Abstract
Because it serves as the cytoplasm of the oocyte and provides a large amount of reserves, the egg yolk has biological significance for developing embryos. The ovary and its hierarchy of follicles are the main reproductive organs responsible for yolk deposition in chickens. However, the genetic architecture underlying the yolk and ovarian follicle weights remains elusive. Here, we measured the yolk weight (YW) at 11 age points from onset of egg laying to 72 weeks of age and measured the follicle weight (FW) and ovary weight (OW) at 73 weeks as part of a comprehensive genome-wide association study (GWAS) in 1,534 F2 hens derived from reciprocal crosses between White Leghorn (WL) and Dongxiang chickens (DX). For all ages, YWs exhibited moderate single nucleotide polymorphism (SNP)-based heritability estimates (0.25–0.38), while the estimates for FW (0.16) and OW (0.20) were relatively low. Independent univariate genome-wide screens for each trait identified 12, 3, and 31 novel significant associations with YW, FW, and OW, respectively. A list of candidate genes such as ZAR1, STARD13, ACER1b, ACSBG2, and DHRS12 were identified for having a plausible function in yolk and follicle development. These genes are important to the initiation of embryogenesis, lipid transport, lipoprotein synthesis, lipid droplet promotion, and steroid hormone metabolism, respectively. Our study provides for the first time a genome-wide association (GWA) analysis for follicle and ovary weight. Identification of the promising loci as well as potential candidate genes will greatly advance our understanding of the genetic basis underlying dynamic yolk weight and ovarian follicle development and has practical significance in breeding programs for the alteration of yolk weight at different age points.
Introduction
Chicken egg yolk is an emulsion of water (48%), lipids (33%), and proteins (17%) [1]. Because it serves as the cytoplasm of the oocyte and provides a large amount of reserves, egg yolk functions biologically to provide the above-mentioned nutrients to the developing embryos [2],and yolk can accumulate significant amounts of IgY immunoglobulins (up to 100 mg per egg) to provide innate immunity to the embryos [3]. Egg yolk is widely used in the food industry for its high nutritional value to humans [4]. Furthermore, the bioactive substances of egg yolk are applied in the pharmaceutical and cosmetics fields for their binding properties, emulsion stability, and natural antioxidants [5–7].
The central area of the chicken ovary is composed of a vascularized medulla and a cortex containing the small follicles that are oocytes covered by follicular epithelium [2], and egg yolk is formed in these ovarian follicles by the consecutive deposition of lipids and proteins [8]. The sequential development of oocytes in ovaries leads to the display of a hierarchy in the follicles with four to six yolky follicles of gradually increasing size at the surface. Yolk precursors, however, are not synthesized in the ovary but are produced by the liver and then transported in the blood to the ovarian oocytes [2, 9]. Vitellogenin, consisting of one phosvitin and two lipovitellins, is the main carrier for protein transportation from the liver to the ovary in the blood [10]. The lipid carrier is very low-density lipoprotein (VLDL), which has a standard structure consisting of a core of triglycerides and cholesterol esters surrounded by a surface layer composed of phospholipids, cholesterol, and apoproteins [11]. Yolk precursors (vitellogenin and VLDL) are transported in the follicular wall and are released near the basolateral membrane of the follicles. Then the penetration of these precursors is ensured through a process of endocytosis induced by the receptor LR8 for the deposition of yolk [12, 13].
Due to the wide utilization of egg yolk, many efforts have been performed to alter egg yolk weight [14]. However, egg yolk weight is a complicated quantitative trait affected by many factors, such as breed and hen age [14]. The yolk weight is increased with the age of the laying birds; for eggs of the same size, old hens produce larger egg yolks than young hens, and the albumen weight is correspondingly decreased [15]. The strategy of identifying the quantitative trait loci (QTLs) or causal genes that are related to yolk formation and ovarian follicle development is a powerful tool to illustrate the genetic control for yolk weight and follicle development. A decade ago, microsatellite markers were employed to detect the causal regions associated with yolk weight, and multiple QTLs were reported [16–19]. Until now, however, only seven QTLs(distributed on chromosomes 4, 6, 9, 11, 15, 22, and 26) that relate to yolk weight had been identified. And these QTLs, which have poor repeatability and wide positional confidence intervals, were deposited in the AnimalQTL Database (http://www.animalgenome.org/cgi-bin/QTLdb/index). In recent years, genome-wide association studies (GWASs) have been utilized to identify the associations between genomic loci and phenotypes with relatively high-density single nucleotide polymorphism (SNP) arrays in chickens [17]. Recently, with the rapid advance in next-generation sequencing technologies, large numbers of SNPs have been discovered in chickens [20]. The development of the Affymetrix 600K Chicken SNP array allows further efficient screening for causal loci and genes with relevance to target traits.
In the current study, we conducted GWA analysis on the yolk weight at 11 time points from the onset of egg laying to 72 weeks utilizing 600K high-density SNP arrays in an F2 chicken population to explore the associated genomic loci and genes that contribute to the dynamic change in the yolk weight with the aging process. Furthermore, the GWA analysis was also performed on ovary and follicle weights at 73weeksof age to detect the causal genes related to follicle and ovary development. This study lays the foundation for an investigation into the genetic control of yolk and follicle development.
Materials and Methods
Ethics statement
All protocols and procedures involving animals were performed in accordance with the Guidelines for the Care and Use of Experimental Animals established by the Ministry of Agriculture of China (Beijing, China). All animal work was approved by the Animal Welfare Committee of China Agricultural University (permit number: SYXK 2007–0023).
Resource population
Reciprocal crosses between standard breed White Leghorns (WL) and Dongxiang chickens (DX), an indigenous Chinese strain, were utilized to develop an F2 resource population. For parents, six WL and six DX males were initially mated with 133 DX and 80 WL females, generating 1,029 and 552 chicks for the F1 generation, respectively. Then 25 males and 407 females from the WL/DX cross and 24 males and 235 females from the DX/WL cross in the F1 generation were used to produce the F2 population, which consisted of 3,749 chicks in a single hatch, originating from 49 half-sib and 590 full-sib families. The hens were kept in individual cages of the same environment with food and water adlibitum at the Jiangsu Institute of Poultry Sciences. Finally, 1,534 hens from 49 half-sib families and 550 full-sib families with sufficient phenotypic and pedigree information were selected for SNP genotyping. All layers were caged individually and subjected to a light/dark cycle of 16h of light and 8h of darkness (16L:8D)and raised in the same environment with feed and water adlibitum.
Phenotypic measurements
Yolk weight (YW) was measured for the first egg of each hen and again at 32, 36, 40, 44, 48, 52, 56, 60, 66, and 72 weeks of age, consisting of 11 age points. Except for the first egg, fresh eggs were collected for four successive days to ensure two eggs per hen at each age point. Eggs were broken to obtain the internal contents, and then the yolk was separated from the albumen and weighed by electronic scale. All hens were humanely sacrificed by 60%–70% carbon dioxide at 73weeks of age, the follicles and ovaries were then separated and weighted for follicle-active hens. FW includes the weight of all follicles that were contained in the ovary, and OW indicates the weight of the follicle-free ovary, which consisted of a highly vascularized central area (medulla) and a peripheral area (cortex). Descriptive statistics were calculated with R project (R version 3.1.2) using all available records. The function of ‘rntransform’ in the GenABEL package of R project was used for the rank-based inverse normal transformations [21].
Genotyping and quality control
Genomic DNA was extracted by the standard phenol/chloroform method and genotyped with the Affymetrix 600K Axiom Chicken Genotyping Array (Affymetrix, Inc. Santa Clara, CA, USA). Affymetrix Power Tools v1.16.0 (APT) software was then used for quality control (QC) and genotype calling. Specifically, only samples with dish quality control (DQC) >0.82 and a call rate>97% were included in the subsequent analyses. An R script supplied by Affymetrix was run to compute the SNP QC metrics and filter out individual SNPs falling below given thresholds. After these QC steps, 1,512 individuals and 532,299 SNPs remained valid. Because the current statistical methods are more powerful for the detection of the associations between phenotypes and autosomal genotypes, all SNPs on sex chromosomes were excluded in the QC process. The PLINK v1.90 package [22] was then used for further quality control to improve the detection power, in which SNPs with a minor allele frequency (MAF)<5% and Hardy—Weinberg equilibrium (HWE) test P< 1 × 10−6 were removed from the downstream analysis. Some sporadic missing genotypes were imputed using the BEAGLE v4.0 procedure [23], and only SNPs with an imputation quality score R 2>0.5 were retained. Ultimately, up to 1,512 individuals and 435,867 SNPs were valid for the GWA analysis.
Estimation of heritability and contribution to phenotypic variance
A univariate restricted maximum likelihood (REML) estimation was implemented in the GCTA v1.24 program [24] to estimate the heritability explained by the eligible SNPs (h 2 snp). We also quantified the pair-wise genetic and phenotypic correlations for each trait at multiple time points with a bivariate mixed model [25]. A genetic relationship matrix (GRM) derived from all genotyped SNPs on autosomes and two linkage groups was created, and the top three PCs calculated by the GCTA tool were included as covariates to account for the potential population structure. We then estimated the contribution to phenotypic variance (CPV)made by these associated loci or genomic regions.
Genome-wide association analysis
The principal component analysis (PCA) implemented in the PLINK package was conducted prior to genome-wide association (GWA) analysis to eliminate spurious associations resulting from the presence of cryptic relatedness or hidden population stratification. To properly decide the thresholds for genome-wide significant/suggestive associations, we used the simpleM method [26] to correct the number of multiple tests. With this approach, an effective number of 59,308 independent tests was suggested; hence, the genome-wide significant and suggestive P values were 8.43 × 10−7 (0.05/59,308) and 1.69 × 10−5 (1.00/59,308), respectively.
A univariate association test equipped with an exact mixed model approach in GEMMA v0.94 software [27] was performed with the valid individuals and SNPs. A centered relatedness matrix was calculated with the independent SNPs, and then the derived Wald test P value was calculated for the significance level between the SNPs and phenotypes. The univariate linear mixed model as follows:
For the equation, y is an n × 1 vector of phenotypic values for n individuals, W is an n × 5 matrix of covariates (fixed effects, i.e., top three PCs, sire and dam effects) including a column vector of 1, α is a 5 × 1 vector of corresponding coefficients including the intercept, x is an n × 1 vector of marker genotypes at the locus tested, β is the corresponding effect size of the marker and all effects are reported for the minor allele in each marker, u is an n × 1 vector of random polygenic effects with a covariance structure as u~ N(0, KVg), where K represents a known n × ngenetic relatedness matrix derived from SNP markers and Vg is the polygenic additive variance, and ε is an n × 1 vector of random residuals with ε ~ N(0, IVe), where I is an n × n identity matrix, and Ve is the residual variance component. We used the Wald test statistic for each SNP to test the null hypothesis β = 0, where the best linear unbiased estimate (BLUE) of β and the corresponding sampling variance are obtained by solving the mixed model equations (MME) based on estimated Vg and Ve.
The Manhattan plots and quantile—quantile (QQ) plots were drawn by the “gap” packages [28] in R project. The genomic inflation factor λ, which is the judgment of the extent of false positive signals, was also calculated with the function of estlambda implemented in the GenABEL package [21] in R project.
Linkage disequilibrium analysis
Notably, many SNPs maybe passively significantly associated with target traits, resulting from their linkage to a strongly causal mutant. In general, GWAS does not distinguish a genuine causal locus from those that are statistically significant loci within a strong linkage disequilibrium (LD) region. Therefore, in order to characterize the potential candidate genes responsible for a trait, we conducted an LD analysis and inferred the haplotype blocks containing peak SNPs by Haploview v4.2 [29]. A block is derived using the solid spine algorithm, and defined as that the first and last SNPs in a region that is strong in LDs (D′ ≥ 0.8) with all intermediate SNPs.
Gene identification
Variant Effect Predictor (VEP) and BioMart tools based on the galGal4 assembly supported by Ensembl were used for the identification of candidate genes in which significant loci were located [30] and for the detection of the genes in a given genomic region [31].
Results
Phenotype description and genetic parameters
Descriptive statistics for egg yolk weights at 11 age points and follicle and ovary weights at 73 weeks were presented in Table 1. We observed a curvilinear increase in yolk weights with advancing hen age from the onset of egg laying to 66 weeks and a slight decrease at 72 weeks. Follicle and ovary weights were measured for 1,429 follicle-active hens (1,508 slaughtered in total) at 73 weeks old. These two traits exhibited large coefficients of variation, which may be due to the different oviposition intervals for old hens. All phenotypic values conformed to the normal distribution after rank-based inverse normal transformation.
Table 1. Descriptive statistics of yolk and ovary traits for the F2 population.
| Trait(weight) | N | Mean (g) | SD | CV(%) | Max | Min |
|---|---|---|---|---|---|---|
| FYW | 1,494 | 9.04 | 0.96 | 10.58 | 13.00 | 6.10 |
| YW32 | 1,473 | 13.75 | 1.04 | 7.57 | 17.10 | 9.40 |
| YW36 | 1,464 | 14.54 | 1.21 | 8.30 | 18.70 | 10.50 |
| YW40 | 1,476 | 15.31 | 1.23 | 8.04 | 19.50 | 11.50 |
| YW44 | 1,420 | 15.82 | 1.40 | 8.86 | 20.70 | 10.50 |
| YW48 | 1,226 | 16.01 | 1.37 | 8.58 | 20.80 | 12.00 |
| YW52 | 1,225 | 16.42 | 1.43 | 8.72 | 21.80 | 11.70 |
| YW56 | 1,348 | 16.65 | 1.58 | 9.52 | 22.90 | 11.30 |
| YW60 | 1,364 | 16.96 | 1.55 | 9.15 | 22.90 | 12.90 |
| YW66 | 1,304 | 16.97 | 1.67 | 9.87 | 24.90 | 11.20 |
| YW72 | 1,253 | 15.62 | 1.73 | 11.08 | 21.80 | 7.30 |
| FW | 1,439 | 29.72 | 10.37 | 35.17 | 60.30 | 2.12 |
| OW | 1,439 | 4.23 | 1.50 | 35.72 | 14.31 | 1.53 |
Abbreviations: FW = follicle weight; FYW = yolk weight of first egg; N = number of samples; OW = ovary weight; SD = standard deviation; YW = yolk weight; YW32, YW36, YW40, YW44, YW48, YW52, YW56, YW60, YW66, and YW72 = yolk weight of 32, 36, 40, 44, 48, 52, 56, 60, 66, and 72 weeks of age, respectively.
We quantified the additive genetic variation in liability to yolk weights at the different ages captured by eligible GWAS markers. Univariate GCTA analyses revealed that all yolk weights had moderate heritable patterns (Table 2), and the highest SNP-based heritability estimate was found in YW40 (h 2 = 0.38). Moreover, bivariate GCTA analyses indicated that yolk weights at the various ages exhibited highly and positively correlation, especially for yolk weights at neighboring time points. At the beginning of the entire laying stage, the yolk weight of the first egg (FYW) showed slightly lower genetic correlations with the yolk weights at the following ages (r g<0.60), compared with those among yolk weights from 32 to 72 weeks of age (r g>0.80). Notably, the follicle weight at 73 weeks had low phenotypic (0.02 to 0.17) and genetic (-0.003 to 0.52) correlation with yolk weights at the 11 age points. Similarly, the ovary weight also poorly correlated with yolk weights, and the phenotypic and genetic correlation coefficients ranged from 0.02 to 0.13 and -0.02 to 0.40, respectively. However, the genetic correlation between ovary weight and follicle weight was relatively high (0.86).
Table 2. Genetic parameters of yolk and ovary weights.
| Traits | FYW | YW32 | YW36 | YW40 | YW44 | YW48 | YW52 | YW56 | YW60 | YW66 | YW72 | FW | OW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F YW | 0.34(0.04) | 0.52(0.09) | 0.58(0.09) | 0.32(0.10) | 0.35(0.11) | 0.52(0.11) | 0.46(0.11) | 0.35(0.12) | 0.44(0.10) | 0.29(0.12) | 0.29(0.11) | -0.003(0.14) | -0.02(0.13) |
| YW32 | 0.28 | 0.34(0.04) | 1.00(0.02) | 0.99(0.02) | 0.97(0.03) | 0.99(0.04) | 0.82(0.07) | 0.81(0.08) | 0.80(0.06) | 0.80(0.07) | 0.80(0.07) | 0.52(0.12) | 0.35(0.12) |
| YW36 | 0.24 | 0.61 | 0.31(0.04) | 1.00(0.02) | 1.00(0.03) | 0.99(0.04) | 0.88(0.06) | 0.87(0.07) | 0.92(0.05) | 0.86(0.06) | 0.85(0.06) | 0.34(0.14) | 0.40(0.12) |
| YW40 | 0.22 | 0.64 | 0.63 | 0.38(0.04) | 0.99(0.02) | 1.00(0.03) | 0.91(0.04) | 0.93(0.05) | 0.96(0.04) | 0.83(0.05) | 0.87(0.05) | 0.34(0.16) | 0.34(0.12) |
| YW44 | 0.18 | 0.56 | 0.56 | 0.65 | 0.31(0.04) | 0.99(0.04) | 0.92(0.05) | 0.95(0.05) | 0.98(0.04) | 0.89(0.05) | 0.86(0.05) | 0.45(0.13) | 0.35(0.12) |
| YW48 | 0.21 | 0.56 | 0.54 | 0.63 | 0.58 | 0.27(0.05) | 0.89(0.05) | 0.95(0.05) | 1.00(0.04) | 0.89(0.05) | 0.83(0.06) | 0.39(0.14) | 0.28(0.14) |
| YW52 | 0.20 | 0.50 | 0.50 | 0.60 | 0.55 | 0.61 | 0.29(0.05) | 1.00(0.04) | 0.97(0.04) | 0.90(0.05) | 0.91(0.05) | 0.22(0.15) | 0.21(0.14) |
| YW56 | 0.18 | 0.47 | 0.45 | 0.57 | 0.53 | 0.55 | 0.61 | 0.25(0.04) | 1.00(0.03) | 0.91(0.05) | 0.97(0.04) | 0.32(0.16) | 0.35(0.14) |
| YW60 | 0.19 | 0.49 | 0.48 | 0.59 | 0.53 | 0.60 | 0.60 | 0.63 | 0.31(0.05) | 0.94(0.04) | 0.95(0.04) | 0.39(0.13) | 0.27(0.13) |
| YW66 | 0.12 | 0.44 | 0.46 | 0.57 | 0.52 | 0.56 | 0.59 | 0.60 | 0.60 | 0.31(0.05) | 0.96(0.03) | 0.15(0.15) | 0.17(0.14) |
| YW72 | 0.12 | 0.41 | 0.38 | 0.50 | 0.46 | 0.50 | 0.50 | 0.55 | 0.54 | 0.58 | 0.35(0.05) | 0.19(0.14) | 0.19(0.13) |
| FW | 0.02 | 0.14 | 0.17 | 0.16 | 0.13 | 0.14 | 0.11 | 0.07 | 0.14 | 0.06 | 0.10 | 0.16(0.04) | 0.86(0.09) |
| OW | 0.02 | 0.12 | 0.09 | 0.06 | 0.08 | 0.04 | 0.08 | 0.07 | 0.05 | 0.03 | 0.05 | 0.13 | 0.20(0.04) |
Diagonal: Heritability estimates. Lower triangle: Phenotypic correlations. Upper triangle: Genetic correlations. Standard errors of the estimates are in parentheses. Abbreviations: FYW = yolk weight of first egg; OW = ovary weight;YW32, YW36, YW40, YW44, YW48, YW52, YW56, YW60, YW66, and YW72 = yolk weight of 32,36,40,44,48,52,56,60,66, and 72 weeks of age, respectively.
Identification of candidate loci by a genome-wide association study
We conducted separate association tests using a univariate model for egg weights at each age point and follicles and ovary weight at 73weeks. The Manhattan plots and the Quantile-Quantile (Q-Q) plots of the yolk weight of first eggs (FYW) are shown as an example in Fig 1, and the other traits are shown in S1–S12 Figs. The detailed messages for genome-wide significant SNPs are listed in Tables 3 and 4, and the descriptions for suggestive significant SNPs are shown in S1 Table. The CPVs of the loci were estimated by a tool of GCTA and ranged from 2.40 to 3.47% for yolk weights (Table 3) and from 2.11 to 3.76% for ovary weights (Table 4).
Fig 1. Manhattan plot (left) and quantile-quantile plot (right) of the observed P values for the yolk weight of first eggs (FYW).
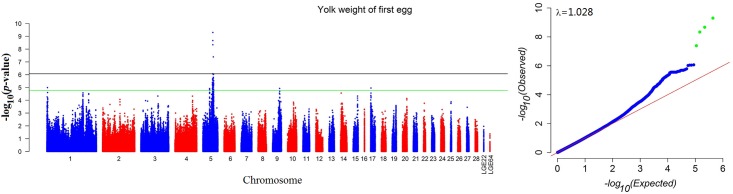
The Manhattan plot indicates the-log10 (observed P values) for genome-wide SNPs (y-axis) plotted against their respective positions on each chromosome (x-axis), and the horizontal black and green lines depict the genome-wide significant (8.43 × 10−7) and suggestive significant (1.69 × 10−5) thresholds, respectively. For the quantile-quantile plot, the x-axis shows the expected-log10-transformed P values, and the y-axis represents the observed-log10-transformed P values. The genomic inflation factors (λ) are shown on the top left in the QQ plots. Green points represent the genome-wide significant associations.
Table 3. Genome-wide association analyses for yolk weight (YW) and follicle weight (FW).
| Traits | SNP ID | Chr | Position | Candidate genes /Nearest Gene | Location | Minor/Majorallele | MAF | N | Effect size a (s.e.m.) | CPV(%) | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FYW | rs313187645 | 5 | 40106943 | DOT1L | intron | A/G | 0.307 | 1,494 | 0.293(0.047) | 3.47 | 5.00E-10 |
| FYW | rs312299419 | 5 | 40167320 | STON2 | intron | T/G | 0.277 | 1,494 | -0.294(0.049) | 3.34 | 2.16E-09 |
| FYW | rs314422825 | 5 | 40194495 | STON2 | exon(missense) | A/G | 0.333 | 1,494 | -0.269(0.046) | 3.18 | 4.61E-09 |
| FYW | rs315568325 | 5 | 42130216 | GALC | downstream_1,172bp | T/C | 0.288 | 1,494 | -0.266(0.048) | 2.80 | 4.08E-08 |
| YW40 | rs16187384 | 23 | 1456741 | RPA2 | Intron | A/G | 0.493 | 1,476 | -0.241(0.044) | 2.77 | 3.76E-08 |
| YW48 | rs313116098 | 1 | 173851584 | BRCA2/ZAR1 | exon/downstream_2,140bp | A/G | 0.298 | 1,226 | 0.242(0.049) | 2.48 | 8.61E-07 |
| YW56 | rs316715137 | 3 | 22975753 | EML4/KCNG3 | intron/downstream_267kb | A/G | 0.291 | 1,348 | -0.263(0.049) | 2.61 | 7.79E-08 |
| YW56 | rs314923267 | 3 | 22971805 | EML4/ KCNG3 | intron/downstream_267kb | T/C | 0.277 | 1,348 | 0.256(0.051) | 2.40 | 6.57E-07 |
| YW66 | rs312474469 | 28 | 3667179 | ELL | intron | T/C | 0.177 | 1,304 | 0.289(0.057) | 2.49 | 4.91E-07 |
| YW66 | rs315213484 | 28 | 3665094 | ELL | upstream_1,134bp | A/C | 0.178 | 1,304 | 0.286(0.057) | 2.47 | 6.95E-07 |
| YW72 | rs312274422 | 3 | 79252286 | ENSGALG00000028314 | downstream_80,003bp | T/G | 0.280 | 1,253 | -0.278(0.050) | 2.79 | 3.53E-08 |
| YW72 | rs312873273 | 17 | 1366852 | RGS3 | intron | C/G | 0.213 | 1,253 | -0.298(0.058) | 2.89 | 3.73E-07 |
| FW | rs14920313 | 1 | 172060693 | DCLK1/SPG20 | intron/upstream_7.3kb | T/C | 0.249 | 1,450 | -0.261(0.051) | 2.60 | 1.09E-07 |
| FW | rs14920355 | 1 | 172084219 | DCLK1 | intron | T/C | 0.246 | 1,450 | -0.257(0.051) | 2.42 | 3.00E-08 |
| FW | rs313140585 | 1 | 172453837 | NBEA/STARD13 | intron/upstream_47.7kb | T/C | 0.277 | 1,450 | -0.242(0.048) | 2.50 | 3.88E-07 |
Abbreviations: Chr = chromosome; CPV = contribution to phenotype variance; FW = follicle weight; FYW = first yolk weight; MAF = minor allele frequency; N = number of samples; SNP = single nucleotide polymorphism; YW40,48,56,66, and 72 = yolk weights of 40,48,56,66,and 72 weeks, respectively.
a Effect size means the effect of minor alleles. Positive/negative effect size value means that the substitution of major allele to minor allele can lead to heavier/lighter yolk or ovary weight.
Table 4. Significant single nucleotide polymorphism (SNP) markers associated with ovary weight (OW).
| SNP | Position | Candidate genes /Nearest Gene | Location | Minor/Majorallele | MAF | N | Effect size a (s.e.m.) | CPV(%) | P value |
|---|---|---|---|---|---|---|---|---|---|
| Chr 1 | |||||||||
| rs317441976 | 169051794 | DLEU7 | upstream(71.3kb) | T/G | 0.412 | 1,450 | -0.278(0.048) | 3.76 | 9.65E-09 |
| rs316815773 | 169670916 | NEK5 | Intron | T/C | 0.489 | 1,450 | 0.247(0.046) | 3.10 | 9.49E-08 |
| rs314051078 | 169046176 | DLEU7 | upstream(76.9kb) | A/G | 0.387 | 1,450 | -0.255(0.048) | 3.24 | 1.16E-07 |
| rs317796161 | 169018288 | DLEU7 | upstream(104.8kb) | C/G | 0.486 | 1,450 | 0.249(0.047) | 2.79 | 1.83E-07 |
| rs15500685 | 169696167 | NEK3 | intron | T/C | 0.477 | 1,450 | 0.240(0.046) | 2.96 | 2.18E-07 |
| rs312737959 | 169709920 | CKAP2 | intron | A/G | 0.443 | 1,450 | -0.250(0.048) | 2.98 | 2.21E-07 |
| rs13972990 | 169562091 | DHRS12 | downstream(6,152bp) | T/C | 0.457 | 1,450 | -0.246(0.048) | 2.93 | 3.96E-07 |
| Chr 28 | |||||||||
| rs315788897 | 1699155 | ACER1b/ACSBG2 | intron/intron | T/C | 0.144 | 1,450 | 0.375(0.059) | 3.18 | 1.70E-10 |
| rs316105069 | 2123244 | SCAMP4 | downstream(493bp) | A/G | 0.136 | 1,450 | 0.382(0.060) | 3.14 | 1.94E-10 |
| rs13663720 | 1788269 | ADAMTS10 | intron | T/C | 0.155 | 1,450 | 0.358(0.059) | 3.11 | 1.89E-09 |
| rs317889060 | 2012197 | DOT1L | intron | T/C | 0.228 | 1,450 | 0.299(0.051) | 2.88 | 7.43E-09 |
| rs318099911 | 2163700 | ABHD17A | upstream(3,437bp) | T/C | 0.143 | 1,450 | 0.341(0.059) | 2.51 | 8.30E-09 |
| rs316358363 | 1626264 | ACSBG2 | upstream(4,223bp) | A/G | 0.161 | 1,450 | 0.320(0.056) | 2.43 | 1.11E-08 |
| rs314735191 | 1514399 | NRTN | upstream(4,727bp) | T/C | 0.186 | 1,450 | 0.298(0.053) | 2.57 | 1.77E-08 |
| rs314625273 | 2230957 | ONECUT3 | downstream(2,970bp) | C/G | 0.128 | 1,450 | 0.350(0.062) | 2.44 | 2.30E-08 |
| rs316444293 | 2342019 | TCF3 | intron | A/C | 0.159 | 1,450 | 0.310(0.057) | 2.39 | 6.18E-08 |
| rs313040427 | 1712749 | ANP32B | intron | T/C | 0.165 | 1,450 | 0.306(0.056) | 2.47 | 6.73E-08 |
| rs314385559 | 2367109 | TCF3 | downstream(1,276bp) | A/G | 0.163 | 1,450 | 0.302(0.057) | 2.3 | 1.11E-07 |
| rs14305841 | 2185417 | REXO1 | intron | T/C | 0.133 | 1,450 | 0.325(0.061) | 2.11 | 1.19E-07 |
| rs318236639 | 1929393 | OAZ | downstream(777bp) | A/G | 0.243 | 1,450 | 0.259(0.049) | 2.45 | 1.27E-07 |
| rs16211139 | 1602011 | RFX2 | downstream(10.9kb) | C/G | 0.239 | 1,450 | 0.262(0.049) | 2.56 | 1.33E-07 |
| rs317858034 | 2042162 | AP3D1 | intron | T/C | 0.212 | 1,450 | 0.277(0.053) | 2.51 | 2.10E-07 |
| rs317537249 | 1843976 | ACTL9 | downstream(2,988bp) | T/C | 0.275 | 1,450 | 0.239(0.046) | 2.2 | 2.18E-07 |
| rs313479236 | 1821752 | ZAP70 | downstream(4,220bp) | T/C | 0.226 | 1,450 | 0.263(0.051) | 2.47 | 2.28E-07 |
| rs316676992 | 1849612 | ACTL9/MUC16 | upstream(1,333 bp)/downstream(2,188 bp) | A/G | 0.275 | 1,450 | 0.236(0.046) | 2.13 | 3.30E-07 |
| rs317641108 | 1926277 | OAZ/C19orf35 | downstream(3,893 bp)/intron | A/G | 0.238 | 1,450 | 0.253(0.049) | 2.32 | 3.51E-07 |
| rs315715629 | 1761595 | ADAMTS10 | intron | T/C | 0.243 | 1,450 | 0.246(0.049) | 2.21 | 6.13E-07 |
| rs313972624 | 1732609 | MYO1F | intron | A/C | 0.241 | 1,450 | 0.245(0.049) | 2.21 | 6.90E-07 |
| rs312587681 | 2335214 | TCF3 | intron | T/C | 0.158 | 1,450 | 0.288(0.058) | 2.03 | 6.99E-07 |
| rs314330209 | 1990610 | DOT1L | intron | A/G | 0.274 | 1,450 | 0.230(0.046) | 2.13 | 7.26E-07 |
| rs14305824 | 2179908 | REXO1 | intron | A/G | 0.176 | 1,450 | 0.269(0.054) | 1.95 | 7.95E-07 |
Abbreviations: Chr = chromosome; CPV = contribution to phenotype variance; MAF = minor allele frequency; N = number of samples; SNP = single nucleotide polymorphism.
a Effect size means the effect of minor alleles. Positive/negative effect size value means that the substitution of major allele to minor allele can lead to heavier/lighter yolk or ovary weight.
Yolk weight
A total of 11 significant loci distributed on chromosome 1, 3, 5, 17, and 28 were associated with YW at different age points including the age of the hen at the first egg(AFE) and 40, 48, 56, 66,and 72 weeks of age (Table 3). Eleven genes, including DOT1L, STON2, GALC, RPA2, BRCA2, ZAR1, EML4, KCNG3, ELL, ENSGALG00000028314 and RGS3F were detected as harboring or being near these significant loci (Table 3).
Follicle weight
Three SNPs located on chromosome 1 (GGA1) were significantly associated with follicle weight (FW). Detailed SNP messages are presented in Table 3. These three significant SNPs were all identified in the intron of annotated genes. Two SNPs, rs14920313 and rs14920355, were located in the doublecortin-like kinase 1 (DCLK1) gene, and rs313140585 was identified in the neurobeachin (NBEA) gene. In addition, two other genes, spastic paraplegia 20 (SPG20) and StAR-related lipid transfer domain containing 13 (STARD13), were found near the significant SNPs.
Ovary weight
Thirty-one SNPs showed significant association with ovary weight (Table 4), seven of which were located within a 700-kb region (169.01–169.71Mb) on GGA1. Furthermore, the other 24 SNPs were located in an 800-kb region spanning from 1.6 to 2.4 Mb on GGA28. Of the loci in GGA1, three were located in annotated genes, including NIMA-related kinase 5(NEK5), NIMA-related kinase 3(NEK3), and cytoskeleton associated protein 2 (CKAP2). Twelve of the 24 SNPs in GGA28 were located in known genes, and the remainders were located 0.4 to 10 kb away from the nearest known genes, which included 19 annotated genes (Table 4).
Considering the large number of significant SNPs (24 loci) in a narrow region (800kb) in GGA28, we speculated that strong LD exists in this region. Hence, the LD analysis was performed in the genomic region from 1.60 to 2.40 Mb, which contained 744 SNPs, and 76 small scale blocks were observed in this region (S2 Table). Eight out of the 24 significant SNPs fell into six blocks, and the remaining 18 SNPs were not located in any of these blocks (Fig 2 and S2 Table). The results indicated that the significant loci in GGA28 were in poor LD with each other and that the loci may play independent roles.
Fig 2. Regional plot for single nucleotide polymorphisms(SNPs) at GGA28 spanning from 1.60 to 2.40 Mb.
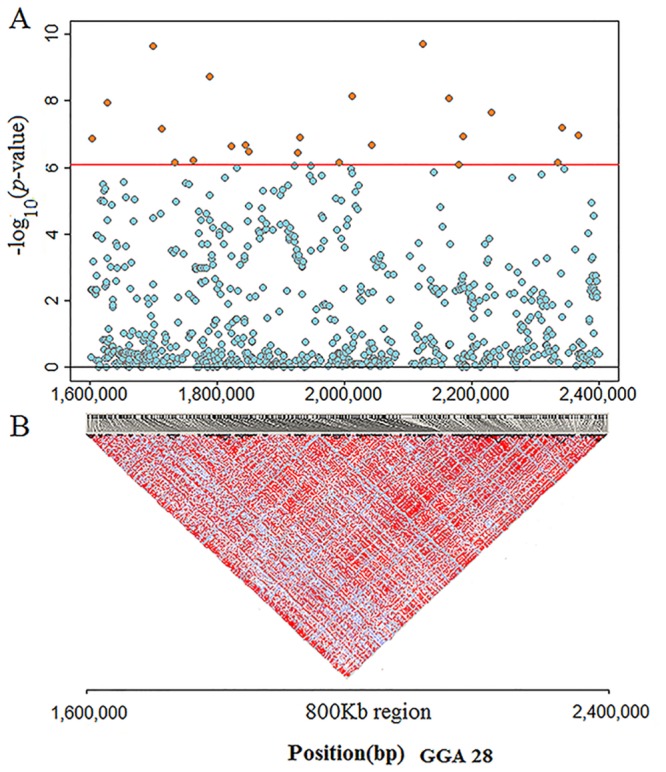
Plot A: In the region of 1.60 to 2.40 Mb,744 SNPs (orange and blue points) were analyzed for their association with ovary weight (OW). The-log10 (observed P values) of the SNPs (y-axis) are presented according to their chromosomal positions (x-axis). Twenty-four SNPs (orange points) reached genome-wide significance level (black line, 8.43 × 10−7). Plot B: Seventy-six small-scale blocks were observed in this region.
Discussion
The population used for this study was anF2 cross population, which could maximize the differences of the traits and increase the power to identify the QTLs for traits that differed between the breeds. Compared with previous association analyses in chickens, our analysis used a higher density (600K) SNP array covering chromosomes 1–28 and two unassigned linkage groups. In addition, this F2 population consisted of 1,512 hens, which is the largest population used for GWA analysis of egg traits so far, and therefore, the novel genomic region and loci revealed by the current study should be accurate and reliable. Several previous studies detected causal genomic regions or genes that controlled yolk weight using GWA analysis [17, 18]. However, these studies utilized phenotypes from limited age points, assuming that the genetic architecture underlying this trait does not vary with the aging process. In the current study, yolk weights of 11 age points from the onset of egg laying to 72 weeks of age were employed to detect the dynamic genetic control for yolk weights. In addition, this study was the first GWA analysis for ovary and follicle weight in chicken.
Yolk weight
By performing GWA analysis, we identified 11 associations with yolk weight at six age points at genome-wide significance levels (P<8.33 × 10−7). These significant loci, which occurred on GGA 1, 3, 5, 17, and 28, had no overlap with previously reported QTLs. This might due to the specially designed F2crosses used in this study. In addition, the significant loci for each age point varied completely, which was notable because it indicated that the genetic control for yolk weight was age-dependent. However, the genetic correlations among yolk weights at each age points were relatively high (Table 2), suggesting certain consistent genetic variants existed to control yolk weight along with the aging process. This contradiction might be explained by the existence of suggestive significant common genomic regions or loci that associated with yolk weight at different age points (S1 Table). In contrast, the 11 genome-wide significant SNPs were considered more powerful and specific loci for yolk weight. These loci were located near or in 16 annotated genes.
One candidate gene with a biologically plausible function is zygote arrest 1(ZAR1), which is downstream (2,140bp) of rs313116098, an SNP significantly associated with YW48.ZAR1 is considered to be an oocyte-specific maternal-effect gene [32] and is thought to function in the initiation of embryogenesis in many vertebrate species including humans, pigs, cattle, sheep, mice, rats, and frogs [33]. Michailidis et al. (2010) revealed that this gene was preferentially expressed in chicken oocytes, ovaries, testes, and embryos during embryonic development [34]. This gene is related to the development of follicular oocytes; however, its involvement in yolk weight is not clear.
Three genes, stonin 2 (STON2), galactosylceramidase(GALC), and potassium channel, voltage-gated modifier subfamily G, member 3 (KCNG3) have functions related to nerve regulation. In fact, nerve networks have close relationships with ovarian follicle development. Specifically, the maturity and ovulation of the yolky follicle is profusely innervated by both adrenergic and cholinergic fibers [35]. STON2encodes a protein that participates in synaptic vesicle recycling through interaction with synaptotagmin 1, which is required for neurotransmission [36]. GALC encodes galactosylceramidase, which is related to globoid cell leukodystrophy in humans [37]. KCNG3encodes for the voltage-gated potassium (Kv) channels, which regulate neurotransmitter release, insulin secretion, and neuronal excitability [38]. Neurons are mainly present within the thecal layers of the largest follicles. These neurons provide numerous neurochemicals (catecholamines, neurotrophins, vasoactive intestinal peptide) to the follicle [39]. Therefore, the differences among yolk weights of first eggs may be partly led by the variable maturity of the sequential follicles that are regulated by the nerve networks. The remaining screened genes have no direct relevance with the yolk deposition orovarian follicle development, and this maybe due to insufficient knowledge about these genes in chickens.
Follicle weight
Follicles that connect to the ovary by pedicles are the sites for yolk formation. Hence, the mass of ovarian follicles represent the capacity of sequential yolk production of laying hens. However, the phenotypic or genetic correlation coefficients between follicle weight and yolk weight at each age point were all very low, even with the yolk weight of 72 weeks of age. This indicated that the genetic architecture might be disparate for single yolk weights and the mass of existing ovarian follicles at a certain age.
Three loci (rs14920313, rs14920355, and rs313140585) in GGA1 were significantly associated with follicle weight, and two candidate genes, SPG20 and STARD13, harbored or were near these loci. SPG20 encodes a protein containing an MIT (microtubule interacting and trafficking molecule) domain and can cause protein translocation to the plasma membrane when stimulated with epidermal growth factor (EGF) [40], whereas EGF, as one of the main intra-ovarian hormones produced by the germinal disc and granulosa cells, can stimulate growth and reduce atresia in follicles [41, 42]. STARD13 encodes a protein possessing C-terminal STAR-related lipid transfer domain, which is a classical type of lipid transporter [43, 44]. In mammals,15proteins, STARD1-STARD15, possess a START domain, and cholesterol, 25-hydroxycholesterol, phosphatidylcholine, phosphatidylethanolamine, and ceramides are ligands for STARD1/STARD3/STARD5, STARD5, STARD2/STARD10, STARD10, and STARD11, respectively [43]. In the current study, the STARD13 gene was identified with follicle weight, indicating its crucial role in lipid metabolism in the process of yolk deposition, an association that needs further investigation.
Ovary weight
The SNPs that were significantly associated with the follicle-free ovary weight were distributed on two chromosomes, GGA1 and GGA28. The loci on GGA1 (169.01–169.70Mb) were upstream from and only 3 Mb away from the loci (172.06–172.45Mb), which were related to follicle weight, whereas the candidate genes in these two regions were completely different. Six genes containing these loci of interest or located near these loci in GGA1 were identified. However, only one gene hasa function plausible for ovarian development. This gene is DHRS12, a gene that encodes the enzyme of the short-chain dehydrogenases/reductases (SDR) family, which metabolizes many different compounds, including steroid hormones [45]. The ovarian steroid hormones include estrogen, testosterone, and progesterone; these are synthesized by follicular interstitial and granulosa cells, and estrogen can stimulate the liver for the synthesis of the yolky lipid and protein precursors [2]. Hence, DHRS12is closely related to ovarian development.
For the loci in GGA 28, the most significant SNP, rs316105069(P = 1.7 × s3-10), was located in two genes, including alkaline ceramidase 1 (ACER1b) andacyl-CoA synthetase bubblegum family member 2 (ACSBG2). These two genes are closely related to the biological process of yolk formation, such as lipoprotein synthesis, lipid droplet promotion, and lipid carrier. ACER1can catalyze the hydrolysis of very long chain ceramides to sphingosine, while sphingosine is absorbed and converted to palmitic acid and then acylated into chylomicron triglycerides (TGs), the materials needed for the synthesis of lipoproteins [46]. Another gene, ACSBG2,encodes proteins belonging to the acyl-CoA synthetase family, which can channel lipids into nascent lipid droplets in mammals [47]. Claire et al. (2013) demonstrated that the ACSBG2 gene is significantly associated with abdominal fat deposition and exhibited functions related to lipid metabolism in chickens [48].
Conclusions
In summary, the current study reports for the first time a GWA analysis on ovary and follicle weights in chickens. Our results revealed 12, 3, and 31 genome-based significant SNPs for yolk, follicle, and ovary weight, respectively, by GWA analysis with a 600K high-density SNP array. The GWA analysis for yolk weights at multiple age points suggested that the genetic control for yolk weight is age-dependent. A list of candidate genes such as ZAR1, STARD13, ACER1b, ACSBG2, and DHRS12 were identified for their plausible function in yolk and follicle developments. Our findings establish a foundation for follow-up studies and create a better understanding of the molecular controls involved in the development of the ovary and its hierarchy of follicles.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(XLSX)
(XLSX)
Acknowledgments
We are grateful to the staff of Jiangsu Institute of Poultry Science for their technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The current research was funded in part by the National Natural Science Foundation of China (31472084), the National High Technology Development Plan of China (2013AA102501), Programs for Changjiang Scholars and Innovative Research in University (IRT1191), and China Agriculture Research Systems (CARS-41).
References
- 1. Burley RW, Vadehra DV. The avian egg: chemistry and biology. New York: John Wiley and Sons; 1989. [Google Scholar]
- 2. Nys Y, Bain M, Van Immerseel F. Improving the safety and quality of eggs and egg products. UK: Woodhead Publishing Limited; 2011. [Google Scholar]
- 3. Mann K, Mann M. The chicken egg yolk plasma and granule proteomes. Proteomics. 2008;8(1):178–191. [DOI] [PubMed] [Google Scholar]
- 4. Kovacs-Nolan J. Advances in the value of eggs and egg components for human health. J Agric Food Chem. 2005;53(22):8421–8431. [DOI] [PubMed] [Google Scholar]
- 5. Hartmann C, Wilhelmson M. The hen's egg yolk: a source of biologically active substances. World Poultry Sci J. 2001;57(1):13–28. [Google Scholar]
- 6. Anton M, Nau F, Nys Y. Bioactive egg components and their potential uses. Proceedings of the XVII European Symposium on the Quality of Poultry Meat and XI European Symposium on the Quality of Eggs and Egg Products, Golden Tulip Parkhotel Doorwerth, Netherlands, 2005 May 23–26. 2005;62(3):429–438. [Google Scholar]
- 7. Mine Y, Kovacs-Nolan J. New insights in biologically active proteins and peptides derived from hen egg. World Poultry Sci J. 2006;62(1):87–95. [Google Scholar]
- 8. Shen X, Steyrer E, Retzek H, Sanders EJ, Schneider WJ. Chicken oocyte growth: receptor-mediated yolk deposition. Cell Tissue Res. 1993;272(3):459–471. [DOI] [PubMed] [Google Scholar]
- 9. George R, Barber DL, Schneider WJ. Characterization of the chicken oocyte receptor for low and very low density lipoproteins. J Biol Chem. 1987;262:16838–16847. [PubMed] [Google Scholar]
- 10. Deeley RG. The effects of estrogen on apolipoprotein synthesis. Can J Biochem Cell Biol. 1985;63(8):882–889. [DOI] [PubMed] [Google Scholar]
- 11. Evans AJ, Burley RW. Proteolysis of apoprotein B during the transfer of very low density lipoprotein from hens' blood to egg yolk. J Biol Chem. 1987;262(2):501–504. [PubMed] [Google Scholar]
- 12. Nimpf J, Schneider W. The VLDL receptor: an LDL receptor relative with eight ligand binding repeats, LR8. Atherosclerosis. 1998;141(2):191–202. [DOI] [PubMed] [Google Scholar]
- 13. Mac Lachlan I, Nimpf J, Schneider WJ. Avian riboflavin binding protein binds to lipoprotein receptors in association with vitellogenin. J Biol Chem. 1994;269(39):24127–24132. [PubMed] [Google Scholar]
- 14. Suk Y, Park C. Effect of breed and age of hens on the yolk to albumen ratio in two different genetic stocks. Poultry Sci. 2001;80(7):855–858. [DOI] [PubMed] [Google Scholar]
- 15. Nangsuay A, Ruangpanit Y, Meijerhof R, Attamangkune S. Yolk absorption and embryo development of small and large eggs originating from young and old breeder hens. Poultry Sci. 2011;90(11):2648–2655. [DOI] [PubMed] [Google Scholar]
- 16. Tuiskula-Haavisto M. Mapping of quantitative trait loci affecting quality and production traits in egg layers. Poultry Sci. 2002;81(7):919–927. [DOI] [PubMed] [Google Scholar]
- 17. Liu W, Li D, Liu J, Chen S, Qu L, Zheng J, et al. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. PloS one. 2011;6(12):1427–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolc A, Arango J, Jankowski T, Dunn I, Settar P, Fulton J, et al. Genome-wide association study for egg production and quality in layer chickens. J Anim Breed Genet. 2014;131(3):173–182. 10.1111/jbg.12086 [DOI] [PubMed] [Google Scholar]
- 19. Sasaki O, Odawara S, Takahashi H, Nirasawa K, Oyamada Y, Yamamoto R, et al. Genetic mapping of quantitative trait loci affecting body weight, egg character and egg production in F2 intercross chickens. Anim Genet. 2004;35(3):188–194. [DOI] [PubMed] [Google Scholar]
- 20. Kranis A, Gheyas AA, Boschiero C, Turner F, Yu L, Smith S, et al. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics. 2013;14:59 10.1186/1471-2164-14-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. [DOI] [PubMed] [Google Scholar]
- 22. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–223. 10.1016/j.ajhg.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SH, DeCandia TR, Ripke S, Yang J, Schizophrenia Psychiatric Genome-Wide Association Study C, International Schizophrenia C, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. 10.1038/ng.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34(1):100–105. 10.1002/gepi.20430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao JH. gap: Genetic Analysis Package. R package version 1.1–12. Available: http://cranr-projectorg/web/packages/gap/indexhtml. 2014.
- 29. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. [DOI] [PubMed] [Google Scholar]
- 30. McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. 10.1093/bioinformatics/btq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford). 2011;2011(1):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu X. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33(2):187–191. [DOI] [PubMed] [Google Scholar]
- 33. Wu X, Wang P, Brown CA, Zilinski CA, Matzuk MM. Zygote arrest 1 (Zar1) is an evolutionarily conserved gene expressed in vertebrate ovaries. Biol Reprod. 2003;69(3):861–867. [DOI] [PubMed] [Google Scholar]
- 34. Michailidis G, Argiriou A, Avdi M. Expression of chicken zygote arrest 1 (Zar1) and Zar1-like genes during sexual maturation and embryogenesis. Vet Res Commun. 2010;34(2):173–184. 10.1007/s11259-010-9343-z [DOI] [PubMed] [Google Scholar]
- 35. Odlind V, Rosén M, Stenbeck M. Female genital organs. Acta Oncologica (Stockholm, Sweden). 1995;34 suppl 4(1):53–59. [DOI] [PubMed] [Google Scholar]
- 36. Maritzen T. Stonins—specialized adaptors for synaptic vesicle recycling and beyond? Traffic. 2010;11(1):8–15. 10.1111/j.1600-0854.2009.00971.x [DOI] [PubMed] [Google Scholar]
- 37. Wenger DA. Molecular genetics of Krabbe disease (globoid cell leukodystrophy): diagnostic and clinical implications. Hum Mutat. 1997;4(4):268–279. [DOI] [PubMed] [Google Scholar]
- 38. Brooke RE. Kv3 voltage-gated potassium channels regulate neurotransmitter release from mouse motor nerve terminals. EurJNeurosci. 2004;20(12):3313–3321. [DOI] [PubMed] [Google Scholar]
- 39. Onagbesan O. Intra-ovarian growth factors regulating ovarian function in avian species: A review. Anim Reprod Sci. 2009;111:121–140. 10.1016/j.anireprosci.2008.09.017 [DOI] [PubMed] [Google Scholar]
- 40. Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol Biol Cell. 2007;18(5):1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y. Epidermal growth factor (EGF) receptor ligands in the chicken ovary: I. Evidence for heparin-binding EGF-like growth factor (HB-EGF) as a potential oocyte-derived signal to control granulosa cell proliferation and HB-EGF and kit ligand expression. Endocrinology. 2007;148(7):3426–3440. [DOI] [PubMed] [Google Scholar]
- 42. Yao HH, Bahr JM. Germinal disc-derived epidermal growth factor: a paracrine factor to stimulate proliferation of granulosa cells. Biol Reprod. 2001;64(1):390–395. [DOI] [PubMed] [Google Scholar]
- 43. Alpy F. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–2801. [DOI] [PubMed] [Google Scholar]
- 44. Soccio RE. StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J Biol Chem. 2003;278(25):22183–22186. [DOI] [PubMed] [Google Scholar]
- 45. Penning TM. Hydroxysteroid dehydrogenases and pre-receptor regulation of steroid hormone action. Hum Reprod Update. 2003;9(3):193–205. [DOI] [PubMed] [Google Scholar]
- 46. Nilsson A, Duan RD. Absorption and lipoprotein transport of sphingomyelin. J Lipid Res. 2006;47(1):154–171. [DOI] [PubMed] [Google Scholar]
- 47. Fujimoto Y. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of Acyl-CoA synthetase. Biol Pharm Bull. 2006;29(11):2174–2180. [DOI] [PubMed] [Google Scholar]
- 48. Claire D’Andre H, Paul W, Shen X, Jia X, Zhang R, Sun L, et al. Identification and characterization of genes that control fat deposition in chickens. J Anim Sci Biotechnol. 2013;4(1):22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


