Abstract
Cytotoxic T lymphocyte–associated antigen-4 (CTLA-4) blockade can promote antitumor T cell immunity and clinical responses. The mechanism by which anti–CTLA-4 antibodies induces antitumor responses is controversial. To determine the effects of CTLA-4 blockade on the T cell repertoire, we used next-generation deep sequencing to measure the frequency of individual rearranged T cell receptor β (TCRβ) genes, thereby characterizing the diversity of rearrangements, known as T cell clonotypes. CTLA-4 blockade in patients with metastatic castration-resistant prostate cancer and metastatic melanoma resulted in both expansion and loss of T cell clonotypes, consistent with a global turnover of the T cell repertoire. Overall, this treatment increased TCR diversity as reflected in the number of unique TCR clonotypes. The repertoire of clonotypes continued to evolve over subsequent months of treatment. Whereas the number of clonotypes that increased with treatment was not associated with clinical outcome, improved overall survival was associated with maintenance of high-frequency clones at baseline. In contrast, the highest-frequency clonotypes fell with treatment in patients with short overall survival. Stably maintained clonotypes included T cells having high-avidity TCR such as virus-reactive T cells. Together, these results suggest that CTLA-4 blockade induces T cell repertoire evolution and diversification. Moreover, improved clinical outcomes are associated with less clonotype loss, consistent with the maintenance of high-frequency TCR clonotypes during treatment. These clones may represent the presence of preexisting high-avidity T cells that may be relevant in the antitumor response.
INTRODUCTION
Cytotoxic T lymphocyte–associated antigen-4 (CTLA-4) is a co-inhibitory receptor that controls T cell activation during initiation and maintenance of adaptive immune responses. CTLA-4 binds to B7 ligands expressed on antigen-presenting cells (APCs) with higher affinity than the costimulatory molecule CD28, and both its gene and surface expression are induced during T cell activation upon APC interaction (1). By competing for and binding to B7 ligands, CTLA-4 inhibits T cell proliferation and cytokine expansion. Monoclonal antibodies (mAbs) that block CTLA-4 interactions with B7 may enhance effector T cell (Teff) function (2) and may also inhibit regulatory T cell (Treg) activity (3, 4), leading to regression of established tumors in mouse models (5). Because CTLA-4 is constitutively expressed on Tregs, antibodies that bind CTLA-4 have also been recently reported to operate independently of CTLA-4–B7 interactions by triggering antibody-dependent cell-mediated cytotoxicity (ADCC) and Fc receptor–mediated elimination of Tregs within tumors in mouse models (6–8).
Two fully human mAbs to CTLA-4, ipilimumab and tremelimumab, have undergone phase 3 studies in human studies (9, 10), with the former being U.S. Food and Drug Administration–approved in the treatment of metastatic melanoma. Both antibodies induce tumor response patterns that manifest as disease stabilization and/or delayed objective responses. These mAbs are also associated with toxicities attributable to inflammation and breaking of self-tolerance in multiple organs. In a randomized phase 3 trial, ipilimumab extended overall survival in patients with previously treated metastatic or unresectable melanoma and, in a subset of patients, produced durable responses (11). Ipilimumab can also induce clinical responses in patients with metastatic castration-resistant prostate cancer (CRPC) (12, 13). Anti–CTLA-4 mAbs have been combined with other agents with complementary immunomodulatory properties, including cytokines such as granulocyte-macrophage colony-stimulating factor (GM-CSF) that expand circulating APCs and thus may promote antigen presentation of endogenous tumor antigens and/or ADCC (14, 15).
In humans, the mechanism of antitumor activity is not fully understood. Disruption of CTLA-4 and B7 interactions by mAbs with ipilimumab or tremelimumab enhances both Teff and Treg proliferation, leading to suggestions that a ratio favoring Teffs over Tregs would promote tumor regression (4, 16). The potential importance of baseline T cell fitness is underscored by factors that have been associated with clinical benefit from ipilimumab and are suggestive of T cell activation and/or proliferation upon treatment with CTLA-4 blockade: elevated absolute lymphocyte counts (17), expression of inflammatory immune-related markers (18), preexisting responses to tumor antigen (19), and increased immune cell infiltration of tumors (20, 21). Notably, high baseline frequency of CTLA-4–expressing T cells may also be associated with clinical benefit to ipilimumab (22). These observations suggest that potential responders to treatment may have preexisting, rather than de novo, tumor-specific T cell clones that have been primed by APC with tumor antigens but are attenuated by subsequent CTLA-4 expression and signaling. Because CTLA-4 blockade may lower the threshold of T cell receptor (TCR) signaling to activate a T cell, one consequence of treatment with blocking antibodies would be to increase the diversity of T cell clones by expanding a range of T cells bearing low-affinity TCRs. However, CTLA-4 surface expression also correlates with strong TCR signal strength, likely by high-affinity interactions with corresponding major histocompatibility complex (MHC) ligands or by ligand density (23). Because T cells are selectively enriched for high-affinity TCR-ligand interactions during the normal development of a T cell response, CTLA-4 may preferentially restrict the expansion of cells with stronger TCR affinities, promoting a diverse population of antigen-specific T cells (24). CTLA-4 blockade could reduce the diversity of responding T cells by narrowing the scope of reactive clones to those bearing high-affinity TCRs for their antigens.
To assess whether CTLA-4 blockade promotes global T cell expansion and/or loss of T cell clonotype diversity, we used next-generation sequencing to assess the impact of CTLA-4 blockade on changes to the T cell repertoire. This method can potentially identify the millions of individual clonotypes by high-throughput sequencing of the repertoire of rearranged TCRβ chains in individual patients, using primers designed to read the variable (V), diversity (D), joining (J), and constant (C) gene segments (25). The number of unique VDJ reads determines the frequency of each clonotype. All the distinct TCRβ sequences can be followed serially over time or quantitatively compared between individuals and cohorts (26, 27). We show that anti–CTLA-4 mAb therapy induces immediate and substantial effects on the size and diversity of the T cell repertoire in both CRPC and metastatic melanoma patients, and that these changes continue to evolve with repeated treatment. Although individual patients can have varying degrees of change to their T cell repertoire, we found that those patients with improved overall survival maintained their baseline high-frequency clones.
RESULTS
CTLA-4 blockade remodels the TCR repertoire
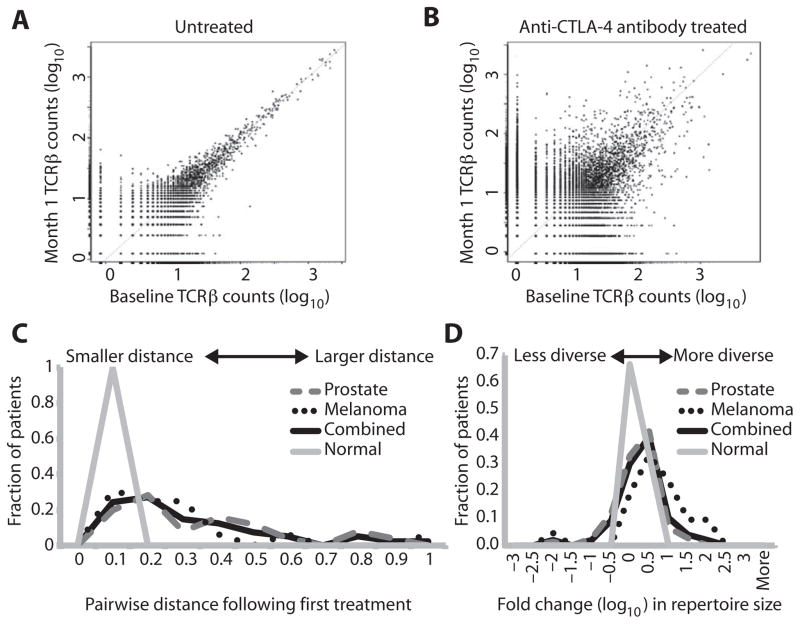
Studies in T cell responses to anti–CTLA-4 mAbs have focused on either specific antigen-directed responses or immunophenotypic changes to T cell subpopulations, rather than on the entire repertoire of T cell clones. To determine the effects of CTLA-4 blockade on the T cell repertoire, we initially assessed whether anti–CTLA-4 blocking antibodies demonstrated changes in abundance in unique clonotypes. We sequenced the TCRβ genes from T cells contained in peripheral blood mononuclear cells (PBMCs) from 25 metastatic CRPC patients treated with ipilimumab and GM-CSF (14), 21 metastatic melanoma patients treated with tremelimumab (28), and 9 untreated healthy control subjects. Prostate cancer patients were treated every 4 weeks in a phase 1/2 trial, and the melanoma patients were treated every 12 weeks in a phase 2 trial. We evaluated the absolute counts at baseline and at 4 weeks of all clonotypes identified by repertoire sequencing. The assay is sensitive to detect rare clonotype frequencies less than 10−6 (25). Clones that do not change in abundance are found on the x=y diagonal, and those that change are found above or below the diagonal. With this approach, untreated control subjects demonstrated minimal variation in individual T cell clonotype abundance from month to month (Fig. 1A). In contrast, every patient who received one treatment of anti–CTLA-4 mAbs developed increases and decreases in absolute clonotype counts (Fig. 1B). To assess the degree of change, we quantified the difference between baseline and month 1 (posttreatment) samples by applying Morisita’s distance to clone count distributions, scaled from 0 to 1 to indicate minimal and maximal distance, respectively (29). This metric is an inverse measure of overlap between two populations (baseline and 4 weeks after treatment). Whereas untreated subjects consistently showed the greatest overlap (minimal travel distance) in repertoires before and 4 weeks after treatment, both prostate cancer and melanoma patients showed a wide distribution of pairwise travel distance, extending out to maximum distance with respect to repertoire change (Fig. 1C). The median distance between untreated samples was 0.039 versus 0.197 for anti–CTLA-4–treated samples (P = 0.0005, Mann-Whitney). These results indicate that anti–CTLA-4 mAbs induce significant changes in clonotype frequencies consistent with T cell repertoire turnover.
Fig. 1. Evolution of T cell repertoire with anti–CTLA-4 antibodies.
(A and B) TCRβ clonotype frequency plots are depicted for a representative untreated individual (A) and for a patient treated with anti–CTLA-4 antibody (B). Each point on the scatter plots represents a single clonotype with normalized log10 clone count graphed at baseline (x axis) and after 1 month (y axis). The increased variance of the low-abundance clones is due to Poisson sampling effects. Clones that are present in only one sample are assigned an arbitrary count of 1 in the sample from which they are missing to permit plotting. (C) The difference between pre- and posttreatment samples (and untreated, sequential, normal samples) was quantified by applying Morisita’s distance to clone count distributions, with 0 indicating minimal distance, and 1 indicating maximal distance. The distribution of distance values was plotted. (D) Repertoire size (that is, sample diversity) was quantified by counting the number of unique clonotypes comprising the top 25th percentile of clones after sorting by abundance. Fold change in repertoire size was calculated, and the distribution of fold change values was plotted on a logarithmic scale.
CTLA-4 blockade influences TCR repertoire diversity
To assess whether anti–CTLA-4 influenced the diversity of the repertoire, we used the metric of repertoire size as a measure of sample diversity. We counted the number of individual clonotypes represented in the top 25th percentile by ranked molecule count after sorting by abundance. Fold changes were then determined after first treatment. This metric is not strongly influenced by rare clonotypes and is therefore relatively stable to sequencing depth differences and different input cell amounts. The metric was calculated for paired pre- and post-treatment patient samples separated by 1 month, as well as untreated control samples, separated by the same time interval. In comparison with untreated subjects, who maintained stable diversity over 1 month, cancer patients treated with anti–CTLA-4 displayed increases in repertoire size beyond the range observed in untreated pairs (Fig. 1D). Thirty-four (45%) of 76 paired CRPC samples and 12 (57%) of 21 paired melanoma samples had more than twofold changes in TCR diversity. Overall, 46 (47%) of all 97 paired samples across prostate and melanoma patients had changes in diversity more than twofold in either direction (Table 1). By comparison, none of the nine untreated sample pairs underwent more than twofold change in diversity (P = 0.005, Fisher’s exact test, two-tailed). The greatest fold increases were observed in melanoma patients, with 43% showing ≥10-fold increases, whereas 9% of all paired CRPC samples demonstrated similar changes (Fig. 1D).
Table 1. CTLA-4 blockade influences T cell diversity.
nd, sample was not evaluated.
| Number of pre/post pairs with fold change in diversity* >2-fold in either direction
|
|||||
|---|---|---|---|---|---|
| Treatment 1 | Treatment 2 | Treatment 3 | Overall | Total pairs | |
| Prostate | 13 | 12 | 9 | 34 | 76 |
| Melanoma | 12 | nd | nd | 12 | 21 |
| Combined | 25 | 12 | 9 | 46† | 97 |
| Untreated controls | 0 | 0 | 0 | 0† | 9 |
Diversity metric is the number of clones represented in the top 25% by ranked molecule count. The metric was determined pre-post for three sequential treatments (1, 2, and 3) separated by 1-month intervals.
Overall anti–CTLA-4–treated versus normal, P = 0.005 (Fisher’s exact test, two-tailed).
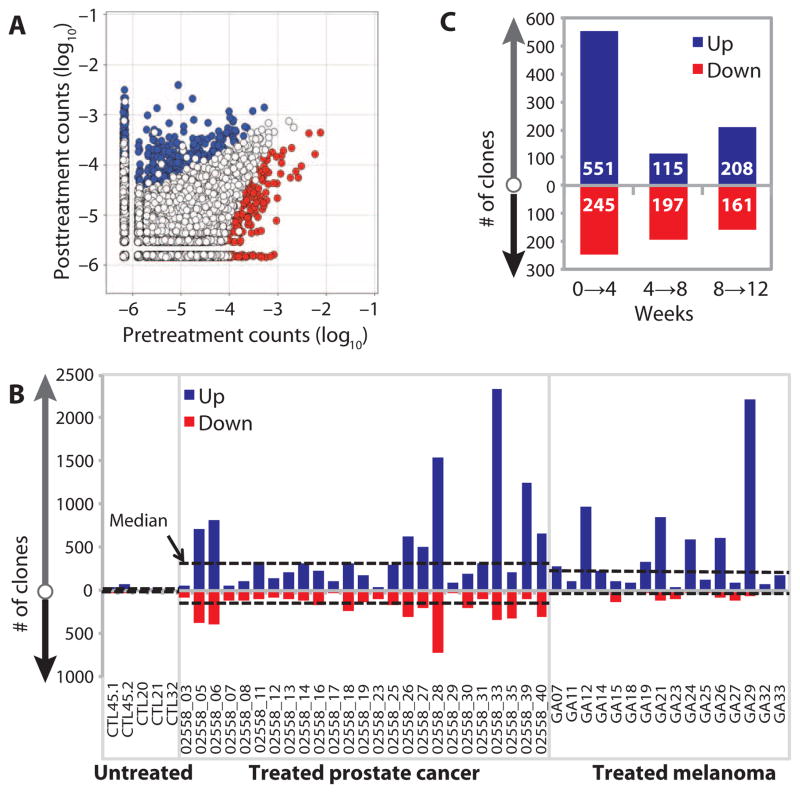
We next assessed the number of clonotypes with significant pairwise differences in abundance from baseline to 4 weeks after treatment, using a DESeq algorithm modified to account for normal variation in the repertoire over time and to compensate for the lack of replicates in the experimental design (Fig. 2A) (30). Of the differentially abundant clonotypes identified in prostate cancer patients, a median of 297 clonotypes showed significant increases in abundance, and a median of 134 showed significant decreases (Fig. 2B and Table 2). In the melanoma patients, we found more clonotype gain than loss (Fig. 2B and Table 2; median, 213 versus 25). The imbalance in differentially abundant clonotypes may account for the increased diversity observed in most melanoma patients (Fig. 1D). Overall, the bidirectional changes in clonotype abundance after one cycle of anti–CTLA-4 treatment occurred at levels greater than observed in untreated control subjects, providing further evidence for anti–CTLA-4–mediated TCR repertoire turnover. These data also suggest that repertoire diversification is driven by the overall contribution of clonotype gain versus clonotype loss.
Fig. 2. Accelerated repertoire turnover with multiple anti–CTLA-4 treatments.
(A) Representative frequency plot, showing the number of clones with significantly increased (blue) or decreased (red) abundance, using DESeq with a median dispersion model fitted to untreated normal samples separated by 1 month. (B) The numbers of clones with significantly changed abundance 1 month after first treatment are plotted for each sample, with increased abundance clones (blue, “Up”) plotted above the axis, and reduced abundance clones (red, “Down”) plotted as negative values. Median values for untreated control, prostate, and melanoma groups are plotted as dashed lines. (C) The number of significantly changing clones was determined for three sequential treatments. Increased abundance (blue, “Up”) and decreased abundance (red, “Down”) are indicated. Values are median values from 13 assessed patients within the prostate cancer cohort.
Table 2. Median values for significantly changed clones after first treatment with anti–CTLA-4 antibodies.
Clones with significantly different abundance separated by 1 month were identified as described in Fig. 2.
| Number of clones with increased abundance | Number of clones with decreased abundance | |
|---|---|---|
| Prostate | 297 | 134 |
| Melanoma | 213 | 25 |
| Combined | 239 | 99 |
| Untreated controls | 20 | 2 |
Clonotype evolution continues with repeated treatment
To assess whether subsequent treatments with anti–CTLA-4 mAbs had similar effects on the T cell repertoire, we determined the changes in clonotype abundance in patients who had received multiple cycles of treatment. We analyzed samples from the 13 prostate cancer patients who received at least three consecutive cycles of treatment given every 4 weeks, determining the number of significantly changed clonotypes after each treatment (Fig. 2C). We consistently observed significant changes in both directions with respect to unique clonotype abundance. To distinguish whether the repertoire change is affected by repeat dosing or the result of just one dose, we assessed a patient who received a single dose of ipilimumab. We found that repertoire change, as measured by Morisita’s distance in that individual over 3 months, is much less than a patient receiving repeated dosing (fig. S1). These results suggest that repeated dosing of anti–CTLA-4 antibody leads to continuous remodeling of the TCR repertoires over time.
Improved overall survival is associated with maintenance of high-frequency T cell clones
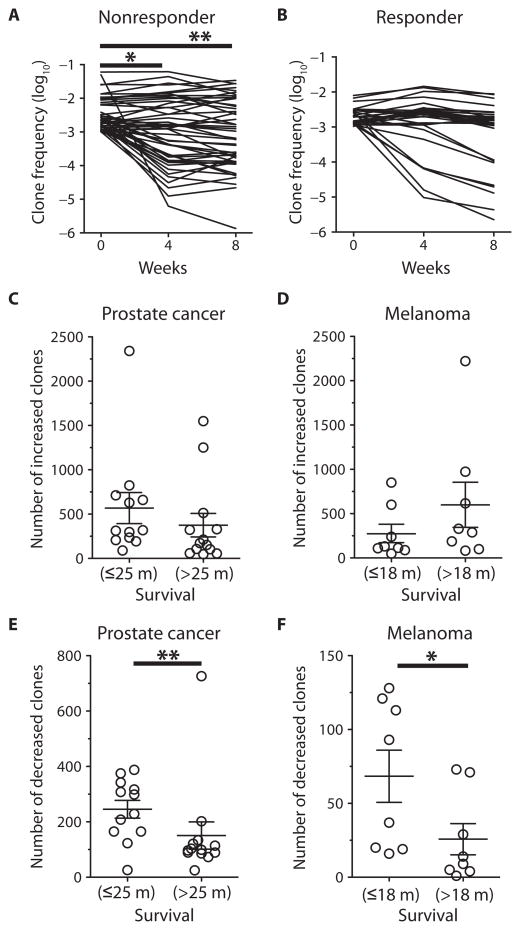
We wished to assess whether the changes seen with repertoire changes were associated with clinical outcome. Objective clinical responses using conventional response criteria are relatively uncommon with anti–CTLA-4, with 5 to 11% of patients treated at active dose levels (11, 31). With the similar low rates of response in our cohorts, we found no association between changes in T cell repertoire and clinical response [Response Evaluation Criteria in Solid Tumors (RECIST) in melanoma patients (32), and PSAWG2 in prostate cancer patients (33)]. We then examined whether an association existed between clonotypic changes and overall survival. We selected preexisting high-frequency clonotypes that were present at or above a frequency of 10−3 in the baseline sample and determined their kinetics over time. Most patients, which include short-term survivors, demonstrated a mixed pattern of clonotype evolution after initial treatment (Fig. 3A, representative CRPC patient with disease progression and 5-month survival). Changes between baseline versus weeks 4 and 8 were statistically significant in the patient with progressive disease (P = 0.013 and P = 0.004, respectively, Mann-Whitney). In contrast, we observed no statistically significant changes in a patient who clinically responded, raising the hypothesis that the high-frequency clonotypes were maintained in patients with good clinical outcome (Fig. 3B, representative CRPC patient with objective response and ongoing 96-month survival; P = 0.695 and P = 0.096, respectively, Mann-Whitney). This lack of significant changes was seen in two additional patients with the longest survivals after treatment (fig. S2).
Fig. 3. Association between clonotype changes and clinical outcome.
(A and B) Representative clonotype frequencies that exist at ≥10−3 at baseline for a clinical nonresponder (A) and clinical responder (B) are presented over time. *P = 0.013; **P = 0.004, two-sided Mann-Whitney test. (C to F) The numbers of significantly increased clones were plotted for metastatic CRPC patients (C) and metastatic melanoma patients (D), and the numbers of significantly decreased clones for metastatic CRPC patients (E) and metastatic melanoma patients (F), with each cohort divided by median survival (m, months). Mean values with error bars as SEM are plotted. P values were calculated using a two-sided Mann-Whitney test: *P = 0.0281; **P = 0.006.
Using median survival (25 months for prostate cancer patients, 18 months for melanoma patients) as a cutoff to classify patients into short survival versus long survival, we next examined whether improved survival was associated with changes in TCR clonotypes. We found no significant differences between the number of clones whose frequencies increased significantly after treatment and of patients above and below the median survival for metastatic CRPC (Fig. 3C) and metastatic melanoma (Fig. 3D). Rather, we observed that patients with survival above the median had significantly lower numbers of clones that significantly decreased in frequency, whereas patients with shorter survival had more frequent declines in their high-frequency clones. This association held for both CRPC (Fig. 3E; P = 0.006, Mann-Whitney) and metastatic melanoma (Fig. 3F; P = 0.0281, Mann-Whitney) cohorts.
Preexisting primed CD8 T cells are maintained in long-term survivors
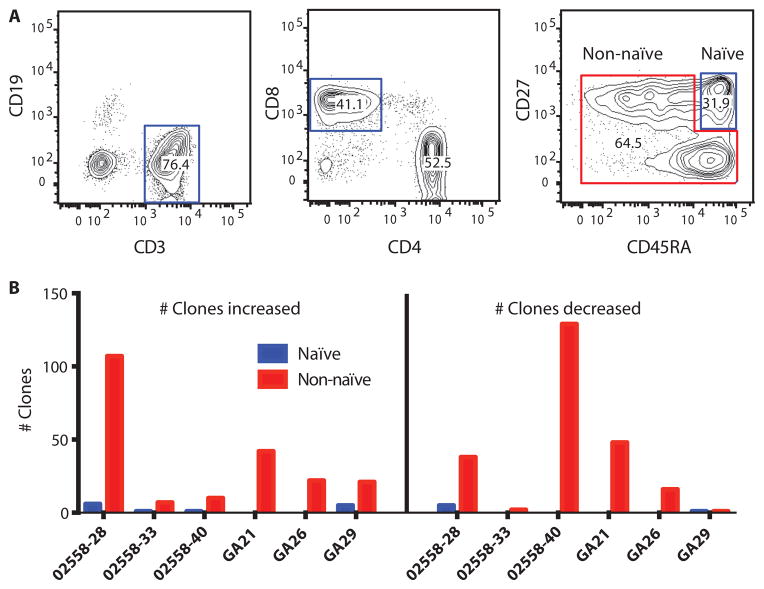
We further sequenced all rearranged TCRβ chains of sorted naïve CD27+CD45RA+ CD8 T cells and non-naïve (primed effector or memory) CD8 cells (Fig. 4A) and compared their contributions to clonotype change. Very few differentially abundant clones, as determined by DESeq, were classified as naïve CD8 T cells (Fig. 4B). Instead, baseline clones that changed in abundance with treatment were non-naïve T cells (Fig. 4B). The evidence, thus, suggests that patients who respond would have a pre-primed T cell response, not a naïve response that converts to an effector response.
Fig. 4. Clonotype change in naïve and effector CD8 T cells.
(A) Gated populations indicate sorting scheme for naïve (CD3+CD8+CD27+CD45RA+) and non-naïve CD8+ T cells. (B) Naïve (blue) or non-naïve (red) T cells sorted before treatment were then sequenced to track clonotypic changes with treatment. The numbers of clones that increased (left panel) and decreased (right panel) after anti–CTLA-4 treatment are shown for three prostate cancer patients and three melanoma patients.
Stability of antigen-specific clonotypes can reflect TCR affinity
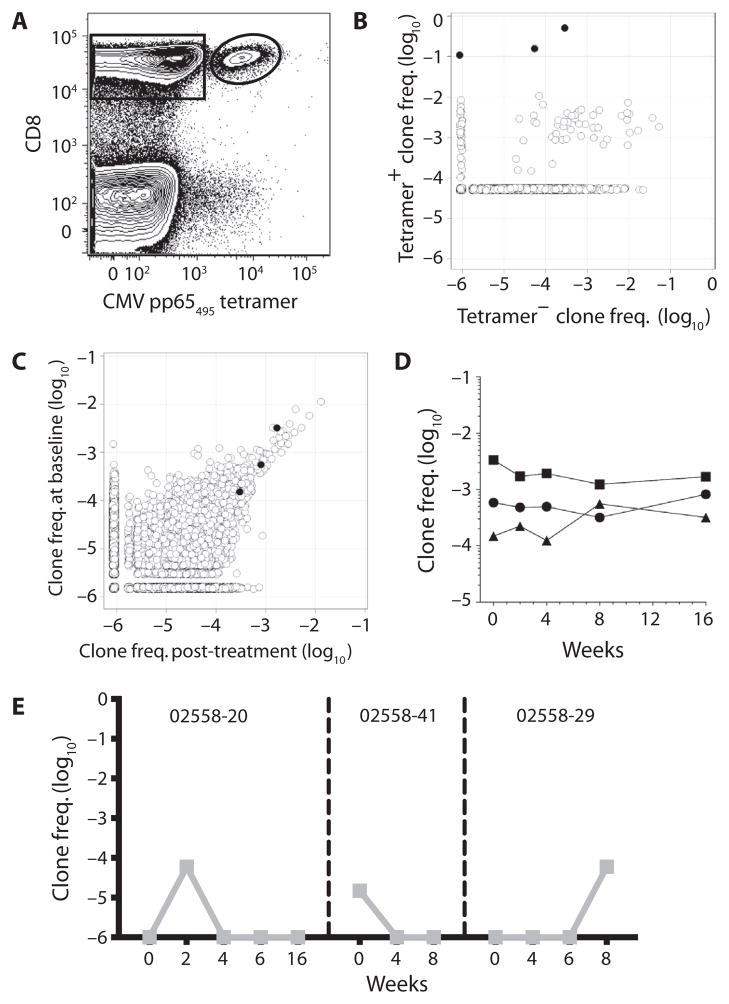
To further elucidate the nature of these preexisting high-frequency clonotypes on anti–CTLA-4 treatment, we used MHC/peptide tetramers to isolate and examine the evolution of T cell responses to specific antigens. Virus-specific T cells, which typically possess high-affinity TCR, can be frequently identified with this approach. Indeed, cytomegalovirus (CMV)–reactive clones could be detected using HLA-A*0201/pp65 peptide tetramers (Fig. 5A). Tetramer+ and tetramer− CD8+ T cells were then sorted and sequenced for TCRβ VDJ regions (Fig. 5B) to identify the CMV pp65-specific TCRβ clones for specific patients. From one clinical responder with CRPC (partial response, 56-month survival), we identified three unique clonotypes that accounted for 80% of tetramer+ sorted cells (Fig. 5B). We then assessed the overall frequency of these clonotypes within the repertoire over time (Fig. 5C). At baseline, these three clonotypes were present at high frequencies (≥~10−3) or at moderately high levels (10−4). Frequencies were stable after initial treatment (Fig. 5C) and maintained over the four cycles (Fig. 5D). The predicted CDR3 amino acid sequence from the most dominant clone (DSAMYRCASSLAPGTTNEKLFFGSGTQLSV) matched a previously published pp65-specific clonotype (34), consistent with CMV-reactive repertoires dominated by a few shared clonotypes with high antigen affinity/avidity across MHC-matched individuals.
Fig. 5. Antigen specificity of specific T cell clonotypes.
(A) Gated populations indicate CD8+ CMV pp65(495–404) tetramer− (rectangle) and tetramer+ (oval) populations sorted by flow cytometry and assessed for TCRβ repertoire sequencing from a clinical responder. (B) Frequency (log10) of each clone identified after TCRβ repertoire sequencing of the sorted tetramer+ and tetramer− CD8+ T cells. The three clones indicated in black are those deemed antigen-specific based on fold enrichment in tetramer+ versus tetramer− T cells and absolute frequency in tetramer+ cells. (C) TCRβ clone frequencies (log10) at baseline (week 0) and after treatment (week 16). The three clones identified in (B) are indicated in black. (D) Frequencies of the three CMV-specific clones identified in (B) and (C) at baseline (week 0) and at various time points after anti–CTLA-4 treatment. (E) Frequencies of p1182-specific clones (left, center) and p1183-specific clone (right) at baseline and at various time points after anti–CTLA-4 treatment for three different CRPC patients.
We assessed HLA-A*0201+ patients to a panel of tetramers for self-antigens because these antigen-specific T cells would be expected to have low-affinity TCR. Using two different HLA-A*0201/self-peptide (p1182 and p1183) tetramers bearing Pak6, we identified two distinct p1182-specific CD8+ TCRβ clones and one p1183-specific clone. Pak6 is a self-antigen expressed in the testis and prostate to which we could detect antibodies in CRPC patients (35, 36). Using the same approach, we found that the presence of these antigen-specific clonotypes was very transient (Fig. 5E). We did assess and, where possible, sort the HLA*0201 patients with tetramers to previously described tumor-associated antigens, including MART1(26–35), tyrosinase(368–376), gp100(209–217), PSA, and PAP (37). We were, however, not able to isolate tetramer+ T cell clones to these tumor-associated antigens.
DISCUSSION
Two models have been proposed for the potential effects of CTLA-4 blockade on Teffs (24): (i) the threshold model whereby CTLA-4 blockade would allow low-avidity T cells to expand and diversity to increase, and (ii) the attenuation model whereby CTLA-4 blockade would promote the preferential expansion of high-avidity T cells and narrowing of the repertoire. Our studies show that anti–CTLA-4 treatment induces diversification, profound turnover, and large-scale remodeling of the entire human T cell repertoire over time. This effect was evident in both CRPC and metastatic melanoma patients, as well as with two different human mAbs. Although these findings would be consistent with the threshold model, we provide evidence that patients with improved survival have lower numbers of preexisting high-frequency T cells that significantly decline in frequency after treatment compared with those who had shorter survival. Moreover, the bulk of changes occur in the effector/ memory T cell compartment rather than in the naïve T cell pool. These results therefore do not cleanly conform to either of the proposed models but suggest that patients with improved outcomes may have meaningful preexisting T cell responses. Nevertheless, the study has implications not only regarding the mechanism of action of anti–CTLA-4 mAbs in human malignancies but also in the usage of repertoire sequencing to identify patterns that may predict clinical benefit to anti–CTLA-4 therapies.
Diverse repertoires are important in limiting the magnitude of immune escape (38, 39) but may also promote self-reactive clones and induce host inflammation. Because anti–CTLA-4 mAbs induce global T cell proliferation and activation, an increasingly diverse repertoire may represent biologic effects of treatment. Conversely, the selection of high-avidity clonotypes, particularly in the setting of chronic antigen stimulation (40–42) or in metastatic melanoma (43, 44), leads to a hierarchy of high-frequency T cell clones dominating the immune repertoire. We show a similar stability in the order and abundance of high-frequency CMV-specific T cell clonotypes over time, suggesting that other high-avidity T cell clonotypes that have been previously selected in the context of persistent antigen stimulation are not influenced by subsequent anti–CTLA-4 mAbs. This stands in contrast to rarer self-reactive T cells, which are present only briefly during the course of CTLA-4 blockade. The lower avidity of these clones may hinder their selection during T cell activation and expansion. We could not isolate tetramer+ T cells reactive to described tumor-associated antigens [for example, MART1, gp100, and tyrosinase for melanoma (37); PAP and PSA for prostate cancer]. These results would support the notion that cancer patients develop individual-specific or “private” immune responses to their tumors (19) that may include T cell responses to tumor-specific mutations (45, 46) rather than developing responses to these shared antigens.
Similarly, the association between the reduced number of high-frequency clonotypes that significantly decline with treatment and over-all survival may reflect a Teff pool that can react strongly to tumor antigens but are inhibited by checkpoint blockade. This model is consistent with previous evidence showing modulation of preexisting antigen-specific responses with respect to anti–CTLA-4 mAbs in humans (19). In contrast, patients who fare worse have more of their high-frequency clonotypes decline with treatment. Clonotype contraction may signify failed selection of clonotypes because of lower TCR affinity, inappropriate antigen selectivity, poor antigen-presenting capacity, or T cell exhaustion and eventual disappearance over time (47).
We noted that repertoire evolution in patients with melanoma treated with tremelimumab favored clonotypes with significant increases in abundance, whereas the numbers of clonotypes with directional changes were more evenly distributed in prostate cancer patients who received ipilimumab. This difference could reflect a disparity in the patients because the prostate cancer cohort is older than the melanoma cohort, or biologic differences in the tumors. Melanomas have a higher mutational frequency than prostate cancer, which could result in a larger pool of neoantigens (48). Recently, it has been proposed that anti–CTLA-4 antibodies may deplete Tregs at tumor sites by ADCC, the absence of which strongly impairs antitumor activity (6–8). As an immunoglobulin G2 (IgG2) antibody, tremelimumab may have less ADCC activity than IgG1 ipilimumab and may be less likely to reduce CTLA-4–expressing clones by ADCC. We did not see a class effect of selective Treg depletion, supporting the notion that lost clonotypes are selected out of the effector pool.
Improved overall survival could, however, alternatively reflect the natural history of disease (such as indolent disease and/or subsequent therapies) rather than direct treatment effect. Other conditions may affect repertoire change, such as corticosteroid use for management of treatment-related immune toxicities. Clonotype maintenance is likely not the only variable affecting survival. Nevertheless, we observed the same association in CRPC and metastatic melanoma, cancers with very different disease courses, and underlying biologies. Moreover, we noted that the reduction in clonotype loss is most marked in patients who had a combined objective partial response with far superior survival (Fig. 3B). Larger sample sizes and prospective validation are needed to determine whether this maintenance in TCR clonotypes can serve as a prognostic and/or predictive biomarker, and whether there are other potential confounding variables. Recent works have shown that tumor-reactive neoantigen-specific T cells can be identified from tumor samples by exome analysis (45, 46). Tumor biopsies were not available to examine this question for the study. Even so, these findings are important in that changes of circulating T cell repertoire, as assessed by a blood sample rather than a tumor biopsy, may have clinical relevance. Finally, the technology used is limited by the inability to determine antigen specificity based solely on TCR-rearranged sequences. Assays that would include epitope mapping or antigen discovery would be best paired with repertoire sequencing in both blood and tumor to provide a more complete picture of structural diversity.
Despite these limitations, the analyses presented here suggest that maintained high-frequency T cell clones present at the start of treatment may be immunologically and clinically relevant for mediating tumor response and/or prolonged survival. The presence of these clones could reflect the immunogenicity of the patient’s tumor, the capacity of human leukocyte antigen (HLA) alleles to present antigen, and/or stochastic rearrangements in developing the TCR repertoire. The outcomes of enhancing global repertoire diversity versus a focus on pre-existing tumor antigen–specific T cells are not necessarily mutually exclusive. We propose that for most long-term survivors, high-avidity clones present at baseline are readily available after checkpoint inhibition to recognize relevant tumor antigens and mediate durable responses. For others, an increasingly diverse and evolving TCR repertoire allows for discovery and expansion of subdominant tumor-reactive clones. Because only a proportion of patients obtain long-term benefit, identification of these preferential clonotypes could improve patient selection for patients receiving single-agent CTLA-4 blocking antibodies. Another challenge with ipilimumab treatment is the phenomena of pseudoprogression, where patients appear to be progressing on scans but later respond. This type of assay could be used to identify patients who may benefit from continued treatment. For most patients who would not derive benefit, determining antigen specificity for these persistent clonotypes could inform complementary strategies, including potential vaccination strategies in combination or in sequence with CTLA-4 blockade to enhance clinical benefit. This study shows that next-generation sequencing with its extraordinary depth of coverage can be used to track the evolution of rearranged T cell clones and reveal further insight into T cell immunity modulated with immunotherapies.
MATERIALS AND METHODS
Study design
PBMCs were cryopreserved from 25 CRPC patients treated with anti–CTLA-4 (ipilimumab; Bristol-Myers Squibb) and GM-CSF (sargramostim; Sanofi) concurrently in a single-center phase 1/2 clinical trial at University of California, San Francisco (ClinicalTrials.gov identifier: NCT00064129) as previously described (14). Patients were treated with up to four doses of ipilimumab ranging from 1.5 to 10 mg/kg and GM-CSF at 250 μg/m2 per day. Anti–CTLA-4 antibody was administered every 4 weeks with GM-CSF given daily on the first 2 weeks of these cycles. Patient characteristics from the phase 1 study were previously described (14). The 21 assessed melanoma patients were enrolled in a phase 2 clinical trial of single-agent tremelimumab at 15 mg/kg administered every 3 months at University of California, Los Angeles (ClinicalTrials.gov identifier: NCT00471887) and were previously characterized (28, 49). Samples from these patients were available at baseline and 1 month after treatment. Patients were not restricted by HLA alleles. Informed consent was obtained for all investigations. PBMCs from untreated controls were obtained from Cellular Technology Limited (25).
TCRβ amplification and sequencing
The amplification and sequencing of TCRβ repertoire were previously described (25). Briefly, RNA was isolated from cells using AllPrep DNA/RNA mini and/or micro kits, according to the manufacturer’s instructions (Qiagen). RNA was reverse-transcribed to complementary DNA (cDNA) using Vilo kits (Life Technologies). cDNA was amplified using locus-specific primer sets for TCRβ. This amplification reaction reproducibly amplified all possible RNA transcripts found in the sample containing the rearranged TCRβ locus regardless of which variable (V) segment and which common constant (C) region allele each rearranged molecule had while appending the necessary sequences for cluster formation and sample indexing. Ultimately, 115 base pairs (bp) were sequenced from the C side sufficient to sequence through the junctional sequence from C to V. In addition, 95 bp was obtained from the V-to-C direction, providing ample sequence to map the V segment accurately.
Clonotype identification and enumeration
This procedure has been previously described in detail (25). Briefly, all reads are mapped to V and J segments. Identical sequences of successfully mapped reads were grouped in clonotypes. The frequency of each clonotype in a sample was determined by calculating the number of sequencing reads for each clonotype divided by the total number of passed sequencing reads in the sample.
Determination of repertoire diversity and repertoire change
Diversity of individual sample repertoires was assessed by calculating the number of unique clonotypes comprising the top 25th percentile of cumulative reads after sorting by clone abundance. Because it is not strongly influenced by rare clonotypes, this metric is stable to sequencing depth differences and amenable to comparisons between experiments.
Repertoire change between sequential experiments was measured using Morisita’s distance, which is not significantly influenced by sample size (29, 50). Numbers are reported here as distances, with 1 indicating maximal dissimilarity.
Identification of differentially abundant clonotypes
Identification of differentially abundant clonotypes in pairwise experiments using the DESeq R package (30) with modifications is described in Supplementary Methods.
T cell sorting
Cryopreserved PBMCs were washed and labeled with fluorescently labeled antibodies to CD3, CD8, CD14, and CD19 (BioLegend). For HLA-A*0201+ PBMCs, cells were labeled with HLA-A*0201/peptide tetramers with CMV pp65(495–404) (NLVPMVATV), Pak6 p1182(405–413) (LLLDSYVKI), and Pak6 p1183(84–92) (GLLNDIQKL) (NIH Tetramer Core) and sorted (FACSAria, BD Biosciences). Labeled CD8+ tetramer+ T cells were then sequenced as above by high-throughput sequencing (25, 40). These clonotypes have substantial fold enrichment (~1000-fold) in tetramer+ cells versus tetramer− cells and high absolute frequency in the tetramer+ population (25).
Statistical analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software). Specific statistical tests were two-sided Fisher’s exact test and two-sided Mann-Whitney tests.
Supplementary Material
Acknowledgments
We thank K. Kong for her assistance in data analysis.
Funding: Supported by the Peter Michael Foundation, 1R01 CA136753, and 1R01 CA163012. MHC tetramers were supplied by the NIH Tetramer Core.
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/6/238/238ra70/DC1
Methods
Fig. S1. Clonotype frequencies over time after single treatment with anti–CTLA-4.
Fig. S2. Clonotype kinetics in additional patients with improved survival.
Author contributions: L.F., M.K., and M.F. conceived the experimental plan. L.F. and A.R. provided clinical samples and annotation. Y.H. and M.K. performed the experiments. E.C., Y.H., M.K., C.C., M.F., and L.F. analyzed the data and interpreted the results.
Competing interests: M.K., C.C., and M.F. are employees of and own stock in Sequenta. Sequenta has pending patent applications on the TCR sequencing platform. E.C., Y.H., A.R., and L.F. declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Kwek SS, Cha E, Fong L. Unmasking the immune recognition of prostate cancer with CTLA4 blockade. Nat Rev Cancer. 2012;12:289–297. doi: 10.1038/nrc3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med. 1996;183:2533–2540. doi: 10.1084/jem.183.6.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Read S, Malmström V, Powrie F. Cytotoxic T lymphocyte–associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quezada SA, Peggs KS, Curran MA, Allison JP. CTLA4 blockade and GM-CSF combination immunotherapy alters the intratumor balance of effector and regulatory T cells. J Clin Invest. 2006;116:1935–1945. doi: 10.1172/JCI27745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 6.Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, Korman AJ. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res. 2013;1:32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- 7.Bulliard Y, Jolicoeur R, Windman M, Rue SM, Ettenberg S, Knee DA, Wilson NS, Dranoff G, Brogdon JL. Activating Fc γ receptors contribute to the antitumor activities of immunoregulatory receptor-targeting antibodies. J Exp Med. 2013;210:1685–1693. doi: 10.1084/jem.20130573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD, Peggs KS, Ravetch JV, Allison JP, Quezada SA. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti–CTLA-4 therapy against melanoma. J Exp Med. 2013;210:1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolchok JD, Hodi FS, Weber JS, Allison JP, Urba WJ, Robert C, O’Day SJ, Hoos A, Humphrey R, Berman DM, Lonberg N, Korman AJ. Development of ipilimumab: A novel immunotherapeutic approach for the treatment of advanced melanoma. Ann N Y Acad Sci. 2013;1291:1–13. doi: 10.1111/nyas.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribas A, Hanson DC, Noe DA, Millham R, Guyot DJ, Bernstein SH, Canniff PC, Sharma A, Gomez-Navarro J. Tremelimumab (CP-675,206), a cytotoxic T lymphocyte associated antigen 4 blocking monoclonal antibody in clinical development for patients with cancer. Oncologist. 2007;12:873–883. doi: 10.1634/theoncologist.12-7-873. [DOI] [PubMed] [Google Scholar]
- 11.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJM, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small EJ, Tchekmedyian NS, Rini BI, Fong L, Lowy I, Allison JP. A pilot trial of CTLA-4 blockade with human anti-CTLA-4 in patients with hormone-refractory prostate cancer. Clin Cancer Res. 2007;13:1810–1815. doi: 10.1158/1078-0432.CCR-06-2318. [DOI] [PubMed] [Google Scholar]
- 13.Slovin SF, Higano CS, Hamid O, Tejwani S, Harzstark A, Alumkal JJ, Scher HI, Chin K, Gagnier P, McHenry MB, Beer TM. Ipilimumab alone or in combination with radiotherapy in metastatic castration-resistant prostate cancer: Results from an open-label, multicenter phase I/II study. Ann Oncol. 2013;24:1813–1821. doi: 10.1093/annonc/mdt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong L, Kwek SS, O’Brien S, Kavanagh B, McNeel DG, Weinberg V, Lin AM, Rosenberg J, Ryan CJ, Rini BI, Small EJ. Potentiating endogenous antitumor immunity to prostate cancer through combination immunotherapy with CTLA4 blockade and GM-CSF. Cancer Res. 2009;69:609–615. doi: 10.1158/0008-5472.CAN-08-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rini BI, Fong L, Weinberg V, Kavanaugh B, Small EJ. Clinical and immunological characteristics of patients with serologic progression of prostate cancer achieving long-term disease control with granulocyte-macrophage colony-stimulating factor. J Urol. 2006;175:2087–2091. doi: 10.1016/S0022-5347(06)00261-8. [DOI] [PubMed] [Google Scholar]
- 16.Kavanagh B, O’Brien S, Lee D, Hou Y, Weinberg V, Rini B, Allison JP, Small EJ, Fong L. CTLA4 blockade expands FoxP3+ regulatory and activated effector CD4+ T cells in a dose-dependent fashion. Blood. 2008;112:1175–1183. doi: 10.1182/blood-2007-11-125435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ku GY, Yuan J, Page DB, Schroeder SEA, Panageas KS, Carvajal RD, Chapman PB, Schwartz GK, Allison JP, Wolchok JD. Single-institution experience with ipilimumab in advanced melanoma patients in the compassionate use setting: Lymphocyte count after 2 doses correlates with survival. Cancer. 2010;116:1767–1775. doi: 10.1002/cncr.24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Yu D, Sarnaik AA, Yu B, Hall M, Morelli D, Zhang Y, Zhao X, Weber JS. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J Transl Med. 2012;10:146. doi: 10.1186/1479-5876-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan J, Adamow M, Ginsberg BA, Rasalan TS, Ritter E, Gallardo HF, Xu Y, Pogoriler E, Terzulli SL, Kuk D, Panageas KS, Ritter G, Sznol M, Halaban R, Jungbluth AA, Allison JP, Old LJ, Wolchok JD, Gnjatic S. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical benefit in advanced melanoma patients treated with ipilimumab. Proc Natl Acad Sci USA. 2011;108:16723–16728. doi: 10.1073/pnas.1110814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamid O, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, Guida M, Hyams DM, Gómez H, Bastholt L, Chasalow SD, Berman D. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med. 2011;9:204. doi: 10.1186/1479-5876-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, Jackson JR, Shahabi V. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santegoets SJ, Stam AG, Lougheed SM, Gall H, Scholten PE, Reijm M, Jooss K, Sacks N, Hege K, Lowy I, Cuillerot JM, von Blomberg BM, Scheper RJ, van den Eertwegh AJ, Gerritsen WR, de Gruijl TD. T cell profiling reveals high CD4+CTLA-4+ T cell frequency as dominant predictor for survival after prostate GVAX/ipilimumab treatment. Cancer Immunol Immunother. 2013;62:245–256. doi: 10.1007/s00262-012-1330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egen JG, Allison JP. Cytotoxic T lymphocyte antigen-4 accumulation in the immunological synapse is regulated by TCR signal strength. Immunity. 2002;16:23–35. doi: 10.1016/s1074-7613(01)00259-x. [DOI] [PubMed] [Google Scholar]
- 24.Egen JG, Kuhns MS, Allison JP. CTLA-4: New insights into its biological function and use in tumor immunotherapy. Nat Immunol. 2002;3:611–618. doi: 10.1038/ni0702-611. [DOI] [PubMed] [Google Scholar]
- 25.Klinger M, Kong K, Moorhead M, Weng L, Zheng J, Faham M. Combining next-generation sequencing and immune assays: A novel method for identification of antigen-specific T cells. PLOS One. 2013;8:e74231. doi: 10.1371/journal.pone.0074231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robins H. Immunosequencing: Applications of immune repertoire deep sequencing. Curr Opin Immunol. 2013;25:646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, Warren EH. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Euw E, Chodon T, Attar N, Jalil J, Koya RC, Comin-Anduix B, Ribas A. CTLA4 blockade increases Th17 cells in patients with metastatic melanoma. J Transl Med. 2009;7:35. doi: 10.1186/1479-5876-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morisita M. Measuring of interspecific association and similarity between communities. Mem Fac Sci Kyushu Univ Ser E (Biol) 1959;3:65–80. [Google Scholar]
- 30.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolchok JD, Neyns B, Linette G, Negrier S, Lutzky J, Thomas L, Waterfield W, Schadendorf D, Smylie M, Guthrie T, Jr, Grob JJ, Chesney J, Chin K, Chen K, Hoos A, O’Day SJ, Lebbé C. Ipilimumab monotherapy in patients with pretreated advanced melanoma: A randomised, double-blind, multicentre, phase 2, dose-ranging study. Lancet Oncol. 2010;11:155–164. doi: 10.1016/S1470-2045(09)70334-1. [DOI] [PubMed] [Google Scholar]
- 32.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 33.Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, Eisenberger MA, Higano C, Bubley GJ, Dreicer R, Petrylak D, Kantoff P, Basch E, Kelly WK, Figg WD, Small EJ, Beer TM, Wilding G, Martin A, Hussain M Prostate Cancer Clinical Trials Working Group. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–1159. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux A, Mourin G, Fastenackels S, Almeida JR, Iglesias MC, Boyd A, Gostick E, Larsen M, Price DA, Sacre K, Douek DC, Autran B, Picard C, de Miranda S, Sauce D, Stern M, Appay V. CMV driven CD8+ T-cell activation is associated with acute rejection in lung transplantation. Clin Immunol. 2013;148:16–26. doi: 10.1016/j.clim.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J Biol Chem. 2001;276:15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- 36.Kwek SS, Dao V, Roy R, Hou Y, Alajajian D, Simko JP, Small EJ, Fong L. Diversity of antigen-specific responses induced in vivo with CTLA-4 blockade in prostate cancer patients. J Immunol. 2012;189:3759–3766. doi: 10.4049/jimmunol.1201529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comin-Anduix B, Lee Y, Jalil J, Algazi A, de la Rocha P, Camacho LH, Bozon VA, Bulanhagui CA, Seja E, Villanueva A, Straatsma BR, Gualberto A, Economou JS, Glaspy JA, Gomez-Navarro J, Ribas A. Detailed analysis of immunologic effects of the cytotoxic T lymphocyte-associated antigen 4-blocking monoclonal antibody tremelimumab in peripheral blood of patients with melanoma. J Transl Med. 2008;6:22. doi: 10.1186/1479-5876-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messaoudi I, Guevara Patiño JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 39.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, Bonneville M. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 41.Ladell K, Hashimoto M, Iglesias MC, Wilmann PG, McLaren JE, Gras S, Chikata T, Kuse N, Fastenackels S, Gostick E, Bridgeman JS, Venturi V, Arkoub ZA, Agut H, van Bockel DJ, Almeida JR, Douek DC, Meyer L, Venet A, Takiguchi M, Rossjohn J, Price DA, Appay V. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 42.Miles JJ, Silins SL, Brooks AG, Davis JE, Misko I, Burrows SR. T-cell grit: Large clonal expansions of virus-specific CD8+ T cells can dominate in the peripheral circulation for at least 18 years. Blood. 2005;106:4412–4413. doi: 10.1182/blood-2005-06-2261. [DOI] [PubMed] [Google Scholar]
- 43.Speiser DE, Baumgaertner P, Barbey C, Rubio-Godoy V, Moulin A, Corthesy P, Devevre E, Dietrich PY, Rimoldi D, Liénard D, Cerottini JC, Romero P, Rufer N. A novel approach to characterize clonality and differentiation of human melanoma-specific T cell responses: Spontaneous priming and efficient boosting by vaccination. J Immunol. 2006;177:1338–1348. doi: 10.4049/jimmunol.177.2.1338. [DOI] [PubMed] [Google Scholar]
- 44.Derré L, Bruyninx M, Baumgaertner P, Devevre E, Corthesy P, Touvrey C, Mahnke YD, Pircher H, Voelter V, Romero P, Speiser DE, Rufer N. In vivo persistence of codominant human CD8+ T cell clonotypes is not limited by replicative senescence or functional alteration. J Immunol. 2007;179:2368–2379. doi: 10.4049/jimmunol.179.4.2368. [DOI] [PubMed] [Google Scholar]
- 45.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJA, Behjati S, Hilkmann H, El Atmioui D, Nieuwland M, Stratton MR, Kerkhoven RM, Kesmir C, Haanen JB, Kvistborg P, Schumacher TN. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, Samuels Y, Rosenberg SA. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Day EK, Carmichael AJ, ten Berge IJM, Waller ECP, Sissons JGP, Wills MR. Rapid CD8+ T cell repertoire focusing and selection of high-affinity clones into memory following primary infection with a persistent human virus: Human cytomegalovirus. J Immunol. 2007;179:3203–3213. doi: 10.4049/jimmunol.179.5.3203. [DOI] [PubMed] [Google Scholar]
- 48.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortés ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Lee RS, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CWM, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, Getz G. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comin-Anduix B, Sazegar H, Chodon T, Matsunaga D, Jalil J, von Euw E, Escuin-Ordinas H, Balderas R, Chmielowski B, Gomez-Navarro J, Koya RC, Ribas A. Modulation of cell signaling networks after CTLA4 blockade in patients with metastatic melanoma. PLOS One. 2010;5:e12711. doi: 10.1371/journal.pone.0012711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolda H. Similarity indices, sample size and diversity. Oecologia. 1981;50:296–302. doi: 10.1007/BF00344966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.