SUMMARY
Primary cilia interpret vertebrate Hedgehog (Hh) signals. Why cilia are essential for signaling is unclear. One possibility is that some forms of signaling require a distinct membrane lipid composition, found at cilia. We found that the ciliary membrane contains a particular phosphoinositide, PI(4)P, whereas a different phosphoinositide, PI(4,5)P2, is restricted to the membrane of the ciliary base. This distribution is created by Inpp5e, a ciliary phosphoinositide 5-phosphatase. Without Inpp5e, ciliary PI(4,5)P2 levels are elevated and Hh signaling is disrupted. Inpp5e limits the ciliary levels of inhibitors of Hh signaling, including Gpr161 and the PI(4,5)P2-binding protein Tulp3. Increasing ciliary PI(4,5)P2 levels or conferring the ability to bind PI(4)P on Tulp3 increases the ciliary localization of Tulp3. Lowering Tulp3 in cells lacking Inpp5e reduces ciliary Gpr161 levels and restores Hh signaling. Therefore, Inpp5e regulates ciliary membrane phosphoinositide composition, and Tulp3 reads out ciliary phosphoinositides to control ciliary protein localization, enabling Hh signaling.
INTRODUCTION
Primary cilia are sensory organelles whose malfunction causes human diseases known as ciliopathies (Hildebrandt et al. 2011). Cilia are also required to interpret vertebrate Hedgehog (Hh) signals, patterning cues vital for embryonic development and adult tissue homeostasis (Goetz and Anderson 2012). How cilia transduce Hh signals is imperfectly understood, but involves the coordinated trafficking of proteins into and out of cilia. In the absence of Hh signals, such as Sonic hedgehog (Shh), the G protein-coupled receptor (GPCR) Gpr161 localizes to cilia and keeps the downstream Gli transcription factors in their repressor forms (Mukhopadhyay et al. 2013). Binding of Shh to its receptor Patched1 (Ptch1) causes the ciliary accumulation of Smoothened (Smo) and the ciliary exit of Gpr161, thereby inducing the activation of Gli transcription factors, which move from the cilium to the nucleus and induce the Hh transcriptional program. Ciliary localization of Gpr161 requires a ciliary trafficking complex that includes Tulp3 and the intraflagellar transport complex A (IFT-A) (Mukhopadhyay et al. 2010 and 2013; Norman et al. 2009; Patterson et al. 2009; Cameron et al. 2009; Qin et al. 2011; Tran et al. 2008; Liem Jr. et al. 2012).
Although it is clear that the protein composition of the ciliary membrane is distinct from that of the surrounding, contiguous plasma membrane, it has been less clear how the lipid composition of the ciliary membrane differs from that of other cellular membranes. In Paramecia, the ciliary membrane is enriched in sphingolipids and a mutation that alters ciliary lipid composition affects ciliary channel activity, suggesting that the ciliary lipid composition is critical for its function (Kaneshiro et al. 1984; Andrews and Nelson 1979; Forte et al. 1981). Similarly, in Tetrahymena and Chlamydomonas certain lipids are enriched in their cilia or flagella (Kennedy and Thompson Jr 1970; Jonah and Erwin 1971; Smith et al. 1970; Gealt et al. 1981; Bloodgood et al. 1985). In Trypanosomes, the flagellum possesses high levels of sterols and saturated fatty acids and shows a high degree of lipid organization (Tyler et al. 2009; Souto-Padron and de Souza 1983). A region of high lipid organization also exists at the base of vertebrate epithelial cilia, indicating that subdomains within the cilium may differ in their lipid composition. (Vieira et al. 2006; Montesano 1979).
Different forms of another class of lipids, the phosphoinositides, help to define different cellular membranes (Di Paolo and De Camilli 2006; Roth 2004; Sasaki et al. 2009). In C. elegans, a phosphoinositide 5-phosphatase, CIL-1, controls PI(3)P levels and the ciliary localization of PKD-2, suggesting that phosphoinositides can participate in ciliary protein trafficking (Bae et al. 2009). In mammals, three phosphoinositide 5-phosphatases, Ocrl, Inpp5b and Inpp5e, can localize to cilia (Jacoby et al. 2009; Bielas et al. 2009; Luo et al. 2012 and 2013). Mutations in human INPP5E can cause the ciliopathy Joubert syndrome, and knockout of mouse Inpp5e results in phenotypes characteristic of ciliopathies, including cystic kidneys and polydactyly (Jacoby et al. 2009; Bielas et al. 2009). Therefore, we investigated whether distinct phosphoinositides were present in the ciliary membrane and whether they participate in the unique signaling functions of vertebrate cilia.
RESULTS
PI(4)P and PI(4,5)P2 localize to distinct ciliary compartments
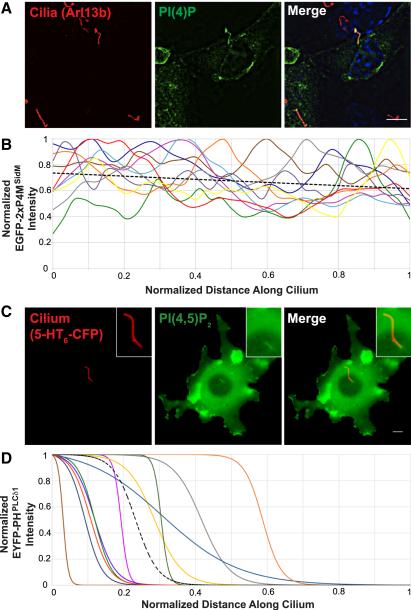
To investigate whether specific phosphoinositides localize to cilia, we expressed specific phosphoinositide-binding domains fused to fluorescent proteins in ciliated cells (Hammond and Balla 2015). Unlike sensors for PI(3)P, PI(5)P, PI(3,4)P2, PI(3,5)P2 and PIP3, a PI(4)P-specific sensor, EGFP-2×P4MSidM (Hammond et al. 2014), was enriched in cilia (Fig.1A and data not included). EGFP-2×P4MSidM was present in 74±4% of IMCD3 cilia. Linescans of ten such cilia reflect the presence of PI(4)P throughout the ciliary membrane (Fig.1B). Antibody staining confirmed the enrichment of PI(4)P within IMCD3 cilia (Fig.S1A). Immunofluorescence also revealed that sea urchin cilia have abundant PI(4)P, indicating that PI(4)P is a component of the ciliary membrane in evolutionarily distant animals (Fig.S1B).
Fig. 1. PI(4)P and PI(4,5)P2 are present in distinct ciliary compartments.
(A) IMCD3 cells transfected with EGFP-2×P4MSidM, a PI(4)P sensor, were stained with antibodies against the ciliary protein Arl13b (red) and EGFP (green). Nuclei were marked by DAPI (blue). (B) Normalized EGFP-2×P4MSidM intensity for ten IMCD3 cilia was plotted against normalized distance along the cilium. The black dotted line is the linear regression of all cilia. (C) Cilia of live NIH-3T3 cells were visualized by 5HT6-CFP fluorescence (false colored red). PHPLCd1- EYFP, a PI(4,5)P2 sensor, accumulated in the proximal ciliary region (false colored green). Scale bar, 5μm. (D) Normalized EYFP-PHPLCd1 intensity for eleven NIH-3T3 cilia was plotted against normalized distance along the cilium and the data fitted to sigmoidal curves. The black dotted line is the average of all curves.
Whereas PI(4)P was present along the length of cilia, a PI(4,5)P2 sensor (EYFP-PHPLCδ1) (Stauffer et al. 1998) localized to the proximal end of NIH-3T3 and IMCD3 cilia (Fig.1C and Fig.S1C-D). EYFP-PHPLCδ1 fluorescence ceased at a sharp boundary near the ciliary base (Fig. 1D). To confirm that EYFP-PHPLCδ1 fluorescence reflected PI(4,5)P2 distribution, we targeted Inp54p, a yeast enzyme that specifically converts PI(4,5)P2 to PI(4)P, to cilia by fusing it to a ciliary GPCR (Serotonin Receptor 6, 5HT6) (Lin et al. 2013; Johnson et al. 2008; Tsujishita et al. 2001; Suh et al. 2006). Coexpression of 5HT6-EYFP-Inp54p with a PI(4,5)P2 sensor (mCerulean3-PHPLCδ1) reduced mCerulean fluorescence at the ciliary base (Fig.S1E-F). A catalytically inactive version, 5HT6-EYFP-Inp54p(D281A), did not affect mCerulean3-PHPLCδ1 localization (Fig.S1E-F) (Suh et al. 2006). Conversely, targeting PI(4)P 5-kinase, type Iγ (PIPK) to cilia by fusing it to 5HT6 expanded mCerulean3-PHPLCδ1 localization to the length of the cilium (Fig.S1E-F) (Suh et al. 2006; Ueno et al. 2011). A catalytically inactive version, 5HT6-EYFP-PIPK(D253A), had no effect on mCerulean3-PHPLCδ1 localization (Fig.S1E-F) (Ueno et al. 2011). Together, these data indicate that the ciliary membrane contains PI(4)P along its length, and PI(4,5)P2 proximally.
Inpp5e generates ciliary PI(4)P and restricts ciliary PI(4,5)P2
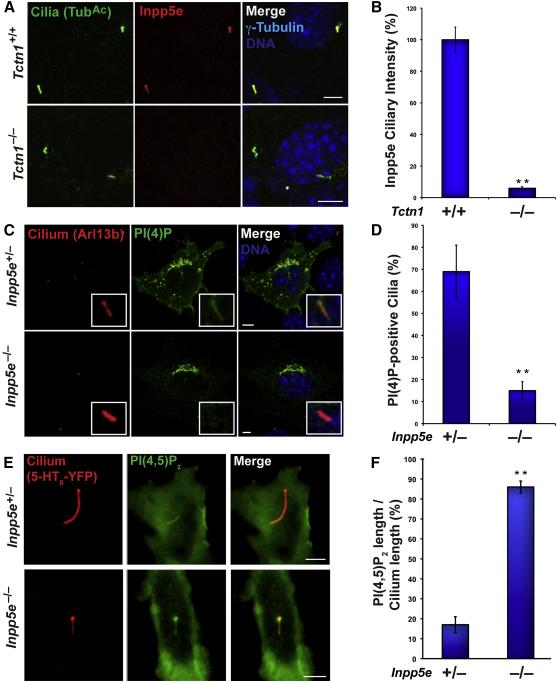
The transition zone, a region of the ciliary base, participates in protein localization to cilia (Czarnecki and Shah 2012; Garcia-Gonzalo and Reiter 2012). We found that Tctn1, a transition zone protein essential for vertebrate Hh signaling (Garcia-Gonzalo et al. 2011; Roberson et al. 2015), is required for the ciliary localization of Inpp5e (Fig.2A-B). Because Inpp5e can convert PI(4,5)P2 into PI(4)P, we hypothesized that Inpp5e affects the relative levels of these lipids in the ciliary membrane. To test this hypothesis, we derived mouse embryonic fibroblasts (MEFs) from Inpp5e+/− and Inpp5e−/− embryos (Jacoby et al. 2009). As expected, Inpp5e was present in the cilia of Inpp5e+/− but not Inpp5e−/− MEFs (Fig.S2A). Consistent with published data (Jacoby et al. 2009), Inpp5e+/− and Inpp5e−/− MEFs ciliated to equal extents (Fig.S2B-C). To assess whether Inpp5e affects PI(4)P distribution, we examined the localization of the PI(4)P probe, EGFP-2×P4MSidM, in Inpp5e+/− and Inpp5e−/− MEFs. EGFP-2×P4MSidM localization to cilia was severely reduced in Inpp5e−/− MEFs, further suggesting that EGFP-2×P4MSidM distribution accurately reflects PI(4)P distribution and indicating that Inpp5e is important for generating ciliary PI(4)P (Fig.2C-D).
Fig. 2. The ciliopathy protein Inpp5e controls ciliary phosphoinositide levels.
(A) Wild type and Tctn1−/− MEFs were stained with antibodies for the ciliary protein acetylated tubulin (TubAc, green), Inpp5e (red), and the basal body protein γ-Tubulin (cyan). Nuclei were marked by DAPI (blue). (B) Quantitation of the amount of Inpp5e fluorescence intensity at cilia of Tctn1−/− MEFs relative to the cilia of wild type MEFs. (C) Inpp5e+/− and Inpp5e−/− MEFs expressing the PI(4)P sensor EGFP-2×P4MSidM were stained for Arl13b (red) and EGFP (green). Nuclei were marked by DAPI (blue). (D) Quantitation of the proportion of Inpp5e+/− and Inpp5e−/− MEF cilia that display ciliary localization of EGFP-2×P4MSidM. (E) Cilia of live Inpp5e+/− and Inpp5e−/− MEFs were visualized by 5HT6-YFP fluorescence (false colored red). The PI(4,5)P2 sensor PHPLCd1-mCerulean3 (false colored green) localized to a restricted proximal domain of cilia of Inpp5e+/− MEFs but localized throughout cilia of Inpp5e−/− MEFs. (F) Quantitation of the extent of the ciliary PHPLCd1-mCerulean3 fluorescence (PI(4,5)P2 length) relative to the extent of 5HT6-YFP fluorescence (Cilium length) in Inpp5e+/− and Inpp5e−/− MEFs. All scale bars are 5 m. Data are shown as means±SEM. Asterisks indicate p<0.01 in unpaired t-tests.
As Inpp5e has the ability to remove the 5-phosphate from PI(4,5)P2, we hypothesized that it regulates ciliary PI(4,5)P2,levels. To test this, we expressed mCerulean3-PHPLCδ1, a PI(4,5)P2 probe, in Inpp5e+/− and Inpp5e−/− MEFs. Similarly to NIH-3T3 and IMCD3 cells, Inpp5e+/− MEFs localized mCerulean3-PHPLCδ1 at the proximal cilium (Fig.2E-F). In contrast, Inpp5e−/− MEFs localized mCerulean3-PHPLCδ1 along the entire length of the ciliary membrane (Fig.2E-F). Thus, Inpp5e limits ciliary PI(4,5)P2 and generates ciliary PI(4)P, consistent with a role in converting PI(4,5)P2 to PI(4)P within cilia.
To test whether the expanded ciliary PI(4,5)P2 in Inpp5e−/− MEFs reflects loss of ciliary PI(4,5)P2 5-phosphatase activity, we examined whether targeting the yeast PI(4,5)P2 5-phosphatase Inp54p to cilia lacking Inpp5e restored normal ciliary PI(4,5)P2 levels. 5HT6-EYFP-Inp54p, but not the catalytically inactive 5HT6-EYFP-Inp54p(D281A), restored the ciliary exclusion of mCerulean3-PHPLCδ1 in Inpp5e−/− MEFs (Fig.S2D). These results further suggest that Inpp5e dephosphorylates ciliary PI(4,5)P2 to restrict it to the proximal cilium.
As Tctn1 is required to localize Inpp5e to cilia, we hypothesized that, like Inpp5e−/− MEFs, Tctn1−/− MEFs would demonstrate altered ciliary phosphoinositide composition. As with other cell types, the PI(4,5)P2 probe mCerulean3-PHPLCδ1, was restricted to the base of Tctn1+/+ MEF cilia. In contrast to the wild type MEFs but like Inpp5e−/− MEFs, mCerulean3-PHPLCδ1 localized along the full length of Tctn1−/− MEF cilia (Fig.S2E-F).
Inpp5e promotes Hedgehog signaling and limits Gpr161 ciliary localization
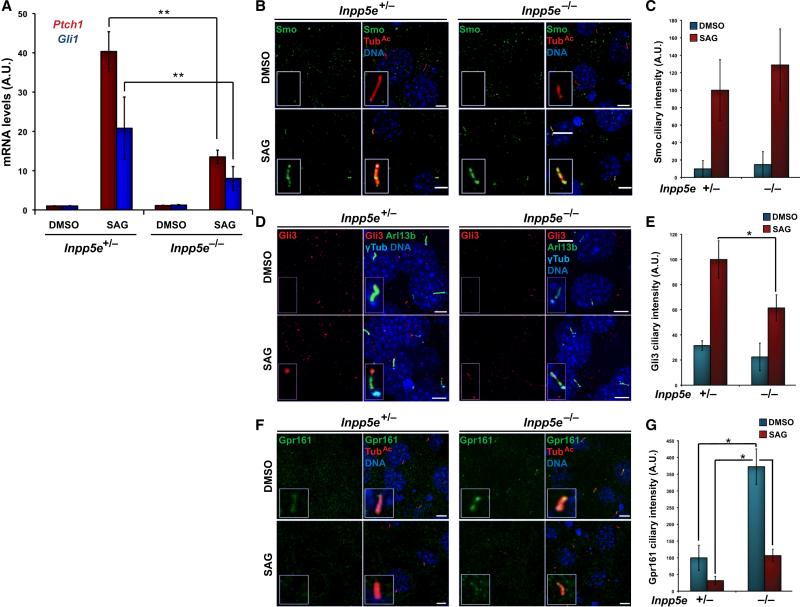
Because cilia are required for vertebrate cells to respond to Hh signals, we examined whether Inpp5e is required for Hh signal transduction. Stimulating control Inpp5e+/− MEFs with either the N-terminal signaling portion of Shh, ShhN, or a pharmacological agonist of Smo, SAG, robustly increased transcription of Gli1 and Ptch1, two Hh target genes (Fig.3A and Fig.S3A-C) (Chen et al. 2002). These responses were abrogated in mutant Inpp5e−/− MEFs, revealing a role for Inpp5e in promoting Hh signaling (Fig.3A and Fig.S3A-C).
Fig. 3. Inpp5e promotes Hh signaling and limits Gpr161 localization to cilia.
(A) Inpp5e+/− and Inpp5e−/− MEFs were treated with SAG or vehicle (DMSO) and expression of Hh target genes Ptch1 and Gli1 was measured by qRT-PCR in arbitrary units (A.U.). Error bars represent standard deviations of six independent experiments. Asterisks indicate p<0.01 in unpaired t-tests. (B) Inpp5e+/− and Inpp5e−/− MEFs were treated with SAG or vehicle (DMSO) and stained for Smo (green), and TubAc (red). (C) Quantitation of Smo ciliary intensity in DMSO or SAG-treated Inpp5e+/− and Inpp5e−/− MEFs. (D) Inpp5e+/− and Inpp5e−/− MEFs were treated with SAG or vehicle (DMSO) and stained for Gli3 (red), Arl13b (green), and γ-Tubulin (γTub, cyan). (E) Quantitation of Gli3 levels at the ciliary tip in DMSO or SAG-treated Inpp5e+/− and Inpp5e−/− MEFs. (F) Inpp5e+/− and Inpp5e−/− MEFs were treated with SAG or vehicle (DMSO) and stained for Gpr161 (green) and TubAc (red). (G) Quantitation of ciliary Gpr161 levels in DMSO or SAG-treated Inpp5e+/− and Inpp5e−/− MEFs. Error bars represent standard deviations. Asterisks indicate p<0.05. Scale bars, 5μm.
To investigate how the Inpp5e-mediated control of ciliary phosphoinositide levels participates in Hh signaling, we examined the subcellular localization of Hh pathway components in control and Inpp5e−/− MEFs. Because Inpp5e is required for cells to respond fully to the Smo agonist SAG, (Fig.3A), ciliary phosphoinositides are likely to participate in Hh signaling at the level of, or downstream of, Smo. As expected, Ptch1 localized normally to cilia in Inpp5e−/− MEF (Fig.S3D) (Rohatgi et al. 2007). As CIL-1, a C. elegans homolog of Inpp5e, helps control the ciliary localization of PKD-2, another ciliary membrane protein and the ortholog of mammalian Polycystin-2 (Bae et al. 2009), we examined whether Inpp5e also affects the ciliary localization of Polycystin-2. Unlike nematodes, mouse Inpp5e is dispensable for the ciliary localization of Polycystin-2 (Fig.S3E). Similarly, Inpp5e was not required either for the exclusion of Smo from cilia in the absence of pathway activation or for its ciliary localization upon stimulation with SAG (Fig.3B-C and Fig.S3F). In contrast, the SAG-dependent accumulation of Gli3 at the ciliary tip was reduced in Inpp5e−/− cells (Fig.3D-E and Fig.S3G). Therefore, Inpp5e promotes Hh signal transduction at a step subsequent to Smo ciliary localization and prior to Gli3 accumulation at the ciliary tip.
Recently, Gpr161 was identified as a negative regulator of Hh signaling that functions upstream of Gli transcription factor accumulation in cilia (Mukhopadhyay et al. 2013). Remarkably, we observed that ciliary levels of Gpr161 in Inpp5e−/− MEFs were greater than in Inpp5e+/− MEFs (Fig.3F-G). SAG treatment lowered ciliary Gpr161 levels approximately threefold in both cell types relative to unstimulated cells, as a result of which Gpr161 levels remained higher in Inpp5e−/− MEFs after SAG treatment (Fig.3G and Fig.S3H). Thus, Inpp5e limits ciliary Gpr161 levels but does not prevent the Hh-dependent ciliary exit of Gpr161.
Inpp5e limits the ciliary localization of Tulp3 and IFT-A
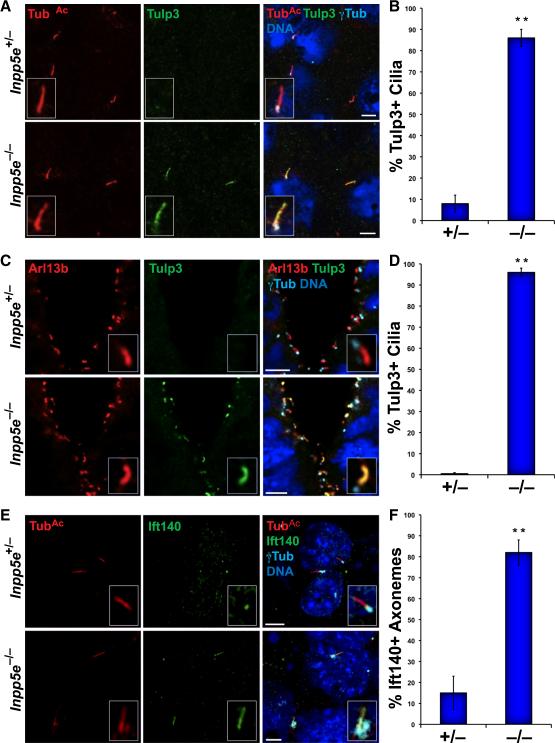
Because the ciliary localization of Gpr161 requires Tulp3, a protein that binds PI(4,5)P2 and that, like Gpr161, restrains vertebrate Hh signaling (Mukhopadhyay et al. 2010 and 2013), we examined whether Inpp5e affects Tulp3 localization. Consistent with a previous report (Norman et al. 2009), Tulp3 was only occasionally observed within control cilia (Fig.4A-B). In contrast, Tulp3 robustly accumulated along the full length of Inpp5e−/− MEF cilia (Fig.4A-B). Tulp3 was also present at abnormally high levels in the primary cilia of Inpp5e−/− E9.5 neural tubes and other tissues (Fig.4C-D and data not shown). Immunoblot analysis revealed that Tulp3 levels are equivalent in Inpp5e+/− and Inpp5e−/− MEFs (Fig.S4A), suggesting that ciliary phosphoinositides affect Tulp3 localization to cilia but not its stability.
Fig. 4. Inpp5e limits the ciliary levels of Tulp3 and IFT-A.
(A) Inpp5e+/− and Inpp5e−/− MEFs were stained for TubAc (red), Tulp3 (green) and γTub (cyan). Nuclei were marked by DAPI in all panels (blue). (B) Quantitation of the percentage of Inpp5e+/− and Inpp5e−/− MEF cilia containing Tulp3 along their length. (C) Staining of E9.5 Inpp5e+/− and Inpp5e−/− neural tubes for Arl13b (red), Tulp3 (green) and γTub (cyan). (D) Quantitation of the percentage of Tulp3-containing cilia in Inpp5e+/− and Inpp5e−/− neural tubes. (E) Inpp5e+/− and Inpp5e−/− MEFs were stained for TubAc (red), Ift140 (green) and γTub (cyan). (F) Quantitation of the percentage of Inpp5e+/− and Inpp5e−/− MEF cilia containing Ift140 beyond the basal body. Data are means±SEM. Asterisks indicate p<0.01 in unpaired t-tests.
Tulp3 interacts with the IFT-A complex, and IFT-A components such as Ift139 (also called Thm1 or Ttc21b) restrain vertebrate Hh signaling, similar to Tulp3 and Gpr161 (Qin et al. 2011; Tran et al. 2008; Liem Jr. et al. 2012). Therefore, we investigated whether Inpp5e also restricts the ciliary localization of IFT-A components. Like Tulp3, IFT-A components Ift139 and Ift140 overaccumulated in the cilia of Inpp5e−/− MEFs (Fig.4E-F and Fig.S4B). In contrast, loss of Inpp5e did not affect the ciliary levels of IFT-B component Ift88 (Fig.S4C). Thus, in addition to promoting Hh signaling, Inpp5e limits the ciliary localization of the Hh signaling negative regulators Ift139, Ift140, Tulp3 and Gpr161. As these proteins interact with each other, we hypothesized that the phosphoinositide levels controlled by Inpp5e limit their localization to cilia.
Since Tctn1−/− MEF cilia, like those of Inpp5e−/− MEFs, lack Inpp5e and accumulate PI(4,5)P2, we tested whether they also accumulate Tulp3 and Gpr161. Ciliary levels of Tulp3 in Tctn1−/− MEFs were not increased (Fig.S4D), and ciliary levels of Gpr161 were decreased in Tctn1−/− MEFs (Fig.S4E). Given that many other membrane-associated proteins, such as Polycystin-2 and Smo, fail to localize to cilia in Tctn1−/− MEFs but localize normally to the cilia of Inpp5e−/− MEFs (Garcia-Gonzalo et al. 2011 and this work), we conclude that the disruption of the ciliary gate at the transition zone caused by loss of Tctn1 leads to a more pervasive defect in localizing ciliary membrane-associated proteins than absence of ciliary Inpp5e.
Inpp5e limits ciliary Tulp3 and IFT-A levels by restricting ciliary PI(4,5)P 2
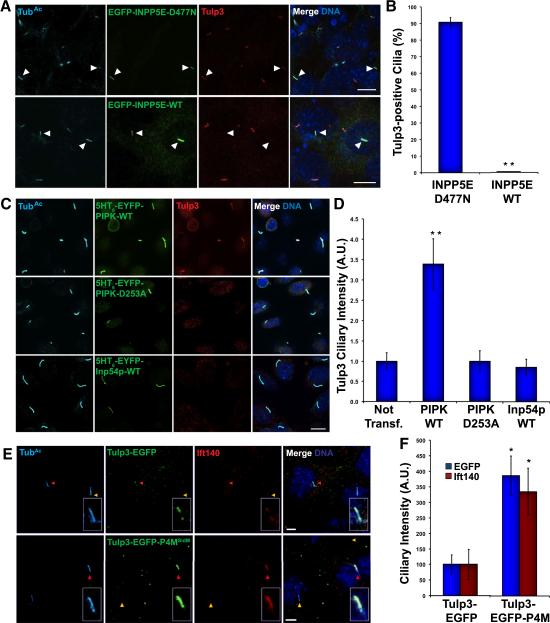
To determine whether the ciliary buildup of Tulp3 in Inpp5e−/− cells was due to the elevated ciliary PI(4,5)P2 levels, we tested whether expression of wild type or catalytically dead Inpp5e affected ciliary Tulp3 levels. Expression of EGFP-INPP5E-WT in Inpp5e−/− cilia removed Tulp3 from cilia, whereas catalytically inactive EGFP-INPP5E-D477N had no effect, consistent with a phosphatase-dependent role for Inpp5e in controlling ciliary Tulp3 levels (Fig.5A-B).
Fig. 5. PI(4,5)P2 recruits Tulp3 and IFT-A to cilia.
(A) Inpp5e−/− MEFs expressing EGFP-INPP5E-D477N or EGFP-INPP5E-WT were stained for TubAc (cyan), EGFP (green) and Tulp3 (red). Nuclei were stained with DAPI in all panels. (B) Quantitation of the percentage of EGFP-INPP5E-D477N- or EGFP-INPP5E-WT-expressing Inpp5e−/− MEF cilia positive for Tulp3. (C) IMCD3 cells expressing 5-HT6-EYFP-PIPK, 5-HT6-EYFP-PIPK(D253A) or 5-HT6-EYFP-Inp54p were stained for TubAc (cyan), EYFP (green) and Tulp3 (red). (D) Quantification of the fluorescence intensity of Tulp3 in cilia of untransfected cells, and those expressing 5-HT6-EYFP-PIPK, 5-HT6-EYFP-PIPK(D253A) or 5-HT6-EYFP-Inp54p. (E) Inpp5e+/− MEFs transfected with expression plasmids for Tulp3-EGFP or Tulp3-EGFP-P4MSidM were stained for TubAc (cyan), EGFP (green), and Ift140 (red). Cilia of transfected and untransfected cells are indicated by red and yellow arrowheads, respectively. (F) Quantification of the fluorescence intensity of the EGFP of Tulp3-EGFP or Tulp3-EGFP-P4MSidM in cilia of transfected Inpp5e+/− MEFs, and of Ift140 in these same cilia. Data are means±SEM. One asterisk indicates p<0.05 and two p<0.01 in unpaired t-tests.
To assess whether PI(4,5)P2 is sufficient to cause Tulp3 to accumulate in cilia, we targeted a PI(4)P 5-kinase (5HT6-EYFP-PIPK) to the cilia of wild type IMCD3 cells. As shown previously, 5HT6-EYFP-PIPK increases ciliary PI(4,5)P2 levels (Fig.S1E-F). In addition to increasing ciliary PI(4,5)P2 levels, 5HT6-EYFP-PIPK increased ciliary Tulp3 levels (Fig.5C-D). In contrast, the inactive mutant 5HT6-EYFP-PIPK-D253A or 5HT6-EYFP-Inp54p had no such effect (Fig.5C-D).
As Tulp3 is required for the ciliary delivery of Gpr161, we tested whether increasing ciliary PI(4,5)P2 levels also increased ciliary Gpr161 localization. Expression of 5HT6-EYFP-PIPK caused a modest but significant increase in ciliary Gpr161 levels as compared to expression of 5HT6-EYFP-PIPK-D253A (Fig.S5A-B). As increasing ciliary PI(4,5)P2 levels, either by removing Inpp5e or by increasing ciliary PI(4)P 5-kinase activity, increased ciliary levels of Tulp3 and Gpr161, we conclude that maintaining low ciliary PI(4,5)P2 levels is critical for restraining ciliary Tulp3 and Gpr161 localization.
Tulp3 binds PI(4,5)P2 but not PI(4)P (Mukhopadhyay et al. 2010). We hypothesized that this phosphoinositide binding specificity accounted for the limited localization of Tulp3 to cilia of wild type cells and the increased localization of Tulp3 to cilia with increased PI(4,5)P2 levels. To test this hypothesis, we generated a Tulp3 with altered phosphoinositide binding specificity by fusing Tulp3-EGFP to P4MSidM, a PI(4)P-binding domain of a Legionella protein (Hammond et al. 2014). Whereas, like endogenous Tulp3, Tulp3-EGFP weakly localized to cilia, Tulp3-EGFP-P4MSidM, like Tulp3 in cilia with elevated PI(4,5)P2 levels, robustly localized cilia (Fig.5E-F). These results suggest that persistent interaction with phosphoinositides of the ciliary membrane is sufficient to increase the ciliary localization of Tulp3, and that increased ciliary PI(4,5)P2 levels are sufficient to increase ciliary localization of Tulp3.
To assess whether altered ciliary Tulp3-phosphoinositide interactions are sufficient to affect the localization of Tulp3-interacting proteins, we examined whether Tulp3-EGFP-P4MSidM affected the ciliary localization of Ift140. Expression of Tulp3-EGFP-P4MSidM caused Ift140 to accumulate in cilia (Fig.5E-F), similar to its behavior in Inpp5e−/− cells, whereas expression of Tulp3-EGFP had not effect on ciliary Ift140. These results are consistent with differential Tulp3 interaction with PI(4,5)P2 in the membrane of the ciliary base and PI(4)P in the more distal ciliary membrane being critical to limit the ciliary localization of both Tulp3 and its interacting protein, Ift140.
Inhibition of Tulp3 or Gpr161 increases Hh signaling in Inpp5e −/− MEFs
We hypothesized that the Hh signaling defects caused by loss of Inpp5e were due to the PI(4,5)P2-dependent increase in ciliary Tulp3 and its associated proteins including Gpr161. We reduced Tulp3 and Gpr161 levels in Inpp5e−/− MEFs using siRNA-mediated knockdown (Fig.S6A-C). Reducing Tulp3 reduced the ciliary levels of Gpr161, consistent with a role for Tulp3 in delivering Gpr161 to the cilium (Fig.S6A,C). In contrast, reducing Gpr161 had no effect on the ciliary levels of Tulp3, indicating that there is no reciprocal requirement (Fig.S6A,C).
In addition to decreasing ciliary Gpr161 levels, depletion of Tulp3 increased SAG-activated Hh signaling in Inpp5e−/− MEFs to a level indistinguishable from that of control cells (Fig.S6D). Depletion of Gpr161 in Inpp5e−/− MEFs led to a more modest but still significant increase in SAG-activated Hh signaling (Fig.S6D,E). We conclude, therefore, that limiting ciliary PI(4,5)P2 levels is critical for restricting the ciliary localization of Tulp3 and other negative regulators of Hh signaling (Fig.6A). Defects in maintaining the normal distribution of ciliary phosphoinositides alters ciliary trafficking of Tulp3, IFT-A and Gpr161, disrupting ciliary Hh signaling.
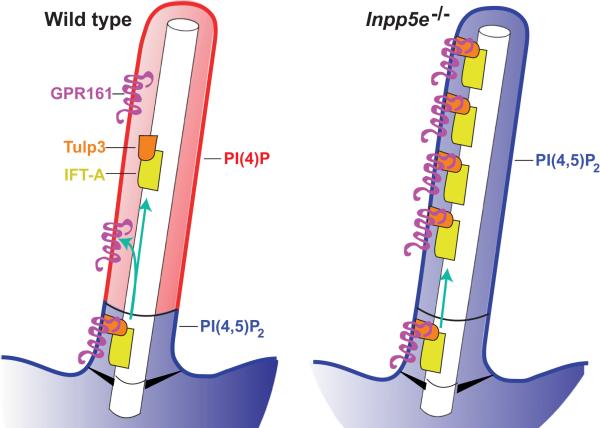
Fig. 6. Model of the role of ciliary phosphoinositides in Hh signaling.
Inpp5e restricts PI(4,5)P2 levels in the ciliary membrane. The ability of Tulp3 to interact with PI(4,5)P2 but not PI(4)P is critical for limiting its accumulation and that of its interactors IFT-A and Gpr161 within the cilium. In the absence of Inpp5e, ciliary PI(4,5)P2 levels increase, increasing the amount of negative regulators of Hh signaling, Tulp3, IFT-A and Gpr161, within the cilium and restricting the ability of the cilium to transduce Hh signals.
DISCUSSION
Phosphoinositides confer identity to organelles. For example, endosome and Golgi membranes possess PI(3)P and PI(4)P, respectively (Di Paolo and De Camilli 2006; Roth 2004), whereas the plasma membrane possesses both PI(4)P and PI(4,5)P2 (Hammond et al. 2012 and 2014). Physical separation of these membrane-bound organelles makes their distinct lipid compositions possible. In contrast to most organelles, the cilium is not membrane-bounded. Despite this difference, we found a sharp boundary at the ciliary base that separates the ciliary membrane into a distal PI(4)P-containing domain and a proximal PI(4,5)P2-containing domain.
Creating this phosphoinositide boundary depends on the Tctn1 complex of the transition zone to confine the Inpp5e phosphatase within the cilium. We previously showed that Tctn1 and other components of the transition zone MKS complex are critical for localizing the small GTPase Arl13b to the cilium (Garcia-Gonzalo et al. 2011). Arl13b is itself critical for the ciliary localization of Inpp5e (Humbert et al. 2012). Thus, it is likely that Tctn1 and the transition zone MKS complex localize Inpp5e to cilia through their effects on Arl13b.
In the absence of Inpp5e, ciliary levels of PI(4)P decrease and PI(4,5)P2 increase, suggesting that Inpp5e is critical for generating the observed distribution of ciliary phosphoinositides. It will be interesting to determine whether Inpp5e shares this function with other ciliary phosphoinositide phosphatases, such as Ocrl or Inpp5b (Luo et al. 2012 and 2013).
Perturbing the ciliary levels of ciliary PI(4)P and PI(4,5)P2, either by removing Inpp5e or by targeting the phosphoinositide 5-kinase PIPK to cilia, is sufficient to increase the ciliary localization of Tulp3. As Tulp3 interacts with PI(4,5)P2 but not PI(4)P (Mukhopadhyay et al. 2010), it is an excellent candidate for distinguishing between these two domains within the cilium. Tulp3 binds to the IFT-A complex to deliver GPCRs such as Gpr161 to cilia (Mukhopadhyay et al. 2010 and 2013). Consistent with a critical role for Tulp3-mediated sensing of phosphoinositides in this process, disruption of the ciliary phosphoinositide distribution not only increases ciliary Tulp3, but it also increases the ciliary levels of IFT-A components and Gpr161. Although Tulp3 is one key interpreter of ciliary phosphoinositides, it may not be the sole one. Other Tubby family proteins may share this role. Mutation of either Tubby or Tulp1 cause retinal degeneration, a phenotype commonly observed in ciliopathies (Mukhopadhyay and Jackson 2011). By analogy to our findings with Tulp3 and Gpr161, it is possible that photoreceptor phosphoinositides are read out by Tubby and Tulp1 to control the delivery of Rhodopsin to the outer segment, defects in which cause photoreceptor loss.
Tulp3, IFT-A components such as Ift139, and Gpr161 are all negative regulators of ciliary Hh signaling (Mukhopadhyay et al. 2013; Norman et al. 2009; Patterson et al. 2009; Cameron et al. 2009; Qin et al. 2011; Tran et al. 2008; Liem Jr. et al. 2012). In addition to limiting ciliary PI(4,5)P2 levels, Inpp5e limits the ciliary localization of these negative regulators and is required for normal Hh signal transduction. Reducing Tulp3 levels in cilia using an siRNA restores Hh signaling in MEFs lacking Inpp5e, suggesting that overaccumulation of Tulp3 in cilia is a key factor in the observed signaling defects. The effect of the Tulp3 siRNA on Hh signaling is consistent with the effect of loss-of-function mutations in Tulp3 on developmental Hh signaling, suggesting that the siRNA effect is on target, but it will be of interest to confirm this epistasis using genetic tools (Norman et al. 2009; Patterson et al. 2009; Cameron et al. 2009). Reducing Gpr161 levels also increased Hh signaling in Inpp5e−/− MEFs, suggesting that it is also sufficient to limit normal Hh signaling. The more modest effect of reducing Gpr161 levels, reflected by the fact that the fold induction of Ptch1 by SAG was unaffected by Gpr161 depletion, raises the possibility that overaccumulation of Tulp3 in cilia also causes the ciliary accumulation of additional negative regulators of Hh signaling. Our finding that reducing Tulp3 or Gpr161 levels increases Hh response even in the presence of increased ciliary PI(4,5)P2 indicates that control of these two protein levels is a critical role for ciliary phosphoinositides in Hh signaling.
In mammalian cells, Tulp3 interprets ciliary phosphoinositides to control the localization of interactors such as IFT-A components and Gpr161. Increasing the ciliary levels of PI(4,5)P2 (by loss of Inpp5e or by expressing a ciliary phosphoinositide 5-kinase) or fusing Tulp3 with a PI(4)P-binding domain, increases the ciliary localization of Tulp3 and IFT-A components. Therefore, restricting ciliary PI(4,5)P2 levels is critical to limiting the ciliary localization of Tulp3 and its interactors. Thus, the interplay between proteins and lipids helps generate the specialized subcellular environment present in the primary cilium and critical for its ability to transduce signals.
In most Drosophila cells, the Hh pathway functions independently of cilia, yet PI(4)P is required for Drosophila Hh signaling (Yavari et al. 2010). Although the cellular contexts may be different, a membrane domain high in PI(4)P and low in PI(4,5)P2 may represent a fundamental requirement for the cilium-dependent and cilium-independent transduction of Hh signals. We did not successfully observe rescue of Hh signaling by overexpressing Inpp5e in Inpp5e−/− MEFs (data not shown). This is likely due to inefficient transfection of Inpp5e−/− MEFs, but also raises the possibility that overexpressed Inpp5e inhibits other aspects of ciliary function, perhaps by altering phosphoinositide pools recognized by FAPP2, a Golgi protein involved in ciliogenesis, or the BBSome (Vieira et al. 2006; Jin et al. 2010).
Just as PI(4)P may have functions in Hh signaling that are independent of cilia, it is possible that PI(4,5)P2 may have functions in cilia that predate the origin of Hh signaling.
Although the ciliary phosphoinositide composition of unicellular organisms has not yet been determined, the wide phylogenetic distribution of Tubby family proteins hints at a broad and evolutionarily ancient role for PI(4,5)P2 in ciliary biology. For example, Tubby family members are present in many ciliated organisms, such as Chlamydomonas, Tetrahymena, and Paramecia. Whether Tubby proteins interpret ciliary phosphoinositides to regulate ciliary protein delivery in these organisms will help determine the evolutionary origins of the role of phosphoinositides in ciliary function.
We have shown that the ciliary membrane has a distinct phosphoinositide composition that is critical for regulating ciliary protein trafficking and, thus, Hh signal transduction. Importantly, this role does not exclude additional roles for ciliary phosphoinositides, including as cofactors for ciliary protein function, determinants of ciliary membrane viscosity, or contributors to membrane surface charge or ion-binding capacity. The specialized compositions of ciliary proteins and lipids depend on each other and are essential for generating the specialized environment required to make the cilium a unique signaling organelle.
Methods
Animal models and cell lines
Inpp5e−/− mice have been described previously, as have Tctn1−/− mice and MEFs (Jacoby et al. 2009; Garcia-Gonzalo et al. 2011; Reiter and Skarnes 2006). We derived MEFs from littermate E19.5 Inpp5e+/− and Inpp5e−/− embryonic tails. Briefly, tails were dissected, rinsed in Dulbecco's PBS containing penicillin and streptomycin (PenStrep), digested for 10 minutes in 0.05% Trypsin-EDTA, disagreggated by pipetting and plated on DMEM medium supplemented with 20%FBS and PenStrep. These primary MEFs were maintained in DMEM, 15%FBS and PenStrep and immortalized by infection with a lentivirus expressing SV40 large T antigen. All MEF experiments in this study were performed using immortalized MEFs grown in DMEM and 10%FBS.
For cilia isolation from sea urchin embryos, eggs and sperm from adult Strongylocentrotus purpuratus (Kerckhoff Marine Lab, Corona del Mar, CA) were collected, combined and cultured for 2 days in natural seawater at ~10,000 embryos/mL at 12°C with constant stirring. Midgastrula embryos were collected, concentrated by centrifugation and washed 3 times with natural seawater. Pelleted embryos were resuspended in natural seawater+0.5M NaCl to amputate cilia. Deciliated embryos were removed from the sample by centrifugation at 400×g for 5 minutes. Cilia were then pelleted from the supernatant by centrifugation at 10,000×g, 20 minutes, 4°C.
Immunofluorescence
Antibodies used in this study were: goat anti-γ-Tubulin (Santa Cruz, sc-7396), chicken anti-EGFP (Aves Labs, GFP-1020), mouse anti-acetylated tubulin (Sigma, 6-11B-1), anti-Arl13b (NeuroMab, 75-287), anti-Gli3 (Gli3N-6F5) (Wen et al. 2010), anti-PI4P (Echelon Bioscience, Z-P004), rabbit anti-Inpp5e (Jacoby et al. 2009), anti-Arl13b (Caspary et al. 2007), anti-Smoothened (Abcam, ab38686), anti-Patched1 (Rohatgi et al. 2007), anti-Tulp3 (Norman et al. 2010), anti-Ift140 (Proteintech, 17460-1-AP), and anti-Gpr161 (Proteintech, 13398-1-AP and Mukhopadhyay et al. 2013). For immunostaining of MEFs, cells were grown on coverslips and fixed with 4%PFA in PBS for 5-10 minutes at RT followed by 3 minutes at −20°C in cold methanol. PFA fixation was omitted when staining for Ift140. PI4P antibody was used according to the manufacturer's instructions. For Tulp3 staining, cells were fixed for 10 minutes at 4°C in acetone. After fixation, cells were blocked in PBS containing 0.1% Triton X-100 and 2% donkey serum. For chicken EGFP staining, cells were also blocked in BlokHen reagent (Aves Labs) diluted in PBS. After blocking, cells were incubated in block containing primary antibodies for 1 hour at 37°C, 3 hours at RT or overnight at 4°C. Coverslips were then rinsed twice in PBS, incubated with secondary antibodies and Hoechst 33342 or DAPI (30min at 37°C or 1h at RT), rinsed twice in milliQ water, and mounted in gelvatol. For mouse embryo analyses, we fixed dissected embryos for 1-2 hours in 4%PFA in PBS, washed three times in PBS and sunk overnight in 30% sucrose. Embryos were then equilibrated and frozen in OCT compound (Sakura) using a dry ice-ethanol bath. For staining, cryosections were thawed and washed thrice in PBST (PBS with 0.1%Triton), encircled with an ImmEdge hydrophobic pen, blocked and stained with antibodies as above.
Live cell imaging
NIH-3T3, mIMCD3 and MEFs were cultured in DMEM (Gibco) supplemented with 10% FBS. For all transient transfections, cells were transfected with the respective DNA constructs by plating them directly in a transfection solution containing DNA plasmid and Xtremegene 9 (Roche). Cells were plated on poly(D-lysine)-coated borosilicate glass Lab-Tek 8-well chambers (Thermo Scientific). Ciliogenesis was induced by serum starvation for 24 h. Live cell imaging was mostly performed using an IX-71 (Olympus) microscope with a 40× oil objective (Olympus) (with additional 1.6× optical zoom) and a CoolSNAP HQ CCD camera (Photometrics). Micrographs were taken using Metamorph 7.5 imaging software (Molecular Devices).
DNA constructs
To construct the 5HT6-EYFP expression plasmid, DNA encoding 5HT6 flanked by 5’ and 3’ AgeI cleavage sites was amplified by PCR from 5HT6-EGFP (Berbari et al. 2007) and subcloned into pEYFP-C1 (Clontech). To construct the 5HT6-EYFP-PIPK, 5HT6-EYFP-PIPK(D253A), 5HT6-EYFP-Inp54p, and 5HT6-EYFP-Inp54p(D281A) expression plasmids, DNAs encoding the wild type or mutant forms of PIPK or Inp54p were digested from CFPFKBP-PIPK or CFP-FKBP-Inp54p (Suh et al. 2006) with 5’ EcoRI and 3’ BamHI cleavage sites and subcloned into the 5HT6-EYFP expression plasmid.
Hh signal transduction assay
MEFs were plated on 12-well plates in full medium and allowed to reach confluency. The medium was replaced by OptiMEM containing either vehicle, 200nM SAG (Cayman Chemicals), or 1μg/ml mouse recombinant ShhN-C25II (R&D Systems). Cells were then incubated for 24 hours and RNA extracted (Qiagen RNeasy kit). For each sample, 1μg RNA was used to make cDNA (Invitrogen Superscript III reverse transcriptase kit). The cDNAs were then analyzed by QPCR using Express SYBR GreenER Supermix with premixed ROX (Invitrogen) and primers for Gli1, Ptch1 and β-actin (Santos et al. 2014). For siRNA transfections, 400,000 cells/well were reverse transfected in 12-well plates using Lipofectamine-RNAiMAX (Invitrogen). Transfection media were replaced 24 hours later with full medium and cells incubated for an additional 24 hours, when Hh agonist-containing media were added, as described above.
Quantitations and statistical tests
Metamorph image analysis software was used to quantitate the relative extension of the PI(4,5)P2 ciliary compartment. Only cilia that lied flat on the cell surface were analyzed to ensure accuracy. Cilia length was measured by tracing a line along the ciliary marker signal. PH-PLCδ signal length was measured analogously and expressed as percentage relative to cilia length. For phosphoinositide sensor signal linescans, a line was traced along the length of each cilium (Sensorcilium) and a second identical line (Sensorbackground) was traced in close proximity to the first line within the cell area. Normalized values of (Sensorcilium – Sensorbackground) were plotted against relative distance along the cilium and fitted to sigmoid curves if appropriate.
Quantitation of ciliary signal intensities was carried out using Image J software. Each experiment was performed at least three times. Z-stacks of seven fields of cells were acquired from each condition using a 63X objective and 1.5× digital zoom in a Leica TCS SPE confocal microscope. For each field, a maximal z-projection was created and cilia identified using Arl13b or TubAc and γ-Tub as markers. Each cilium (or its tip for Gli3) was outlined with the polygon tool and the mean intensity of the desired channel inside the area measured in an 8-bit scale (0-255). These values were then background-subtracted and averaged for each field, which typically contained ten or more cilia. Means and SEMs were then calculated for each condition and significance assessed using unpaired, one-tailed homoscedastic Student's t-tests. In cases where cilia clearly fell into positive and negative categories, cilia were counted visually and data treated as above. For Tulp3 and Ift140 in MEFs, cilia were only counted as positive if the marker was present inside the cilium, not just at the transition zone.
For Hh assays, QPCR data were analyzed using the ΔΔCt method. Means and SEMs of at least three independent experiments, each run in triplicate, were calculated, plotted and significance assessed by Student's t-tests as above.
Supplementary Material
Highlights.
The ciliary membrane contains different phosphoinositides than that of its base
The ciliopathy-associated protein Inpp5e generates the phosphoinositide distribution
Ciliary phosphoinositides are required for normal Hedgehog (Hh) signaling
Tulp3 senses phosphoinositides to limit ciliary Gpr161, an inhibitor of Hh signaling
Acknowledgments
This work was supported by grants from the NIH (AR054396 and GM095941 to J.F.R. and DK102910 to T.I.) and from the Burroughs Wellcome Fund, the Packard Foundation, and the Sandler Family Supporting Foundation to J.F.R. S.C.P. is supported by the Agency for Science, Technology and Research in Singapore.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
F.R.G, S.P, T.I. and J.F.R. devised all the experiments, most of which were executed by F.R.G and S.P. Western blots and SIM imaging were performed by E.C.R. and G.G., respectively. Sea urchin flagella and Inpp5e mutant mice were generated by M.A and S.S., respectively. F.R.G, S.P, T.I. and J.F.R. wrote the manuscript. T.I. and J.F.R. supervised the work.
The authors declare no competing interests.
References
- Andrews D, Nelson DL. Biochemical studies of the excitable membrane of Paramecium tetraurelia. II. Phospholipids of ciliary and other membranes. Biochim Biophys Acta. 1979;550:174–87. doi: 10.1016/0005-2736(79)90205-0. [DOI] [PubMed] [Google Scholar]
- Bae YK, Kim E, L'hernault SW, Barr MM. The CIL-1 PI 5-phosphatase localizes TRP Polycystins to cilia and activates sperm in C. elegans. Curr Biol. 2009;19:1599–607. doi: 10.1016/j.cub.2009.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidylinositol signaling to the ciliopathies. Nat. Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood RA, Woodward MP, Young WW., Jr Unusual distribution of a glycolipid antigen in the flagella of Chlamydomonas. Protoplasma. 1985;185:123–130. [Google Scholar]
- Cameron DA, Pennimpede T, Petkovich M. Tulp3 is a critical repressor of mouse hedgehog signaling. Dev. Dyn. 2009;238:1140–1149. doi: 10.1002/dvdy.21926. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki PG, Shah JV. The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 2012;22:201–210. doi: 10.1016/j.tcb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Forte M, Satow Y, Nelson D, Kung C. Mutational alteration of membrane phospholipid composition and voltage-sensitive ion channel function in paramecium. Proc Natl Acad Sci U S A. 1981;78:7195–9. doi: 10.1073/pnas.78.11.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J. Cell Biol. 2012;197:697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gealt MA, Adler JH, Nes WR. The sterols and fatty acids from purified flagella of Chlamydomonas reinhardtii. Lipids. 1981;16:133–136. [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Balla T. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim. Biophys. Acta. 2015;1851:746–758. doi: 10.1016/j.bbalip.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, Balla T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 2014;205:113–126. doi: 10.1083/jcb.201312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, et al. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N. Engl. J. Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci U S A. 2012;109:19691–6. doi: 10.1073/pnas.1210916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby M, et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 2009;41:1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan FP, Nachury MV. The conserved Bardet-Biedl Syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–19. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Chichili GR, Rodgers W. Compartmentalization of phosphatidylinositol 4,5-bisphosphate signaling evidenced using targeted phosphatases. J Biol Chem. 2008;283:29920–29928. doi: 10.1074/jbc.M805921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonah M, Erwin JA. The lipids of membraneous cell organelles isolated from the ciliate, Tetrahymena pyriformis. Biochim Biophys Acta. 1971;231:80–92. doi: 10.1016/0005-2760(71)90256-6. [DOI] [PubMed] [Google Scholar]
- Kaneshiro ES, Matesic DF, Jayasimhulu K. Characterizations of six ethanolamine sphingophospholipids from Paramecium cells and cilia. J Lipid Res. 1984;25:369–77. [PubMed] [Google Scholar]
- Kennedy KE, Thompson GA., Jr Phosphonolipids: localization in surface membranes of Tetrahymena. Science. 1970;168:989–91. doi: 10.1126/science.168.3934.989. [DOI] [PubMed] [Google Scholar]
- Liem KF, Jr., et al. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J. Cell Biol. 2012;197:789–800. doi: 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, et al. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat. Chem. Biol. 2013;9:437–443. doi: 10.1038/nchembio.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, et al. OCRL localizes to the primary cilium: a new role for cilia in Lowe syndrome. Hum. Mol. Genet. 2012;21:3333–3344. doi: 10.1093/hmg/dds163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, et al. Compensatory role of inositol 5-phosphatase INPP5B to OCRL in primary cilia formation in oculocerebrorenal syndrome of Lowe. PLoS One. 2013;8:e66727. doi: 10.1371/journal.pone.0066727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano R. Inhomogeneous distribution of filipin-sterol complexes in the ciliary membrane of rat tracheal epithelium. Am J Anat. 1979;156:139–45. doi: 10.1002/aja.1001560115. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Jackson PJ. The Tubby family proteins. Genome Biol. 2011;12:225. doi: 10.1186/gb-2011-12-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, et al. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes. Dev. 2010;24:2180–2193. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RX, et al. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum. Mol. Genet. 2009;18:1740–1754. doi: 10.1093/hmg/ddp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson VL, et al. Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects and polydactyly: identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Hum. Mol. Genet. 2009;18:1719–1739. doi: 10.1093/hmg/ddp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Lin Y, Norman RX, Ko HW, Eggenschwiler JT. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc. Natl. Acad. Sci. USA. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Skarnes WC. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 2006;20:22–27. doi: 10.1101/gad.1363606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson EC, et al. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol. 2015;209:129–142. doi: 10.1083/jcb.201411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- Santos N, Reiter JF. A central region of Gli2 regulates its localization to the primary cilium and transcriptional activity. J. Cell Sci. 2014;127:1500–1510. doi: 10.1242/jcs.139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, et al. Mammalian phosphoinositide kinases and phosphatases. Prog Lipid Res. 2009;48:307–343. doi: 10.1016/j.plipres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Smith JD, Snyder WR, Law JH. Phosphonolipids in Tetrahymena cilia. Biochem Biophys Res Commun. 1970;39:1163–9. doi: 10.1016/0006-291x(70)90682-0. [DOI] [PubMed] [Google Scholar]
- Souto-Padron T, de Souza W. Freeze-fracture localization of filipin-cholesterol complexes in the plasma membrane of Trypanosoma cruzi. J Parasitol. 1983;69:129–37. [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. Receptor-induced Transient Reduction in Plasma Membrane Phosphatidylinositol-4,5 Biphosphate Concentration Monitored in Living Cells. Current Biology. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujishita Y, Guo S, Stolz LE, York JD, Hurley JH. Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell. 2001;105:379–389. doi: 10.1016/s0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- Tyler KM, Fridberg A, Toriello KM, Olson CL, Cieslak JA, Hazlett TL, Engman DM. Flagellar membrane localization via association with lipid rafts. J Cell Sci. 2009;122:859–66. doi: 10.1242/jcs.037721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Falkenburger BH, Pohlmeyer C, Inoue T. Triggering actin comets versus membrane ruffles: distinctive effects of phosphoinositides on actin reorganization. Sci Signal. 2011;4:ra87. doi: 10.1126/scisignal.2002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Gaus K, Verkade P, Fullekrug J, Vaz WL, Simons K. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin-Darby canine kidney (MDCK) cells. Proc Natl Acad Sci U S A. 2006;103:18556–61. doi: 10.1073/pnas.0608291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, et al. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell. Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavari A, et al. Role of lipid metabolism in smoothened derepression in hedgehog signaling. Dev Cell. 2010;19:54–65. doi: 10.1016/j.devcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.