Abstract
Background
Glomerular podocytes are highly differentiated cells that are key components of the kidney filtration units. The podocyte cytoskeleton builds the basis for the dynamic podocyte cytoarchitecture and plays a central role for proper podocyte function. Recent studies implicate that immunosuppressive agents including the mTOR-inhibitor everolimus have a protective role directly on the stability of the podocyte actin cytoskeleton. In contrast, a potential stabilization of microtubules by everolimus has not been studied so far.
Methods
To elucidate mechanisms underlying mTOR-inhibitor mediated cytoskeletal rearrangements, we carried out microarray gene expression studies to identify target genes and corresponding pathways in response to everolimus. We analyzed the effect of everolimus in a puromycin aminonucleoside experimental in vitro model of podocyte injury.
Results
Upon treatment with puromycin aminonucleoside, microarray analysis revealed gene clusters involved in cytoskeletal reorganization, cell adhesion, migration and extracellular matrix composition to be affected. Everolimus was capable of protecting podocytes from injury, both on transcriptional and protein level. Rescued genes included tubulin beta 2B class IIb (TUBB2B) and doublecortin domain containing 2 (DCDC2), both involved in microtubule structure formation in neuronal cells but not identified in podocytes so far. Validating gene expression data, Western-blot analysis in cultured podocytes demonstrated an increase of TUBB2B and DCDC2 protein after everolimus treatment, and immunohistochemistry in healthy control kidneys confirmed a podocyte-specific expression. Interestingly, Tubb2b brdp/brdp mice revealed a delay in glomerular podocyte development as showed by podocyte-specific markers Wilm’s tumour 1, Podocin, Nephrin and Synaptopodin.
Conclusions
Taken together, our study suggests that off-target, non-immune mediated effects of the mTOR-inhibitor everolimus on the podocyte cytoskeleton might involve regulation of microtubules, revealing a potential novel role of TUBB2B and DCDC2 in glomerular podocyte development.
Introduction
Podocytes are highly differentiated renal glomerular visceral epithelial cells that cover the outer layer of the glomerular basement membrane playing a crucial role in the regulation of glomerular function [1]. These specialized cells show a complex cellular organization consisting of a cell body, thick primary foot processes, and thin secondary foot processes, which are linked by the glomerular slit diaphragms (SDs) [2]. The sophisticated cell shape of podocytes is maintained by the coordinated intracellular filamentous network of cytoskeletal elements, including microtubules (MTs), intermediate filaments (IFs) and actin filaments (AFs). Physiological podocyte function mainly depends on the dynamic regulation of complex cellular structures, in particular the foot processes. MTs and the actin cytoskeleton seem to coordinately control formation of podocyte foot processes [3]. In particular, MT-rich primary foot processes extending from the cell body split into secondary foot processes containing a dynamic actin meshwork that interacts with the secondary foot processes of neighboring podocytes via Nephrin-linked SDs [3]. Over the past few years, the importance of cytoskeletal components for proper podocyte morphology and glomerular function has emerged from a body of functional data. Genetic studies in glomerular disorders identified several mutated genes encoding proteins associated with the podocyte cytoskeleton such as Nephrin, Podocin, CD2AP, Synaptopodin, alpha-Actinin-4, Inverted formin 2 and TRPC6 [4]. Nonetheless, molecular mechanisms regulating podocyte foot process formation are still poorly understood. Among various intracellular signals, multiple actin based cytoskeletal responses have been established to be mediated by the Rho family small GTPases [5]. In particular, Rac1 and Cdc42 stimulate dynamic protrusions, whereas RhoA together with its effector ROCK control formation of contractile actin-myosin stress fibers [6]. Interestingly, our recent publication revealed that this pathway also mediates the cytoskeletal stabilizing effects of the mTOR inhibitor everolimus (EV) [7].
In contrast to the actin cytoskeleton, the role of MTs in podocyte architecture is still insufficiently studied. Taking advantages of an immortalized murine cell line, Kobayashi and colleagues have previously shown that morphogenesis of podocytes requires proper assembly of MTs as well as their transport by a MT-based motor protein, and is regulated by the extracellular matrix [8,9]. Emerging data from multiple different cell systems suggest a reciprocal crosstalk between the actin regulatory signal transduction pathways and MT-dynamics [10,11]. First, MT polymerization has been associated with activation of the Rho GTPase Rac1 promoting dynamic cell protrusions called lamellipodia [12]. On the other hand, MT de-polymerization activates the RhoA-ROCK pathway via release of the guanine nucleotide exchange factor GEF-H1 [13,14]. In turn, RhoA mediated stress fiber contractility is critically involved in the dynamics of cell-substrate contacts which themselves have been suggested to capture MTs in distinct cellular regions [15,16]. Despite the fact that morphogenesis of podocytes strongly depends on MTs [8,17,18], it is not well understood, how MTs might be coordinated with the actin cytoskeleton to control podocyte behavior and how MT dynamics is affected during podocyte injury.
In the present study we performed microarray analysis using cultured human podocytes treated with EV in a puromycin aminonucleoside (PAN) experimental model of podocyte injury in order to define genes that are strongly associated with cytoskeletal damage. We revealed strong association of tubulin beta 2B class IIb (TUBB2B) and doublecortin domain containing 2 (DCDC2) with proper podocyte function, two proteins involved in MT formation in neuronal cells but not identified in podocytes so far. We provide direct evidence that PAN-induced disorganization of the actin cytoskeleton and microtubules is associated with downregulation of TUBB2B and DCDC2 in human cultured podocytes and podocyte dysfunction. EV ameliorated podocyte damage by preventing these deregulations. Recently, Stottmann and coworkers described neurodevelopmental defects in mice with a homozygous missense mutation in Tubb2b. These homozygous mutants do not survive past birth. They have a profoundly affected brain with significant cortical thinning, largely due to massively increased cell death and in addition show an abnormal proliferation of the basal progenitors [19]. Kidneys of these mutant mice have not been studied so far. Here, we present first data demonstrating a specific expression of TUBB2b in podocytes and a significant delay in glomerular podocyte development in Tubb2b brdp/brdp mice underscoring a role for TUBB2B in proper podocyte function.
Materials and Methods
Cell culture
Conditionally immortalized human podocytes were generated by Prof. Dr. Moin A. Saleem (University of Bristol, South Mead Hospital, Bristol, UK) [20]. Culture conditions were described previously [20].
Animals and kidney preparation
Tubb2b mice were maintained as previously indicated [19]. Kidneys were dissected at embryonic day (E) 18.5, 1 day before birth, because homozygous mutant mice did not survive past birth. Tissues were formalin-fixed and paraffin-embedded for immunocytochemistry or glutaraldehyde-fixed for transmission electron microscopy. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the Cincinnati Children’s Hospital Medical Center (protocol numbers 1D05052 and IACUC2013-0068). All euthanasia and embryo harvests were performed after isoflurane sedation to minimize animal suffering and discomfort. Animal euthanasia was via cervical dislocation and thoracotomy.
Human tissue samples
Formalin-fixed, paraffin-embedded and snap-frozen specimens from normal control kidney sections were included in this study. Kidneys were obtained from medical nephrectomy specimens. Written informed consent from the donors were obtained for the use of these samples in research. Kidney tissues were used following the guidelines of the local ethics committee of the Medical Faculty of Essen, Germany (protocol number 09–3954), which specifically approved this study. The tissue samples were not procured from a tissue bank or donation center.
Experimental design and drug treatment
Podocytes were treated with EV in a PAN-model of podocyte injury as described previously [7].
RNA isolation from cells
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions including DNase digestion.
RNA and protein isolation from snap-frozen tissues
Total RNA and protein were isolated using the AllPrep DNA / RNA / Protein Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations using the TissueLyser (Qiagen, Hilden, Germany) for homogenization and including DNase digestion for RNA-isolation.
Real-time RT-PCR analysis
1–2 μg of total RNA was reverse-transcribed with random hexamers and SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s instructions.
Quantitative real-time RT-PCR (qPCR) was performed in triplets in 20 μl with 20 ng cDNA, 10 μl SYBR Green JumpStart Taq ready Mix (Sigma-Aldrich, Munich, Germany) and 2.5 pmol of sequence specific primers obtained from biomers.net GmbH, Ulm, Germany. qPCR was performed with StepOnePlus engine (Applied Biosystems, Darmstadt, Germany) and the following conditions: 95°C for 2 min; 40 cycles: 95°C for 30 s; 60°C for 1 min; 72°C for 1 min; followed by melting curve analysis.
qPCR for the endogenous control 18S rRNA was performed according to the manufacturer’s instructions using the TaqMan Gene Expression Assay Hs99999901_s1 (Applied Biosystems, Darmstadt, Germany) in combination with the TaqMan Fast Universal PCR Master Mix (Applied Biosystems, Darmstadt, Germany).
Relative gene expression was calculated by the 2−ΔΔCT method. Primer sequences are listed in Table 1.
Table 1. Primer used for qPCR.
| Gene Symbol | Gene Title | Sequence forward 5'-3' | Sequence reverse 5'-3' |
|---|---|---|---|
| COL4A3 | Collagen, type IV, alpha 3 | GGATTGCCAGGATTTTCTGGT | TGGTACACCGACAAGTCCGTA |
| DCDC2 | Doublecortin domain containing 2 | CCAGTCGATCAGAGGCCAG | CTTCAAGGTCACCATTCATTCCT |
| FN1 | Fibronectin 1 | CGGTGGCTGTCAGTCAAAG | AAACCTCGGCTTCCTCCATAA |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | ACAACTTTGGTATCGTGGAAGG | GCCATCACGCCACAGTTTC |
| LPXN | Leupaxin | CCAACGACTACCACCAACTTT | GCCAGGTCTGGTTCATTGCT |
| MMP24 | Matrix metallopeptidase 24 | GCCGGGCAGAACTGGTTAAA | CCCGTAAAACTGCTGCATAGT |
| MYLIP | Myosin regulatory light chain interacting protein | GCAGGCGACTGGGAATCATAG | CGGTTTCTCAGGTTTAGCCAT |
| MYO5C | Myosin VC | TCGTGGGCGAGAATGACCT | GGCAACTGCTTGTAAGGATTCA |
| NRG1 | Neuregulin 1 | AGAGCCTGTTAAGAAACTCGC | GTCCACTTCCAATCTGTTAGCA |
| PCDH10 | Protocadherin 10 | TGGATGGTGGAAGGAGTCTTT | TTCAGCGATATTCCCCACGAA |
| PCDH17 | Protocadherin 17 | TGATTGACTCCAACGACAACAG | TGAAAGAGTAGAGCACTTCACCA |
| PCDH9 | Protocadherin 9 | AACAGATCCTGACACAGGCTT | TCCAGTCCAAAAACACTCTGC |
| SYNPO2 | Synaptopodin 2 | CTCGCCCCTGTCAAGACTG | CCAGGCTGTACCGCTTCTA |
| TGFB1I1 | Transforming growth factor beta 1 induced transcript 1 | TACAGCACGGTATGCAAGCC | GCAACCGATCTAGCTCACAGAG |
| THBS1 | Thrombospondin 1 | TGCTATCACAACGGAGTTCAGT | GCAGGACACCTTTTTGCAGATG |
| TUBA1A | Tubulin, alpha 1a | TCGATATTGAGCGTCCAACCT | CAAAGGCACGTTTGGCATACA |
| TUBB | Tubulin, beta class I | ACCAACCTACGGGGATCTGAA | TTGACTGCCAACTTGCGGA |
| TUBB2B | tubulin, beta 2B class 2b | GGCACGATGGATTCGGTTAGG | ACACGAAATTGTCTGGTCTGAAG |
Affymetrix expression analysis
Target preparation was performed with 200ng RNA essentially as described in the GeneChip 3’ IVT Express Kit user manual (Affymetrix, Santa Clara, USA). Hybridization, washing and staining of the arrays were done according to the GeneChip Hybridization, Wash, and Stain Kit on the GeneChip Scanner 3000 with G7 update.
Following fragmentation, cRNA was hybridized on Affymetrix GeneChips Human Genome U133_Plus_2.0. GeneChips were washed and stained in the Affymetrix Fluidics Station 450 using the Affymetrix Hybridization, Wash and Stain Kit. Hybridization signals were detected with Affymetrix Gene Chip Scanner 3000 and global scaling was performed by Affymetrix GeneChipOperatingSoftware 1.4 using the MAS5 algorithm. Microarray data of all groups have been published in Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO accession number GSE66107.
For data interpretation we only took those gene products into account that provided present hybridization signals in at least one of the two groups compared. Furthermore, only gene products (probe sets) with fold changes (FC) ≤0.5 or ≥2.0 were considered as significantly, differentially expressed.
To identify differentially expressed targets, we performed an ANOVA test study, in which experimental samples were analyzed against the respective baseline (control) samples using the Partek GS ANOVA algorithm including multiple testing correction.
Western-blot analysis
Cells were harvested using CelLytic MT-buffer (Sigma-Aldrich, Munich, Germany) according to the manufacturer’s instructions. Lysis-buffer was supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich, Munich, Germany), 10 μg / ml aprotinin (Roche, Mannheim, Germany), 10 μg / ml leupeptin (Roche, Mannheim, Germany) and 2 mM phenylmethanesulfonylfluoride (PMSF, Sigma, Munich, Germany). Isolation was performed at 4°C. Total protein content was measured by Bio-Rad protein assay (Bio.Rad, Munich, Germany). Samples (each > 10 μg protein) were supplemented with Laemmli sample buffer (Bio-Rad, Munich, Germany) and boiled for 10 min at 95°C.
Protein extracts from tissues were obtained as described above. 20 μl of each sample in buffer ALO was boiled for 10 min at 95°C.
Proteins were separated using 10% or 12% Mini-PROTEAN TGX Precast Gels (Bio-Rad, Munich, Germany) and transferred on 0.45 μm PVDF Transfer Membranes (Thermo Scientific, Schwerte, Germany) with a MiniProtean Tetra Cell electrophoresis system (Bio-Rad, Munich, Germany) and a Biometra fastblot B34 blotting device (Biometra, Göttingen, Germany). 15 μl Precision Plus Protein All Blue Standard (Bio-Rad, Munich, Germany) were used as marker. Membranes were incubated with primary antibodies against TUBB2B (1:500; AM09375PU-N, Acris, Herford, Germany), THBS1 (1:500; SAB1401385, Sigma, Munich, Germany), DCDC2 (1:500; SAB2500298, Sigma, Munich, Germany), DCDC2 (1:250; HPA031582, Sigma, Munich, Germany), COL4A3 (1:500; HPA042064, Sigma, Munich, Germany), SYNPO2 (1:500; SAB3500586, Sigma, Munich, Germany), PCDH9 (1:500; HPA015581, Sigma, Munich, Germany) or GAPDH (1:10000; G8795, Sigma, Munich, Germany). Secondary antibodies used were horseradish peroxidase-conjugated goat anti-rabbit IgG (Santa Cruz, Heidelberg, Germany; 1:10000 against COL4A3, SYNPO2 and PCDH9) donkey anti-goat IgG (Santa Cruz, Heidelberg, Germany; 1:10000 against DCDC2) and goat anti-mouse IgG (Santa Cruz, Heidelberg, Germany; 1:20000 against GAPDH, 1:10000 against TUBB2B and THBS1). Signal detection was performed with SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific, Schwerte, Germany) and visualized by the FUSION FX7 chemiluminescence-system (PEQLAB, Erlangen, Germany) und Fusion-software (PEQLAB, Erlangen, Germany). Intensity of signals was determined using ImageJ software (http://rsbweb.nih.gov/ij/index.html). Densitometric data were normalized to GAPDH-loading control.
Immunofluorescence and cell imaging of podocytes in vitro
For immunofluorescence, podocytes were plated on glass coverslips. After treatment, cells were fixed with 100% icecold methanol (Sigma, Munich, Germany) for 10 min at 4°C, washed with PBS and permeabilized with PBS / 0.5% Triton X-100 / 3% BSA / 3% chicken serum / 3% goat serum for 45 min at room temperature. After washing with blocking buffer (PBS / 0.5% BSA), cells were incubated with the appropriate antibody dilutions. Primary antibodies used were: alpha-Tubulin, rabbit polyclonal antibody ab18251 (1:100; Abcam, Cambridge, UK); TUBB2B, mouse polyclonal antibody AM09375PU-N (1:100; Acris, Herford, Germany) and DCDC2, goat polyclonal antibody SAB2500298 (1:100; Sigma, Munich, Germany).
For staining, cells were incubated with the primary antibody at 4°C overnight followed by the corresponding Alexa Fluor 488 / 594 chicken anti-rabbit IgG (H+L) (alpha-Tubulin), Alexa Fluor 488 / 594 goat anti-mouse IgG (H+L) (TUBB2B) or Alexa Fluor 488 / 594 chicken anti-goat IgG (H+L) (DCDC2) (all 1:1000, Invitrogen, Karlsruhe, Germany) in blocking buffer for 1 h at room temperature. In parallel, nuclei were stained with 4.6-diamidino-2-phenylindole dihydrochloride (DAPI, Sigma, Munich, Germany). For actin staining cells were incubated with phalloidin-TRITC (1:1000, Sigma, Munich, Germany).
Negative controls were performed without primary antibodies.
Fluorescence imaging was performed on a fully automated Nikon ECLIPSE Ti microscope controlled by NIS Elements AR 3.2 software (Nikon, Düsseldorf, Germany) equipped with a CoolSNAP HQ2 camera (Photometrics, Tucson, Arizona). Images were acquired using a 20x phase contrast objective with appropriate filter sets. Image processing and analysis was performed with ImageJ (http://rsbweb.nih.gov/ij/index.html) and Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Immunohistochemistry staining of tissues
Immunohistochemistry on formalin-fixed, paraffin-embedded tissues was performed using the Labeled StreptAvidin Biotin kit from DAKO according to the manufacturer’s recommendations. Briefly, paraffin-embedded tissues were sectioned at 5 μm. Sections were deparaffinized in xylene and rehydrated in graded ethanols. After blocking endogenous peroxidase by incubation in 3% hydrogen peroxide (DAKO, Hamburg, Germany) antigen demasking procedure was done with Heat Induced Epitope Retrieval (HIER) in Target retrieval solution pH 6 (DAKO, Hamburg, Germany), using a pressure boiler as heat source for 10 min (TUBB2B, DCDC2, NPHS2, NPHS1 and SYNPO) or with 1 mg / ml protease at 38°C for 10 min (WT1). After blocking of unspecific sites (0,1% Avidin, 0,01% Biotin and Protein Block Serum-free; all obtained from DAKO, Hamburg, Germany), sections were stained with the first antibody for 2 h diluted in Antibody Diluent with Background Reducing Components (DAKO, Hamburg, Germany) followed by incubation with the biotinylated secondary antibody (30 min; DAKO, Hamburg, Germany) and the streptavidin-peroxidase reagent (30 min, DAKO, Hamburg, Germany) and finally 3,3'-diaminobenzidine (DAB; DAKO, Hamburg, Germany) used as the chromogen. Sections were counterstained with Mayer’s Hematoxylin (DAKO, Hamburg, Germany), mounted with Immu-Mount (Thermo Scientific, Schwerte, Germany), and coverslipped. Negative controls consisted of consecutive tissue sections of each case in which the primary antibody was omitted.
Primary antibodies were as follows: TUBB2B, mouse polyclonal antibody AM09375PU-N (1:100; Acris, Herford, Germany); DCDC2, rabbit polyclonal antibody HPA031582 (1:200; Sigma, Munich, Germany); WT1, mouse monoclonal antibody Clone 6F-H2 (1:50; DAKO, Hamburg, Germany); NPHS2, rabbit polyclonal antibody ab93650 (1:200; Abcam, Cambringe, UK); NPHS1, rabbit polyclonal antibody SAB3500770 (1:200; Sigma, Munich, Germany) and SYNPO, goat polyclonal antibody sc-21537 (1:100; Santa Cruz, Heidelberg, Germany).
In addition, periodic acid-Schiff (PAS) staining (Sigma, Munich, Germany) was performed according to the manufacturer’s instructions.
Imaging was performed on a Leica DM RBE microscope (Leica Microsystems, Wetzlar, Germany) equipped with an Olympus DP73 camera (Olympus, Hamburg, Germany) using cellSens Dimension 1.7 software (Olympus Soft Imaging Solutions, Münster, Germany). Images were acquired with a 10x, 20x or 40x PL APO objective. Image processing and analysis was performed using ImageJ (http://rsbweb.nih.gov/ij/index.html) and Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Tissue Sampling and Processing for Electron Microscopy
Upon initial dissection at embryonic day (E) 18.5, 1 day before birth, mice kidneys were fixed at room temperature in 2.5% glutaraldehyde in PBS. Repositioning was performed in 2.5% glutaraldehyde in 0.19 M cacodylate buffer at pH 7.4. Samples were postfixed in 1% Osmium tetroxide in 0.19 M cacodylate buffer, pH 7.4, for 2 hours. Tissues were stained en bloc in 2% uranyl acetate in 0.05 M maleate buffer, pH 4.6. Specimens then were dehydrated in graded ethanols and embedded in epoxy resin [21]. Toluidine-blue stained semithin sections were used to search for glomeruli. Ultrathin sections were picked up onto Formvar carbon–coated grids, stained with lead citrate, and analyzed with a transmission electron microscope (TEM 910; Zeiss, Oberkochen, Germany) equipped with a cooled CCD digital camera (TRS, Moorenweis, Germany). During TEM studies, 4–11 glomeruli were captured randomly in each of two different wild types and Tubb2b brdp/brdp kidneys.
Statistical analysis
Values from multiple experiments were expressed as means ± SD. Statistical analysis was assessed using Student’s T-Test. Statistical significance was defined as p < 0.05.
Results
EV treatment modulates the transcriptome of injured human podocytes
To identify molecules involved in mechanisms underlying EV-mediated protective activity during podocyte injury, we first generated microarray expression data from MeOH (solvent for EV), EV, PAN+MeOH and PAN+EV treated human differentiated podocytes. The Affymetrix GeneChip Human Genome U133_Plus_2.0 was used for transcriptomic analyses with three biological replicates for each treatment. Overall, PAN treatment (PAN+MeOH) caused differential regulation of a substantial number of genes as compared to solvent control (MeOH). Expression of a large portion of these transcripts (195 differentially expressed genes) was partially rescued in the combination with EV (PAN+EV) with more than two-fold changes between the two given treatments (S1 Table). We identified 73 transcripts decreased after PAN treatment and increased in the combination with EV at the same time as well as 122 transcripts increased in PAN+MeOH and decreased in PAN+EV (fold change > 2.0; p-value < 0.05). Critically, among those transcripts, an accumulation of genes involved in the regulation of microtubules was detected.
Table 2 lists selected results from the microarray analysis of podocyte gene expression in PAN+MeOH treated cells compared to solvent control (MeOH) and PAN+EV samples compared to PAN+MeOH. The top of Table 2 contains genes with a significant decrease in expression following PAN+MeOH treatment when compared with solvent only (MeOH). Interestingly, expression of several genes in this group is substantially elevated upon treatment with PAN in combination with EV (PAN+EV). Several of those genes encode proteins playing important roles in cytoskeletal organization and stability, including TUBB2B, DCDC2, MYO5C, TUBB and TUBA1A. Others that were elevated with EV encode proteins involved in cell-cell adhesion (THBS1, TGFB1I1, NRG1, FN1), migration (DCDC2, MMP24, FN1) and extracellular matrix composition (COL4A3). Interestingly, the expression of the three major components of MTs tubulin beta class I (TUBB), tubulin beta 2B class IIb (TUBB2B) and tubulin alpha 1a (TUBA1A) was decreased in PAN+MeOH samples as compared to MeOH alone but substantially increased in the presence of EV. The bottom of Table 2 depicts genes that were up-regulated with PAN+MeOH and significant down-regulated in PAN+EV treated cells. In particular, we found accumulation of genes that encode proteins associated with cytoskeletal organization and stability such as SYNPO2 and MYLIP as well as cell adhesion including LPXN, PCDH9, PCDH10 and PCDH17. Strikingly, the expression of PCDH9, -10 and -17 (protocadherin 9, -10, -17) was increased up to tenfold when treated with PAN+MeOH as compared to MeOH alone and strongly decreased in combination with EV, the largest fold-changes measured. These genes are known to encode adhesion proteins that mediate cell-cell adhesion in neural tissues [22,23]. Thus, such strong differential regulation of expression levels suggests a potential critical role in podocyte adhesion as well.
Table 2. Affymetrix gene expression data selected according to function.
| Probeset | Gene Symbol | Gene Title | PAN+MeOH vs. MeOH | PAN+EV vs. PAN+MeOH | Function | ||
|---|---|---|---|---|---|---|---|
| p < 0.05 | FC > 1.5 | p < 0.05 | FC > 1.5 | ||||
| Decreased in PAN+MeOH / Increased in PAN+EV | |||||||
| 214023_x_at | TUBB2B | tubulin, beta 2B | 1.83E-08 | -19.00 | 1.45E-05 | 4.23 | cytoskeletal organization / stability |
| 201109_s_at | THBS1 | thrombospondin 1 | 3.98E-07 | -13.51 | 1.17E-03 | 2.53 | adhesion |
| 222925_at | DCDC2 | doublecortin domain containing 2 | 3.71E-09 | -6.59 | 4.71E-06 | 2.28 | cytoskeletal organization / stability, migration |
| 218966_at | MYO5C | myosin VC | 3.05E-07 | -5.80 | 2.65E-04 | 2.25 | cytoskeletal organization / stability |
| 212320_at | TUBB | tubulin, beta | 6.07E-09 | -5.47 | 8.81E-06 | 2.05 | cytoskeletal organization / stability |
| 209651_at | TGFB1I1 | transforming growth factor beta 1 induced transcript 1 | 2.95E-08 | -5.06 | 4.58E-05 | 1.95 | adhesion |
| 222073_at | COL4A3 | collagen, type IV, alpha 3 | 3.06E-06 | -4.80 | 1.67E-04 | 3.35 | extracellular matrix composition |
| 206343_s_at | NRG1 | neuregulin 1 | 6.12E-07 | -3.18 | 9.39E-05 | 2.14 | adhesion |
| 209118_s_at | TUBA1A | tubulin, alpha 1a | 7.33E-09 | -2.86 | 2.52E-06 | 1.98 | cytoskeletal organization / stability |
| 213171_s_at | MMP24 | matrix metallopeptidase 24 (membrane-inserted) | 3.36E-06 | -2.42 | 1.41E-03 | 1.56 | migration |
| 214702_at | FN1 | fibronectin 1 | 2.14E-05 | -1.78 | 1.23E-03 | 1.50 | adhesion, migration |
| Increased in PAN+MeOH / Decreased in PAN+EV | |||||||
| 227662_at | SYNPO2 | synaptopodin 2 | 6.66E-05 | 2.11 | 9.69E-05 | -2.96 | cytoskeletal organization / stability |
| 216250_s_at | LPXN | leupaxin | 3.30E-06 | 2.40 | 8.17E-05 | -2.23 | adhesion, migration |
| 223130_s_at | MYLIP | myosin regulatory light chain interacting protein | 3.77E-08 | 4.86 | 1.18E-05 | -2.57 | cytoskeletal organization / stability |
| 219737_s_at | PCDH9 | protocadherin 9 | 5.41E-08 | 9.46 | 6.24E-06 | -5.69 | adhesion |
| 228635_at | PCDH10 | protocadherin 10 | 1.06E-06 | 13.92 | 2.12E-04 | -4.94 | adhesion |
| 228863_at | PCDH17 | protocadherin 17 | 2.30E-08 | 17.66 | 2.61E-06 | -10.52 | adhesion |
Genes were selected based on their putative roles their products may play in stabilizing podocyte cytoskeleton or adhesion and on the intensity of the EV rescue effect. PAN = puromycin aminonucleoside. EV = everolimus. MeOH = methanol, solvent for EV. p = p-value. FC = fold change.
Quantitative RT-PCR (qPCR) was initially used to verify selected microarray results. We selected all 17 genes of Table 2 on the basis of their possible contribution to the functional endpoint of the study (i.e. increased cytoskeletal stability and cell adhesion with EV). These included 11 upregulated and 6 downregulated genes in the presence of EV. Table 3 lists the qPCR results with PAN+EV treated cells compared to PAN+MeOH. In agreement with the microarray, 16 genes were differentially regulated as measured with qPCR from which 12 reached statistical significance.
Table 3. qPCR analysis of human podocyte gene expression: Verification of selected microarray results.
| Gene Symbol | Gene Title | PAN+MeOH vs. MeOH | PAN+EV vs. PAN+MeOH | Function | ||||
|---|---|---|---|---|---|---|---|---|
| FC | p-value | FC Array* | FC | p-value | FC Array* | |||
| COL4A3 | Collagen, type IV, alpha 3 | -1.3 | ns | -4.8 | 2.6 | 0.002 | 3.3 | extracellular matrix composition |
| DCDC2 | Doublecortin domain containing 2 | -3.9 | 0.001 | -6.6 | 2.4 | 0.004 | 2.3 | cytoskeletal organization / stability, migration |
| FN1 | Fibronectin 1 | -2.0 | 0.03 | -1.8 | -1.0 | ns | 1.5 | adhesion, migration |
| LPXN | Leupaxin | 5.6 | 0.01 | 2.4 | -2.2 | 0.03 | -2.2 | adhesion, migration |
| MMP24 | Matrix metallopeptidase 24 | -1.6 | 0.01 | -2.4 | 3.0 | ns | 1.6 | migration |
| MYLIP | Myosin regulatory light chain interacting protein | 15.7 | 0.01 | 5.3 | -2.3 | 0.04 | -2.6 | cytoskeletal organization / stability |
| MYO5C | Myosin VC | -4.1 | 0.002 | -5.8 | 2.8 | 0.009 | 2.3 | cytoskeletal organization / stability |
| NRG1 | Neuregulin 1 | -1.6 | ns | -3.2 | 1.2 | ns | 2.1 | adhesion |
| PCDH10 | Protocadherin 10 | 71.1 | 0.009 | 13.9 | -4.6 | 0.01 | -4.9 | adhesion |
| PCDH17 | Protocadherin 17 | 371.4 | 0.005 | 17.7 | -12.0 | 0.006 | -10.5 | adhesion |
| PCDH9 | Protocadherin 9 | 32.7 | 0.02 | 9.5 | -6.7 | 0.02 | -5.7 | adhesion |
| SYNPO2 | Synaptopodin 2 | 1.9 | ns | 2.1 | -2.4 | 0.04 | -3.0 | cytoskeletal organization / stability |
| TGFB1I1 | Transforming growth factor beta 1 induced transcript 1 | -1.7 | 0.05 | -5.1 | 2.9 | 0.008 | 2.0 | adhesion |
| THBS1 | Thrombospondin 1 | -10.8 | 0.0003 | -13.5 | 2.5 | 0.03 | 2.5 | adhesion |
| TUBA1A | Tubulin, alpha 1a | -3.8 | 0.0001 | -2.9 | 1.6 | 0.03 | 2.0 | cytoskeletal organization / stability |
| TUBB | Tubulin, beta class I | -3.9 | 0.0007 | -5.5 | 2.2 | 0.004 | 2.0 | cytoskeletal organization / stability |
| TUBB2B | tubulin, beta 2B class 2b | -13.5 | 0.00008 | -19.0 | 5.8 | 0.003 | 4.2 | cytoskeletal organization / stability |
PAN = puromycin aminonucleoside. EV = everolimus. MeOH = methanol, solvent for EV. p = p-value. FC = fold change.
* = p < 0.05.
EV protects components of cytoskeletal stability and podocyte adhesion
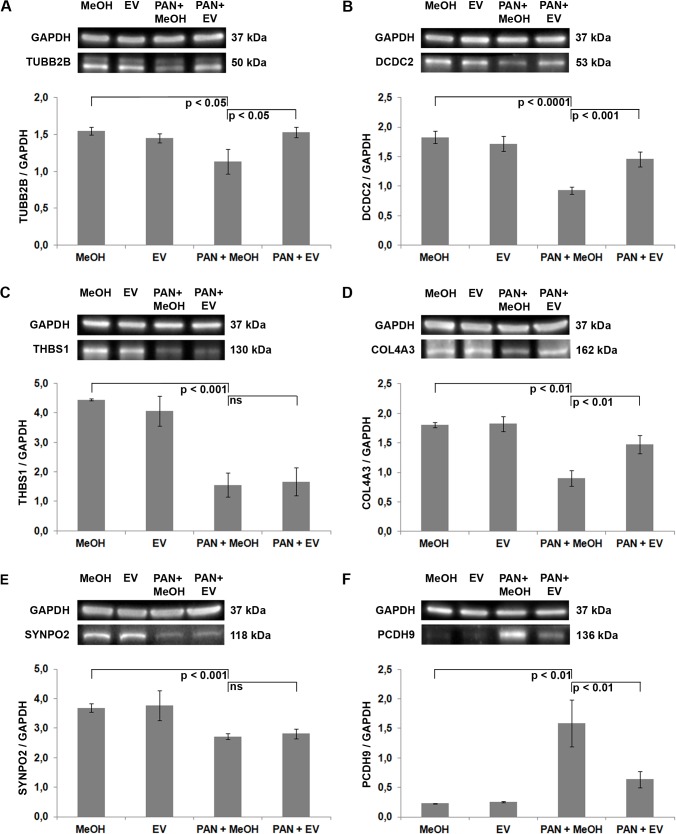
To further confirm the results of the microarray analysis, we performed Western-blot analysis of TUBB2B, DCDC2, THBS1, COL4A3, SYNPO2 and PCDH9 in cultured podocytes selected due to their possible roles in podocytes (Fig 1). We measured the levels of these differentially expressed gene products after PAN treatment and in the presence of EV. PAN dramatically reduced the amount of TUBB2B, DCDC2, THBS1, COL4A3 and SYNPO2 protein (Fig 1A–1E). Addition of EV significantly restored the protein levels of TUBB2B, DCDC2 and COL4A3, without altering THBS1 and SYNPO2 expression. In contrast, PAN lead to an elevated PCDH9 expression, whereas EV tempered the increase of PCDH9 (Fig 1F). Taken together, with the exception of SYNPO2 protein expression, these findings confirm the microarray results.
Fig 1. Western-blot analysis of human podocytes: Verification of selected microarray results.
Quantification of total TUBB2B (A), DCDC2 (B), THBS1 (C), COL4A3 (D), SYNPO2 (E) and PCDH9 (F) protein. GAPDH = loading control. For quantification, total protein was normalized with respect to GAPDH (representative example from 3 independent experiments). PAN = puromycin aminonucleoside. EV = everolimus. MeOH = methanol, solvent for EV. p = p-value. Data are means ± SD. ns = not significant.
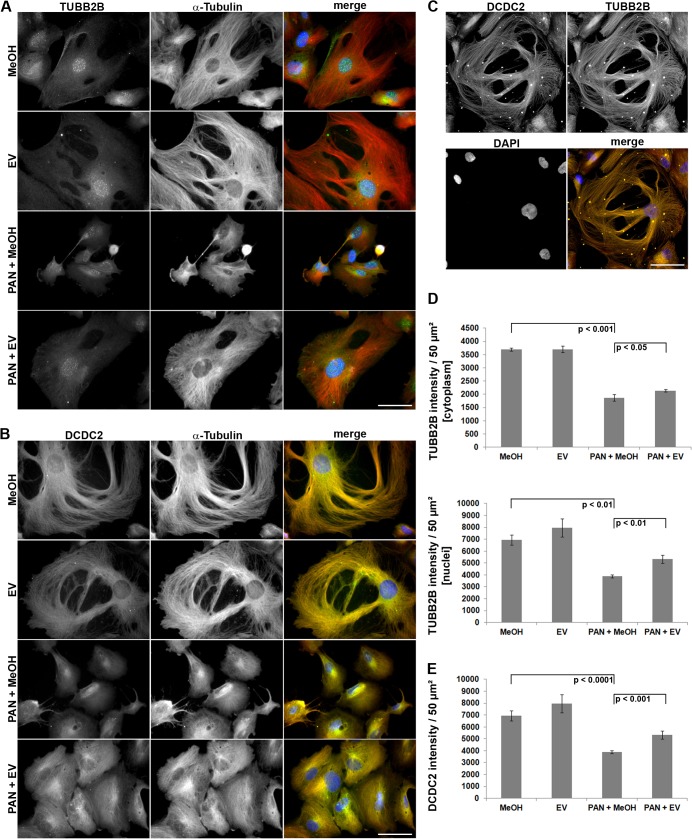
The subcellular localization of tubulin beta 2B class IIb (TUBB2B) and doublecortin domain containing 2 (DCDC2), both known to be involved in MT assembly, was subsequently assessed by immunocytochemical stainings in podocytes (Fig 2). We characterized TUBB2B protein localization both in the cytoplasm and in the nucleus of differentiated podocytes (Fig 2A). The nuclear pattern of TUBB2B appeared punctate whereas the cytoplasmic protein colocalized with the alpha-tubulin staining. DCDC2 protein was restricted to the cytoplasm and also colocalized with alpha-tubulin (Fig 2B). Immunostaining of TUBB2B and DCDC2 revealed a colocalization of the two proteins on filamentous structures reminiscent of microtubules (Fig 2C).
Fig 2. Immunofluorescence analysis of human podocytes: Verification of selected microarray results.
(A+B) alpha-tubulin, TUBB2B and DCDC2 images are presented in gray scale for maximum contrast. The merge image depicts TUBB2B and DCDC2 in green and alpha-tubulin in red. DAPI was used to visualize nuclei (blue). PAN-treated cells are smaller compared to control cells, due to alterations of the actin cytoskeleton and cell-substrate adhesion dynamics [7]. (C) Double staining of TUBB2B (red) and DCDC2 (green) of untreated control cells (D+E) Quantification of TUBB2B- and DCDC2 staining intensity with Image J software (n = 3 experiments, ≥ 10 cells per condition). PAN = puromycin aminonucleoside. EV = everolimus. MeOH = methanol, solvent for EV. p = p-value. Data are means ± SD. Scale bar = 50 μm.
In addition to the cellular localization we quantified the staining intensity of TUBB2B and DCDC2 in all treatment groups. This data revealed a reduced expression of TUBB2B in PAN-treated podocytes when compared to MeOH-treated cells, for both the nuclear and the cytoplasmic pattern. Addition of EV significantly restored the staining intensity of TUBB2B in the cytoplasm and to a greater extend in the nuclei (Fig 2D). In line with this data, the expression of DCDC2 was also significantly restored by EV (Fig 2E).
TUBB2B and DCDC2 are expressed in human kidney tissues
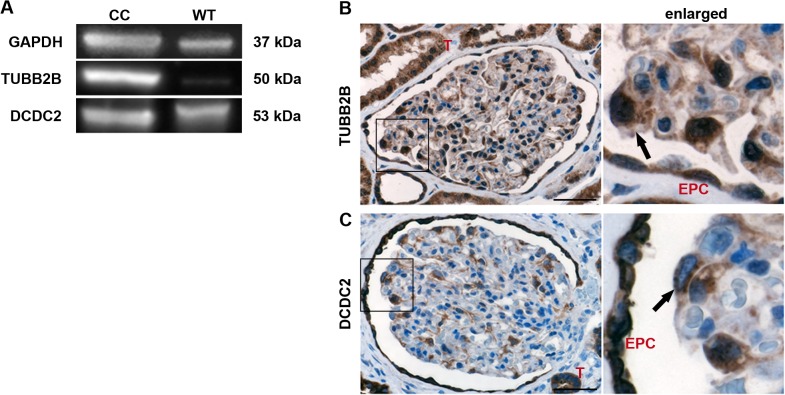
To confirm the presence of TUBB2B and DCDC2 in vivo in normal human kidney tissues we performed qPCR and Western-blot analyses from extracts received from total kidneys. Both were present on RNA level (data not shown) as well as on protein level (Fig 3A).
Fig 3. TUBB2B and DCDC2 are expressed in human kidneys.
(A) Western-blot analysis to measure the existence of TUBB2B and DCDC2 in human kidneys. GAPDH = loading control. CC = Podocyte cell culture. WT = Wild type kidney. (B+C) Immunohistochemical staining of TUBB2B and DCDC2 in healthy human kidneys. 3,3'-diaminobenzidine (DAB) was used as chromogen (brown staining) and nuclei were stained with hematoxylin (blue). The black arrows mark a nuclear as well as cytoplasmic staining of TUBB2B and a cytoplasmic staining of DCDC2 in podocytes. A negative control was performed without the primary antibody (S1 Fig). T = tubule. EPC = parietal epithelial cell. Scale bar = 50 μm.
To evaluate the localization of both proteins within the kidney in vivo, immunohistochemical stainings for TUBB2B and DCDC2 were performed in healthy control human kidneys (Fig 3B+3C). TUBB2B showed distinct cytoplasmic and nuclear staining in tubules, podocytes and epithelial parietal cells (EPCs) (Fig 3B). Pronounced cytoplasmic DCDC2 expression was observed in EPCs, whereas lower levels of DCDC2 were found in podocytes (Fig 3C).
Delay in glomerular development in mutant Tubb2b mice
Recently, Stottmann et al. described a homozygous mouse model expressing a missense mutation in Tubb2b [19]. To define the effect of Tubb2b brdp/brdp on glomerular ultrastucture we compared kidneys from E18.5 Tubb2b brdp/brdp and Tubb2b +/+ mice by light microscopy and by TEM studies.
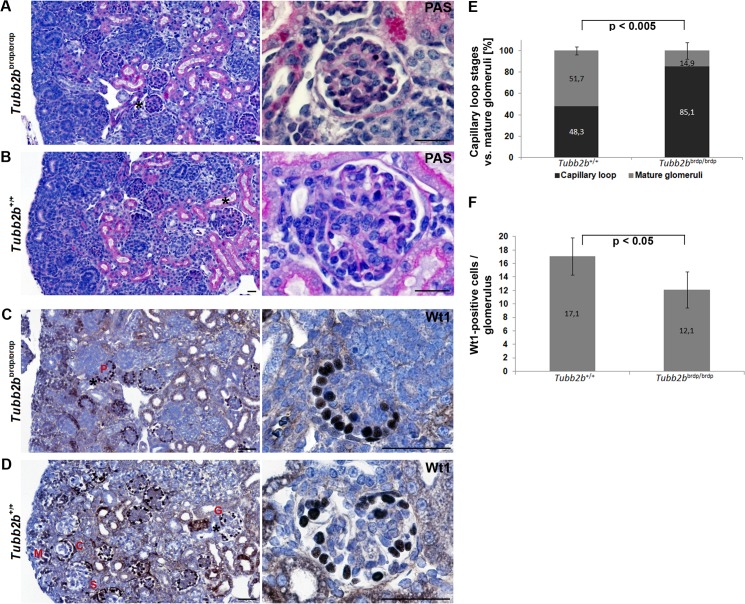
Light microscopy examination of Tubb2b brdp/brdp glomeruli revealed a significant delay in glomerular tuft and capillary lumen development (Fig 4A), whereas wild type Tubb2b +/+ glomeruli had age-dependent normally developed glomeruli with open capillary loops (Fig 4B).
Fig 4. Periodic acid-Schiff and Wilm’s tumour 1 staining of wild type and Tubb2b brdp/brdp mouse kidneys (E18.5).
(A+B) Glomeruli were stained with periodic acid-Schiff to highlight basement membranes of glomerular capillary loops and tubular epithelium. (A) Tubb2b brdp/brdp mice often show a lack in glomerular tuft and capillary lumen development. (B) The capillary loops of the wild type glomeruli are well-defined and thin. Scale bars = 25 μm. (C+D) Wilm’s tumour 1 (Wt1) expression was detectable by immunohistochemistry (dark-brown nuclear staining results from 3,3'-diaminobenzidine). Nuclei were stained with hematoxylin (blue). (C) Tubb2b brdp/brdp kidneys show a specific Wt1 staining in podocytes (P) arranged in a string of pearls-like pattern at the periphery of the glomerulus, characteristic of an early developmental stage. (D) In the developing wild type kidney, Wt1 is expressed in mesenchymal cells that are starting the mesenchymal-to-epithelial transition (condensing metanephric mesenchyme (M)), in early epithelial structures (comma- (C) and S-shaped (S) bodies) and in fully differentiated epithelial cells (glomerular podocytes (G)). Black asterisks: Enlarged section areas on the right. Scale bars = 50 μm. (E) Percentage of capillary loop stages versus mature glomeruli in 3 mutant and 4 wild type animals. For each animal, 15 to 30 capillary loop stages / mature glomeruli were counted. Data are means ± SD. (F) Average number of Wt1-positive cells per glomerulus in 3 mutant and 4 wild type animals. 15 to 30 glomeruli / animal were counted. Data are means ± SD.
To further explore the cellular alterations underlying the glomerular phenotype observed in Tubb2b brdp/brdp mice, we examined the expression of the podocyte-specific protein Wilm’s tumour 1 (Wt1). The expression pattern of Wt1 in Tubb2b brdp/brdp kidneys revealed a delay in podocyte maturation with podocytes arranged in a string of pearls-like pattern at the periphery of the glomerulus (Fig 4C). In the developing wild type kidney, Wt1 expression is detectable in mesenchymal cells that are starting the mesenchymal-to-epithelial transition (condensing metanephric mesenchyme), in early epithelial structures (comma- and S-shaped bodies) as well as in differentiated epithelial cells (glomerular podocytes) (Fig 4D). To confirm the podocyte maturation defect in Tubb2b brdp/brdp kidneys the percentage of capillary loop stages versus mature glomeruli and the average number of Wt1-positive cells per glomerulus were analyzed. Indeed, a significantly increased number of capillary loop stages in Tubb2b brdp/brdp kidneys compared to mature glomeruli was found (Fig 4E). In addition, significantly less Wt1-positive cells per glomerulus were detected in Tubb2b brdp/brdp kidneys as compared to wild type mice (Fig 4F).
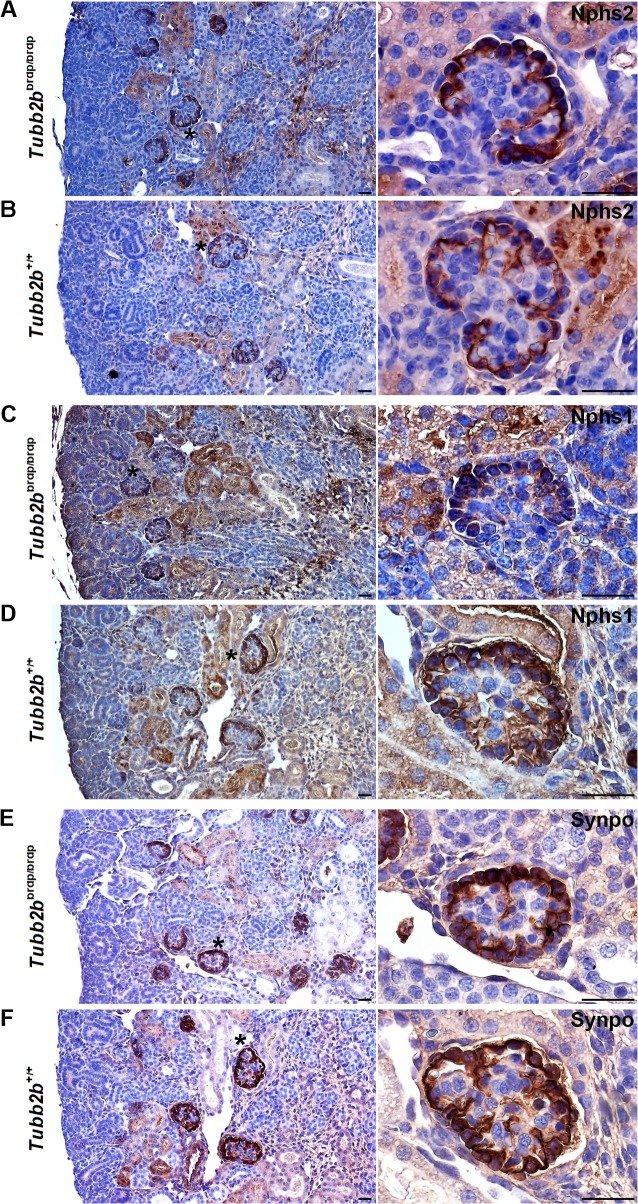
To underscore these observed defects in podocyte development we analyzed the expression of the podocyte-specific proteins Podocin (Nphs2), Nephrin (Nphs1) and Synaptopodin (Synpo) by immunohistochemistry (Fig 5). Nphs1 and Nphs2 expression in Tubb2b brdp/brdp kidneys was found to be elevated and localized to podocytes arranged like a row of pearls at the periphery of the glomerulus, further supporting a delay in podocyte maturation (Fig 5A+5C). In the developing wild type kidney Nphs2 and Nphs1 were detected in podocytes from the early capillary loop stage in the developing nephrons, and in mature glomeruli (Fig 5B+5D).
Fig 5. Nphs2-, Nphs1- and Synpo staining of wild type and Tubb2b brdp/brdp mouse kidneys (E18.5).
Immunohistochemical staining of mice kidneys (dark-brown Nphs2, Nphs1 and Synpo stainings result from 3,3'-diaminobenzidine. Nuclei were stained with hematoxylin (blue). (A+B) Nphs2 staining. (A) Tubb2b brdp/brdp kidneys show a specific Nphs2 staining in podocytes arranged like a row of pearls at the periphery of the glomerulus. (B) Within the developing wild type kidney early capillary loop stages and maturing glomeruli are labeled. (C+D) Nphs1 staining. (E+F) Synpo staining. (C-F) Both, Nphs1 and Synpo patterns are comparable to the Nphs2 staining. Black asterisks: Enlarged section areas on the right. Scale bars = 50μm.
The Synpo expression pattern in the Tubb2b brdp/brdp kidneys was found to be similar to the Wt1, Nphs2 and Nphs1 staining, confirming the developmental defects seen in the mutant mice. (Fig 5E). As shown in Fig 5F, Synpo was observed in the wild type kidneys in the early capillary loop stage glomeruli and in glomeruli of later developmental stages.
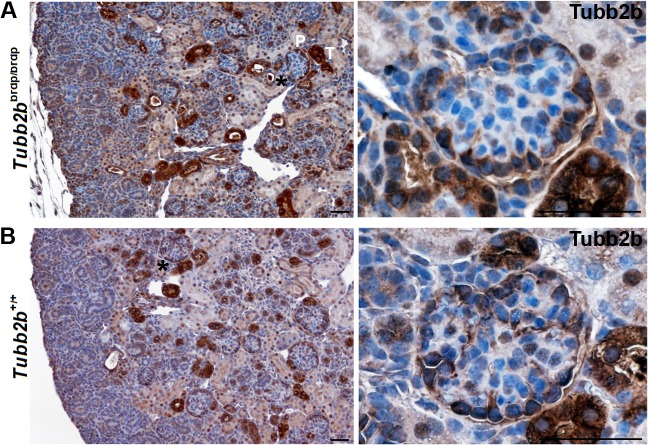
Similar to Wt1, Nphs2, Nphs1 and Synpo, expression of Tubb2b can be found in podocytes organized like pearls at the periphery of the glomerulus, further establishing the developmental defects in Tubb2b brdp/brdp mice (Fig 6A). In wild type kidneys, Tubb2b is not expressed at early stages of podocyte development but restricted to mature podocytes (Fig 6B).
Fig 6. Tubb2b staining of wild type and Tubb2b brdp/brdp mouse kidneys (E18.5).
Immunohistochemical staining of mice kidneys (dark-brown Tubb2b staining results from 3,3'-diaminobenzidine. Nuclei were stained with hematoxylin (blue). (A) Tubb2b brdp/brdp kidneys show a specific cytoplasmic Tubb2b expression in tubuli (T) and in podocytes (P), confirming the developmental defects seen with the Wt1, Nphs2, Nphs1 and Synpo stainings. (B) Tubb2b expression in wild type kidneys is restricted to the mature podocytes. Interestingly in the developing wild type kidney Tubb2b is not expressed in the early developmental stages of maturing podocytes. Note, that in murine podocytes nuclear Tubb2b seems much less expressed compared to human kidneys. Black asterisks: Enlarged section areas on the right. Scale bars = 50 μm.
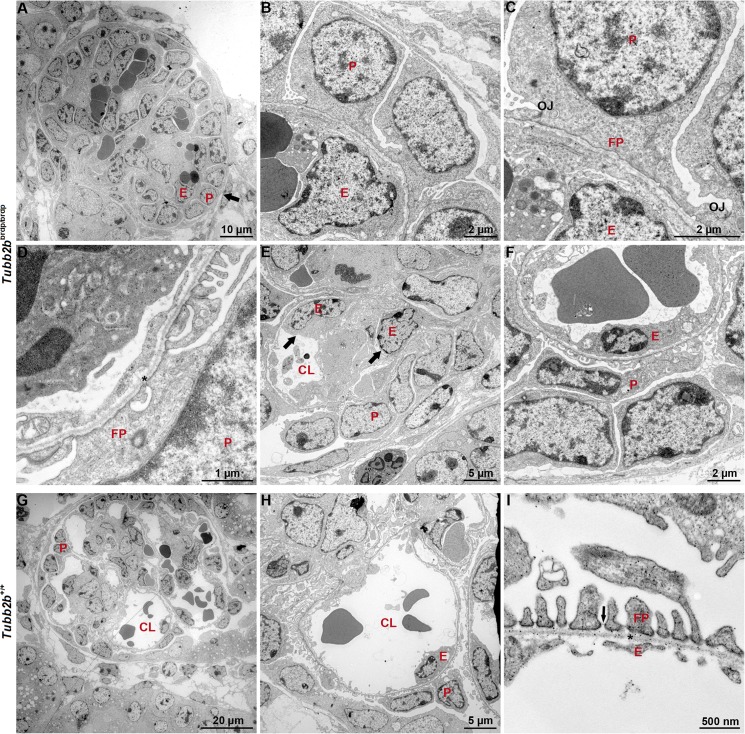
On examination with TEM, significant morphologic changes of foot process and glomerular endothelium development were observed in Tubb2b brdp/brdp glomeruli (Fig 7). Some glomeruli from Tubb2b brdp/brdp mice had immature, cuboidal podocytes (Fig 7A+7B). In some areas, podocyte foot processes appeared incompletely differentiated. They were extremely wide and linked by occludens junctions (OJs) rather than SDs (Fig 7C+7D). Often, there was no visible capillary lumen in Tubb2b brdp/brdp glomeruli, even in most mature glomeruli, and multiple endothelial cells were visualized within the capillary loops (Fig 7A+7E). In addition, glomerular endothelial cells were swollen and vacuolated, with decreased fenestrations (Fig 7F). The glomerular basement membrane in Tubb2b brdp/brdp mice appeared morphologically similar to that seen in controls (Fig 7D). Glomeruli of wild type Tubb2b +/+ mice had, according to age, open glomerular capillaries with fenestrated endothelium, differentiated podocyte foot processes linked by SDs and a normal glomerular basement membrane (Fig 7G–7I).
Fig 7. Transmission electron micrographs of wild type and Tubb2b brdp/brdp mouse kidneys (E18.5).
(A-F) With TEM, a delay in glomerular endothelial (E) and podocyte (P) development could be observed in Tubb2b brdp/brdp mice. (A+B) Glomerulus with immature, cuboidal podocytes (black arrow). (C+D) Incompletely differentiated podocyte foot processes (FPs); some are extremely wide and linked by occludens junctions (OJs). (D) The glomerular basement membrane (asterisk) appears to be normal. (A+E) Glomeruli with no or small visible capillary lumen (CL) and multiple endothelial cells within the capillary loops (black arrows). (F) Swollen and vacuolated glomerular endothelial cells with decreased fenestrations. (G-I) Glomeruli of wild type Tubb2b mice had open glomerular capillaries with fenestrated endothelium, differentiated podocyte foot processes linked by SDs (black arrow) and a normal glomerular basement membrane.
Collectively, the observed delay in podocyte differentiation following alteration of Tubb2b function underscores a role of the MT protein TUBB2B during kidney organogenesis. The immature podocyte phenotype seems not to be due to a prolonged proliferation of the Wt1-positive podocyte progenitors at the comma- and S-shaped body stage as no difference in the expression of the proliferation marker MKI67 could be observed (data not shown). These findings and the delay in capillary development remains to be studied more precisely in future experiments.
Discussion
The immense variation in podocyte cell shape required for proper podocyte function depends on the underlying network of dynamic and interconnected actin and MT polymers. Glomerular diseases are often associated with foot process effacement and podocytopenia leading to severe proteinuria, and are closely linked to disaggregation and altered distribution of AFs [24]. Podocytes in vivo display typical arborized morphology with two types of processes–thick primary foot processes containing MTs as well as IFs [25] and thin secondary foot processes in which AFs represent the core cytoskeletal elements [26]. The molecular mechanisms of the reorganization of the podocyte actin cytoskeleton have been well studied over the last years whereas the function of the primary foot processes as well as the role of MTs and IFs is still poorly understood. Previous experiments, both in vivo and in vitro, showed that MTs are essential not only for formation but also for maintenance of intact primary podocyte foot processes [17,18,27].
In the present study, we identified by differential gene expression analysis of EV treated podocytes in an experimental model of podocyte injury a large number of genes being involved in cytoskeletal organization and stability comprising MT and actin-associated genes. In particular we observed a significant increase in the expression of TUBB, TUBB2B and TUBA1A, three major components of MTs. Interestingly a podocyte-specific expression of TUBB2B was observed in human and mouse podocytes.
MTs are produced by polymerized heterodimers of alpha- and beta-tubulin, encoded by tubulin genes which share substantial levels of sequence homology and are regulated spatially and temporally [28,29]. The function of MTs has been extensively studied in neurons. Here, they are crucial for neurite initiation, cortical development, cortical malformations and neuronal migration [30–32]. Functional studies suggest that abnormal neuronal migration would result from defective interactions in the tubulin heterodimer assembly and altered three-dimensional conformation of the tubulins, with compromised interaction with MT associated proteins or MT motor proteins [33]. Recently, mutations in neuronal tubulin genes (e.g. TUBA1A, TUBB2B, TUBB3 and TUBA8) have been identified in patients suffering from nervous system disorders, underlining the importance of tubulin isotypes in MT dynamics during brain development and axon guidance [34,35]. Overall, the identified TUBB2B mutations are suggested to alter the proper functions of MTs but the precise molecular function of TUBB2B in cortical development is still unclear. Stottmann and coworkers describe neurodevelopmental defects in mice with a homozygous missense mutation in Tubb2b due to a major increase in apoptosis and abnormal proliferation of the basal progenitors [19]. Besides these well-studied effects in the brain, podocyte-specific functions of TUBB2B have not been analyzed yet.
In podocytes, MTs are the scaffold of primary foot processes and the central cell body [25,36–40]. Disruption of MT elongation with vinblastine resulted in severe damage of primary foot processes in vivo [17,37,41] and blocked foot process formation in vitro [18].
In the present study, analyses of kidneys from Tubb2b brdp/brdp mice by the use of the podocyte-specific markers Wt1, Nphs2, Nphs1 and Synpo together with the Tubb2b expression pattern showed an abnormal podocyte morphology consistent with a delay in kidney development. A MT associated migratory defect of premature glomerular and perhaps endothelial cells might underlie this phenomenon, which is a focus of ongoing experimental studies.
Besides TUBB2B the MT associated protein DCDC2 was identified as a novel gene in podocytes, differentially regulated by mTOR inhibition. DCDC2 belongs to the superfamily of doublecortin (DCX) domain containing proteins. It regulates cytoskeletal dynamics by binding to and stabilizing MTs by linking adjacent tubulin protofilaments and has been identified in primary cilia of neuronal cells [42–50]. Kobayashi and colleagues demonstrated that MT associated proteins cause MT-based foot process formation in vitro via enhancement of MT assembly as well as an increase in MT stability [51]. DCX-microtubule effects have been extensively studied in neurons, including recent studies revealing cooperative binding effects, changes in MT structure, and regulation of molecular motors [48,49,52–54]. Mutations in the human DCX gene, the first characterized gene of the family, result in abnormal neuronal migration, epilepsy, mental retardation and cause double cortex syndrome and lissencephaly in humans [43]. Downregulation of DCX by RNA interference in animal models leads to neuronal migration disorders similar to those seen in the brains of dyslexic individuals [55,56]. Very recently, a novel clinical entity has been linked to recessive human DCDC2 mutations with disruption of Wnt signaling causing a form of renal hepatic nephronophthisis-related ciliopathy [57]. In this work of Schueler et al., DCDC2 protein has been identified to colocalize with alpha-tubulin to the axoneme of primary cilia of human renal tubule cells and cholangiocytes in human liver and to multiciliated ependymal cells and pia mater cells in mouse brain. This is the first description of DCDC2 expression in the kidney, however the cellular function of DCDC2 in podocytes remains unclear. Interestingly, a homozygous human mutation in another ciliary gene TTC21B has lately been implicated in a familial form of focal-segmental glomerulosclerosis, clinically characterized by nephrotic-range proteinuria and severe podocyte damage [58]. In this work, the TTC21B gene product IFT139 was redistributed along the microtubule network in mature podocytes. These findings link defective functions of MTs and associated ciliary proteins to clinically relevant proteinuric disease.
Overall, this data is in line with the results of the present study demonstrating an up-regulation of TUBB2B and the MT associated protein DCDC2 as a consequence of mTORi-related rescue in damaged human podocytes. We here report for the first time an expression of TUBB2B and DCDC2 in human and mouse podocytes highlighting their potential relevance for the MT network and the actin cytoskeleton. Further studies will evaluate the role of TUBB2B and DCDC2 in human proteinuric disease.
Supporting Information
Negative control for TUBB2B and DCDC2 antibodies is performed without the primary antibody. 3,3'-diaminobenzidine (DAB) was used as chromogen (brown staining) and nuclei were stained with hematoxylin (blue). Scale bar = 50 μm.
(TIF)
Fold change (FC) > 2.0 and p-value (p) < 0.05 for both comparisons. Not included data: Unknown gene annotations, chromosome open reading frames, family with sequence similarities, hypothetical proteins, genes with the same fold change tendency for both comparisons. PAN = puromycin aminonucleoside. EV = everolimus. MeOH = methanol, solvent for EV.
(DOCX)
Acknowledgments
Anja K. Büscher and Stefanie Weber received financial support from the Deutsche Forschungsgemeinschaft (BU 2602/3-1) and Stefanie Jeruschke from the Forschungsunterstützungskreis Kindernephrologie e.V. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors thank Moin Ahson Saleem for providing the immortalized human podocyte cell line, Wilhelm Kriz and Jan Ulrich Becker for evaluation of histologic results and Heike Rekasi for excellent technical support.
Data Availability
Microarray data of all groups have been published in Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO accession number GSE66107.
Funding Statement
AKB and SW received financial support from the Deutsche Forschungsgemeinschaft (BU 2602/3-1) and Stefanie Jeruschke from the Forschungsunterstützungskreis Kindernephrologie e.V. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mundel P, Kriz W. Structure and function of podocytes: an update. Anat Embryol (Berl). 1995; 192(5): 385–397. [DOI] [PubMed] [Google Scholar]
- 2. Pavenstadt H, Kriz W, Kretzler M. Cell biology of the glomerular podocyte. Physiol Rev. 2003; 83(1): 253–307. [DOI] [PubMed] [Google Scholar]
- 3. Yuan H, Takeuchi E, Salant DJ. Podocyte slit-diaphragm protein nephrin is linked to the actin cytoskeleton. Am J Physiol Renal Physiol. 2002; 282(4): F585–F591. [DOI] [PubMed] [Google Scholar]
- 4. Büscher AK, Weber S. Educational paper: the podocytopathies. Eur J Pediatr. 2012; 171(8): 1151–1160. 10.1007/s00431-011-1668-2 [DOI] [PubMed] [Google Scholar]
- 5. Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001; 81(1): 153–207. [DOI] [PubMed] [Google Scholar]
- 6. Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998; 279(5350): 509–514. [DOI] [PubMed] [Google Scholar]
- 7. Jeruschke S, Büscher AK, Oh J, Saleem MA, Hoyer PF, Weber S, et al. Protective effects of the mTOR inhibitior everolimus on cytoskeletal injury in human podocytes are mediated by RhoA signaling. PLoS One. 2013; 8(2): e55980 10.1371/journal.pone.0055980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kobayashi N, Reiser J, Kriz W, Kuriyama R, Mundel P. Nonuniform microtubular polarity established by CHO1/MKLP1 motor protein is necessary for process formation of podocytes. J Cell Biol. 1998; 143(7): 1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kobayashi N, Mominoki K, Wakisaka H, Shimazaki Y, Matsuda S. Morphogenetic activity of extracellular matrices on cultured podocytes. Laminin accelerates podocyte process formation in vitro. Ital J Anat Embryol. 2001; 106(2 Suppl 1): 423–430. [PubMed] [Google Scholar]
- 10. Hirose M, Ishizaki T, Watanabe N, Uehata M, Kranenburg O, Moolenaar WH, et al. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J Cell Biol. 1998; 141(7): 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scaife RM, Job D, Langdon WY. Rapid microtubule-dependent induction of neurite-like extensions in NIH 3T3 fibroblasts by inhibition of ROCK and Cbl. Mol Biol Cell. 2003; 14(11): 4605–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999; 1(1): 45–50. [DOI] [PubMed] [Google Scholar]
- 13. Chang YC, Nalbant P, Birkenfeld J, Chang ZF, Bokoch GM. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol Biol Cell. 2008; 19(5): 2147–2153. 10.1091/mbc.E07-12-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nalbant P, Chang YC, Birkenfeld J, Chang ZF, Bokoch GM. Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol Biol Cell. 2009; 20(18): 4070–4082. 10.1091/mbc.E09-01-0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rottner K, Hall A, Small JV. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol. 1999; 9(12): 640–648. [DOI] [PubMed] [Google Scholar]
- 16. Kaverina I, Rottner K, Small JV. Targeting, capture, and stabilization of microtubules at early focal adhesions. J Cell Biol. 1998; 142(1): 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andrews PM. The effect of vinblastine-induced microtubule loss on kidney podocyte morphology. Am J Anat. 1977; 150(1): 53–61. [DOI] [PubMed] [Google Scholar]
- 18. Kobayashi N, Reiser J, Schwarz K, Sakai T, Kriz W, Mundel P. Process formation of podocytes: Morphogenetic activity of microtubules and regulation by protein serine/threonine phosphatase PP2A. Histochem Cell Biol. 2001; 115(3): 255–266. [DOI] [PubMed] [Google Scholar]
- 19. Stottmann RW, Donlin M, Hafner A, Bernard A, Sinclair DA, Beier DR. A mutation in Tubb2b, a human polymicrogyria gene, leads to lethality and abnormal cortical development in the mouse. Hum Mol Genet. 2013; 22(20): 4053–4063. 10.1093/hmg/ddt255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saleem MA, O'Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002; 13(3): 630–638. [DOI] [PubMed] [Google Scholar]
- 21. Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969; 26(1): 31–43. [DOI] [PubMed] [Google Scholar]
- 22. Weiner JA, Jontes JD. Protocadherins, not prototypical: a complex tale of their interactions, expression, and functions. Front Mol Neurosci. 2013; 6: 4 10.3389/fnmol.2013.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adh Migr. 2011; 5(2): 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whiteside CI, Cameron R, Munk S, Levy J. Podocytic cytoskeletal disaggregation and basement-membrane detachment in puromycin aminonucleoside nephrosis. Am J Pathol. 1993; 142(5): 1641–1653. [PMC free article] [PubMed] [Google Scholar]
- 25. Kobayashi N, Mundel P. A role of microtubules during the formation of cell processes in neuronal and non-neuronal cells. Cell Tissue Res. 1998; 291(2): 163–174. [DOI] [PubMed] [Google Scholar]
- 26. Ichimura K, Kurihara H, Sakai T. Actin filament organization of foot processes in rat podocytes. J Histochem Cytochem. 2003; 51(12): 1589–1600. [DOI] [PubMed] [Google Scholar]
- 27. Kerjaschki D. Polycation-induced dislocation of slit diaphragms and formation of cell junctions in rat kidney glomeruli. The effects of low temperature, divalent cations, colchicine, and cytochalasin B. Lab Invest. 1978; 39(5): 430–440. [PubMed] [Google Scholar]
- 28. Villasante A, Wang D, Dobner P, Dolph P, Lewis SA, Cowan NJ. Six mouse alpha-tubulin mRNAs encode five distinct isotypes: testisspecific expression of two sister genes. Mol Cell Biol. 1986; 6(7): 2409–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang D, Villasante A, Lewis SA, Cowan NJ. The mammalian beta-tubulin repertoire: hematopoietic expression of a novel, heterologous beta-tubulin isotype. J Cell Biol. 1986; 103(5): 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dehmelt L, Nalbant P, Steffen W, Halpain S. A microtubule-based, dynein-dependent force induces local cell protrusions: Implications for neurite initiation. Brain Cell Biol. 2006; 35(1): 39–56. [DOI] [PubMed] [Google Scholar]
- 31. Breuss M, Keays DA. Microtubules and neurodevelopmental disease: the movers and the makers. Adv Exp Med Biol. 2014; 800: 75–96. 10.1007/978-94-007-7687-6_5 [DOI] [PubMed] [Google Scholar]
- 32. Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009; 41(6): 746–752. 10.1038/ng.380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Friocourt G, Marcorelles P, Saugier-Veber P, Quille ML, Marret S, Laquerrière A. Role of cytoskeletal abnormalities in the neuropathology and pathophysiology of type I lissencephaly. Acta Neuropathol. 2011; 121(2): 149–170. 10.1007/s00401-010-0768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jaglin XH, Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 2009; 25(12): 555–566. 10.1016/j.tig.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 35. Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010; 140(1): 74–87. 10.1016/j.cell.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Drenckhahn D, Franke RP. Ultrastructural organization of contractile and cytoskeletal proteins in glomerular podocytes of chicken, rat, and man. Lab Invest. 1988; 59(5): 673–682. [PubMed] [Google Scholar]
- 37. Andrews PM. Investigations of cytoplasmic contractile and cytoskeletal elements in the kidney glomerulus. Kidney Int. 1981; 20(5): 549–562. [DOI] [PubMed] [Google Scholar]
- 38. Cortez P, Méndez M, Riser BL, Guérin CJ, Rodríguez-Barbero A, Hassett C, et al. F-actin fiber distribution in glomerular cells: structural and functional implications. Kidney Int. 2000; 5(6): 2452–2461. [DOI] [PubMed] [Google Scholar]
- 39. Farquhar MG, Vernier RL, Good RA. An electron microscopic study of the glomerulus in nephrosis, glomerulonephritis and lupus erythematosus. J Exp Med. 1957; 106(5): 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vasmant D, Maurice M, Feldmann G. Cytoskeleton ultrastructure of podocytes and glomerular endothelial cells in man and in the rat. Anat Rec. 1984; 210(1): 17–24. [DOI] [PubMed] [Google Scholar]
- 41. Tyson GE. Scanning electron microscopic study of the effect of vinblastine on podocytes of rat kidney. Virchows Arch B Cell Pathol. 1977; 25(2): 105–115. [DOI] [PubMed] [Google Scholar]
- 42. Coquelle FM, Levy T, Bergmann S, Wolf SG, Bar-El D, Sapir T, et al. Common and divergent roles for members of the mouse DCX superfamily. Cell Cycle. 2006; 5(9): 976–983. [DOI] [PubMed] [Google Scholar]
- 43. Reiner O, Coquelle FM, Peter B, Levy T, Kaplan A, Sapir T, et al. The evolving doublecortin (DCX) superfamily. BMC Genomics. 2006; 7: 188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim MH, Cierpicki T, Derewenda U, Krowarsch D, Feng Y, Devedjiev Y, et al. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat Struct Biol. 2003; 10(5): 324–333. [DOI] [PubMed] [Google Scholar]
- 45. Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998; 92(1): 63–72. [DOI] [PubMed] [Google Scholar]
- 46. Francis F, Koulakoff A, Boucher D, Chafey P, Schaar B, Vinet MC, et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999; 23(2): 247–256. [DOI] [PubMed] [Google Scholar]
- 47. Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999; 23(2): 257–271. [DOI] [PubMed] [Google Scholar]
- 48. Moores CA, Perderiset M, Francis F, Chelly J, Houdusse A, Milligan RA. Mechanism of microtubule stabilization by Doublecortin. Mol Cell. 2004; 14(6): 833–839. [DOI] [PubMed] [Google Scholar]
- 49. Moores CA, Perderiset M, Kappeler C, Kain S, Drummond D, Perkins SJ, et al. Distinct roles of Doublecortin modulating the microtubule cytoskeleton. EMBO J. 2006; 25(19): 4448–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Massinen S, Hokkanen ME, Matsson H, Tammimies K, Tapia-Páez I, Dahlström-Heuser V, et al. Increased expression of the dyslexia candidate gene DCDC2 affects length and signaling of primary cilia in neurons. PLoS One. 2011; 6(6): e20580 10.1371/journal.pone.0020580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kobayashi N, Heid H, Sakai T, Kriz W, Huber G, Mundel P. Molecular characterization reveals the identity of microtubule associated proteins MAP3 and MAP4. Biochem Biophys Res Commun. 2000; 268(2): 306–309. [DOI] [PubMed] [Google Scholar]
- 52. Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999; 23(2): 257–271. [DOI] [PubMed] [Google Scholar]
- 53. Bechstedt S, Brouhard GJ. Doublecortin recognizes the 13-protofilament microtubule cooperatively and tracks microtubule ends. Dev Cell. 2012; 23(1): 181–192. 10.1016/j.devcel.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu JS, Schubert CR, Fu X, Fourniol FJ, Jaiswal JK, Houdusse A, et al. Molecular basis for specific regulation of neuronal kinesin-3 motors by doublecortin family Proteins. Mol Cell. 2012; 47(5): 707–721. 10.1016/j.molcel.2012.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meng HY, Smith SD, Hager K, Held M, Liu J, Olson RK, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA. 2005; 102(47): 17053–17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Burbridge TJ, Wang Y, Volz AJ, Peschansky VJ, Lisann L, Galaburda AM, et al. Postnatal analysis of the effect of embryonic knockdown and overexpression of candidate dyslexia susceptibility gene homolog Dcdc2 in the rat. Neuroscience. 2008; 152(3): 723–733. 10.1016/j.neuroscience.2008.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schueler M, Braun DA, Chandrasekar G, Gee HJ, Klasson TD, Halbritter J, et al. DCDC2 Mutations Cause a Renal-Hepatic Ciliopathy by Disrupting Wnt Signaling. Am J Hum Genet. 2015; 96(1): 81–92. 10.1016/j.ajhg.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cong EH, Bizet AA, Boyer O, Woerner S, Gribouval O, Filhol E, et al. A homozygous missense mutation in the ciliary gene TTC21B causes familial FSGS. J Am Soc Nephrol. 2014; 25(11): 2435–2443. 10.1681/ASN.2013101126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Negative control for TUBB2B and DCDC2 antibodies is performed without the primary antibody. 3,3'-diaminobenzidine (DAB) was used as chromogen (brown staining) and nuclei were stained with hematoxylin (blue). Scale bar = 50 μm.
(TIF)
Fold change (FC) > 2.0 and p-value (p) < 0.05 for both comparisons. Not included data: Unknown gene annotations, chromosome open reading frames, family with sequence similarities, hypothetical proteins, genes with the same fold change tendency for both comparisons. PAN = puromycin aminonucleoside. EV = everolimus. MeOH = methanol, solvent for EV.
(DOCX)
Data Availability Statement
Microarray data of all groups have been published in Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO accession number GSE66107.