially as two of the three children not available for this study had received a diagnosis of autism or PDD-NOS. Consequently, we believe it is imperative for future studies to recruit and evaluate other individuals with invdupdel(8p), to use CGH microarrays and comprehensive neurobehavioral assessment batteries to elucidate the genotype and phenotype, and to ascertain the genes involved in cognitive function and autism.
Acknowledgment
We thank the Fondation Jérôme Lejeune, Paris, France, for their generous support without which this project could not have been brought to fruition.
Footnotes
We thank Reviewer #4 for this interpretation and analysis.
Contributor Information
Gene S. Fisch, Email: gene.fisch@nyu.edu, Department of Epidemiology and Health Promotion, Colleges of Dentistry and Nursing, New York University, NY, USA.
Ryan Davis, M.I.N.D. Institute, University of California, Davis, CA, USA.
Janey Youngblom, Department of Biological Sciences, California State University, Stanislaus, CA, USA.
Jeff Gregg, M.I.N.D. Institute, University of California, Davis, CA, USA.
References
- Anderlid BM, Schoumans J, Annerén G, Sahlén S, Kyllerman M, Vujic M, Hagberg B, Blennow E, Nordenskjöld M. Subtelomeric rearrangements detected in patients with idiopathic mental retardation. Am J Med Genet. 2002;107:275–284. doi: 10.1002/ajmg.10029. [DOI] [PubMed] [Google Scholar]
- Badaruddin DH, Andrews GL, Bölte S, Schilmoeller KJ, Schilmoeller G, Paul LK, Brown WS. Social and behavioral problems of children with agenesis of the corpus callosum. Child Psychiatry Hum Dev. 2007;38:287–302. doi: 10.1007/s10578-007-0065-6. [DOI] [PubMed] [Google Scholar]
- Barber JC, Maloney VK, Huang S, Bunyan DJ, Cresswell L, Kinning E, Benson A, Cheetham T, Wyllie J, Lynch SA, Zwolinski S, Prescott L, Crow Y, Morgan R, Hobson E. 8p2.3 1 duplication syndrome: a novel genomic condition with unexpected complexity revealed by array CGH. Eur J Hum Genet. 2008;16:18–27. doi: 10.1038/sj.ejhg.5201932. [DOI] [PubMed] [Google Scholar]
- Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Buysse K, Antonacci F, Callewaert B, Loeys B, Fränkel U, Siu V, Mortier G, Speleman F, Menten B. Unusual 8p inverted duplication deletion with telomere capture from 8q. Eur J Med Genet. 2009;52:31–36. doi: 10.1016/j.ejmg.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Chien WH, Gau SS, Wu YY, Huang YS, Fang JS, Chen YJ, Soong WT, Chiu YN, Chen CH. Identification and molecular characterization of two novel chromosomal deletions associated with autism. Clin Genet. 2010 doi: 10.1111/j.1399-0004.2010.01395.x. [DOI] [PubMed] [Google Scholar]
- Claeys I, Holvoet M, Eyskens B, Adriaensens P, Gewillig M, Fryns JP, Devriendt K. A recognisable behavioural phenotype associated with terminal deletions of the short arm of chromosome 8. Am J Med Genet. 1997;74:515–520. doi: 10.1002/(sici)1096-8628(19970919)74:5<515::aid-ajmg12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- de Vries BB, Lees M, Knight SJ, Regan R, Corney D, Flint J, Barnicoat A, Winter RM. Submicroscopic 8pter deletion, mild mental retardation, and behavioral problems caused by a familial t(8;20) (p23;p13) Am J Med Genet. 2001;99:314–319. doi: 10.1002/ajmg.1182. [DOI] [PubMed] [Google Scholar]
- Digilio MC, Marino B, Guccione P, Giannotti A, Mingarelli R, Dallapiccola B. Deletion 8p syndrome. Am J Med Genet. 1998;75:534–536. [PubMed] [Google Scholar]
- Fisch GS, Grossfeld P, Falk R, Battaglia A, Youngblom J, Simensen R. Cognitive-behavioral features of Wolf–Hirschhorn syndrome and other subtelomeric micro deletions. Am J Med Genet C Semin Med Genet. 2010;154C:417–426. doi: 10.1002/ajmg.c.30279. [DOI] [PubMed] [Google Scholar]
- Floridia G, Piantanida M, Minelli A, Dellavecchia C, Bonaglia C, Rossi E, Gimelli G, Croci G, Franchi F, Gilgenkrantz S, Grammatico P, Dalprá L, Wood S, Danesino C, Zuffardi O. The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am J Hum Genet. 1996;58:785–796. [PMC free article] [PubMed] [Google Scholar]
- Fryns JP, Kleczkowska A, Vogels A, Van den Berghe H. Normal phenotype and slight mental retardation in de novo distal 8p deletion (8pter—8p23.1) Ann Genet. 1989;32:171–173. [PubMed] [Google Scholar]
- Gilmore L, Cuskelly M, Jobling A, Smith S. Deletion of 8p: a report of a child with normal intelligence. Dev Med Child Neurol. 2001;43:843–846. doi: 10.1017/s0012162201001530. [DOI] [PubMed] [Google Scholar]
- Glancy M, Barnicoat A, Vijeratnam R, de Souza S, Gilmore J, Huang S, Maloney VK, Thomas NS, Bunyan DJ, Jackson A, Barber JC. Transmitted duplication of 8p23.1–8p23.2 associated with speech delay, autism, and learning difficulties. Eur J Hum Genet. 2009;17:37–43. doi: 10.1038/ejhg.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand M, Gray C, Glew G, Tsuchiya KD. Mild phenotype in a patient with mosaic del(8p)/invdupdel(8p) Am J Med Genet A. 2010 doi: 10.1002/ajmg.a.33669. [DOI] [PubMed] [Google Scholar]
- Hutchinson R, Wilson M, Voullaire L. Distal 8p deletion (8p23.1–8pter): a common deletion? J Med Genet. 1992;29:407–411. doi: 10.1136/jmg.29.6.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Nillesen WM, Versteeg MH, Merkx GF, Knoers NV, Kets M, Vermeer S, van Ravenswaaij CM, de Kovel CG, Brunner HG, Smeets D, de Vries BB, Sistermans EA. Screening for subtelomeric rearrangements in 210 patients with unexplained mental retardation using multiplex ligation dependent probe amplification (MLPA) J Med Genet. 2004;41:892–899. doi: 10.1136/jmg.2004.023671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A, Floridia G, Rossi E, Clementi M, Tenconi R, Camurri L, Bernardi F, Hoeller H, Previde Re C, Maraschio P, Wood S, Zuffardi O, Danesino C. D8S7 is consistently deleted in inverted duplications of the short arm of chromosome 8 (invdup 8p) Hum Genet. 1993;92:391–396. doi: 10.1007/BF01247342. [DOI] [PubMed] [Google Scholar]
- Moog U, Engelen JJ, Albrechts JC, Baars LG, de Die-Smulders CE. Familial dup(8) (p12p21.1): mild phenotypic effect and review of partial 8p duplications. Am J Med Genet. 2000;94:306–310. doi: 10.1002/1096-8628(20001002)94:4<306::aid-ajmg8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Ozgen HM, van Daalen E, Bolton PF, Maloney VK, Huang S, Cresswell L, van den Boogaard MJ, Eleveld MJ, van ‘t Slot R, Hochstenbach R, Beemer FA, Barrow M, Barber JC, Poot M. Copy number changes of the microcephalin 1 gene (MCPH1) in patients with autism spectrum disorders. Clin Genet. 2009;76:348–356. doi: 10.1111/j.1399-0004.2009.01254.x. [DOI] [PubMed] [Google Scholar]
- Papanikolaou K, Paliokosta E, Gyftodimou J, Kolaitis G, Vgenopoulou S, Sarri C, Tsiantis J. A case of partial trisomy of chromosome 8p associated with autism. J Autism Dev Disord. 2006;36:705–709. doi: 10.1007/s10803-006-0104-3. [DOI] [PubMed] [Google Scholar]
- Rooms L, Reyniers E, Kooy RF. Diverse chromosome breakage mechanisms underlie subtelomeric rearrangements, a common cause of mental retardation. Hum Mutat. 2007;28:177–182. doi: 10.1002/humu.20421. [DOI] [PubMed] [Google Scholar]
- Shao L, Shaw CA, Lu X-Y, Sahoo T, Bacino CA, Lalani SR, Stankiewicz P, Yatsenko SA, Li Y, Neill S, Pursley AN, Chinault AC, Patel A, Beaudet AL, Lupski JR, Cheung SW. Identification of Chromosome Abnormalities in Subtelomeric Regions by Microarray Analysis: a Study of 5, 380 Cases. Am J Med Genet A. 2008;146A:2242–2251. doi: 10.1002/ajmg.a.32399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa O, Kurosawa K, Ida T, Harada N, Kondoh T, Miyake N, Yoshiura K, Kishino T, Ohta T, Niikawa N, Matsumoto N. Molecular characterization of invdupdel(8p): analysis of five cases. Am J Med Genet A. 2004;128A:133–137. doi: 10.1002/ajmg.a.30063. [DOI] [PubMed] [Google Scholar]
- Sklower-Brooks SS, Genovese M, Gu H, Duncan CJ, Shanske A, Jenkins EC. Normal adaptive function with learning disability in duplication 8p including band p22. Am J Med Genet. 1998;78:114–117. doi: 10.1002/(sici)1096-8628(19980630)78:2<114::aid-ajmg3>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- Stec I, van Ommen GJ, den Dunnen JT. WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics. 2001;76:5–8. doi: 10.1006/geno.2001.6581. [DOI] [PubMed] [Google Scholar]
- Tabarés-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- Ventola PE, Kleinman J, Pandey J, Barton M, Allen S, Green J, Robins D, Fein D. Agreement among four diagnostic instruments for autism spectrum disorders in toddlers. J Autism Dev Disord. 2006;36:839–847. doi: 10.1007/s10803-006-0128-8. [DOI] [PubMed] [Google Scholar]
- Wu BL, Schneider GH, Sabatino DE, Bozovic LZ, Cao B, Korf BR. Distal 8p deletion (8)(p23.1): an easily missed chromosomal abnormality that may be associated with congenital heart defect and mental retardation. Am J Med Genet. 1996;62:77–83. doi: 10.1002/(SICI)1096-8628(19960301)62:1<77::AID-AJMG16>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang J, Xiao J, Wang H, Li J, Gao Z, Yang Y, Cai B, Wang L, Zhou Z, Tian L, Wang X, Zhong N, Qin J, Wu X, Jiang Y. Submicroscopic subtelomeric aberrations in Chinese patients with unexplained developmental delay/mental retardation. BMC Med Genet. 2010;11:11–72. doi: 10.1186/1471-2350-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenamandra A, Perrone R, McLaughlin J, Mehta L. Inverted duplication/deletion of chromosome 8p: mild clinical phenotype. Am J Med Genet. 1999;82:91–93. doi: 10.1002/(sici)1096-8628(19990101)82:1<91::aid-ajmg19>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Yu S, Fiedler S, Stegner A, Graf WD. Genomic profile of copy number variants on the short arm of human chromosome 8. Eur J Hum Genet. 2010;18:1114–1120. doi: 10.1038/ejhg.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]