Abstract
Background
Equine neuroaxonal dystrophy/equine degenerative myeloencephalopathy (NAD/EDM) is a neurodegenerative disorder affecting young horses of various breeds that resembles ataxia with vitamin E deficiency in humans, an inherited disorder caused by mutations in the alpha-tocopherol transfer protein gene (TTPA). To evaluate variants found upon sequencing TTPA in the horse, the mode of inheritance for NAD/EDM had to be established.
Hypothesis
NAD/EDM in the American Quarter Horse (QH) is caused by a mutation in TTPA.
Animals
88 clinically phenotyped (35 affected [ataxia score ≥2], 53 unaffected) QHs with a diagnosis of NAD/EDM with 6 affected and 4 unaffected cases confirmed at postmortem examination.
Procedures
Pedigrees and genotypes across 54,000 single nucleotide polymorphism (SNP) markers were assessed to determine heritability and mode of inheritance of NAD/EDM. TTPA sequence of exon/intron boundaries was evaluated in 2 affected and 2 control horses. An association analysis was performed by 71 SNPs surrounding TTPA and 8 SNPs within TTPA that were discovered by sequencing. RT-PCR for TTPA was performed on mRNA from the liver of 4 affected and 4 control horses.
Results
Equine NAD/EDM appears to be inherited as a polygenic trait and, within this family of QHs, demonstrates high heritability. Sequencing of TTPA identified 12 variants. No significant association was found using the 79 available variants in and surrounding TTPA. RT-PCR yielded PCR products of equivalent sizes between affected cases and controls.
Conclusions and Clinical Importance
NAD/EDM demonstrates heritability in this family of QHs. Variants in TTPA are not responsible for NAD/EDM in this study population.
Keywords: Alpha-tocopherol, Equine, Genetics, Vitamin E
Equine neuroaxonal dystrophy (NAD) is an inherited neurodegenerative condition that has been described in young horses of various breeds.1–4 The histologic lesions of NAD consist of neuronal degeneration within specific nuclei of the brainstem.1,3,5 Equine degenerative myeloencephalopathy (EDM) is a pathologically more advanced form of NAD that also appears to be inherited. A histologic diagnosis of EDM is made when axonal necrosis and demyelination are found within the dorsal and ventral spinocerebellar tracts and ventromedial funiculi of the cervicothoracic spinal cord.6–10 Both NAD and EDM have been associated with vitamin E deficiency.3,6 In American Quarter Horses (QHs) with histopathologic lesions consistent with EDM, we have demonstrated that there also are lesions consistent with NAD.3 Therefore, the term NAD encompasses the primary disease process, with EDM considered a more severe variant of NAD.
Comparatively, both the clinical and histologic findings in horses with NAD/EDM resemble ataxia with vitamin E deficiency (AVED) in humans. AVED is caused by various mutations in the alpha-tocopherol transfer protein gene (TTPA), which encodes for the protein responsible for vitamin E transport in the liver and incorporation of α-tocopherol, a main component of vitamin E, into very low-density liposomes for transport throughout the body.11,12 Human patients with AVED develop neurologic abnormalities similar to those seen in horses with NAD/EDM and pathologic findings in AVED are similar to those observed in NAD/EDM, including neuroaxonal dystrophy within the gracile and cuneate nuclei.13 Although we have previously demonstrated no difference in expression of TTPA between affected and unaffected horses,3 mutated genes can maintain normal expression levels while altering protein confirmation and their interactions,14 and therefore we aimed to definitively evaluate TTPA as a candidate gene for NAD/EDM.
A mode of inheritance for NAD/EDM is required to determine if variants uncovered in TTPA are putative mutations. Although previous studies have described pedigree analysis15 and a prospective breeding trial was performed in Morgan horses,2 the mode of inheritance for NAD/EDM has been difficult to determine. Either an autosomal dominant mode of inheritance with variable expression or a polygenic mode of inheritance was considered likely based on a breeding trial in Morgan horses.2 It is not known, however, if NAD/EDM is allelic (ie, if specific causal variants are shared) between Morgans and QHs. Using the large group of horses phenotyped in a previous study,3 we aimed to define the mode of inheritance of NAD/EDM in the QH.
Materials and Methods
Pedigree Analysis
Pedigrees were collected from 88 clinically phenotyped QHs, including 35 affected and 53 unaffected horses. Six affected and 4 unaffected controls were confirmed at postmortem examination, with histologic findings of NAD/EDM-affected horses previously described.3 DNA was collected and purified from all of these horses at the time of examination.a All horses originated from 1 breeding farm with a high incidence of NAD/EDM and were classified as affected with a mean ataxia score ≥2 as previously described.3 Horses were classified as unaffected with a mean ataxia score of 0. Attributable to the age of onset of NAD/EDM ranging from 4 to 36 months,2,4 horses were only classified as unaffected if they were confirmed to be neurologically normal at 3 years of age. Ataxia scores are reported as mean ± standard deviation. Serum vitamin E concentrations have been reported previously.3 A complete 5-generation pedigree was obtained from the farm for each horse. Pedigrees were visually inspected to determine mode of inheritance.
Serum α-Tocopherol Concentrations
Serum α-tocopherol concentrations were measured in randomly selected horses during DNA collection and determined by high-performance liquid chromatography with fluorescence detection as previously described.3 Serum α-tocopherol concentrations are described as mean ± standard deviation. Based on the small sample size, a nonparametric Wilcoxon signed rank test was used to compare serum α-tocopherol concentrations between affected (n = 8) and unaffected (n = 5) horses.
Complex Segregation Analysis
A simple segregation analysis could not be performed on this family of horses because families were closely interrelated. To evaluate for a single gene Mendelian effect, complex segregation analysis was performed on the 35 affected and 53 unaffected horses. Complex segregation analysis is intended to integrate Mendelian transmission genetics and models of penetrance with the patterns of covariance expected in polygenic inheritance. A more complete description of complex segregation analysis is available.16
An outline of the criteria that must be satisfied before acceptance of the single major locus model has been provided.17 Adherence to these criteria decreases the number of false positives. Evaluation of the models necessary for complex segregation analysis was conducted with the package iBay.18 The goal of this strategy was to simultaneously estimate the caudal density for a polygenic contribution to disease along with the contributions of a putative Mendelian locus. Specifically, for this mixed-inheritance model, the strategy allowed the evaluation of a polygenic variance component, the additive and dominance contributions of a single locus (the parameters −a, d, and a for the putative major locus genotypes AA, AB, and BB, respectively) and the frequency of allele A of the putative major locus (defined as p). Given our scoring of binary phenotypes, where affected is 1 and normal is scored as 0, the “B” allele represents the putative disease- enhancing allele. The iBay software models the unobservable scale of this threshold trait such that the residual variance is fixed at 1.0 (ie, σe2 = 1).
Creation of the Gibbs sample requires several key assumptions about the behavior of these unknown parameters. Although a variety of models can be considered, all are some variant of the following: with or without sex, as a fixed effect with flat (ie, uniform) prior densities, the polygenic variance component with a flat prior density, as well as flat prior densities for the additive, dominance, and allele frequency parameters. A Gibbs sample of 7,500 was generated, beginning with the creation of 2,000,000 total samples, a “burn-in” of 500,000, and a sampling rate of every 200-th Gibbs value. This process was repeated 2 additional times to create 3 replicate chains. As outlined above, the post-Gibbs analysis was implemented with the packages BOA [Bayesian output analysis]19 and CODA,20 both part of the R-program.21 Convergence of the Gibbs sampling process was performed by contrasting sample means from the first 10% of the sample with the last 50% of the sample.22 Also as outlined above, from the 22,500 Gibbs samples, the mean, standard deviation, and the upper and lower limits of a 95% highest density region (HDR; the Bayesian equivalent of a confidence interval) were computed for each of the unknown parameters with hdrcde.23
Heritability
A subset of 71 horses (33 affected [16 males, 17 females], 38 unaffected [13 males, 25 females]) was genotyped for 54,000 single nucleotide variants (SNPs) by the Illumina EquineSNP50 array.b Total genotyping rate was 98.3% on average across all markers. Pseudoheritability, defined as the additive component of heritability as estimated with the kinship matrix, was calculated using phenotype and SNP marker information by a computer program (EMMAX)c in this subset of horses. The basis for pseudoheritability calculation has been described previously.24 Briefly, a pairwise relatedness matrix from high-density markers is computed, which is used to represent the sample structure. The contribution of the sample structure to the phenotype then is estimated by a variance component model that results in an estimated covariance matrix of phenotypes, modeling the effect of genetic relatedness on the phenotypes. To verify this pseudoheritability calculation, a heritability estimate was generated from only pedigree and phenotype information.
The phenotype of disease is discrete and binary, requiring any analyses to accommodate the distinction between a binary observation and a continuous liability (risk) for disease. This is most often accomplished in threshold models.25 Specifically, we will assume that liability for disease is a normally distributed random variable, such that the ith observation, yi, can be modeled as yi = μ + sexi + ai + ei, where μ is a constant common to all animals; sexi, the contribution to disease risk of the ith sex (i = 1,2); ai, the additive genetic contribution to disease risk; and ei, the unknown residual. In addition, ai was assumed to have originated in a vector, a, of additive genetic values from a multivariate normal density with mean 0 and covariance Aσ2a, with A being the numerator relationship matrix and the residuals to follow from a normal density with mean 0 and variance I σ2e. Given the nature of threshold models, because the liability is an underlying unobservable variable, we fixed the residual variance at σ2e = 1. The objective was to estimate the unknown constant μ, along with the unknown variance σ2a. With that, the heritability of disease liability can be estimated [h2 = σ2a/(σ2a + 1)]. For completeness, we also considered a model without the term for sex, leaving only a constant common to all observations.
A Bayesian framework, a strategy of considerable power and plasticity, was used to evaluate these unknown values.25 The public domain package iBayd was used. The prior distribution for the constant μ and the 2 sex effects was a normal density with null mean and an unknown variance, so as to make the prior uninformative, or “flat”.18 The prior distribution for the variance component (σ2a) was assumed to be an inverse chi-square density, with settings to make it uninformative or “flat”.18 As implied by the use of a Bayesian strategy, estimates of the caudal density for the unknown parameters were generated by a Monte Carlo Markov chain. This was performed with 3 chains, each starting at dispersed values for the unknown variance and constant μ and the 2 sex effects. Convergence of the chains was examined by trace plots making use of the R package CODA20 and computation of the Gelman-Rubin statistic.26 Each chain was run a total of 2,000,000 rounds with a burn-in of 500,000 rounds and a thinning interval of 200 (creating a single chain sample of 7,500 values). In this scenario, both parameters had chains that mixed well (with Gelman-Rubin statistics of 1.00), with the absolute value of all autocorrelations for parameters below 0.05.
TTPA Sequencing
The equine ortholog to human gene TTPA (NM 000370) was identified by the equine BLAT search at the University of California Santa Cruz Genome Bioinformatics website.e Exons 2–5 were identified within the September 2007 2.0 draft assembly of the domestic horse (Equus caballus).f For exons 2–5, genomic DNA from 2 postmortem confirmed NAD/EDM-affected horses (1 horse classified as NAD and the other as the more severe variant, EDM) and 1 postmortem confirmed unaffected horse was chosen for sequencing and the equine reference sequencef was used as a second control sequence. Exon 1 could not to be identified based on a gap in the equine assembly of unknown size from chromosome 9:21665110-21665210. For this region, 1 additional postmortem confirmed unaffected horse was used for sequencing to provide a second control because the reference sequence was not available.
For exons 2–5, polymerase chain reaction (PCR) primers flanking each exon were designed by Primer3 software.g PCR was performed as previously described27 using primer-specific melting temperature (Table 1). For exon 1, primers were designed to flank the gap in the sequence (F-5′AGTACGGAGCAGGGCTCATA3′ and R- 5′GCGAAGGGACAGAACTCAAG3′) and the products were clonedh and sequenced. An additional set of primers (F-5′ AGGGGAGGGAAGGAAGGAA3′ and R-5′ GGGATTTCTCAGAACACCTG3′) was created to fill the remaining gap (700 bp) and the sequence was used to design equine-specific primers for exon 1. Once the complete sequence was obtained, PCR primers were designed to flank exon 1 in genomic DNA (Table 1), a PCR protocol designed for GC-rich regionsi was used and the resulting PCR products sequenced. An open reading frame was hypothesized for the gene by investigating the equine sequence for probable start and stop sites that conferred the most similarity to the human homologous protein sequence. All exon sequences and flanking intron sequences were scanned for variants and the equine reference sequenceg was used as an additional control sample. The equine sequence has been submitted to GenBank (submission #1528593).
Table 1.
Primers sequences, melting temperatures (Tm), and genomic locations used for sequencing TTPA.
| Region | Primers | Tm(°C) | Genomic Position |
|---|---|---|---|
| Exon 1 | F-CCTGGGGGAAGTTTCAGATT | 60 | N/A |
| R-GCTTGCAACTTGGGCTTTC | 60 | N/A | |
| Exon 2 | F-TTGTCTGGATCCCAAAAAGC | 58 | g.21671464-21671483 |
| R-CATCGAGCACCAGAACAGAA | 60 | g.21672427-21672446 | |
| Exon 3 | F-CTTGCTTTTCCCATTTCCAA | 56 | g.21676601-21676620 |
| R-TTATTGGGCAAAGATGATGG | 56 | g.21677569-21677588 | |
| Exon 4 | F-TGTGTTTGTTGTACTCTTTCAACG | 59 | g.21678366-21678389 |
| R-TACAATGCATCCTGCGAATC | 58 | g.21679329-21679348 | |
| Exon 5 | F-TTTTGCTAAGAATCACTTGGACA | 57 | g.21681411-21681433 |
| R-GACACCCACCCAGAATGAGT | 62 | g.21682376-21682395 | |
| 3′UTR | F-CGTGAGTGAGATCCTAATTGGT | 60 | g.21681986-21682007 |
| R-ACAGTACAAACGGCACATTTT | 56 | g.21682946-21682966 |
Association Analysis
SNP markers from the Illumina Equine SNP50 platform that passed quality control settings (minor allele frequency >0.01 and genotyping across individuals >90%) surrounding TTPA were chosen to cover 1.5 Mb 5′ of the gene and 1.5 Mb 3′ of the gene (Table S1). A custom-designed SNP genotyping platform was created by the 12 SNPs uncovered from sequencing of TTPA and genotyping was performed by the iPLEX Gold assayj as previously described28 on the same 33 affected and 38 unaffected horses that had been genotyped on the Illumina EquineSNP50 platform. Surrounding sequence data used to design multiplex primers are listed in Table S2. Two SNPs failed design (TTPA_5UTR_SNP3 and TTPA_Exon1_SNP2) and 2 genotyped poorly (TTPA_Exon1_SNP1 and TTPA_Exon1_SNP4). A casecontrol allelic chi-square association analysis was performed by PLINK softwarek on the 33 affected and 38 unaffected horses with these 79 total SNPs. Permutation testing (100 permutations) was performed to account for multiple testing procedures. To account for a potential recessive mode of inheritance with a high allelic frequency, an additional case-control genotypic chi-square association analysis was performed by permutation testing. Significance was set at P < 0.05.
RT-PCR
Because TTPA is primarily expressed in the liver,29 liver samples from NAD/EDM-affected (n = 4) and unaffected horses (n = 4) were flash-frozen immediately at necropsy and mRNA prepared.l Primers within exon 1 (F5′CTCACCGACTCC TTCTTGCT3′) and exon 5 (R5′GGGAAATGCTGAAGTA AGCTC 3′) were used for reverse transcription polymerase chain reaction (RT-PCR) amplification of mRNA. GAPDH primers F primer (5′AAGATTGTCAGCAATGCCTCC3′) and R primer (5′ CCAGGAAATGAGCTTGACAAA3′) were included to ensure that equivalent amounts of cDNA were added. PCR product size (expected 681 bp) was compared between affected and control cases.
Results
Pedigree Analysis
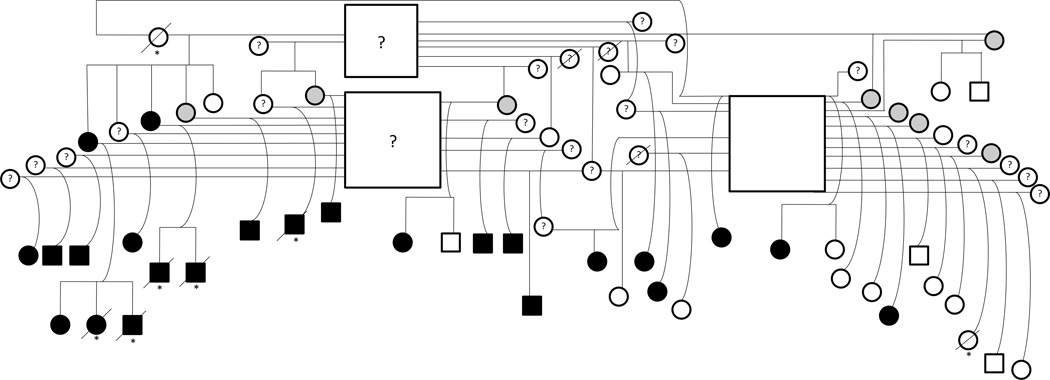
Complete neurologic examinations were performed on 88 horses (81 QHs, 5 Thoroughbred/QH [TB/QH] crosses, 1 Standardbred/QH cross, and 1 TB) from 9 extended families, including 38 males and 50 females. Ten of these horses had complete postmortem examinations performed, confirming the clinical phenotype (6 affected, 4 unaffected). Of all the cases evaluated, 35 were determined to be affected (mean score 2.53 ± 0.48; 18 males, 17 females; 35 QHs) and 53 unaffected (20 males, 33 females; 46 QHs, 5 TB/QH crosses, 1 TB and 1 Standardbred/QH cross). The most complete families were used for assessing inheritance patterns (partial pedigree; Fig 1).
Fig 1.
Partial pedigree of family of Quarter Horses affected with neuraxonal dystrophy/equine degenerative myeloencephalopathy (NAD/EDM). Square = male, circle = female, black = affected (ataxia score ≥2 as previously reported3), white = unaffected (ataxia score of 0), gray = equivocal (ataxia score >0 but <2), ? = unknown phenotype status, / = deceased, * = phenotype confirmed at postmortem.
Several patterns of inheritance were evaluated within families. The pedigrees did not support an X-linked pattern of inheritance, because there was an overall equal sex distribution of affected horses (18 males, 17 females). An autosomal recessive pattern of inheritance could not be definitively ruled out from these families, because 2 affected horses were never bred to evaluate if an unaffected foal was produced. A completely penetrant autosomal dominant mode of inheritance was not supported by 2 breedings (unaffected stallion × 2 unaffected mares producing affected foals).
Serum α-Tocopherol Concentrations
Serum α-tocopherol concentrations were below the reference range (1.5–10 µg/mL) in 7/8 affected horses (mean 0.99 ± 0.46; range, 0.55–1.7 µg/mL) and 4/4 unaffected horses (mean 0.76 ± 0.31; range, 0.3–0.99 µg/mL). There was no significant difference between affected and unaffected horses (P = 0.37).
Complex Segregation Analysis
Table 2 presents results of the complex segregation analysis of our binary measure of NAD. Although several genetic models were evaluated, for brevity, only those results for the model with a constant, a polygenic term and a putative major locus, are presented. In this table, 2 models are considered, one in which the putative major locus is of a general form (ie, a, the additive effect and d, the dominance deviation are estimated) and a second, recessive major locus model (ie, d = −a). In both settings, only the Mendelian transmission of the major alleles is presented. In both analyses, the 95% HDR region for the putative major locus variance overlaps with 0 (0.0), which has the straightforward interpretation of no evidence for a segregating major locus. This conclusion also is illustrated, not independently, by observing that the 95% HDR for the major allele frequency overlap with 1.0.
Table 2.
Mixed-inheritance model parameters for NAD in American Quarter Horses.a
| Polygenic Variance |
Locus Variance |
Additive Effect (a) |
Dominance Deviation (d) |
Frequency (P) | |
|---|---|---|---|---|---|
| Recessive major model, Mendelianbtransmission | |||||
| Mean | 2.61 | 0.89 | 0.88 | – | .55 |
| SD | 0.96 | 1.51 | 0.72 | – | .26 |
| HDR 95% Lowc | 0.79 | 0.00 | 0.00 | – | .11 |
| HDR 95% High | 4.03 | 3.88 | 2.29 | – | 1.00 |
| General major locus model, Mendelian transmission | |||||
| Mean | 2.68 | 0.59 | 0.91 | 0.02 | .56 |
| SD | 0.94 | 0.73 | 0.67 | 0.94 | .28 |
| HDR 95% Low | 0.84 | 0.00 | 0.00 | −1.88 | .07 |
| HDR 95% High | 4.08 | 2.07 | 2.17 | 1.83 | 1.00 |
Estimates are taken from a Gibbs sample of 22,500 values.
Mendelian transmission governs the probability of transmitting the putative major “A” allele. For Mendelian transmission these values are fixed as 1.0, .50, and 0.0 for putative major genotypes AA, AB, and BB, respectively.
Highest density region.
Heritability
When marker information based on genotypes from 49,747 markers was included with pedigree and phenotypic criteria (ataxia score ≥2 classified as affected and a score of 0 classified as unaffected), pseudoheritability was estimated at 0.795. This value then was compared to a true heritability calculation based on pedigree and phenotype information. Table 3 presents mean estimates of the unknown variance and the more easily interpreted statistic of heritability in the model without a term for sex differences in disease risk. The heritability of disease liability has a mean value of 0.70. Table 4 presents results from the Bayesian analysis in a model for disease that includes terms for the possibility of differential disease risk by sex. Although the mean estimates of the unknown polygenic variance and heritability are relatively the same as in the model without sex, there is no significant difference in disease risk by sex. This conclusion arises from the fact that the 95% highest probability density interval easily spans the value of 0.
Table 3.
Mean, standard deviation, and 95% highest probability density for the Gibbs samples of the additive genetic variance and heritability of disease liability in the three combined chains in a model without correction for sex.
| Mean | SD | 2.5% Quantile |
97.5% Quantile |
|
|---|---|---|---|---|
| Additive variance | 2.60 | 0.09 | 0.77 | 3.94 |
| Heritability | 0.70 | 0.10 | 0.44 | 0.80 |
Table 4.
Mean, standard deviation, and 95% highest probability density for the Gibbs samples of the additive genetic variance and heritability of disease liability in the three combined chains in a model including a term for sex.
| Mean | SD | 2.5% Quantile | 97.5% Quantile | |
|---|---|---|---|---|
| Male–female | 0.15 | 0.64 | −1.10 | 1.40 |
| Additive variance | 2.83 | 0.08 | 0.98 | 3.95 |
| Heritability | 0.72 | 0.08 | 0.50 | 0.80 |
TTPA Sequencing
Genomic DNA from 2 postmortem confirmed NAD/EDM-affected horses (age 2 years) and 1 postmortem confirmed unaffected horse (age 28 years) was used for sequencing exon/intron boundaries for exons 2–5. In addition to these horses, 1 additional postmortem confirmed unaffected QH (34 years) had exon 1 sequenced because the reference sequence was not complete. Sequence was obtained 600 bp 5′ of exon 1, ≥100 bp of each exon/intron boundary and 500 bp 3′ to the last exon. A total of 12 variants were identified in the QH DNA samples relative to the published equine genome sequenceg (Table 5). Of the variants, 4/12 were within putative exons (all within exon 1). All 4 exonic variants were synonymous. As NAD/EDM appears to be inherited as a complex trait, an association analysis was performed by the 12 variants discovered from sequencing.
Table 5.
Variants found in sequencing TTPA.
| Region | Polymorphism | Position within Gene | Type of Mutation | Individuals with 1 Minor Allele |
|---|---|---|---|---|
| 5′UTR | UCD_Accession.21665078C>Gb | 179 bp 5′ of exon 1 | SNP: non-coding | 1 affected |
| 2 controls | ||||
| 5′UTR | UCD_Accession.21665177G>Tb | 80 bp 5′ of exon 1 | SNP: non-coding | 1 control |
| 5′UTR | UCD_Accession.21665247A>Cb | 10 bp of 5′ exon 1 | SNP: non-coding | 1 control |
| Exon 1 | UCD_Accession.21665278C>Gb | Exon 1+21 | SNP: synonymous | 1 control |
| Exon 1 | UCD_Accession.21662663C>Gb g.21665331C>Ga | Exon1+106 | SNP: synonymous | 1 control |
| Exon 1 | UCD_Accession.21665441C>Tb g.21665398C>Ta | Exon 1+171 | SNP: synonymous | 1 affected |
| Exon 1 | UCD_Accession.21665466A>Gb g.21665423T>A,Ga | Exon1+196 | SNP: synonymous | 2 controls |
| Intron 3 | g.21677206A>Ga | Intron3+186 | SNP: non-coding | 1 affected |
| Intron 3 | g.21677352A>Ga | Intron3+331 | SNP: non-coding | 1 affected |
| Intron 3 | g.21678485C>Ta | Intron 3: 288 bp 5′ to exon 4 | SNP: non-coding | 1 affected |
| 3′UTR | g.21682198T>Ca | 3′UTR+378 | SNP: non-coding | 1 affected |
| 3′UTR | g.21682218C>Ta | 3′UTR+398 | SNP: non-coding | 1 affected |
Equine genome reference sequence considered wild type.
Reference sequence not available for this region: wild type based on major allele. Genomic location was determined as the position 5′ to Exon 1.
Association Analysis
None of the SNPs either surrounding TTPA or any of the identified 8 variants that were genotyped within TTPA achieved significance after permutation testing using a case-control allelic chi-square association (Table S1) or a genotypic chi-square association.
RT-PCR
RT-PCR of mRNA from 4 NAD/EDM affected (1 [n = 2], 2, and 4 years of age) and 4 unaffected (1, 6, 28, and 34 years of age) was performed to cover exons 1 through 5, and PCR products of equal size (681 bp) and banding intensity were observed between affected cases and controls, thus indicating that, in the liver, no alternative splicing was present in affected animals.
Discussion
This study of NAD/EDM in the QH established a high heritability of 0.70 in this particular study population and investigated the mode of inheritance of NAD/EDM. An X-linked pattern of inheritance was excluded. In addition, we have excluded a fully penetrant autosomal dominant mode of inheritance based on 2 crosses by an unaffected stallion in this report, which is in agreement with the findings in Morgans with NAD by Beech et al.2 However, based on our previous research describing the clinical phenotype of NAD/EDM in the QH,3 including the effect that supplemental vitamin E, specifically α-tocopherol, during the 1st year of life may have on the overall phenotype of affected horses, it is likely that there is a strong environmental component in the development of NAD/EDM and the trait may be incompletely penetrant or polygenic. This theory is supported by the findings in this study. Therefore, a horse may have the putative genetic mutation, but not display a consistent moderate (grade 2) to severe (grade ≥3) ataxia.
Beech et al2 excluded an autosomal recessive mode of inheritance of NAD in the Morgan horse by producing unaffected foals when affected sires were bred to affected mares. In our population of horses, an autosomal recessive mode of inheritance cannot be excluded, as affected horses were not bred to affected horses. Based on the number of horses affected within these families, if the mode of inheritance was autosomal recessive, the carrier frequency for this mutation within this family would be quite high.
Heritability is defined as the proportion of the phenotype that is attributable to genetic variance. Recently, in genetic association studies, new methods that exploit the use of genetic marker data have been used to explain the fraction of phenotypic variance due to relatedness by generating a kinship matrix.30 This calculation is termed pseudoheritability because it resembles the heritability estimated from a pedigree, yet is not directly interchangeable with heritability of the trait because the estimated pairwise relatedness does not correspond exactly to the kinship coefficients. Pseudoheritability was estimated at 0.795 for NAD/EDM in this population of horses. To further evaluate this calculation, we performed an additional analysis, estimating heritability from phenotype and pedigree information alone, and obtained a comparable heritability estimate of 0.70. There was no significant effect of sex on this estimate. Such a value suggests a considerable amount of genetic variation, enough to promote a breeding program designed to substantially change disease risk. Moreover, a value of this magnitude would give support to a search for the genes responsible for this character, the so-called dissection of this complex trait. Ours was a relatively small sample and pedigree, and any values estimated from this sample may vary considerably from another selection of individuals. However, the behavior of this small sample, in the setting of this Bayesian estimation strategy, did yield stable and informative results. Heritability estimates may not be completely reliable in such a population as described here, where all horses were exposed to an environmental variable (ie, low dietary vitamin E) that may have impacted the incidence of NAD/EDM in a genetically susceptible population. Therefore, the high heritability as determined in this study should not be extrapolated to the entire QH population.
Complex segregation analysis determined that there was insufficient evidence for a single gene effect and therefore lends evidence to NAD/EDM being inherited as a complex trait. Polygenic traits may include the effects of more than 1 gene along with environmental influences to determine the phenotype. Susceptibility loci, or “risk alleles”, are base-pair variants that are found in a higher frequency in diseased individuals as compared with healthy individuals. Those individuals with the gene variant are at a higher risk to develop a particular disease, but the presence of the variant does not induce or cause the disease. Susceptibility loci have been associated with a wide variety of diseases in humans, including Alzhiemer disease,31 multiple sclerosis,32 and Parkinson’s disease.33 We propose that the genetic variant associated with NAD/EDM may be a susceptibility locus and environmental effects, specifically the amount of α-tocopherol received by the foal during the 1st year of life, may play a role in determining the overall phenotype. The majority of randomly sampled horses in this study were α-tocopherol deficient, as determined by serum α-tocopherol concentrations, and there was no statistical difference between affected and unaffected horses, as previously reported.9,10
Based on the association analysis performed by SNPs in the region and the direct sequencing of TTPA, it is unlikely that genetic mutations in TTPA are causative for NAD/EDM in the QH. A susceptibility locus for NAD/EDM in TTPA also is highly unlikely because none of the identified variants were significantly associated with the disease phenotype. Although 4 variants were unable to be genotyped on the multiplex assay, including 2 synonymous mutations in exon 1, these would be unlikely to be causative for NAD/EDM because there was no evidence of an association in the adjacent markers. Although potential regulatory mutations in TTPA cannot be completely ruled out based on this study, they are unlikely because of the fact that the cDNA from affected and unaffected horses has the same length (ie, contains the same number of exons). Size changes of 100 bp can be distinguished on a 2% agarose gel and none of the exons in TTPA were <100 BPM in length. In addition, we have previously demonstrated that there is no significant difference in the expression of TTPA between NAD/EDM-affected horses and controls.3
Although TTPA was the strongest candidate gene for NAD/EDM and there are prominent similarities of NAD/EDM to AVED, there are distinct histologic differences between the diseases. Although lesions of AVED include spheroid formation within the gracile and cuneate nuclei of the brainstem, there also is mild Purkinje cell loss and axonal degeneration in the dorsal columns,13 features that are not found in horses with NAD/EDM.5,34 In addition, the axonal degeneration of the lateral and ventromedial funiculi seen in EDM is not observed in human patients with AVED. From a clinical perspective, the most striking distinction between the 2 diseases is that cases of AVED appear to stabilize or improve with supplemental α-tocopherol,35 whereas cases of NAD/EDM do not.3,36,37
A limitation of this study was the characterization of 29 affected NAD/EDM horses based on clinical examination and farm history alone. At this time, a definitive diagnosis of NAD/EDM can only be achieved by postmortem examination with careful histologic evaluation of the brainstem and spinal cord. All horses on this particular farm were vitamin E deficient at the time of diagnosis, which, in conjunction with the degree of relatedness among the horses, supports a likely clinical diagnosis of NAD/EDM as a cause of their ataxia. In addition, by classifying only the most severely affected horses as cases and horses with mild neurologic deficits as equivocal, we have minimized our chances of misphenotyping. The horses used for sequencing of TTPA were postmortem-confirmed cases and controls. Genetic heterogeneity in NAD/EDM within and across breeds is a distinct possibility, but we limited this study population to a small family of QH in an attempt to minimize this effect.
The association between α-tocopherol deficiency and the development of NAD/EDM remains unclear. There have been many reports describing α-tocopherol deficiency in NAD/EDM-affected horses,6,15 whereas other studies demonstrate that an α-tocopherol deficiency does not increase the risk of NAD/EDM development.9,38 There does appear to be strong evidence, however, that supplementation with α-tocopherol decreases the prevalence of NAD/EDM in genetically susceptible horses.3,6,8 In addition to TTPA, there currently are over 30 proteins known to play a role in αtocopherol absorption, transport, and metabolism39 and these genes can be considered additional candidates for NAD/EDM. Of interest, polymorphisms in many of these α-tocopherol-related genes have been associated with variable protective effects of supplemental vitamin E in humans.39
In conclusion, NAD/EDM appears to be inherited as a complex trait and demonstrates an estimated heritability of 0.70 in a high-risk environment (ie, low dietary vitamin E). Variants in TTPA are not causative for NAD/EDM in this family of QHs. A genome-wide association study is necessary to provide insight into the genetic cause of NAD/EDM in QHs and additional breeds.
Supplementary Material
Acknowledgments
This project was supported, in part, by the Center for Equine Health with funds provided by the State of California pari-mutuel fund and contributions by private donors. Additional funding was provided by the Center for Food Animal Health NRSP008 National Animal Genome Research Project and by private donations to support equine neurologic research. Dr. Finno’s graduate work was supported, in part, by an NIH T32 grant (5 T32 DC 8072-3).
Abbreviations
- ACD
acid-citrate-dextrose
- AVED
ataxia with vitamin E deficiency
- DNA
deoxyribonucleic acid
- EDM
equine degenerative myeloencephalopathy
- HDR
highest density region
- NAD
neuroaxonal dystrophy
- QH
American Quarter Horse
- SNP
single nucleotide polymorphism
- SR-BI
scavenger receptor class B, type I
- TAP
tocopherol-associated transfer protein
- TTPA
alpha-tocopherol transport protein
- Tm
melting temperature
Footnotes
Conflict of Interest: Authors disclose no conflict of interest.
Gentra Puregene blood kit, Quiagen, Valencia, CA
Illumina SNP50 Genotyping Beadchip, Illumina, San Diego, CA
EMMAX, available from http://genetics.cs.ucla.edu/emmax/index.html
iBay, version 1.33, Janss Biostatistics, Leiden, The Netherlands
UCSC Genome Browser, available from http://genome.ucsc.edu/
September 2007 2.0 draft assembly Equus caballus, available from http://www.ncbi.nlm.nih.gov/genome/145
Steve Rozen and Helen J. Skaletsky (2000). Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds), Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386. Source code available at http://fokker.wi.mit.edu/primer3/
TOPO cloning kit for sequencing, Invitrogen, Grand Island, NY
AccuPrime GC-rich DNA polymerase, Invitrogen, Grand Island, NY
MassArray® iPlex Gold Sequenom, San Francisco, CA
PLINK Package: PLINK Version 1.06, author: Shaun Purcell, URL: http://pngu.mgh.harvard.edu/purcell/plink/
FastTrack 2.0 mRNA Isolation kit, Invitrogen, Grand Island, NY
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. SNP markers from the Illumina Equine SNP50 platform that passed quality control settings (minor allele frequency > 0.01 and genotyping across individuals >90%) surrounding TTPA were chosen to cover 1.5 Mb 5′ of the gene and 1.5 Mb 3′ of the gene and results of association analysis of all 79 SNPs (71 from Illumina Equine SNP50 platform and 12 from SNP discovery on direct sequencing) with χ2 and permuted P values after 100 permutations.
Table S2. Forward, reverse, and extension primers used for multiplex genotyping of the SNPs discovered during sequencing of TTPA.
References
- 1.Baumgartner W, Frese K, Elmadfa I. Neuroaxonal dystrophy associated with vitamin E deficiency in two Haflinger horses. J Comp Pathol. 1990;103:114–119. [PubMed] [Google Scholar]
- 2.Beech J, Haskins M. Genetic studies of neuraxonal dystrophy in the Morgan. Am J Vet Res. 1987;48:109–113. [PubMed] [Google Scholar]
- 3.Aleman M, Finno CF, Higgins RJ, et al. Evaluation of epidemiological, clinical and pathologic features of neuroaxonal dystrophy in Quarter horses. J Am Vet Med Assoc. 2011;239:623–633. doi: 10.2460/javma.239.6.823. [DOI] [PubMed] [Google Scholar]
- 4.Adams AP, Collatos C, Fuentealba C, Illanes O, Blanchard R. Neuroaxonal dystrophy in a two-year-old quarter horse filly. Can Vet J. 1996;37:43–44. doi: 10.4141/cjas57-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beech J. Neuroaxonal dystrophy of the accessory cuneate nucleus in horses. Vet Pathol. 1984;21:384–393. doi: 10.1177/030098588402100404. [DOI] [PubMed] [Google Scholar]
- 6.Mayhew IG, Brown CM, Stowe HD, et al. Equine degenerative myeloencephalopathy: A vitamin E deficiency that may be familial. J Vet Intern Med. 1987;1:45–50. doi: 10.1111/j.1939-1676.1987.tb01985.x. [DOI] [PubMed] [Google Scholar]
- 7.Blythe LL, Craig AM. Equine degenerative myelencephalopathy: Part I. Clinical signs and pathogenesis. Comp Cont Educ Pract Vet. 1992;14:1215–1221. [Google Scholar]
- 8.Gandini G, Fatzer R, Mariscoli M, et al. Equine degenerative myeloencephalopathy in five Quarter horses: Clinical and neuropathological findings. Equine Vet J. 2004;36:83–85. doi: 10.2746/0425164044864741. [DOI] [PubMed] [Google Scholar]
- 9.Dill SG, Correa MT, Erb HN, et al. Factors associated with the development of equine degenerative myeloencephalopathy. Am J Vet Res. 1990;51:1300–1305. [PubMed] [Google Scholar]
- 10.Finno CJ, Higgins RJ, Aleman M, et al. Equine degenerative myeloencephalopathy in Lusitano horses. J Vet Intern Med. 2011;25:1439–1446. doi: 10.1111/j.1939-1676.2011.00817.x. [DOI] [PubMed] [Google Scholar]
- 11.Ouahchi K, Arita M, Kayden H, et al. Ataxia with isolated vitamin E deficiency is caused by mutations in the alphatocopherol transfer protein. Nat Genet. 1995;9:141–145. doi: 10.1038/ng0295-141. [DOI] [PubMed] [Google Scholar]
- 12.Gotoda T, Arita M, Arai H, et al. Adult-onset spinocerebellar dysfunction caused by a mutation in the gene for the alpha-tocopherol-transfer protein. N Engl J Med. 1995;333:1313–1318. doi: 10.1056/NEJM199511163332003. [DOI] [PubMed] [Google Scholar]
- 13.Yokota T, Uchihara T, Kumagai J, et al. Postmortem study of ataxia with retinitis pigmentosa by mutation of the alpha-tocopherol transfer protein gene. J Neurol Neurosurg Psychiatry. 2000;68:521–525. doi: 10.1136/jnnp.68.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strachan T, Read AP. Instability of the human genome: Mutation and DNA repair. In: Strachan T, Read AP, editors. Human Molecular Genetics. 2nd ed. New York: Wiley-Liss; 1999. pp. 315–349. [Google Scholar]
- 15.Blythe LL, Hultgren BD, Craig AM, et al. Clinical, viral, and genetic evaluation of equine degenerative myeloencephalopathy in a family of Appaloosas. J Am Vet Med Assoc. 1991;198:1005–1013. [PubMed] [Google Scholar]
- 16.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates, Inc.; 1998. [Google Scholar]
- 17.Elston RC, Namboodiri KK, Glueck CJ, et al. Study of the genetic transmission of hypercholesterolemia and hypertriglyceridemia in a 195 member kindred. Ann Hum Genet. 1975;39:67–87. doi: 10.1111/j.1469-1809.1975.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 18.Janss L. iBay Reference Manual, Version 1.33. 2007 http://www.lucjanss.com. [Google Scholar]
- 19.Smith BJ. boa: Bayesian Output Analysis Program (BOA) for MCMC. R package version 1.6.6. 2007 [Google Scholar]
- 20.Plummer M, Best N, Cowles K, Vines K. coda: Output analysis and diagnostics for MCMC. R package version 11-2. 2007 [Google Scholar]
- 21.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: 2005. [Google Scholar]
- 22.Geweke J. Evaluating the accuracy of sampling-based approaches to the calculation of posterior moments. In: Bernardo JM, Berger JO, Dawid AP, et al., editors. Bayesian Statistics 4. Oxford: Oxford University Press; 1992. pp. 169–193. [Google Scholar]
- 23.Hyndman R. hdrcde: Highest density regions and conditional density estimation. R package version 2.06. 2007 [Google Scholar]
- 24.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorensen D, Gianola D. Likelihood, Bayesian and MCMC Methods in Quantitative Genetics. New York: Springer; 2002. [Google Scholar]
- 26.Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Statistical Science. 1994;7:457–472. [Google Scholar]
- 27.Young AE, Bower LP, Affolter VK, et al. Evaluation of FOXC2 as a candidate gene for chronic progressive lymphedema in draft horses. Vet J. 2007;174:397–399. doi: 10.1016/j.tvjl.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Millis MP. Medium-throughput SNP genotyping using mass spectrometry: multiplex SNP genotyping using the iPLEX (R) gold assay. Methods Mol Biol. 2011;700:61–76. doi: 10.1007/978-1-61737-954-3_5. [DOI] [PubMed] [Google Scholar]
- 29.Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. Vitamin E imaging and localization in the neuronal membrane. J Am Chem Soc. 2005;127:12152–12153. doi: 10.1021/ja051223y. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura S, Watanabe T, Mizoshita K, et al. Genome-wide association study identified three major QTL for carcass weight including the PLAG1-CHCHD7 QTN for stature in Japanese Black cattle. BMC Genet. 2012;13:40. doi: 10.1186/1471-2156-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertram L, Tanzi RE. The genetics of Alzheimer’s disease. Prog Mol Biol Transl Sci. 2012;107:79–100. doi: 10.1016/B978-0-12-385883-2.00008-4. [DOI] [PubMed] [Google Scholar]
- 32.Gourrard PA, Harbo HF, Hauser SL, Baranzini SE. The genetics of multiple sclerosis: An up-to-date review. Immunol Rev. 2012;248:87–103. doi: 10.1111/j.1600-065X.2012.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kummar KR, Djarmati-Westenberger A, Grunewald A. Genetics of Parkinson’s disease. Semin Neurol. 2011;31:433–440. doi: 10.1055/s-0031-1299782. [DOI] [PubMed] [Google Scholar]
- 34.Siso S, Ferrer I, Pumarola M. Abnormal synaptic protein expression in two Arabian horses with equine degenerative myeloencephalopathy. Vet J. 2003;166:238–243. doi: 10.1016/s1090-0233(02)00302-7. [DOI] [PubMed] [Google Scholar]
- 35.Doria-Lamba L, De Grandis E, Cristiani E, et al. Efficacious vitamin E treatment in a child with ataxia with isolated vitamin E deficiency. Eur J Pediatr. 2006;165:494–495. doi: 10.1007/s00431-006-0085-4. [DOI] [PubMed] [Google Scholar]
- 36.Miller MM, Collatos C. Equine degenerative myeloencephalopathy. Vet Clin North Am Equine Pract. 1997;13:43–52. doi: 10.1016/s0749-0739(17)30254-7. [DOI] [PubMed] [Google Scholar]
- 37.Beech J. Equine degenerative myeloencephalopathy. Vet Clin North Am Equine Pract. 1987;3:379–383. doi: 10.1016/s0749-0739(17)30680-6. [DOI] [PubMed] [Google Scholar]
- 38.Dill SG, Kallfelz FA, deLahunta A, Waldron CH. Serum vitamin E and blood glutathione peroxidase values of horses with degenerative myeloencephalopathy. Am J Vet Res. 1989;50:166–168. [PubMed] [Google Scholar]
- 39.Zingg JM, Azzi A, Meydani M. Genetic polymorphisms as determinants for disease preventive effects of vitamin E. Nutr Rev. 2008;66:406–414. doi: 10.1111/j.1753-4887.2008.00050.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.