Abstract
TREM2 and TYROBP are causal genes for Nasu–Hakola disease (NHD), a rare autosomal recessive disease characterized by bone lesions and early-onset progressive dementia. TREM2 forms a receptor signaling complex with TYROBP, which triggers the activation of immune responses in macrophages and dendritic cells, and the functional polymorphism of TREM2 is reported to be associated with neurodegenerative disorders such as Alzheimer’s disease (AD). The objective of this study was to reveal the involvement of TYROBP and TREM2 in the pathophysiology of AD and schizophrenia. Methods: We investigated the mRNA expression level of the 2 genes in leukocytes of 26 patients with AD and 24 with schizophrenia in comparison with age-matched controls. Moreover, we performed gene association analysis between these 2 genes and schizophrenia. Results: No differences were found in TYROBP mRNA expression in patients with AD and schizophrenia; however, TREM2 mRNA expression was increased in patients with AD and schizophrenia compared with controls (P < 0.001). There were no genetic associations of either gene with schizophrenia in Japanese patients. Conclusion: TREM2 expression in leukocytes is elevated not only in AD but also in schizophrenia. Inflammatory processes involving TREM2 may occur in schizophrenia, as observed in neurocognitive disorders such as AD. TREM2 expression in leukocytes may be a novel biomarker for neurological and psychiatric disorders.
Introduction
Nasu–Hakola disease (NHD), also called polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL), is an extremely rare autosomal recessive disease characterized by bone lesions and early-onset progressive neurocognitive disorders [1]. NHD is caused by mutations in the triggering receptor expressed on myeloid cell 2 (TREM2) on chromosome 6p21.1 or TYRO protein tyrosine kinase binding protein (TYROBP) on chromosome 19q13.1 [2]. These genes encode different domains of the same receptor signaling protein in the activation of immune response, called the TREM2/TYROBP signaling cascade. Several studies have shown that TREM2/TYROBP signaling is essential for the development of osteoclasts and dendritic cells under homeostatic conditions [3] and that synaptogenesis is deregulated in TYROBP-deficient mice [4, 5]. However, the absence of immunological derangement in NHD remains enigmatic.
TREM2 is also known to be associated with Alzheimer’s disease (AD) and other neurodegenerative diseases. A functional single nucleotide polymorphism (SNP) in TREM2 (rs75932628>T, p.R47H) is associated with AD [6], Parkinson’s disease [7], frontotemporal dementia [8], and amyotrophic lateral sclerosis [9]. Moreover, Lue et al. [10] reported that TREM2 expression is upregulated in the brain of patients with AD. Although a vast amount of clinical data has strongly implicated the role of inflammatory and degenerative processes in the pathophysiology of schizophrenia, TREM2 expression in schizophrenia has not yet been examined. In the present study, we report gene expression and association analyses of both TYROBP and TREM2 in patients with AD and schizophrenia.
Methods
Subjects
Descriptive data for each group of participants are shown in Table 1. All participants in this study were of Japanese origin and unrelated to each other.
Table 1. Demographic data and clinical characteristics of each group.
| A. Expression analysis for Alzheimer’s disease. | |||
| Alzheimer's disease (n = 26) | AD-cnt (n = 26) | P-value | |
| sex (male: female) | 8:18 | 13:13 | 0.26 |
| age(years mean ± SD) | 79.6 ± 4.0 | 77.5 ± 4.1 | 0.07 |
| MMSE score (points mean ± SD) | 18.0 ± 5.6 | 29.7 ± 0.6 | < 0.001 |
| CDR number of each grade (0: 0.5: 1: 2: 3) | 0: 1: 8: 14: 3 | 26: 0: 0: 0: 0 | < 0.001 |
| B. Expression analysis for schizophrenia. | |||
| schizophrenia (n = 24) | Sc-cnt (n = 24) | P-value | |
| Sex (male: female) | 7:17 | 7:17 | 1 |
| age(years mean ± SD) | 54.8 ± 18.0 | 54.8 ± 18.0 | 1 |
| disease duration (years mean ± SD) | 24.2 ± 14.7 | none | ― |
| C. Genotyping analysis. | |||
| schizophrenia (n = 796) | control (n = 510) | P-value | |
| sex (male: female) | 457: 339 | 265: 245 | 0.06 |
| age(years mean ± SD) | 53.0 ± 14.4 | 35.1 ± 14.2 | < 0.001 |
AD-cnt, the controls against Alzheimer’s disease in expression analysis; MMSE, Mini Mental State Examination; CDR, Clinical Dementia Rating. The score of 0–3 shows classification of dementia (0 = none, 0.5 = questionable, 1 = mild, 2 = moderate, 3 = severe); Sc-cnt, the controls against schizophrenia in expression analysis. The p-value was calculated by student T test, Chi-squired test, and Fisher’s exact test.
Participants in AD analysis
We recruited 26 patients with AD [8 males and 18 females, mean age ± standard deviation (SD) = 79.6 ± 4.0 years] from Ehime University Hospital and related community hospitals. AD was diagnosed according to criteria established by the National Institute on Aging and the Alzheimer’s Association and classified as probable AD dementia [11] with bilateral hippocampal atrophy using brain CT or brain MRI findings. Subjects were also assessed by the Mini Mental State Examination (MMSE) [12] and Clinical Dementia Rating (CDR) [13]. The AD control group (AD-cnt) included 22 age-matched elderly participants with normal cognitive function (13 males and 13 females, mean age ± SD = 77.5 ± 4.1 years) who agreed to participate in this study. For inclusion, subjects had to be capable of living independently, have MMSE scores over 28, and be free of cognitive impairment or morphological brain abnormality.
Participants in schizophrenia analysis
We recruited 24 patients with schizophrenia (7 males and 17 females, mean age ± SD = 54.8 ± 18.0 years, disease duration at blood draw = 24.2 ± 14.7 years) as well as 24 age-matched controls (Sc-cnt; 7 males and 17 females, mean age ± SD = 54.8 ± 18.0 years) from Ehime University Hospital and related community hospitals. In addition, we enrolled 796 patients with schizophrenia (457 males, 339 females, age = 53.0 ± 3.4 years, 34 patients did not indicate age) who visited Ehime or Tokushima University Hospitals for a gene association study. Schizophrenia was diagnosed according to the Diagnostic and Statistical Manual of Mental Disorders IV criteria by at least 2 certified psychiatrists on the basis of clinical interviews and review of medical records. The comparison group included 510 healthy volunteers (265 males, 245 females, mean age ± SD = 35.1 ± 14.2 years) without psychiatric signs, psychiatric family history, or past history of mental disorders.
NHD participant
A 42-year-old male NHD patient homozygous for a TYROBP mutation (TYROBP c.141delG; manuscript in preparation) was included as a reference. We obtained his RNA twice, at intervals of 30 months, and confirmed that both samples expressed equivalent levels of TYROBP. We also analyzed his father and mother (aged 72 and 68 years, respectively), both of whom were heterozygous for the TYROBP c.141delG mutation.
Ethical issues
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later revision. This study was approved by the institutional ethics committees of The Ethics Review Committee for Human Genome/Gene Analysis Research in Ehime University Graduate School of Medicine and the University of Tokushima Graduate School as “Genetic Studies on Neuropsychiatric Diseases” (registry number; 25-K4), and written informed consent was confirmed by all participants or their guardians before the acquisition of blood samples.
Blood sample collection
Whole peripheral blood samples were collected for the extraction of total RNA and genomic DNA, according to the standard protocol, in PaxGene Blood RNA Systems tubes (BD, Tokyo, Japan) and potassium EDTA tubes, respectively. Absorption spectrophotometry, using NanoDrop-1000 (Thermo Fisher scientific, Yokohama, Japan), was used to determine RNA concentrations and purity (260/280 ratio above 1.8 was used). RNA (1 μg per sample) was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, USA), in a total reaction volume of 40 μL. Genomic DNA was isolated from whole blood leukocytes using the QIAamp DNA Blood Mini Kit (Qiagen, Tokyo, Japan), according to the manufacturer’s protocol.
Expression analyses
For a quantitative estimate of TYROBP and TREM2 mRNA levels, the StepOnePlus Real-Time PCR System (Applied Biosystems) was used. Specific TaqMan probes were employed (Assay ID: TYROBP; Hs00182426_m1, TREM2; Hs00219132_m1), and the relative expression level of TYROBP or TREM2 mRNA was determined by comparison with the housekeeping gene GAPDH [14–16], serving as an internal standard (all Taqman probes from Applied Biosystems). The final volume reaction was 10 μL using the TaqMan Universal Master Mix (Applied Biosystems). Relative mRNA levels were calculated via the 2−ΔΔCT method [17] using StepOne software (Applied Biosystems). We averaged the fold changes from three wells for each sample (triplicate) and used this average value for statistical analyses. The NHD patient was used for calibration in all experiments to correct the observational error.
SNP analysis
Three SNPs, rs8113524 (NG_009304.1; assay ID: C___2604899_10), rs3817624 (NG_009304.1; assay ID: C__25603557_20), and rs2234256 (NG_011561.1; assay ID: C__15948232_10) (Table 2)were selected using HaploView v4.2 (Cambridge, MA) as tag SNPs, with squared correlation coefficient (r2) between 2 SNPs > 0.8 and minor allele frequency (MAF) > 0.01, on the basis of the current International HapMap project database (http://hapmap.ncbi.nlm.nih.gov/index.html.en). These were analyzed for association study with schizophrenia. Additional SNPs, rs429358 (NG_007084.2; assay ID: C___3084793_20) and rs7412 (NG_007084.2; assay ID: C____904973_10) were used to determine apolipoprotein E (APOE) isoforms using a real-time SNP genotyping system (TaqMan Assays, Applied Biosystems). Following this, 1× TaqMan PCR Master Mix, 1× TaqMan SNP genotyping assay, 10 ng genomic DNA, and ultrapure water to a final reaction volume of 6 μL were mixed in each well of an optical plate. Allelic discrimination was performed using StepOnePlus and analyzed using its software.
Table 2. Characteristics of the selected two tagging SNPs in TYROBP and one tagging SNP in TREM2.
| No. | Gene | SNP(major/minor allele) | Chromosome Location |
|---|---|---|---|
| 1 | TYROBP | rs8113524 (T/C) | 35905035, Chr 19 intron |
| 2 | TYROBP | rs3817624 (C/T) | 36398899, Chr 19 intron |
| 3 | TREM2 | rs2234256 (T/C) | 41126655, Chr 6 ex4 (L211P) |
Statistics
Statistical analyses were performed using SPSS v22 (IBM, Tokyo, Japan). Expression levels of TYROBP and TREM2 were compared using the Mann–Whitney U-test and Student’s t-test. Correlations of gene expression with age, disease duration, and MMSE score were analyzed using the Spearman test. Fisher’s exact test with simulated P-values was used to compare TREM2 genotype distributions between patients with schizophrenia and controls. Linkage disequilibrium (LD), TYROBP haplotypes, genotype distributions, minor allele frequencies, and Hardy–Weinberg equilibrium (HWE) were determined using HaploView v4.2. Statistical power was calculated with G*Power 3 (http://www.gpower.hhu.de/), and statistical significance was defined at the 95% confidence level (P = 0.05). The ratio of successful genotyping was over 99%, and we extrapolated the missing values for genotype analysis. Bonferroni corrections were applied to maintain an overall type I error rate of 0.05, taking multiple comparisons into account.
Results
TYROBP and TREM2 expression in NHD
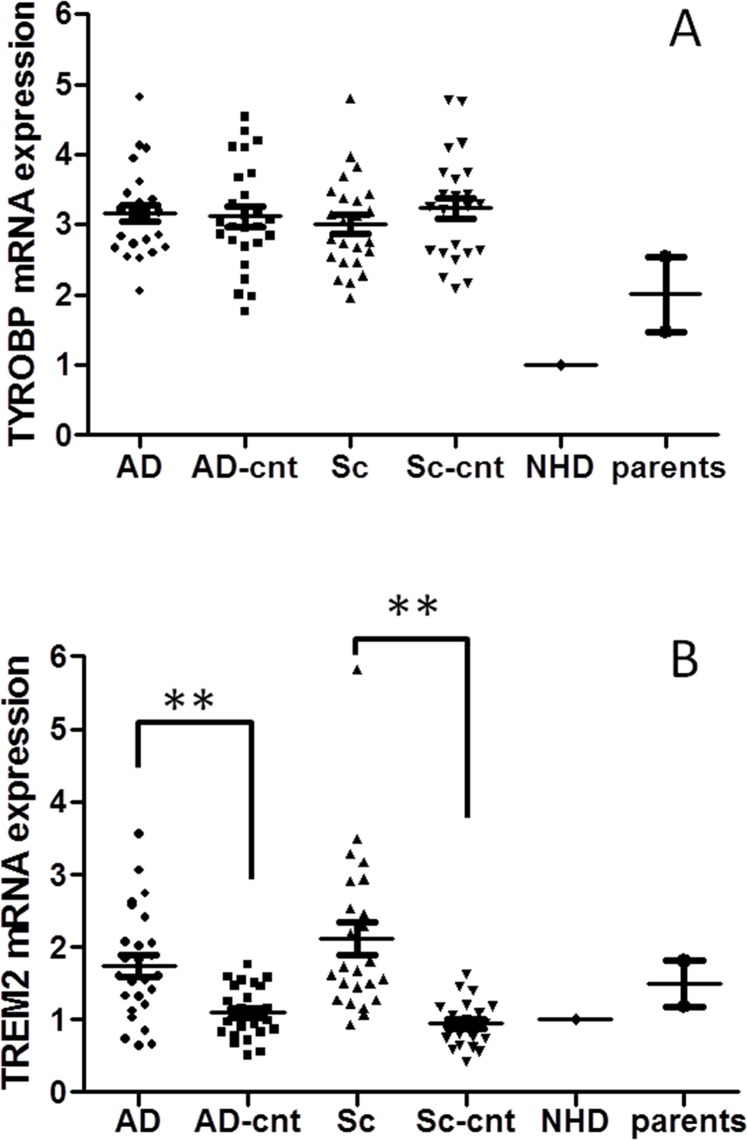
The NHD patient homozygous for the TYROBP c.141delG mutation exhibited the lowest level of TYROBP expression among the study participants, while his parents, heterozygous for the mutation, showed no difference from other groups. In contrast, TREM2 was expressed in the NHD patient and his parents at the same levels as that in control subjects.
TYROBP and TREM2 expression in AD and schizophrenia
Fig 1 shows TYROBP and TREM2 expression in leukocytes of patients with AD or schizophrenia and their controls. TYROBP expression in patients with AD and schizophrenia was similar to that in their respective control groups (P = 0.44 and P = 0.13, respectively). In contrast, TREM2 expression was significantly higher in patients with AD and schizophrenia compared to that in their respective controls (P < 0.001 in both cases). This was also confirmed after Bonferroni corrections. There was no correlation between TREM2 expression and other clinical variables such as age (r = 0.08, P = 0.71), MMSE (r = 0.01, P = 0.95) in AD, and disease duration (r = 0.28, P = 0.19) in schizophrenia.
Fig 1. Expression of TYOBP (1A) and TREM2 (1B) in each group.
TYROBP expression was the lowest in the NHD patient and showed no difference in other groups.TREM2 expression was higher in schizophrenic and AD patients than in the respective controls. P < 0.01 is indicated by ** when the P values were calculated by Mann–Whitney U-test.
TREM2 expression in AD with APOE ɛ4
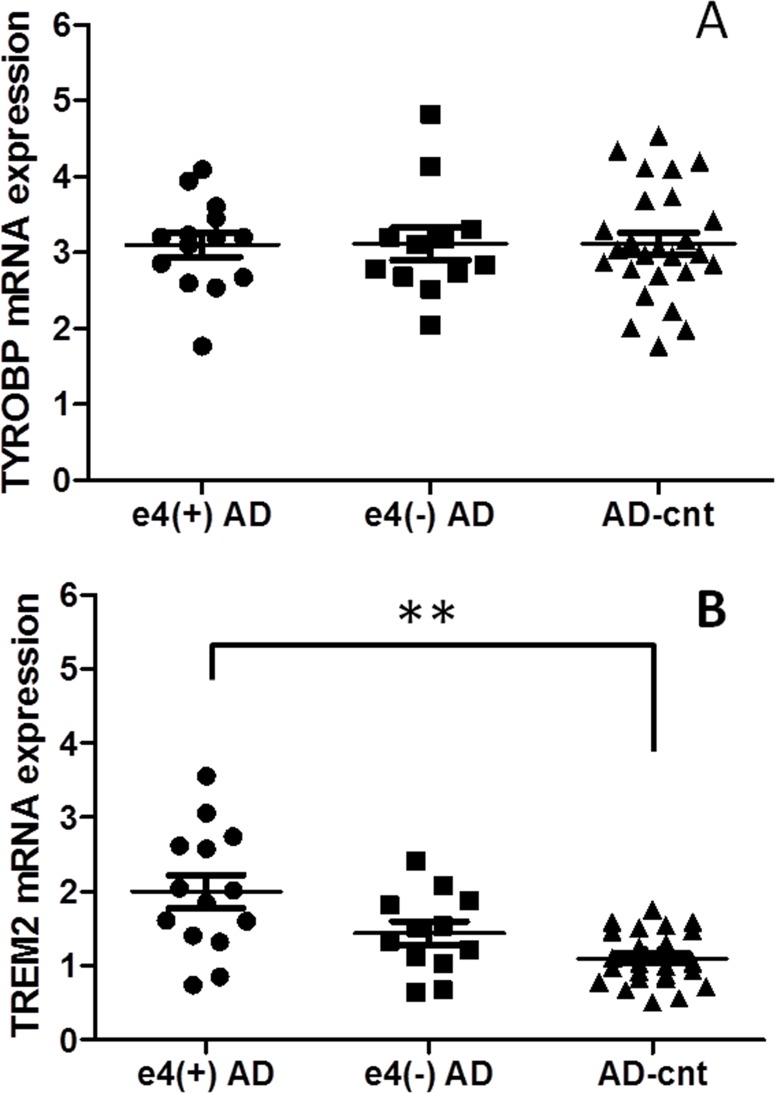
AD patients were typed for the APOE ɛ4 allele (Table 3). There were no differences in age, sex, MMSE score, or CDR between the 14 participants (all were ɛ3/ ɛ4) in the APOE ɛ4-positive AD group [ɛ4(+) AD] and the 12 participants (all were ɛ3/ ɛ3) in the APOE ɛ4-negative AD group [ɛ4(−) AD]. In the AD control group (n = 26), only 2 subjects were positive for the APOE ɛ4 allele. For these 3 groups, TYROBP and TREM2 mRNA expression levels in leukocytes are shown in Fig 2. In ɛ4(+) AD, TREM2 expression was significantly higher than that in AD-cnt (P < 0.001). Although there were no significant differences between ɛ4(+) and ɛ4(−) AD (P = 0.07) or between ɛ4(−) AD and AD-cnt (P = 0.07), the relative expression of both genes tended to decrease across groups, in the order ɛ4(+) AD, ɛ4(−)AD, and AD-cnt.
Table 3. Characteristics of APOE ɛ4 positive and negative group in AD.
| ɛ4-positive (n = 14) | ɛ4-negative (n = 12) | P-value | |
|---|---|---|---|
| sex (male: female) | 5:09 | 3:09 | 0.68 |
| age(years mean ± SD) | 80.1 ± 4.3 | 78.9 ± 3.7 | 0.45 |
| MMSE score (points mean ± SD) | 17.7± 7.1 | 18.2 ± 3.6 | 0.84 |
| CDR number of each grade (0: 0.5: 1: 2: 3) | 0: 0: 5: 7: 2 | 0: 1: 3: 7: 1 | 0.64 |
AD, Alzheimer’s Disease; MMSE, Mini Mental State Examination; CDR, Clinical Dementia Rating. The score of 0–3 shows classification of dementia (0 = none, 0.5 = questionable, 1 = mild, 2 = moderate, 3 = severe). The P-value was calculated by Student t test, Chi-squired test, and Fisher’s exact test.
Fig 2. Expression of TYOBP (2A) and TREM2 (2B) in APOE ɛ4-positive AD [ɛ4(+) AD], APOE ɛ4-negative AD [ɛ4(−) AD], and AD-cnt groups.
TYROBP expression showed no difference. TREM2 expression was significantly higher in ɛ4-positive AD group than in AD-cnt (including 2 ɛ4-positive subjects) group. The expression levels tended to decline in the following order: ɛ4(+) AD, ɛ4(−) AD, and AD-cnt. P < 0.001 after Bonferroni correction is indicated by * * when the P values were calculated by Mann–Whitney U-test.
Case–control association studies in schizophrenia
We performed genotyping of 3 tag SNPs (rs8113524 and rs3817624 in TYROBP and rs2234256 in TREM2) in 796 patients with schizophrenia and 510 controls (unrelated Japanese participants). The results are shown in Table 4. The result of haplotype analysis between TYROBP and schizophrenia is shown in Table 5. In schizophrenia, rs8113524 was not in HWE; however, rs3817624 and rs2234256 were in HWE. No associations between either TYROBP or TREM2 and schizophrenia were revealed by single marker or haplotype analyses.
Table 4. Genotyping results of the selected two tagging SNPs in TYROBP and one tagging SNP in TREM2.
| No. | SNP | n (dd/dD/DD) | HWE P-value | MAF | single marker P-value | |
|---|---|---|---|---|---|---|
| 1 | rs8113524 | cases | 14/129/651 | 0.03 | 0.099 | 0.74 |
| controls | 2/90/407 | 0.29 | 0.095 | |||
| 2 | rs3817624 | cases | 1/59/734 | 1 | 0.038 | 0.86 |
| controls | 1/36/472 | 1 | 0.037 | |||
| 3 | rs2234256 | cases | 1/11/782 | 0.1 | 0.008 | 0.32 |
| controls | 0/5/501 | 1 | 0.005 |
d, minor allele; HWE, Hardy‐Weinberg equilibrium; MAF, Minor Allele Frequency
The single marker p-value is calculated by Chi-squared test with omission the missing values.
Table 5. Result of association study in the TYROBP gene.
| Haplotype | Frequencies | Ratio Counts | Frequencies | P-Value | ||
|---|---|---|---|---|---|---|
| case | control | case | control | |||
| TC | 0.87 | 1370.0: 218.0 | 890.7: 135.3 | 0.863 | 0.868 | 0.69 |
| CC | 0.1 | 157.0: 1431.0 | 97.3: 928.7 | 0.099 | 0.095 | 0.74 |
| TT | 0.04 | 61.0: 1527.0 | 38.0: 988.0 | 0.038 | 0.037 | 0.86 |
P-value is calculated by Chi-squared test with omission the missing values.
Discussion
In NHD, the TREM2/TYROBP signaling pathway is damaged by the mutations in either TYROBP or TREM2, and this has been suggested to cause defects in microglial activity [18]. It is reported that TYROBP expression is normal in the brain of NHD individuals with the TREM2 mutation [2] and that TREM2 mRNA extracted from leukocytes is severely reduced in NHD individuals with this mutation [19]. Our results, showing decreased levels of TYROBP mRNA and normal levels of TREM2 mRNA in leukocytes of an NHD patient with the TYROBP mutation, are consistent with previous reports.
In the present study, we found that TREM2 was more highly expressed in leukocytes of patients with AD and schizophrenia, which were of different disease and age groups, whereas TYROBP expression was not altered in these groups. Our results may be consistent with the previous study that revealed TREM2 mRNA expression in monocytes was elevated in AD [16]. In the brain, TREM2 is primarily expressed in microglia; however, absent or expressed at low levels in granulocytes, monocytes, and macrophages [20]. Kiialainen et al. [21] reported that the microglial expression of Trem2 was high in cultured mouse neuronal cells. The product of TREM2 is highly expressed in microglia of the AD brain [10], and Trem2 expression is particularly high in the amyloid plaques of AD model mice [22]. Higher TREM2 mRNA levels in APOE ɛ4-positive individuals may suggests that TREM2 expression in leukocytes reflects the AD pathology. Although, unexpectedly, we found no differences between the ɛ4(+) AD and ɛ4(−) AD groups or between the ɛ4(−) AD and AD-cnt groups, it is possible that this is a false-negative result and that differences in expression may be confirmed by increasing the number of subjects.
Hu et al. [16] reported that TREM2 mRNA and protein levels were significantly higher in monocytes of AD and that the expression was inversely associated with the MMSE score. TREM2 mRNA was significantly increased in leukocytes of AD in the present study, and we found the tendency to inverse correlation between TREM2 mRNA expression in leukocytes and the MMSE score in APOE ɛ4-negative AD patients (P = 0.08, r = −0.53). We suggest that elevated TREM2 expression in leukocytes can be utilized as a novel biological marker for AD and may associate with severity in APOE ɛ4-negative AD patients.
Surprisingly, TREM2 mRNA expression was also elevated in patients with schizophrenia. To date, this is the first report showing TREM2 upregulation in schizophrenia. Although schizophrenia has been thought of as a non-neurodegenerative disorder, the evidence from several recent reports suggests that schizophrenia may actually be neurodegenerative [23]. Compared to brain tissues, TREM2 is highly expressed in microglia [24] and reduced microglial activity is reported in Trem2-deficient mice [25]. Although we did not examine inflammatory markers such as C-reactive protein and IL-6, it is well known that C-reactive protein [26, 27] and IL-6 [28] are elevated in schizophrenic patients who are free of overt inflammation. Interestingly, increased microglial activity in schizophrenia was also confirmed by both electron microscopy [29] and positron emission tomography [30]. Higher TREM2 mRNA levels in leukocytes of schizophrenia may be associated with peripheral inflammation or microglial involvement. Because inflammation in glial cells may lead to neuronal changes in schizophrenia [31], TREM2 expression in brain samples of patients with schizophrenia should be studied in the future.
TYROBP expression was not changed in either AD or schizophrenia. TREM2 in extracellular regions acts as a receptor to bind the ligand [32], whereas TYROBP is present as a second transmembrane receptor and transmits signal [33]. The difference in the role of each domain in TYROBP/TREM2 signaling may explain the different expression patterns between TYROBP and TREM2 in leukocytes. Further studies of the TREM2/TYROBP signaling should be performed to elucidate its role in neurological and psychiatric diseases.
We could not find significant association between TYROBP and TREM2 gene polymorphisms and schizophrenia. In addition, the allele rs8113524 was not present in HWE in schizophrenic patients; this may be due to small population size and low allele frequency [34]. Recently, an association study of 2190 Japanese patients with late-onset AD revealed that TREM2 variants, including R47H and L211P, were not associated with the disease [35]. Therefore, higher level of TREM2 mRNA in AD and schizophrenia in this study may not be caused by gene polymorphisms but by other factors such as epigenetic modification.
Our study has several limitations that should be considered. All patients with schizophrenia were taking antipsychotics and all patients with AD were taking cholinesterase inhibitors at the time of the blood sampling. Although our NHD patient was also taking antipsychotics and his TREM2 mRNA levels were similar to those of controls, we could not exclude the possibility that these pharmacological treatments affected the expression of both genes. Influences of food or physical condition (e.g., body mass index, smoking, infection, and inflammation) on gene expression in leukocytes were not examined. Moreover, it remains to be confirmed whether TREM2 expression in the brain is in good agreement with that in leukocytes. In addition, future studies should assess the correlation between the severity of inflammation or its markers in central spinal fluid (such as neopterin) and expression of the 2 genes.
In conclusion, we report for the first time that TREM2 expression in leukocytes is elevated not only in AD but also in schizophrenia and may be a clinical biomarker for these diseases. We believe that our study is an important first step in understanding the role of TREM2 in the pathophysiology of AD and schizophrenia.
Acknowledgments
We thank Miss Takako Muneta for her technical assistance and Dr. Win Thiri Kyaw for her proofreading and encouragement. This work was partially supported by a Health and Labor Science Research Grant from the Japanese Ministry of Health, Labour and Welfare; a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology; and the Ehime Graduate University of Medicine’s Good Practice Fund.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was partially supported by a Health and Labor Science Research Grant from the Japanese Ministry of Health, Labour and Welfare; a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology; and the Ehime Graduate University of Medicine’s Good Practice Fund.
References
- 1. Hakola HP. Neuropsychiatric and genetic aspects of a new hereditary disease characterized by progressive dementia and lipomembranous polycystic osteodysplasia. Acta Psychiatr Scand Suppl 1972; 232: 1–173. [PubMed] [Google Scholar]
- 2. Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet 2002; 71: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paradowska-Gorycka A, Jurkowska M. Structure, expression pattern and biological activity of molecular complex TREM-2/DAP12. Human Immunology 2013; 74: 730–737. 10.1016/j.humimm.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 4. Colonna M. DAP12 signaling: from immune cells to bone modeling and brain myelination. Journal of Clinical Investigation 2003; 111: 313–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaifu T, Nakahara J, Inui M, Mishima K, Momiyama T, Kaji M, et al. Osteopetrosis and thalamic hypomyelinosis with synaptic degeneration in DAP12-deficient mice. Journal of Clinical Investigation 2003; 111: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer’s disease. The New England Journal of Medicine 2013; 368: 117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rayaprolu S, Mullen B, Baker M, Lynch T, Finger E, Seeley W, et al. TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson’s disease. Molecular Neurodegeneration 2013; 8: 19 10.1186/1750-1326-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giraldo M, Lopera F, Siniard AL, Corneveaux JJ, Schrauwen I, Carvajal J, et al. Variants in triggering receptor expressed on myeloid cells 2 are associated with both behavioral variant frontotemporal lobar degeneration and Alzheimer’s disease. Neurobiol Aging 2013; 34: 2077.e11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cady J, Koval ED, Benitez BA, Zaidman C, Jockel-Balsarotti J, Allred P, et al. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol 2014; 71: 449–453. 10.1001/jamaneurol.2013.6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lue L, Schmitz C, Sorrano G, Sue L, Beach T, Walker D. TREM2 protein expression changes correlate with Alzheimer’s disease neurodegenerative pathologies in postmortem temporal cortices. Brain Pathol. 2015; 25(4): 469–480 10.1111/bpa.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McKhann G, Knopman D, Chertkow H, Hyman B, Jack C, Kawas C, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer’s & Dementia: The Journal of the Alzheimer's Association 2011; 7: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975; 12: 189–198. [DOI] [PubMed] [Google Scholar]
- 13. Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 1993; 43: 2412–2414. [DOI] [PubMed] [Google Scholar]
- 14. Carrol ED, Salway F, Pepper SD, Saunders E, Mankhambo LA, Ollier WE, et al. Successful downstream application of the Paxgene Blood RNA system from small blood samples in paediatric patients for quantitative PCR analysis. BMC Immunol. 2007; 12: 8:20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watanabe S, Iga J, Ishii K, Numata S, Shimodera S, Fujita H, et al. Biological tests for major depressive disorder that involve leukocyte gene expression assays. J Psychiatr Res. 2015; 66–67: 1–6. 10.1016/j.jpsychires.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 16. Hu N, Tan MS, Yu JT, Sun L, Tan L, Wang YL, et al. Increased expression of TREM2 in peripheral blood of Alzheimer’s disease patients. J Alzheimers Dis 2014; 38: 497–501. 10.3233/JAD-130854 [DOI] [PubMed] [Google Scholar]
- 17. Livak KJ, Schmittgen TD . Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 18. Takahashi K, Rochford C, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. The J Exp Med. 2005; 201(4): 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chouery E, Delague V, Bergougnoux A, Koussa S, Serre JL, Mégarbané A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Human Mutation 2008; 29: E194–204. 10.1002/humu.20836 [DOI] [PubMed] [Google Scholar]
- 20. Neumann H, Takahashi K. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol 2007; 184: 92–99. [DOI] [PubMed] [Google Scholar]
- 21. Kiialainen A, Hovanes K, Paloneva J, Kopra O, Peltonen L. Dap12 and Trem2, molecules involved in innate immunity and neurodegeneration, are co-expressed in the CNS. Neurobiology of Disease 2005; 18: 314–322. [DOI] [PubMed] [Google Scholar]
- 22. Frank S, Burbach G, Bonin M, Walter M, Streit W, Bechmann I, et al. TREM2 is upregulated in amyloid plaque‐associated microglia in aged APP23 transgenic mice. Glia 2008; 56: 1438–1447. 10.1002/glia.20710 [DOI] [PubMed] [Google Scholar]
- 23. Monji A, Kato T, Mizoguchi Y, Horikawa H, Seki Y, Kasai M, et al. Neuroinflammation in schizophrenia especially focused on the role of microglia. Progress in Neuro-Psychopharmacology & Biological Psychiatry 2013; 42: 115–121. [DOI] [PubMed] [Google Scholar]
- 24. Hickman S, Khoury J. TREM2 and the neuroimmunology of Alzheimer’s disease. Biochem Pharmacol. 2014; 88(4): 495–498. 10.1016/j.bcp.2013.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cantoni C, Bollman B, Licastro D, Xie M, Mikesell R, Schmidt R, et al. TREM2 regulates microglial cell activation in response to demyelination in vivo. Acta Neuropathol 2015; 129: 429–447. 10.1007/s00401-015-1388-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Avramopoulos D, Pearce BD, McGrath J, Wolyniec P, Wang R, Eckart N, et al. Infection and inflammation in schizophrenia and bipolar disorder: a genome wide study for interactions with genetic variation. PloS one. 2015; 10.1371/journal.pone.0116696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dickerson F, Stallings C, Origoni A, Vaughan C, Khushalani S, Yang S, et al. C-reactive protein is elevated in schizophrenia. Schizophr Res. 2013; 143(1): 198–202. 10.1016/j.schres.2012.10.041 [DOI] [PubMed] [Google Scholar]
- 28. Al-Hakeim HK, Al-Rammahi AD, Al-Dujaili AH. IL-6, IL-18, sIL-2R, and TNFα proinflammatory markers in depression and schizophrenia patients who are free of overt inflammation. J Affect Disord. 2015; 10.1016/j.jad.2015.04.044 [DOI] [PubMed] [Google Scholar]
- 29. Uranova NA, Vikhreva OV, Rachmanova VI, Orlovskaya DD. Ultrastructural alterations of myelinated fibers and oligodendrocytes in the prefrontal cortex in schizophrenia: a postmortem morphometric study. Schizophr Res Treatment 2011; 2011: 325789 10.1155/2011/325789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doorduin J, Vries E, Willemsen A, Groot J, Dierckx R, Klein H. Neuroinflammation in schizophrenia-related psychosis: a PET study. Journal of Nuclear Medicine: Official Publication, Society of Nuclear Medicine 2009; 50: 1801–1807. [DOI] [PubMed] [Google Scholar]
- 31. Takahashi N, Sakurai T. Roles of glial cells in schizophrenia: possible targets for therapeutic approaches. Neurobiology of Disease 2013; 53: 49–60. 10.1016/j.nbd.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 32. Daws M, Lanier L, Seaman W, Ryan J. Cloning and characterization of a novel mouse myeloid DAP12‐associated receptor family. Eur J Immunol 2001; 31: 783–791. [DOI] [PubMed] [Google Scholar]
- 33. Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 1998; 391: 703–707. [DOI] [PubMed] [Google Scholar]
- 34. Masel J. Genetic drift. Curr Biol. 2011; 21(20): R837–838. 10.1016/j.cub.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 35. Miyashita A, Wen Y, Kitamura N, Matsubara E, Kawarabayashi T, Shoji M, et al. Lack of genetic association between TREM2 and late-onset Alzheimer’s disease in a Japanese population. J Alzheimers Dis 2014; 41: 1031–1038. 10.3233/JAD-140225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.