Abstract
Constitutive activation of the EGFR is common in cancer due to EGFR wild type (EGFRwt) overexpression or the presence of mutant EGFR. Signaling by constitutively active NSCLC EGFR mutants or the EGFRvIII mutant in glioblastoma has been studied intensively and the downstream signals are known. Normally, the EGFRwt is activated when it is exposed to ligand resulting in activation of canonical signals such as ERK and Akt. The EGFRwt also becomes tyrosine phosphorylated and constitutively activated without ligand when it is overexpressed, but downstream signals are unclear. Recent studies have identified a non-canonical form of signaling triggered by EGFRwt exclusively in the absence of ligand that does not involve ERK or Akt activation but, instead, results in activation of the transcription factor IRF3. Addition of ligand turns off IRF3 dependent transcription and activates ERK and Akt. Thus, the EGFR triggers distinct and mutually exclusive signaling networks depending on the presence of ligand. Furthermore, non-canonical EGFRwt signaling may influence response to treatment in cancer. Also, there are reports of both synergistic and antagonistic interactions between ligand-dependent EGFRwt and EGFRvIII signaling, Here, we discuss ligand-independent EGFR signal transduction by oncogenic EGFR mutants and EGFRwt, and review the interplay between EGFRwt and EGFRvIII.
Keywords: EGFR, constitutive signaling, EGFR wild type, EGFRvIII, lung cancer, EGFR mutants, non-canonical EGFR signaling, glioblastoma, GBM
Epidermal growth factor receptor (EGFR) is a transmembrane receptor for members of the epidermal growth factor family (EGF-family) of extracellular protein ligands (1). The EGFR plays an important role in regulating various cellular functions such as proliferation, motility and differentiation. The binding of a ligand to EGFR causes dimerization, followed by autophosphorylation of the EGFR and activation of downstream signaling pathways (1). Activation of the EGFR triggers multiple signaling cascades within the cell culminating in gene transcription and a biological response.
Constitutive or ligand-independent EGFR signaling has been intensively studied in EGFR mutants expressed in lung cancer and glioblastoma (GBM). In NSCLC (non-small cell lung cancer), EGFR overexpression is common and EGFR mutations are detected in 10–20% of patients being more common in Asian patients (2, 3) (4). EGFR mutations in NSCLC have generated intense interest because patients with these mutations in their tumors have an increased responsiveness to EGFR tyrosine kinase inhibitors (5, 6). The most common NSCLC EGFR mutations are an exon 19 deletion and a point mutation of L858R accounting for 90% of all EGFR activating mutations, and are referred to ‘classical’ activating mutations, may facilitate dimerization (7), leading to ligand-independent activation of the EGFR (7, 8). Thus, EGFR mutations in NSCLC alter responsiveness to treatment and are biologically significant. A number of excellent reviews have discussed signal transduction by mutant EGFR in NSCLC (3, 9). In this review, our focus is on EGFR signaling in GBM.
Constitutive or ligand-independent signaling has largely been considered a property of oncogenic EGFR mutants (10), even though it is known that EGFRwt overexpression also results in tyrosine phosphorylation and presumably a constitutive activation of the EGFRwt (11). However, the downstream signals triggered by constitutively activated EGFRwt remained undefined and the biological role of constitutive EGFRwt activation remains unknown. As discussed below, recent studies have identified downstream signals triggered by constitutive EGFRwt activation that involve activation of the transcription factor IRF3 (12). This ligand-independent and constitutive EGFRwt signaling is termed non-canonical EGFR signaling because it does not involve activation of canonical EGFR signals such as ERK and Akt (12). Non-canonical EGFR signaling may be important because of the frequency of EGFRwt overexpression in cancer and because in cancers like GBM, lung, and breast cancer, the EGFR may be commonly overexpressed without significant co-expression of ligand (12–16). In addition, constitutive signaling by EGFR mutants may render cells more sensitive to EGFR inhibition as seen in lung cancer EGFR mutants, or resistant to EGFR inhibition as in the case of EGFRvIII. The differences in sensitivity to inhibition between the different EGFR mutants may reflect differential receptor binding and occupancy resulting from altered confirmation of the mutant receptors (17, 18).
The EGFRwt is often co-expressed with oncogenic EGFR mutants. In the case of GBM, a cancer in which the frequency of EGFR gene amplification is estimated to be 40–50%, the most common oncogenic EGFR mutant, EGFRvIII, is detected almost exclusively in tumors expressing EGFRwt (19, 20). EGFRvIII may have a more focal or limited distribution compared to EGFRwt in GBM (21, 22). However, recent studies suggest that EGFRwt is usually co-expressed in EGFRvIII expressing tumor cells. Fan et al, showed the co-localization of EGFRwt and EGFRvIII in individual tumor cells by immunohistochemistry in GBM samples with antibodies specific for EGFR and EGFRvIII (23), while Puliyapaddamba et al., showed that clones derived from single cells from primary GBM cultures expressed both EGFRwt and EGFRvIII (24). The co-expression of both EGFRwt and EGFRvIII within the same tumor raises the possibility of interactions between the receptors and a number of studies have investigated EGFRwt-EGFRvIII interactions as outlined below. In addition to EGFRvIII, other less common constitutively active EGFR mutants in GBM include the deletion mutant EGFRvIV, a tandem kinase domain duplication in EGFR (TKD-EGFR) and a series of point mutations in the EGFR extracellular domain (10, 20, 25).
Constitute activation of EGFR wild-type
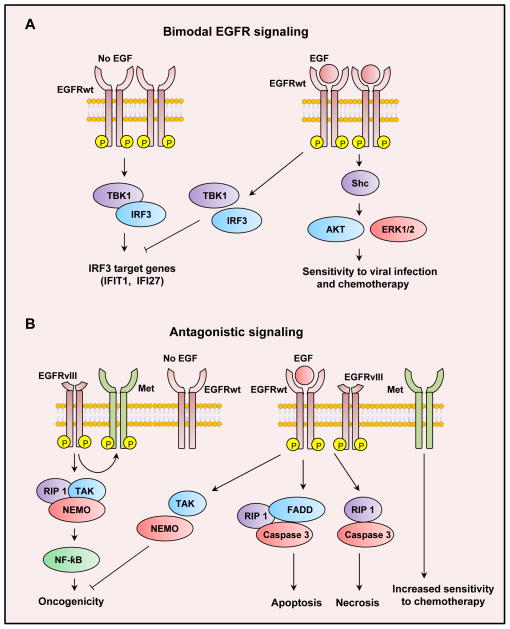
While studies have reported that EGFRwt overexpression results in persistent tyrosine phosphorylation and a constitutive activation of the receptor in the absence of ligand (11), the downstream signals triggered by this constitutive EGFRwt activation have never been clearly described, and the general assumption may have been that constitutive and ligand-induced signals are similar. Ramnarain et al., reported that conditional expression of EGFRwt in glioma cells leads to increased expression of mRNA for 66 genes in the absence of exogenous ligand (26), providing evidence for a constitutive EGFRwt signaling program. In a subsequent study constitutive vs. ligand induced EGFR signaling was studied in detail, in glioma and breast cancer cells (12). In the absence of ligand, increased expression of the EGFRwt resulted in tyrosine phosphorylation of the EGFR, but did not lead to activation of canonical signals such as ERK and Akt. In addition, ligand-independent EGFR signaling did not result in expression of immediate early genes such as EGR1 and EGR2. Instead, ligand-independent EGFR signaling triggers a non-canonical EGFR pathway that is regulated by activation of the transcription factor IRF3. Thus, EGFR overexpression leads to phosphorylation and activation of IRF3 and transcription of IRF3-dependent downstream genes such as IFIT1, IFI27 and TRAIL. An autocrine loop was excluded by the use of Cetuximab, which failed to prevent ligand-independent EGFR signaling, and the use of a non-ligand binding EGFR mutant, EGFRvIII, which also triggered activation of IRF3 dependent target genes, thus excluding an intracellular autocrine activation of the EGFRwt (12). Furthermore, expression of a kinase inactive EGFRwt failed to induce IRF3 activation, and the use of Erlotinib blocked ligand-independent EGFR signaling, demonstrating that the kinase activity of the EGFR is required. Interestingly, addition of EGF resulted in a termination of the IRF3-transcriptional program, as demonstrated by a loss of IRF3 phosphorylation, loss of IRF3 transcriptional activity and downregulation of IRF3 dependent downstream genes IFIT1, IRF3 and TRAIL, with a concomitant activation of canonical ERK and Akt signals and induction of immediate early genes EGR1 and EGR2 (12). These data challenge the a priori view that ligand-independent and ligand-dependent EGFR signals are similar and demonstrate that the downstream signals activated by ligand-independent EGFR signaling are non-canonical and distinct from ligand-activated EGFR signaling. Furthermore, this study suggested that ligand-independent and ligand-activated signals are mutually exclusive. These data together suggest that the overexpressed EGFRwt oscillates between two distinct and mutually exclusive modes of signaling, depending on the presence of ligand (Figure 1A).
Figure 1.
A. Bimodal EGFR signaling: When the EGFR is overexpressed it becomes tyrosine phosphorylated and constitutively activated forming a complex with TBK1 and IRF3 resulting in activation of IRF3. When EGF is added, this complex dissociates, IRF3 activation is lost and the ligand-activated EGFR activates ERK and Akt and cells become more sensitive to chemotherapy. B. Ligand-activated EGFRwt antagonizes EGFRvIII mediated activation of NF-kappaB and Met.
The underlying mechanism may be a switch of EGFR associated proteins with ligand (12) (Figure 1A). Thus, in the absence of ligand, the EGFR forms a ternary complex that includes IRF3 and its upstream kinase TBK1. It was proposed that IRF3 activation resulted from the increased association of IRF3 and TBK1 induced by the presence of EGFRwt. Addition of EGF resulted in a dissolution of the EGFR-IRF3-TBK1 complex and a loss of IRF3 transcriptional activity. The ligand-activated EGFR now forms a complex with Shc. The changes in EGFR-associated proteins in response to ligand are likely a consequence of the altered conformation of the ligand-activated receptor, but this awaits experimental confirmation. Initial studies indicate that cells overexpressing EGFR are more resistant to virus induced cell death in the absence of ligand (12), raising the possibility that ligand-independent EGFR signaling may confer a survival advantage during the clonal evolution of tumors. Similarly, EGFR overexpressing glioma cells are more resistant to chemotherapy with temozolomide in the absence of ligand (12).
Constitutive EGFR signaling may be activated in cancer. A recent study found that about 60 percent of glioblastoma (GBM) tumors that express high levels of the EGFR had low levels of TGFα (12). Interestingly, there is a statistically significant inverse correlation between expression of TGFα and levels of IRF3-dependent genes IFI27 and IFIT1, suggesting that in EGFR overexpressing tumors with high levels of TGFα and hence ligand-activation of EGFR, IRF3 is not activated. Conversely, in EGFR overexpressing tumors in which TGFα was low, levels of IFIT1 and IFI27 were high suggesting that non-canonical EGFR signaling is activated. Other studies have also suggested EGFR overexpression may occur without TGFα expression in GBMs (13) or reported that a significant subset of EGFR overexpressing GBMs do not have a high level of EGFR phosphorylation suggesting a paucity of ligand in GBM (27). Similar results have been found in lung cancer in which estimates of low or absent ligand expression in high EGFR expressing cancers range from 25–32% (14, 15). Similarly in a breast cancer study, about 48% of EGFR positive tumors were positive for TGFα (16). However, while TGFα is a major EGFR ligand in cancer, there are a number of other known EGFR ligands. These include EGF, HB-EGF, Amphiregulin, Betacellulin, and Epigen (28). Thus, determination of the ligand status of EGFR overexpressing tumors will require an assessment of multiple ligands.
EGFRvIII is a constitutively active mutant in GBM
EGFR gene amplification and overexpression are a striking feature of GBM, observed in about 40–50% of GBMs (20), but are rare in low grade gliomas suggesting a causal role for aberrant EGFR signaling in the pathogenesis of GBM. A specific EGFR mutant (EGFR Type III, EGFRvIII, de2–7, ΔEGFR) can be detected in about one third of GBMs (20). EGFRvIII is generated from a deletion of exons 2–7 of the EGFR gene, which results in an in frame deletion of 267 amino acids from the extracellular domain of the receptor. EGFRvIII is unable to bind ligand and signals constitutively.
EGFRvIII has a greater oncogenic potential compared to EGFRwt (29) and a significant effort has been focused on investigating what makes EGFRvIII more tumorigenic compared to EGFRwt. A detailed discussion of EGFRvIII downstream signaling can be found in previous reviews (19, 20). EGFRvIII downstream signaling is clearly distinct from EGFRwt signaling in a number of ways. Firstly, EGFRvIII expression is sufficient to induce a ligand-independent continuous activation of Ras (30), ERK (31) and PI3K-Akt (20, 32). EGFRwt expression fails to activate these canonical signals unless ligand is added, even when the EGFR is overexpressed, and ligand-activated signals are limited in time. Expression of EGFRvIII in U87MG cells leads to an increase in Bcl-XL and resistance to apoptotic cell death in response to chemotherapy (33). Other studies have reported an important role for JNK activation in EGFRvIII signaling (34). Additional effector mechanisms used by EGFRvIII include Dock190-Rac1 activation (35), downregulation of miR-9 and upregulation of its target FOXP1 (36), and alternative splicing of Max leading to glycolytic growth of tumors (37).
Aberrant EGFR signaling is an important mechanism of NF-κB activation in GBM (38, 39). EGFRvIII is reported to induce NF--κB activation and higher expression levels of the pro-angiogenic factor interleukin IL-8 in glioma (40). There may be multiple mechanisms used by EGFRvIII to activate NF--κB. EGFRvIII has been reported to activate NF--κB via mTORC2 kinase (41). Another study reported that EGFRvIII activated NF--κB by a RIP1 kinase dependent mechanism that includes formation of a signaling platform including EGFRvIII, RIP1, NEMO, and TAK1 (24). Importantly, several studies have indicated that NF--κB activation is required for EGFRvIII-mediated oncogenicity in glioma models (24, 40, 41).
EGFRvIII activates the receptor tyrosine kinase Met and combined inhibition of EGFRvIII and Met resulted in enhanced cytotoxicity of EGFRvIII overexpresing compared with inhibition of either receptor alone, indicating a synergistic effect of EGFRvIII and Met (42–44).
Interactions between constitutively activated and ligand activated EGF receptors
EGFRvIII is usually co-expressed with EGFRvIII in GBM and the question of whether EGFRwt influences EGFRvIII signaling and vice versa have been the subject of a number of studies.
Synergistic Interactions
Luwor et al., showed that expression of EGFRvIII in BaF/3 cells promoted their proliferation and found that co-expression of EGFRwt enhanced this effect. They proposed that EGFRvIII heterodimerizes with EGFRwt resulting in transphosphorylation of EGFRwt (45), but did not find increased ligand-activated EGFRwt mediated transphosphorylation of EGFRvIII when both receptors were overexpressed (45). Recently, two studies proposed a role for EGFRwt in the activation of EGFRvIII. Li et al., proposed that EGFRwt played a key role in the constitutive activation of EGFRvIII by forming a complex with EGFRvIII and promoting its dimerization. Thus, increasing EGFRwt levels increased EGFRvIII mediated oncogenicity in an orthotopic model, while decreasing EGFRwt significantly attenuated the oncogenicity of EGFRvIII (46). The authors did not find evidence for a direct transphosphorylation of EGFRvIII by EGFRwt and, in agreement with a previous study (47), found that EGFRwt failed to phosphorylate a kinase dead EGFRvIII mutant, with or without ligand (46). Li et al., suggested that EGFRwt constitutively facilitated the dimerization of EGFRvIII leading to autophosphorylation and activation of EGFRvIII (46). EGFRvIII induced the expression of HB-EGF, a ligand for EGFRwt, suggesting a role for EGFRvIII in the activation of EGFRwt (26, 46). In another study, Fan et al also found that co-expression of EGFRwt and EGFRvIII promoted tumor growth in vivo. They proposed a direct transphosphorylation of EGFRvIII by ligand-activated EGFRwt, leading to an enhanced activation of downstream STAT signaling and increased tumorigenicity (23). The EGFR kinase domain is allosterically activated in an asymmetric dimer (48), with one monomer acting as the activator kinase and another as the receiver kinase. Fan et al. used an elegant experimental approach by generating receiver impaired and activator impaired EGFRwt and EGFRvIII mutants to demonstrate that EGFRvIII is a substrate for EGFRwt (23). Although there are some differences in the proposed mechanisms by which EGFRwt mediates activation of EGFRvIII, which may reflect differences in the experimental approach, EGFRwt levels and/or cell type specific differences, the data suggest that EGFRwt plays an important role in EGFRvIII activation. In addition to these interactions, synergism between EGFRvIII and EGFRwt was suggested by an earlier study that found an EGFRvIII specific induction of an eight gene signature in glioma cells. Three of these genes were ligands for EGFRwt suggesting that EGFRvIII signaling generated an autocrine loop in these cells (26). EGFRvIII induced expression of HB-EGF is biologically significant, and HB-EGF overexpression was shown to accelerate EGFRvIII induced tumorigenicity while HB-EGF silencing attenuated EGFRvIII-mediated tumorigenicity in an orthotopic model (46). Another study has reported a biologically significant paracrine loop triggered by EGFRvIII that activates EGFRwt via induction of IL-6 (22).
Antagonistic interactions
There are two reports of antagonistic interactions between EGFRwt and EGFRvIII (Figure 1B). EGFRvIII-mediated activation of NF-kB in glioma cells is abolished by treating cells with EGF. Thus, it was demonstrated that inducible or constitutive stable expression of EGFRvIII induced NF-kB activation that was abolished by activation of co-expressed EGFRwt receptor with ligand (24). Overexpression of EGFRwt was not required for this antagonistic effect. EGFRvIII induces the K63-linked ubiquitination of RIP1 and formation of a EGFRvIII-RIP1-NEMO-TAK1 signaling complex that mediates NF-kB activation and promotes proliferation of tumor cells in vivo (24). Addition of EGF results in a loss of this signaling platform and a loss of NF-kB activity. RIP1 is known to have a role in both cell survival as well as cell death (49). In the context of EGFRvIII signaling, activation of EGFRwt results in a loss of RIP1 ubiquitination and it’s dissociation from EGFRvIII, NEMO and TAK1. RIP1 now becomes associated with FADD and Caspase-8 and leads to an EGF and RIP1 dependent death of glioma cells (24) (Figure 1B). EGFRwt activation has also been reported to antagonize EGFRvIII-mediated activation of Met in glioma cells. Thus, EGFRvIII expression results in Met phosphorylation and activation. Addition of EGF to EGFRvIII expressing cells results in activation of co-expressed EGFRwt and a rapid loss of Met phosphorylation and activation (50) (Figure 1B). EGFRvIII becomes associated with Met and addition of EGF leads to a loss of the EGFRvIII-Met association suggesting a possible mechanism for the loss of EGFRvIII-induced Met activation (50). Furthermore, the loss of Met activation may sensitize cells to chemotherapy (50)
These studies suggest that interactions between the ligand-dependent EGFRwt receptor and the ligand-independent EGFRvIII receptor are multinodal, frequent and biologically significant.
Concluding comments
The finding that constitutive and ligand-dependent EGFR signaling are distinct and that EGFR signaling in cancer is bimodal may prove relevant to clinical practice. The observation that EGFR overexpressing cells are more sensitive to chemotherapy in the presence of ligand is potentially important in stratification of patients with EGFR overexpressing tumors. The presence or level of expression of EGFR ligand(s) in a particular tumor has been largely been ignored in clinical practice. If EGFR signaling in cancer is indeed bimodal, determination of the EGFR ligand status of a particular tumor may help to predict response to chemotherapy. The newly identified signaling networks that are triggered exclusively by ligand-independent EGFRwt signaling may provide new targets for treatment. In addition, new information about interactions between ligand-independent EGFR mutants and ligand-activated EGFRwt may identify new approaches and new targets for combinatorial treatment. An intriguing example is the finding that ligand-activated EGFRwt can direct the RIP1 kinase cell death switch. Thus, when EGFRvIII is expressed, RIP1 is ubiquitinated and acts in a prosurvival role by activating NF--κB. When the EGFRwt is overexpressed in the same cells and activated with ligand, the RIP1 switch is turned into a cell death mode (24). Thus, there are data that ligand-activated EGFR may render the cell more vulnerable to cell death, an observation that could be exploited for treatment. Future studies will examine the biological significance and therapeutic implications of the new insights into EGFR signaling in cancer.
Acknowledgments
This work was supported in part by a Merit Review Award from the Departments of Veterans Affairs (1I01BX002559-01) and by NIH grant R01 NS062080 (to AH). The contents do not represent the views of the Department of Veterans Affairs or the United States Government.
References
- 1.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. Journal of the National Cancer Institute. 2005;97(5):339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 3.Siegelin MD, Borczuk AC. Epidermal growth factor receptor mutations in lung adenocarcinoma. Laboratory investigation; a journal of technical methods and pathology. 2014;94(2):129–37. doi: 10.1038/labinvest.2013.147. [DOI] [PubMed] [Google Scholar]
- 4.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Critical reviews in oncology/hematology. 1995;19(3):183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Okabe T, Okamoto I, Tamura K, Terashima M, Yoshida T, Satoh T, et al. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res. 2007;67(5):2046–53. doi: 10.1158/0008-5472.CAN-06-3339. [DOI] [PubMed] [Google Scholar]
- 8.Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2(11):e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 10.Pines G, Kostler WJ, Yarden Y. Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS Lett. 2010;584(12):2699–706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152(3):543–56. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty S, Li L, Puliyappadamba VT, Guo G, Hatanpaa KJ, Mickey B, et al. Constitutive and ligand-induced EGFR signalling triggers distinct and mutually exclusive downstream signalling networks. Nat Commun. 2014;5:5811. doi: 10.1038/ncomms6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maruno M, Kovach JS, Kelly PJ, Yanagihara T. Transforming growth factor-alpha, epidermal growth factor receptor, and proliferating potential in benign and malignant gliomas. Journal of neurosurgery. 1991;75(1):97–102. doi: 10.3171/jns.1991.75.1.0097. [DOI] [PubMed] [Google Scholar]
- 14.Rusch V, Klimstra D, Venkatraman E, Pisters PW, Langenfeld J, Dmitrovsky E. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin Cancer Res. 1997;3(4):515–22. [PubMed] [Google Scholar]
- 15.Volante M, Saviozzi S, Rapa I, Ceppi P, Cappia S, Calogero R, et al. Epidermal growth factor ligand/receptor loop and downstream signaling activation pattern in completely resected nonsmall cell lung cancer. Cancer. 2007;110(6):1321–8. doi: 10.1002/cncr.22903. [DOI] [PubMed] [Google Scholar]
- 16.de Jong JS, van Diest PJ, van der Valk P, Baak JP. Expression of growth factors, growth inhibiting factors, and their receptors in invasive breast cancer. I: An inventory in search of autocrine and paracrine loops. The Journal of pathology. 1998;184(1):44–52. doi: 10.1002/(SICI)1096-9896(199801)184:1<44::AID-PATH984>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 17.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2(5):458–71. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barkovich KJ, Hariono S, Garske AL, Zhang J, Blair JA, Fan QW, et al. Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer Discov. 2012;2(5):450–7. doi: 10.1158/2159-8290.CD-11-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor (EGFR) in glioma: Signal transduction, neuropathology, imaging and radioresistance. Neoplasia. 2010;12(9):675–84. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2(87):re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 21.Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21(2):53–6. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- 22.Inda MD, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–45. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24(4):438–49. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puliyappadamba VT, Chakraborty S, Chauncey SS, Li L, Hatanpaa KJ, Mickey B, et al. Opposing Effect of EGFRWT on EGFRvIII-Mediated NF-kappaB Activation with RIP1 as a Cell Death Switch. Cell Rep. 2013;4(4):764–75. doi: 10.1016/j.celrep.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ymer SI, Greenall SA, Cvrljevic A, Cao DX, Donoghue JF, Epa VC, et al. Glioma Specific Extracellular Missense Mutations in the First Cysteine Rich Region of Epidermal Growth Factor Receptor (EGFR) Initiate Ligand Independent Activation. Cancers. 2011;3(2):2032–49. doi: 10.3390/cancers3022032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66(2):867–74. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- 27.Talasila KM, Soentgerath A, Euskirchen P, Rosland GV, Wang J, Huszthy PC, et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta neuropathologica. 2013;125(5):683–98. doi: 10.1007/s00401-013-1101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Exp Cell Res. 2003;284(1):2–13. doi: 10.1016/s0014-4827(02)00105-2. [DOI] [PubMed] [Google Scholar]
- 29.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91(16):7727–31. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prigent SA, Nagane M, Lin H, Huvar I, Boss GR, Feramisco JR, et al. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J Biol Chem. 1996;271(41):25639–45. doi: 10.1074/jbc.271.41.25639. [DOI] [PubMed] [Google Scholar]
- 31.Lorimer IA, Lavictoire SJ. Activation of extracellular-regulated kinases by normal and mutant EGF receptors. Biochim Biophys Acta. 2001;1538(1):1–9. doi: 10.1016/s0167-4889(00)00129-4. [DOI] [PubMed] [Google Scholar]
- 32.Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273(1):200–6. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 33.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci U S A. 1998;95(10):5724–9. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonyak MA, Kenyon LC, Godwin AK, James DC, Emlet DR, Okamoto I, et al. Elevated JNK activation contributes to the pathogenesis of human brain tumors. Oncogene. 2002;21(33):5038–46. doi: 10.1038/sj.onc.1205593. [DOI] [PubMed] [Google Scholar]
- 35.Feng H, Hu B, Vuori K, Sarkaria JN, Furnari FB, Cavenee WK, et al. EGFRvIII stimulates glioma growth and invasion through PKA-dependent serine phosphorylation of Dock180. Oncogene. 2014;33(19):2504–12. doi: 10.1038/onc.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez GG, Volinia S, Croce CM, Zanca C, Li M, Emnett R, et al. Suppression of microRNA-9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity. Cancer Res. 2014;74(5):1429–39. doi: 10.1158/0008-5472.CAN-13-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17(6):1000–8. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364(7):627–37. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park S, Hatanpaa KJ, Xie Y, Mickey BE, Madden CJ, Raisanen JM, et al. The receptor interacting protein 1 inhibits p53 induction through NF-kappaB activation and confers a worse prognosis in glioblastoma. Cancer Res. 2009;69(7):2809–16. doi: 10.1158/0008-5472.CAN-08-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonavia R, Inda MM, Vandenberg S, Cheng SY, Nagane M, Hadwiger P, et al. EGFRvIII promotes glioma angiogenesis and growth through the NF-kappaB, interleukin-8 pathway. Oncogene. 2012;31(36):4054–66. doi: 10.1038/onc.2011.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1(6):524–38. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A. 2007;104(31):12867–72. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Puliyappadamba VT, Chakraborty S, Rehman A, Vemireddy V, Saha D, et al. EGFR wild type antagonizes EGFRvIII-mediated activation of Met in glioblastoma. Oncogene. 2013 doi: 10.1038/onc.2013.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 45.Luwor RB, Zhu HJ, Walker F, Vitali AA, Perera RM, Burgess AW, et al. The tumor-specific de2–7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene. 2004;23(36):6095–104. doi: 10.1038/sj.onc.1207870. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Chakraborty S, Yang CR, Hatanpaa KJ, Cipher DJ, Puliyappadamba VT, et al. An EGFR wild type-EGFRvIII-HB-EGF feed-forward loop regulates the activation of EGFRvIII. Oncogene. 2014;33(33):4253–64. doi: 10.1038/onc.2013.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272(5):2927–35. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–49. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol. 2013;14(11):727–36. doi: 10.1038/nrm3683. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Puliyappadamba VT, Chakraborty S, Rehman A, Vemireddy V, Saha D, et al. EGFR wild type antagonizes EGFRvIII-mediated activation of Met in glioblastoma. Oncogene. 2015;34(1):129–34. doi: 10.1038/onc.2013.534. [DOI] [PMC free article] [PubMed] [Google Scholar]