Abstract
Regulation of the Hedgehog (Hh) pathway relies on an interaction of two receptors that is not fully understood. Patched1 (Ptch1) binds the Hh ligand, but is also a negative regulator of pathway activity. Binding of Hh ligand to Ptch1 leads to the relocation of the activating receptor Smoothened (Smo) to the primary cilium, which is required for the transcriptional Hh response. Besides the transcriptional response, Hh can also induce chemotaxis, and we assessed the effects of defective ciliary localization of Smo on its subcellular itineraries and chemotactic signaling capacity. We find that defective ciliary localization of Smo results in markedly different intracellular trafficking of Smo to sites not involving the primary cilium. These itineraries correlate with a decreased transcriptional signaling capacity, and an enhanced chemotactic responsiveness. These data imply that the ciliary localization machinery functions to transport Smo to sites where it can mediate transcriptional signaling, and away from locations where it can mediate chemotactic signaling. The subcellular localization of Smo is thus a crucial determinant of its signaling characteristics and implies the existence of pool of Smo dedicated to chemotaxis.
INTRODUCTION
The Hedgehog (Hh) pathway is involved in many inductive events in the developing embryo, the maintenance of tissue integrity in adult organisms, and tumorigenesis (1). In amniotes, there are three related ligands that can activate this pathway, Sonic (Shh), Desert, (Dhh) and Indian (Ihh) Hedgehog. Of these, Shh has been studied the most. The contemporary working model for Hh pathway activation in vertebrates holds that the binding of Shh ligand to Patched1 (Ptch1) results in the localization of Smoothened (Smo) to the primary cilium, a cellular appendage shaped by the microtubule cytoskeleton (2–5). Additional proteins involved in mediating the response downstream of Smo localize to the primary cilium as well, and the trafficking to and from the cilium is correlated with the Gli-mediated response, together indicating that the primary cilium is critical for this Shh response. This is further supported by the observations that loss of machinery involved in transport to and from the primary cilium is associated with an attenuation or loss of the Gli-mediated response to Shh (2, 4, 6). Several mouse mutants deficient for cytoskeleton-associated proteins necessary for primary cilium formation show a range of Shh-related phenotypes (7–9), generally consisting of a loss of ligand-mediated modulation of pathway activity through the Gli transcription factors (9). It is hypothesized that in such mice Smo cannot function normally to activate the Gli transcription factors.
Besides activating a transcriptional response, Shh can also act as a cellular chemoattractant (10, 11). Pathfinding of commissural axons and retinal ganglion axons is mediated by a chemotactic Shh response as well (12–16). The chemotactic Shh response does not require de novo transcription or translation, nor does it require the function of Gli proteins (11, 17–19). The chemotactic Shh response, however, does require Smo (13, 20). The requirement for Smo in both the transcriptional and chemotactic responses suggests that a bifurcation of the Shh response into a transcriptional and a chemotactic branch occurs at, or downstream of Smo, but upstream of Gli activation. A requirement for Shh-induced localization of Smo to the primary cilium preceding the alteration of the cytoskeleton at the site of Shh reception seems improbable.
We have shown before that the binding of Shh to Ptch1 causes a redistribution of activated Smo to different intracellular locations not involving the primary cilium (21, 22). However, we did not address the signaling capacities of Smo outside of the primary cilium. Here we report that in addition to the ciliary localization phenotype, intracellular trafficking of Smo not involving the primary cilium was greatly changed as a consequence of defective ciliary localization motifs in Smo, or defective ciliogenesis. These altered itineraries correlated with a much-decreased transcriptional signaling capacity, but unexpectedly, a strongly enhanced chemotactic responsiveness. This suggests that the ciliary localization machinery plays a role in the transport of Smo to sites where it can mediate transcriptional signaling, and away from sites where it can only mediate chemotactic signaling. From this we can deduct a model in which Smo localization is a crucial determinant in discerning between a transcriptional and a chemotactic response to Shh.
RESULTS
Kif3A affects Smo localization outside the primary cilium
The activation of Smo either by Shh or by small molecule agonists results in, but also relies on, the localization dynamics of Smo to and from the primary cilium (5, 23, 24). Several Smo mutants have previously been described that fail to localize to the primary cilium in response to Shh, and these mutants cannot mediate the Gli-mediated Shh response. In Smo−/− immortalized mouse embryo fibroblasts (MEFs) wild type Smo (SmoWT) localized to the primary cilium (Fig. S1A), consistent with earlier observations (24). The SmoΔCLD mutant contains a small mutation in the intracellular C-terminal ciliary localization domain, and the SmoC151Y mutant has a point mutation in the aminoterminal extracellular cysteine rich domain (2, 25). Both mutants showed an impaired localization to the primary cilium (Fig. S1B, C). An oncogenic form of human SMO (SMOM2) has been described to induce a transcriptional Shh response independent of ligand (26, 27) and localized to the primary cilium of Smo−/− MEFs (Fig. S1D). All mutants were expressed at similar quantities after transfection (Fig. S1E). These Smo mutants provide us with the appropriate tools to investigate the effects of defective ciliary localization on subcellular localization of Smo and Shh responsiveness.
Although the staging points of Smo trafficking towards the cilium remain unresolved (23), these events probably initiate outside the primary cilium. As a consequence, the critical interactions between Smo and the trafficking machinery responsible for transport into the cilium likely occur outside the cilium as well. Furthermore, the observation that relatively small mutations, such as found in SmoΔCLD or SmoC151Y, prevent Smo localization to the primary cilium (or at least affect cycling of Smo through the cilium to such an extent that at any fixed timepoint no Smo appears to be in the cilium) indicates there are relatively specific interactions between Smo and its trafficking machinery. To test the relationship between ciliary trafficking machinery and the ciliary localization domain of Smo, we used MEFs from mice mutant for the kinesin II motor protein Kif3A. These cells lack well-organized primary cilia, and Kif3A−/− embryos display a phenotype consistent with an incomplete transcriptional response to Shh (8, 28, 29).
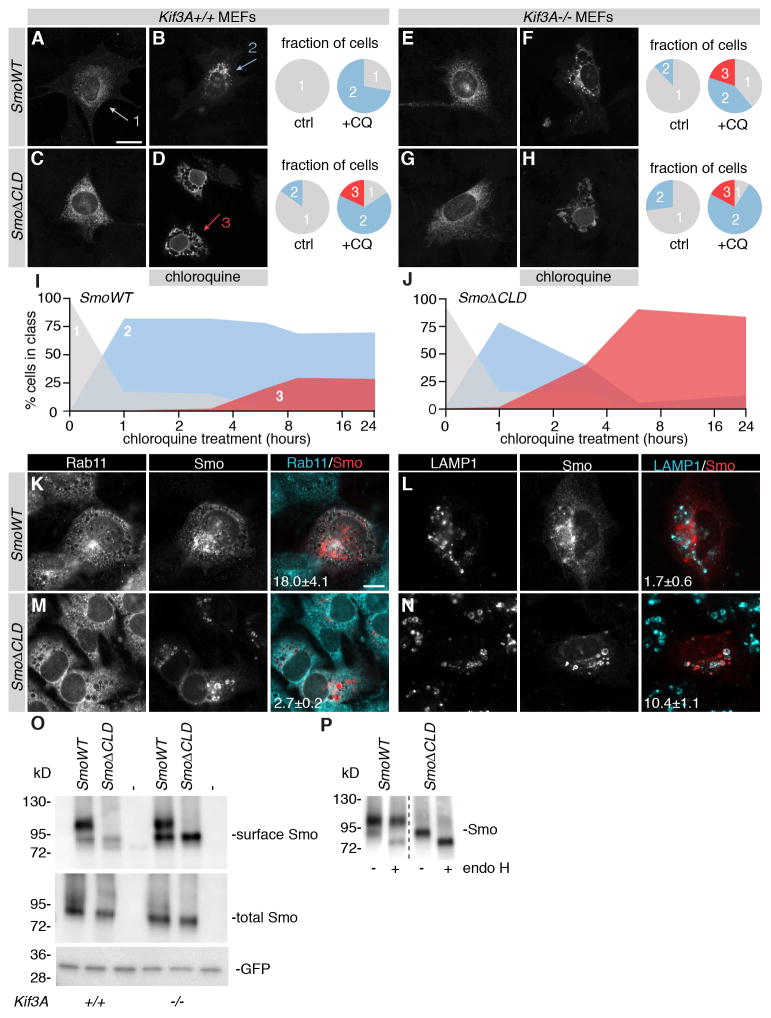
Wild type (Kif3A+/+) and Kif3A−/− MEFs were transfected with SmoWT or SmoΔCLD, and treated with cycloheximide and chloroquine (Fig. 1A–H). Chloroquine prevents acidification of endosomes, thus preventing early to late endosome maturation and this compound has been used previously to study interaction dynamics of Ptch1 and Smo, and trafficking into late endosomes (21, 30). Blocking translation with cycloheximide prevents newly synthesized protein from obscuring the recycling fraction of proteins. The changed Smo localization in response to chloroquine represents the fraction of protein to traffic away from the plasma membrane, and this was quantified by classifying the different intracellular localization patterns observed for Smo (examples of typical localizations indicated by arrows, quantification shown in pie charts in corresponding colors). In the presence of Kif3A, chloroquine induced a moderate perinuclear localization of SmoWT (Fig. 1B, blue arrow ‘2’). However, SmoΔCLD was detected in large vesicles in a fraction of the cells analyzed following chloroquine treatment (Fig. 1D, red arrow ‘3’). In the absence of Kif3A, the distribution of SmoWT was similar to that of SmoΔCLD (Fig. 1F). The absence of Kif3A did not affect the distribution of SmoΔCLD (Fig. 2H). This suggests that both Kif3A and the CLD on Smo regulate the events that underlie trafficking of Smo from the plasma membrane to endosomes. To determine how penetrant the localization phenotype of ciliary localization-defective Smo was, longer chloroquine incubations were performed. In these assays, SmoWT showed vesicular localization in only a small percentage of cells measured (Fig. 1I), whereas the vesicular localization of SmoΔCLD under these conditions was near complete (Fig. 1J).
Fig. 1.
Reduced ciliary localization machinery correlates with altered intracellular itineraries of Smoothened. (A–D) Kif3A+/+ and Kif3A−/− MEFs (E–H) were transfected with SmoWT or SmoΔCLD and treated with 1 μg/ml cycloheximide and 100 μM chloroquine for 3 hours. Smo was visualized by immunofluorescence, and its localization divided in three classes; unperturbed localization to cytosol and membrane, indicated by grey arrow and grey fill in graphs; moderate perinuclear accumulation indicated in blue; widespread vesicular localization indicated in red. Fraction of cells with different Smo localization was quantified. Shown is mean of >60 cells in 3 independent experiments. Scale bar, 20 μm. (I–J) As for panels A–D using wild type MEFs, treatment for times indicated. (K–N) As for panels A–D. Immunofluorescence for indicated proteins was performed. Colocalization of Smo and organelle staining was analyzed and mean percentage of colocalizing pixels is indicated, ± SEM. (O) MEFs were transfected with SmoWT or SmoΔCLD and GFP, and surface biotinylation was performed. Shown is Western blot for surface labeled Smo, and Smo and GFP in total lysates. (P) As for panel O, lysates were treated wit Endo H to assess glycosylation status. Shown is Western blot for surface labeled Smo.
Fig. 2.
Defective ciliary localization enhances chemotactic signaling by Smoothened. (A) MEFs were transfected with indicated forms of Smo and Gli-luciferase reporter. Luciferase activity was measured and corrected for co-transfected Renilla luciferase following 2 μM purmorphamine or control stimulation for 16 hours. Shown is mean fraction of vector, ± SEM, n=6. (B) MEFs were transfected, and transferred to a modified Boyden chamber, using 2 μM purmorphamine or 2 nM ShhN as attractant. Solvent control migration was subtracted to establish net chemotaxis. RFU, relative fluorescence unit; 1 cycle, 2 min. (C) Shown is average migration from 3 experiments as for panel B, using Smo−/− MEFs transfected with vector or indicated mutant forms of Smo, ± SEM, n≥3. For quantitative details, see Methods section. (D) As for panel C, using 2mM recombinant ShhN; (E) 500 nM SAG; (F) As for panel A, using Smo−/− MEFs transfected with mouse (M. musculus), zebrafish (D. rerio), or fruitfly (D. melanogaster) Smo. (G) As for panel C, using indicated constructs. (H) Smo−/− MEFs were transfected with SmoWT, pretreated with the indicated inhibitors for 10 min, and chemotaxis to purmorphamine was assessed. Concentrations used; cyclopamine, 5 μM; pertussis toxin (PTX), 1 μM; MK-571, 5 μM, PP1 and PP2, 10μM.
To more accurately define the different intracellular localizations observed for the different forms of Smo, stainings for markers of distinct intracellular sorting routes were performed. Following treatment with chloroquine, we observed co-labeling of Rab11 and SmoWT (Fig. 1K), suggesting that SmoWT is trafficking via a Rab11-positive compartment (31). Although a subpopulation of SmoWT was localized to LAMP1-positive vesicles (which are late endosomes and lysosomes) as well (Fig. 1L), this population appeared to be much smaller. We did not observe significant co-labeling of SmoΔCLD and Rab11 (Fig. 2M), but instead found SmoΔCLD to accumulate in vesicles containing LAMP1, indicating that SmoΔCLD preferentially traffics to the late endosomes and lysosomes (Fig. 2N).
Rab11 has been implicated in trafficking towards the basal body of the primary cilium (32), and our results suggest that SmoWT enters the cilium from a Rab11-positive vesicle, an itinerary not taken by SmoΔCLD. Consistent with this idea, we found that stable knockdown of Rab11 resulted in a diminished ciliary localization of SmoWT (Fig. S2). However, this effect was not absolute, suggesting that other proteins are able to mediate trafficking of Smo to the cilium, or that the knockdown was incomplete. In any case, these results imply that the CLD of Smo is somehow required for its loading onto Rab11-positive vesicles and that the failure of SmoΔCLD to localize to the primary cilium is caused by altered trafficking at locations in the cell other than the cilium. The chloroquine-induced accumulation of SmoΔCLD in endosomes suggests that its itinerary includes the plasma membrane, and we tested by cell surface biotinylation if Smo localization to the plasma membrane is affected by Kif3A and the CLD. We observed two distinct forms of Smo at the plasma membrane, with apparent Mw of 110kD and 95kD. SmoWT was present at the surface predominantly as an 110kD product, whereas SmoΔCLD was predominantly detected as a 95kD product (Fig. 1O). Endoglycosidase H (Endo H) treatment showed these size differences to be caused by differential glycosylation (Fig. 1P). The 95kD product was sensitive to Endo H treatment, which decreased the apparent Mw to 84 kD, the predicted size of the protein backbone. However, the more extensively glycosylated 110 kD form of SmoWT was largely Endo H resistant. Endo H-sensitive forms of Smo were previously suggested to be ER-localized (33), but our results indicate that this form is also localized at the cell surface, perhaps indicative of altered trafficking of SmoΔCLD (and SmoWT in the absence of Kif3A) through the Golgi. These results demonstrate that the consequences of the loss of the CLD on Smo or Kif3A extend beyond the primary cilium.
Smo was detected at the cell surface regardless of the presence or absence of Kif3A (Fig. 1O). In Kif3A−/− MEFs, the 95kD SmoWT product was enriched about 3-fold relative to the 110kD form. The 95kD product is the predominant surface form of SmoΔCLD regardless of the presence of Kif3A, supporting the general observation that the behavior of SmoWT in Kif3A−/− cells is similar to that of SmoΔCLD. This suggests that the amount of glycosylated Smo at the cell surface involves a role of Kif3A acting on the CLD of Smo. In other words, a mutation in either Kif3A or the CLD will allow the presence of non-fully glycosylated Smo at the cell surface. Taken together, these results show that the loss of Kif3A as well as mutations of the CLD of Smo alters the intracellular itinerary of Smo in a similar manner. We propose that the altered localization and intracellular itineraries of Smo are the reason why in Kif3A embryos the transcriptional response is affected, and why SmoΔCLD is unable to mediate this response.
For the SmoC151Y mutant, we observed a different phenotype altogether. Following chloroquine treatment, SmoC151Y distribution changed rapidly (Fig. S3A), but this distribution was not affected by the absence of Kif3A (Fig. S3B) and like SmoWT, SmoC151Y colocalized with Rab11, but not LAMP-1 (Fig. S3C, D). Surface biotinylation experiments revealed SmoC151Y to be present at the membrane at the same glycosylation status as SmoΔCLD in both Kif3A+/+ and Kif3A−/− MEFs (Fig. S3E), and this form was found to be Endo H sensitive (Fig. S3F). However, we also found much increased total amounts of SmoC151Y in Kif3A−/− MEFs relative to wild type MEFs (Fig. S3E). This implies that in Kif3A−/− MEFs, a relatively small fraction of SmoC151Y makes it to the cell surface. These data indicate that Kif3A facilitates trafficking of non-fully glycosylated forms of Smo to the plasma membrane.
Smo mutants unable to localize to the primary cilium preferentially mediate Hedgehog chemotaxis
To establish a functional role of the observed changes in intracellular trafficking of Smo, we measured the transcriptional response to agonist mediated by the different forms of Smo. SmoWT restored the transcriptional response of Smo−/− MEFs to the Smo agonist purmorphamine (34), as measured using a Gli-luciferase reporter (35) (Fig. 2A). SmoΔCLD was unable to restore a transcriptional Shh response (Fig. 2A). The finding that SmoΔCLD is inactive and unresponsive in this assay argues against overexpression artifacts driving responses that would normally not be mediated by Smo. Similarly, the SmoC151Y mutant could also not restore the transcriptional response to Smo−/− MEFs (Fig. 2A and (25)). On the other hand, SMOM2 showed the highest induction of the transcriptional response independent of agonist (Fig. 2A). SMOM1 was found to confer a higher basal pathway activity than SmoWT, but was unresponsive to agonist. These results are in line with the previously established correlation between ciliary localization of Smo and its ability to activate the transcriptional response (2).
To test the requirement for ciliary localization of Smo in the non-transcriptional Shh response, we assessed the ability of cells transfected with SmoΔCLD and SmoC151Y to migrate towards sources of Shh. As a measure for chemotactic signaling, we assessed cell migration in a modified Boyden camber (11). Transfection of wild SmoWT restored the ability of Smo−/− MEFs to migrate towards Shh and purmorphamine (Fig. 2B). Dose-response experiments showed half-maximum migration (EC50) at approximately 200 nM purmorphamine, and 20 nM SAG, a Smo agonist (33). These EC50s are in the same order of magnitude as those observed for the transcriptional response (33, 36). Transfection of Smo−/− MEFs with SmoΔCLD and SmoC151Y restored chemotaxis to purmorphamine (Fig. 2C). Interestingly, the Smo mutants unable to localize to the primary cilium mediated Shh chemotaxis much more efficiently than SmoWT. This demonstrates that the mechanisms responsible for intracellular trafficking events underlying the ciliary localization of Smo are not required for Shh chemotaxis, and that the altered intracellular localization of these mutants enhances the chemotactic Shh response. Migration to FCS was determined as a control for non-specific cell motility defects and was unaffected by the presence or absence of Smo. Inclusion of purmorphamine in both chambers, thus eliminating its gradient, did not result in net cell movement of cells transfected with any form of Smo (Fig. 2C). The enhanced chemotactic responsiveness of cells transfected with SmoΔCLD relative to those transfected with SmoWT was confirmed in assays using recombinant ShhN (Fig. 2D), and SAG (Fig. 2E).
SMOM1 and SMOM2 transfected cells showed a chemotactic response similar to that mediated by SmoWT, demonstrating that these mutants can be activated by purmorphamine to mediate chemotaxis. This is in apparent contradiction with the idea that these alleles are constitutively active (26, 27), and it would be interesting to see to what extent the activity of activated SMO in other experimental models can be modulated by Shh ligand. The observation that there is no balance between chemotactic versus transcriptional signaling for the SMOM2 mutant demonstrates that these distinct responses are not necessarily mutually exclusive.
To test if activating mutations in Smo are dominant over a defective CLD, the mouse equivalent of the M2 mutation, A1 (26), was introduced in SmoΔCLD, yielding SmoA1-CLD (Fig. S4A). The A1 mutation was unable to rescue the defective ciliary localization and pathway activity caused by a mutated (Fig. S4B–E). It also did not diminish the enhanced chemotactic signaling capacity conferred by the mutated CLD (Fig. S4F).
The ability of transcriptionally unresponsive mutant forms of Smo to mediate chemotaxis suggests that forms of Smo from species, or even phyla other than mouse might also be able to mediate chemotactic signaling in a mammalian background. To assess the inter-species specificity for chemotactic Shh signaling by Smo, Smo−/− MEFs were transfected with zebrafish (Danio rerio), and fruit fly (Drosophila melanogaster) Smo, and pathway activity was assessed. Both these forms of Smo were not able to restore pathway activity as mouse Smo did (Fig. 2F), but surprisingly, both proteins efficiently mediated chemotactic signaling, and the fruit fly more so than mouse Smo (Fig. 2G). This exchangeability implies that chemotactic signaling by Smo is either more robust, or perhaps more evolutionarily conserved, than transcriptional signaling.
To assess if previously described signaling mediators between Smo and the cytoskeleton were responsible for the observed chemotaxis, we performed inhibitor experiments on Smo−/− MEFs transfected with SmoWT. The alkaloid cyclopamine directly binds to and inhibits Smo (37). Pretreatment with this inhibitor diminished migration to purmorphamine, confirming a requirement for Smo (Fig. 2H). Signaling downstream of Smo to the cytoskeleton has previously been described to rely on Smo-mediated activation of G proteins (38). Specifically, it was shown that pertussis toxin (PTX) sensitive G proteins are responsible for this, and indeed we found a marked reduction in migration to purmorphamine in the presence of PTX (Fig. 2H). Further downstream signaling components that have been shown to mediate Shh chemotaxis involve the leukotriene synthesis machinery (20). In agreement, we found the CysLT inhibitor MK-571 to be a very efficient inhibitor of chemotaxis to purmorphamine (Fig. 2H). This suggests some autocrine leukotriene signaling loop to be important for Shh chemotaxis, although the exact mechanism and implications of this remain elusive.
Shh-mediated axon guidance has been described to rely on the activation of Src family kinases (16). The evidence for this comes from the use of the inhibitor PP2. The migration of MEFs to purmorphamine could not be inhibited by the addition of PP2 or a related Src family kinase inhibitor PP1 (Fig. 2H). In addition, we only observed phosphorylation of Src in cells transfected with SmoΔCLD (Fig. S5), suggesting that for Shh chemotaxis in fibroblasts, phosphorylation of Src is not required, and only achieved by very high levels of chemotactic Smo signaling.
Cells without primary cilia retain their chemotactic response to Shh
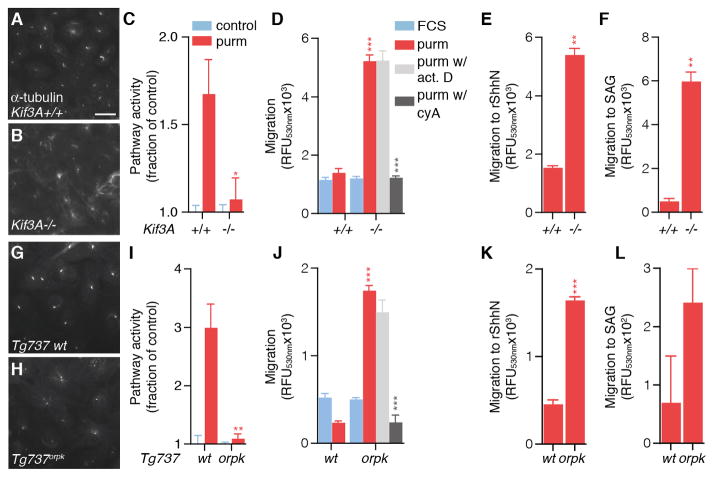
The enhanced chemotactic responsiveness found in cells transfected with ciliary defective forms of Smo raises the question if cells without a primary cilium can still respond to Shh by chemotaxis. We assessed the consequences for transcriptional and chemotactic Shh signaling of genetic perturbations resulting in defective ciliary formation. Staining Kif3A−/− MEFs for acetylated α-tubulin showed that these cells did not have an obvious primary cilium (Fig. 3A, B). Although many cells had distinct puncta of acetylated α-tubulin we never observed the rod-like appearance typical for primary cilia. Similar to SmoΔCLD transfected Smo−/− MEFs, Kif3A−/− MEFs were significantly impaired in their transcriptional response to purmorphamine (Fig. 3C). Nevertheless, Kif3A−/− MEFs displayed a chemotactic response to purmorphamine, which exceeded that of wild type MEFs (Fig. 3D). The chemotactic response to purmorphamine was resistant to actinomycin D (and thus transcription-independent), but sensitive to cyclopamine and therefore dependent on Smo. An enhanced chemotactic response of Kif3A−/− MEFs was also found for recombinant ShhN (Fig. 3E) and SAG (Fig. 3F). These results are consistent with the described requirement of the primary cilium for the transcriptional response, but also demonstrate that the intracellular distribution of Smo following ablation of the primary cilium favors Shh chemotaxis apparently at the expense of the transcriptional response.
Fig. 3.
Cells with defective ciliary function show enhanced chemotactic signaling by Smoothened. (A) Primary cilia on Kif3A+/+ and Kif3A−/− MEFs (B) were visualized by immunofluorescence for acetylated α-tubulin. Scale bar, 20 μm. (C) Kif3A MEFs were transfected with Gli-luciferase reporter and stimulated as for Fig. 2A. Shown is the mean ± SEM, n=4. P-value indicated is compared to Kif3A+/+ MEFs. (D) Chemotaxis to purmorphamine or FCS of Kif3A MEFs was assessed and shown as mean fluorescence ± SEM, n=4. Pretreatment with 500 ng/mL actinomycin D or 10 μM cyclopamine was for 10 min. P-value indicated by lines is compared to wild type MEFs. P-value indicated by grey symbols is compared to purmorphamine stimulated (no inhibitors). (E) As for panel D, using recombinant ShhN; (F) using SAG. (G, H) Primary cilia on wild type or Tg737orpk MEFs were visualized (I) Tg737 MEFs were transfected with Gli-luciferase reporter, stimulated, and analyzed as for panel C. (J) As for panel E using Tg737orpk or wild type MEFs. (K) As for panel J, using recombinant ShhN; (L) using SAG.
Tg737orpk is a hypomorphic allele of the gene coding for the Ift88 protein. Cells homozygous for this allele have severely shortened primary cilia (39), and in mouse models, this results in impaired Shh-mediated patterning of the developing neural tube (8, 40). Consistent with these observations we found that immortalized Tg737orpk MEFs have greatly shortened cilia compared to wildtype MEFs (Fig. 3G, H), and showed a reduced transcriptional response to purmorphamine (Fig. 3I). Similar to the Kif3A−/− MEFs, Tg737orpk MEFs were significantly more efficient in Shh chemotaxis than their wildtype counterparts (Fig. 3J–L). The enhanced chemotactic response observed for these mutant MEFs was not to due to a general increase in migratory capacity, as the migration towards FCS was unaffected by the loss of the primary cilium (Fig. 3D, J).
Concluding, we find a remarkable congruence in the biological consequences between the inability of a cell to make a primary cilium, and the inability of Smo to effectively localize to the primary cilium. The cilium-independent mechanism of Shh chemotaxis appears to be negatively affected when Smo can localize to the primary cilium, indicating that these distinct responses are mediated by different fractions of Smo. The recruitment of Smo into the pool that mediates the cilium-dependent transcriptional response is at the expense of the cilium-independent Smo fraction mediating chemotaxis. Based on this observation we tested if introduction of cilia localization-defective Smo into neurons enhanced their ability to extend neurites.
Ciliary localization-impaired Smo enhances neurite outgrowth
Axonal pathfinding and cell migration are related events that share molecular mechanisms and guidance molecules (41). An in vitro model for neurite extension as a proxy for axonal pathfinding comes from embryonic stem (ES) cells, which by virtue of their pluripotency and sensitivity to differentiation clues can mimic in vivo differentiation programs including neural differentiation. In neuralized, ES cell-derived embryoid bodies (EBs), exposure to Shh results in the upregulation of Isl1/2, an established marker for motor neuron (MN) differentiation (42). In addition to assessing Shh-driven differentiation, staining for class III β-tubulin (Tuj-1) enables us to evaluate the projection of neurites in neuralized EBs. We have previously shown that in neuralized EBs, the induction of motor neurons is a consequence of activating the transcriptional Shh response, whereas the Shh-induced projection of neurites from neurons is independent of Gli activity and transcription, and represents a (not necessarily directional) cytoskeletal Shh response (20). A relative measure of neurite network density is obtained by quantifying the distance between crossing neurites. This distance is negatively correlated to network density and allows for quantification of neurite outgrowth in very dense reticula where tracing individual neurite projections is impossible (20). Thus, using neuralized EBs allows us to study both the transcriptional and the cytoskeletal Shh responses simultaneously, as well as to manipulate them independently.
EBs derived from Smo−/− ES cells did not show a robust ventral neuralization in response to retinoic acid (RA) and SAG (42, 43) as measured by neurite length (Fig. 4A–C) or Isl1/2 (Fig. 4D). We were unable to confer additional responsiveness to these cells by transfecting Smo and for further experiments, EBs were derived from Smo+/− ES cells transfected with vector, SmoWT, or SmoΔCLD. Transfected Smo constructs were expressed at similar amounts. Vector-transfected ES cells showed a moderate increase in the length and number of neurite projections in response to SAG (Fig. 4E–G), as well as an increase in motor neuron differentiation (Fig. 4H). Both the induction of differentiation as well as the projection of neurites in response to SAG was enhanced by transfection of SmoWT, confirming that Smo is important for both these responses (Fig. 4I–L). Consistent with the enhanced ability of SmoΔCLD to mediate non-transcriptional Shh signaling in fibroblasts, we found that ES cells transfected with SmoΔCLD showed a very strong induction of neurite outgrowth in response to SAG (Fig. 4M–O). These cells did not show an increase in the number of motor neurons compared to vector transfected EBs (Fig. 4P). This indicates that SmoΔCLD can mediate SAG-induced neurite outgrowth, but cannot enhance the SAG-induced transcriptional response.
Fig. 4.
Smoothened localized outside the primary cilium enhances neurite outgrowth. (A) Smo−/− ES cells were transfected with vector. After 1 days, cells were grown into embryoid bodies (EBs) and after 3 days replated without inducers, or neuralized and ventralized by addition of 1 μM retinoic acid (RA) and 200 nM SAG (B). After an additional 4 days in culture, class III β-tubulin-positive neurites were visualized. (C) Distance between crossing neurites was measured and frequency distribution analysis was performed. Over 100 measurements were made. (D) Induction of motor neurons was quantified by staining for Isl1/2. Number of Isl1/2-positive cells per EB was counted, and shown is mean ± SEM, n≥100 over 2 experiments. Scale bar, 100 μm. (E–H) As for panels A–D, using Smo+/− ES cells transfected with vector; (I–L) SmoWT; (M–P) SmoΔCLD. P-value indicated is compared to vector. (Q) Cells were transfected with SmoΔCLD and EBs were formed in the presence of RA and SAG, and the leukotriene inhibitor MK-886 (μM indicated). After 4d, EBs were stained for Isl1 and class III β-tubulin. Shown is mean ± SEM, n≥50.
We have previously shown that neurite extension and motor neuron differentiation are both Shh-dependent, but can be uncoupled pharmacologically using leukotriene inhibitors (20). We used this same method to confirm that the neurite outgrowth in transfected Smo+/− EBs is uncoupled from differentiation and that they represent independent read-outs for the different Shh responses. Treatment of Smo+/− EBs transfected with SmoΔCLD with MK-886, a 5-lipoxygenase inhibitor specifically disturbed neurite extension while leaving the differentiation of cells intact (Fig. 4Q). This underscores that Isl1 and Tuj-1 stainings specifically represent the transcriptional or the chemotactic Shh response in these EBs. Together, the data show that defective ciliary localization of Smo enhances neurite outgrowth, and at least suggest that Smo localized outside the primary cilium can mediate this process.
DISCUSSION
In the presented work, we describe the impact of subcellular localization of Smo on its signaling ability and unravel a negative correlation between its ciliary localization and its chemotactic signaling capacity. The observation that small mutations in Smo (or the absence of proteins involved in ciliary function) that cause it to no longer localize at the primary cilium cause an enhanced chemotactic response indicates that 1) ciliary localization of Smo is not required for chemotactic signaling through Smo, from which we can infer that 2) much of the decisions on Smo signaling and thus the nature of the Shh response (i.e. chemotactic or transcriptional) are made prior to localization to, or cycliung through, the primary cilium.
Although the cilium is enriched for Smo, it is unlikely to contain the majority of Smo present in a cell. Thus, observing a dramatic enhancement of chemotactic signaling by defective localization of Smo to the primary cilium cannot be explained by the fact that under normal circumstances all the Smo resides in the cilium, unavailable for chemotactic signaling. Instead, it is much more likely that the changes in the nature of Smo signaling output are caused by shifts in subcellular localization independent of the primary cilium. In other words, the consequences of Smo activation are dependent on its subcellular localization. Whereas the trafficking events culminating in localization of activated Smo to the primary cilium favor the Gli-mediated response, localization of activated Smo to other sites in a cell results in local rearrangement of the cytoskeleton underlying chemotaxis.
In addition, the notion that the primary cilium would be the location where Shh chemotaxis is initiated is hard to reconcile with the consideration that pathway components, when concentrated in a single cellular structure like the primary cilium, cannot easily sense Shh concentration differences along the cell surface. For this, distribution of receptors over a larger domain of the cell membrane would be needed, although the migratory response to PDGF has been described to depend on the primary cilium (39). Another argument against the primary cilium mediating Shh chemotaxis comes from the well-established role for the primary cilium in mediating Shh signaling to the downstream transcriptional mediators, which have been shown to be dispensable for Shh chemotaxis (19). Furthermore, transcriptional events in the nucleus are not likely to retain the directional information that is needed for chemotaxis, nor is this signaling compatible with the timescale on which Shh chemotaxis is observed.
The observation that the increased chemotactic signaling capacity of Kif3A−/− MEFs and cells transfected with SmoΔCLD correlates with differential Smo glycosylation at the cell surface suggests that the different itineraries of Smo split either in the late Golgi, or soon thereafter. Although our data do not show if this is causal or merely correlative to the enhanced chemotactic signaling capacity, the fact that we observe partially glycosylated SmoΔCLD and SmoC151Y at the cell surface, where it is predicted to mediate Shh chemotaxis, at least suggests that complete glycosylation of Smo is not strictly required for chemotactic signaling. We have not been able to directly address the requirement for Smo glycosylation in Shh chemotaxis, but the surface biotinylation experiments seem to suggest that this requirement is not absolute.
The transcriptional Shh response appears to be more ‘fragile’ than the chemotactic Shh response. Genetic loss of the primary cilium, as well as forms of Smo that show impaired localization to the cilium, result in an inability to respond to Shh by transcription of target genes. However, in all these conditions the chemotactic response remains intact. This implies that chemotaxis is a more general Shh response, while under more restricted conditions, Shh can be interpreted by Smo at subcellular localizations like the primary cilium resulting in activation of the transcriptional response. It is tempting to speculate that the easy uncoupling of the two responses is employed in vivo to allow Shh chemotaxis or pathfinding without activating the transcriptional response, although the presented study does not formally prove this notion, and further in vivo experiments should establish the relevance of non-ciliary Shh chemotaxis in development. Another fascinating hypothesis is that the chemotactic Shh response precedes the transcriptional response in evolution, explaining its relative robustness.
A role for primary cilia as the location favoring the transcriptional over a non-transcriptional response is reminiscent of the related Wnt pathway, in which the absence of a primary cilium has been shown to favor signaling through the β-catenin-mediated transcriptional pathway (44). Although the mechanisms underlying the switching event are presumably very different from the one described here, it is interesting to see how separate developmental pathways use the primary cilium as a determinant for qualitative differences in the responses to their inducers.
MATERIALS AND METHODS
Materials
Cycloheximide, chloroquine, NDGA, actinomycin D, chloral hydrate, MK-886 and MTT were from Sigma (St. Louis, MO). Cyclopamine was from Biomol (Plymouth Meeting, PA). Purmorphamine and SAG were from EMD Biochemicals (Darmstadt, Germany). Cell Tracker Green CMFDA (5-chloromethylfluorescein diacetate) was from Invitrogen (Carlsbad, CA). Recombinant ShhN was from R&D Systems (Minneapolis, MN).
Constructs
The SmoWT, SmoΔCLD, SmoC151Y in pCS107 constructs were a kind gift from Dr. Reiter (25). SMOM2 and SMOM1 were from Genentech (South San Francisco, CA). pcDNA3.1 vector was obtained from Invitrogen. The Gli-luciferase reporter and the Renilla control were a kind gift from Dr. H. Sasaki (35). pEGFP-N1 and the NF-κB reporter were from Clontech (Mountain View, CA). Zebrafish Smo was in pCS107 (Dr. J. Chen). Fruit fly Smo in pAC (Dr. S. Ogden) was cloned into pCS107 using HindIII and XhoI.
Cell culture
All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal calf serum (FCS, Invitrogen). Shh-LIGHT II cells (ATCC (26)) were grown in medium supplemented with 400 μg/ml neomycin and 150 μg/ml zeocin.
Transfections
All transfections were performed using Effectene (Qiagen, Hilden, Germany). For transfections in 6-well plates, 2 μg DNA was used at a 1:15 ratio of DNA:Effectene. For 12-well plates, 1 μg DNA was used. Cells were incubated with transfection complexes for 16 hours.
Luciferase assay
Cells grown to 70% confluence in a 12-wells plate were transfected as described above with a Firefly Gli-reporter construct and a CMV-driven Renilla luciferase control. Cells were grown to confluence and medium with 0.5% FCS was added. After stimulation, luciferase activity was measured using the Dual-Glo Luciferase Assay System from Promega (Madison, WI). Raw relative luminescence units (RLU) were corrected for their Renilla control.
Modified Boyden chamber migration assay
Cell migration assays were performed as described earlier (11). Cells were labeled with 10 μM CellTracker Green (Invitrogen) according to manufacturer’s protocol. After labeling, cells were detached with 5mM EDTA, and cells were transferred into FluoroBlok Transwell inserts (BD Falcon) at 5×104 cells per insert. The bottom compartments of the Transwell setups contained the chemoattractants of choice. GFP-spectrum fluorescence in the bottom compartment was measured in a Victor3 plate reader (PerkinElmer, Waltham, MA) every 2 min for 99 cycles (approximately 3 hours), after which background fluorescence (medium without cells) and a no-attractant control was subtracted from each time point. Starting points of migration were set to 0. For Smo−/− MEFs, the number of cells per RFU was approximately 5. This gives a number of migrated cells for the SmoWT transfected MEFs to purmorphamine of 1.4×104 (28% of input), whereas 0.8×104 (16% of input) cells migrated to the bottom well in the absence of attractant.
Fluorescence microscopy
Cells grown on cover slips were washed with PBS and fixed with 4% formaldehyde in PBS for 20 min. Tubulin cytoskeleton was stained with 1:1,000 anti-acetylated-α-tubulin (Sigma) in 10% NGS in PBS-T. For transfected Smo, 9B11 anti-Myc (Cell Signaling) was used at 1:5,000. SMOM2 was visualized using anti-gD (LP14 supernatant) at 1:100. anti-Rab-11 antibody (BD Biosciences) was used at 1:500. ID4B anti-LAMP1 (DSHB) was used at 1:100. Appropriate secondary antibodies were used at 1:500 (Invitrogen). After staining, cells were mounted in ProLong Antifade mounting medium (Invitrogen) and examined on a Zeiss Observer Z1 epifluorescence microscope (Wetzlar, Germany). Colocalization was analyzed in ImageJ.
Western blotting
Cells were lysed using LDS sample buffer (Invitrogen) and subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes, blocked with 5% milk in Tris-buffered saline with 0.1% Tween-20 (TBS-T), and incubated in 9B11 anti-Myc at 1:2,000, anti-β-tubulin at 1:2,000, or anti-GFP at 1:1,000. Appropriate HRP-conjugated secondary antibodies were used at 1:5,000. Proteins were visualized using a FujiFilm LAS 3000 imager.
Embryoid body neuralization
EB differentiation was performed according to Wichterle et al. 2002. EBs stained with 1:500 Tuj1 primary antibody (Covance, Princeton, NJ) or anti-Isl1/2 antibody at 1:1,000 and imaged using a fluorescence microscope. Network density was quantified as described previously (20). Neurite length between crossing neurites (nodes) was measured in Zeiss Axiovision software. Frequency distribution analysis of the measured lengths was analyzed using Graphpad Prism.
Neural explants
Dorsal neural tube explants were isolated from stage 21 chick embryos as described previously (45). Explants were transfected 24 hours after isolation, and immunofluorescence was performed 48 hours after the transfection. Transfection, immunofluorescence, and quantifications were performed as described above.
Surface biotinylation
Cells in 100mm dishes were transfected with 1 μg pEGFP-N1 and 3 μg SmoWT, SmoΔCLD, or SmoC151Y in (both in pCS107). Following surface biotinylation and lysis according to the manufacturer’s protocol (Cell Surface Protein Isolation Kit; Pierce, Rockford, IL), a fraction of the lysate was not precipitated on column, but immunoblotted for GFP to correct for transfection efficiency. Precipitated protein was immunoblotted for Smo. For Fig. 1P and Fig. S3F, lysates were treated with 1,500 units of Endo H (New England Biolabs, Ipswich, MA) for 1 hour at 37°C prior to Western blot analysis.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 4.0. The significance of difference was tested by a two-sided Student’s t-test. Indicated p-values are; *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Fig. S1. Smo mutants fail to localize to the primary cilium.
Fig. S2. Rab11 functions to traffic Smo to the primary cilium.
Fig. S3. Intracellular itineraries of SmoC151Y differ from those of SmoWT and SmoΔCLD.
Fig. S4. Activating mutations in Smo do not rescue the defective CLD phenotype.
Fig. S5. Activation of non-ciliary Smo results in activation of Src.
Summary.
The intracellular trafficking machinery that mediates ciliary localization of Smoothened determines if pathway activation results in the transcription of target genes, or chemotaxis.
Acknowledgments
We thank Dr. J. Reiter for the mouse Smo mutants, the Smo antibody, and the Kif3A cells. We also thank Dr. J. Taipale for Smo−/− cells, Dr. F. De Sauvage for SMOM2, Dr. S. Ogden for fruitfly Smo, Dr. J. Chen for zebrafish Smo, Dr. S. Christensen for the Tg737 MEFs, Dr. H. Sasaki for the 3′GliBS reporter, Dr. for the LP14 antibody, and Dr. L. Kwong for helpful discussions.
Funding: This work was supported by R01GM097035 to HR and a KWF Dutch Cancer Society Fellowship to MFB (no grant number).
Footnotes
Author contributions: M.F.B. and H.D. performed the experiments and analyzed the data. M.F.B., H.D. and H.R. designed the experiments. M.F.B. and H.R. wrote the paper.
Competing interests: The authors have no conflict of interest to declare.
References
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 3.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 5.Wilson CW, Chen MH, Chuang PT. Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium. PLoS One. 2009;4:e5182. doi: 10.1371/journal.pone.0005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorivodsky M, Mukhopadhyay M, Wilsch-Braeuninger M, Phillips M, Teufel A, Kim C, Malik N, Huttner W, Westphal H. Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain. Dev Biol. 2009;325:24–32. doi: 10.1016/j.ydbio.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 9.May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 10.Angot E, Loulier K, Nguyen-Ba-Charvet KT, Gadeau AP, Ruat M, Traiffort E. Chemoattractive activity of sonic hedgehog in the adult subventricular zone modulates the number of neural precursors reaching the olfactory bulb. Stem Cells. 2008;26:2311–2320. doi: 10.1634/stemcells.2008-0297. [DOI] [PubMed] [Google Scholar]
- 11.Bijlsma MF, Borensztajn KS, Roelink H, Peppelenbosch MP, Spek CA. Sonic hedgehog induces transcription-independent cytoskeletal rearrangement and migration regulated by arachidonate metabolites. Cell Signal. 2007;19:2596–2604. doi: 10.1016/j.cellsig.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Bourikas D, Pekarik V, Baeriswyl T, Grunditz A, Sadhu R, Nardo M, Stoeckli ET. Sonic hedgehog guides commissural axons along the longitudinal axis of the spinal cord. Nat Neurosci. 2005;8:297–304. doi: 10.1038/nn1396. [DOI] [PubMed] [Google Scholar]
- 13.Charron F, Stein E, Jeong J, McMahon AP, Tessier-Lavigne M. The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell. 2003;113:11–23. doi: 10.1016/s0092-8674(03)00199-5. [DOI] [PubMed] [Google Scholar]
- 14.Kolpak A, Zhang J, Bao ZZ. Sonic hedgehog has a dual effect on the growth of retinal ganglion axons depending on its concentration. J Neurosci. 2005;25:3432–3441. doi: 10.1523/JNEUROSCI.4938-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trousse F, Marti E, Gruss P, Torres M, Bovolenta P. Control of retinal ganglion cell axon growth: a new role for Sonic hedgehog. Development. 2001;128:3927–3936. doi: 10.1242/dev.128.20.3927. [DOI] [PubMed] [Google Scholar]
- 16.Yam PT, Langlois SD, Morin S, Charron F. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron. 2009;62:349–362. doi: 10.1016/j.neuron.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Chinchilla P, Xiao L, Kazanietz MG, Riobo NA. Hedgehog proteins activate pro-angiogenic responses in endothelial cells through non-canonical signaling pathways. Cell Cycle. 2010;9:570–579. doi: 10.4161/cc.9.3.10591. [DOI] [PubMed] [Google Scholar]
- 18.Jenkins D. Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal. 2009;21:1023–1034. doi: 10.1016/j.cellsig.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Lipinski RJ, Bijlsma MF, Gipp JJ, Podhaizer DJ, Bushman W. Establishment and characterization of immortalized Gli-null mouse embryonic fibroblast cell lines. BMC cell biology. 2008;9:49. doi: 10.1186/1471-2121-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bijlsma MF, Peppelenbosch MP, Spek CA, Roelink H. Leukotriene synthesis is required for hedgehog-dependent neurite projection in neuralized embryoid bodies but not for motor neuron differentiation. Stem Cells. 2008;26:1138–1145. doi: 10.1634/stemcells.2007-0841. [DOI] [PubMed] [Google Scholar]
- 21.Incardona JP, Gruenberg J, Roelink H. Sonic hedgehog induces the segregation of patched and smoothened in endosomes. Curr Biol. 2002;12:983–995. doi: 10.1016/s0960-9822(02)00895-3. [DOI] [PubMed] [Google Scholar]
- 22.Incardona JP, Lee JH, Robertson CP, Enga K, Kapur RP, Roelink H. Receptor-mediated endocytosis of soluble and membrane-tethered sonic hedgehog by patched-1. Proc Natl Acad Sci U S A. 2000;97:12044–12049. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milenkovic L, Scott MP, Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci U S A. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aanstad P, Santos N, Corbit KC, Scherz PJ, Trinh le A, Salvenmoser W, Huisken J, Reiter JF, Stainier DY. The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr Biol. 2009;19:1034–1039. doi: 10.1016/j.cub.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 27.Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr, de Sauvage FJ. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 28.Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, Schneider-Maunoury S, Alvarez-Buylla A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 29.Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, Ros MR, Garcia-Verdugo JM, Alvarez-Buylla A. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Duve C, de Barsy T, Poole B, Trouet A, Tulkens P, Van Hoof F. Lysosomotropic agents. Biochem Pharmacol. 1974;23:2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- 31.Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem. 2004;279:33430–33437. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- 32.Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, Sheffield VC, Scheller RH, Jackson PK. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci U S A. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha S, Chen JK. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem Biol. 2006;2:29–30. doi: 10.1038/nchembio753. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Walker J, Zhang J, Ding S, Schultz PG. Purmorphamine induces osteogenesis by activation of the hedgehog signaling pathway. Chem Biol. 2004;11:1229–1238. doi: 10.1016/j.chembiol.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polizio AH, Chinchilla P, Chen L, Kim S, Manning DR, Riobo NA. Heterotrimeric Gi Proteins link Hedgehog signaling to activation of Rho small GTPases to promote fibroblast migration. J Biol Chem. 2011 doi: 10.1074/jbc.M110.197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 41.de Castro F. Chemotropic molecules: guides for axonal pathfinding and cell migration during CNS development. News Physiol Sci. 2003;18:130–136. doi: 10.1152/nips.01414.2002. [DOI] [PubMed] [Google Scholar]
- 42.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 43.Frank-Kamenetsky M, Zhang XM, Bottega S, Guicherit O, Wichterle H, Dudek H, Bumcrot D, Wang FY, Jones S, Shulok J, Rubin LL, Porter JA. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 45.Yamada T, Pfaff SL, Edlund T, Jessell TM. Control of cell pattern in the neural tube: motor neuron induction by diffusible factors from notochord and floor plate. Cell. 1993;73:673–686. doi: 10.1016/0092-8674(93)90248-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Smo mutants fail to localize to the primary cilium.
Fig. S2. Rab11 functions to traffic Smo to the primary cilium.
Fig. S3. Intracellular itineraries of SmoC151Y differ from those of SmoWT and SmoΔCLD.
Fig. S4. Activating mutations in Smo do not rescue the defective CLD phenotype.
Fig. S5. Activation of non-ciliary Smo results in activation of Src.