Abstract
Purpose
Blocking the immunosuppressive PD-1/PD-L1 pathway has anti-tumor activity in multiple cancer types, and PD-L1 expression on tumor cells and infiltrating myeloid cells correlates with the likelihood of response. We previously found that IFNG (interferon-gamma) was over-expressed by TILs in PD-L1+ vs. PD-L1(−) melanomas, creating adaptive immune resistance by promoting PD-L1 display. The current study was undertaken to identify additional factors in the PD-L1+ melanoma microenvironment coordinately contributing to immunosuppression.
Experimental design
Archived, formalin-fixed paraffin-embedded melanoma specimens were assessed for PD-L1 protein expression at the tumor cell surface with immunohistochemistry (IHC). Whole genome expression analysis, quantitative (q)RT-PCR, immunohistochemistry, and functional in vitro validation studies were employed to assess factors differentially expressed in PD-L1+ versus PD-L1(−) melanomas.
Results
Functional annotation clustering based on whole genome expression profiling revealed pathways up-regulated in PD-L1+ melanomas, involving immune cell activation, inflammation, and antigen processing and presentation. Analysis by qRT-PCR demonstrated over-expression of functionally related genes in PD-L1+ melanomas, involved in CD8+ T cell activation (CD8A, IFNG, PRF1, CCL5), antigen presentation (CD163, TLR3, CXCL1, LYZ), and immunosuppression [PDCD1 (PD-1), CD274(PD-L1), LAG3, IL10]. Functional studies demonstrated that some factors, including IL-10 and IL-32-gamma, induced PD-L1 expression on monocytes but not tumor cells.
Conclusions
These studies elucidate the complexity of immune checkpoint regulation in the tumor microenvironment, identifying multiple factors likely contributing to coordinated immunosuppression. These factors may provide tumor escape mechanisms from anti-PD-1/PD-L1 therapy, and should be considered for co-targeting in combinatorial immunomodulation treatment strategies.
Keywords: PD-L1, PD-1, melanoma, immunotherapy
INTRODUCTION
Programmed death ligand 1 (PD-L1, B7-H1) expression by antigen presenting cells (APCs) is a normal feedback mechanism for terminating immune responses appropriately and maintaining self-tolerance.1 Aberrant PD-L1 expression in cancers co-opts this mechanism, facilitating escape from immune attack. PD-1, the dominant receptor for PD-L1, is found on activated T, B and NK cells in the tumor microenvironment (TME). Its ligation by PD-L1 down-modulates anti-tumor immune effector functions. Antibodies (mAbs) interrupting the PD-1 pathway, blocking either PD-1 or PD-L1, have durable efficacy in patients with advanced melanoma and other cancers, further highlighting the key role of this pathway in local tumor immunosuppression.2, 3
Multiple studies using different detection methods and analytic criteria have demonstrated that PD-L1 expression on tumor cells and/or leukocytes in the TME may predict response to PD-1 pathway blockade. In some cancers, such as MSI colon cancer, PD-L1 is expressed predominantly on tumor-infiltrating monocytic cells rather than on tumor cells themselves,4 whereas we reported that PD-L1+ melanomas and head and neck cancers express PD-L1 on both tumor and monocytic cells.5, 6 A recent study suggests that leukocyte expression of PD-L1 is most predictive of response to an anti-PD-L1 antibody.7 Therefore, understanding TME factors that coordinately influence PD-L1 expression on tumor cells and/or leukocytes is essential to augmenting the clinical impact of anti-PD-1/PD-L1 therapies. Such factors may warrant further study as candidate biomarkers of clinical outcomes to PD-1 blockade, or as co-targets for developing synergistic combination therapies with anti-PD-1/PD-L1.
METHODS
Melanoma specimens
Forty-nine formalin-fixed paraffin-embedded (FFPE) melanoma specimens were characterized for PD-L1 expression by immunohistochemistry (IHC) as described,5 including 4 primary and 45 metastatic lesions. “PD-L1+” was defined as ≥5% of tumor cells showing cell surface staining with the murine anti-human PD-L1 mAb 5H1 (Lieping Chen, Yale University). The geographic association of PD-L1 expression with the presence of TILs was also noted, and TILs were scored as none (0), mild (1), moderate (2), or severe (3) in intensity, as previously described.5 Eleven specimens were subjected to laser capture microdissection (LCM) followed by cDNA-mediated Annealing, Selection, extension and Ligation (DASL) microarray to profile differential gene expression between 5 PD-L1+ and 6 PD-L1(−) melanomas. Another set of eleven specimens, including 4 specimens previously assessed with microarray and 7 new cases, was used to validate differential expression of candidate genes with quantitative (q)RT-PCR, including 6 PD-L1+ cases and 5 PD-L1(−) specimens with TIL intensities similar to the original set (detailed in Supplementary Methods). A separate cohort of 8 lymph node metastases was used to develop amplified in situ hybridization (ISH) detection methods for LAG3 expression, and 25 lymph node metastases were used to investigate geographic relationships between tumor cell PD-L1 expression and TIL LAG-3 expression with IHC. Six specimens were each included in two cohorts. Studies were approved by the Institutional Review Board at the Johns Hopkins University School of Medicine.
Laser capture microdissection (LCM) and RNA isolation
Tumor cells and neighboring immune infiltrates (lymphocytes and macrophages) were excised from FFPE melanoma specimens with LCM, avoiding necrotic areas. RNA was isolated as previously described using the High Pure RNA Paraffin Kit (Roche Diagnostics, Indianapolis, IN).5 For PD-L1+ tumors, IHC on neighboring tissue sections was used to identify areas of PD-L1 expression for excision. For PD-L1(−)tumors, regions of tumor and associated infiltrating immune cells were sampled.
Whole genome microarray analysis
Gene expression was detected by DASL assays arrayed on the Illumina Human HT-12 WG-DASL V4.0 R2 expression bead chip (GEO platform GPL14951), per the manufacturer's specifications. This platform detects 29,377 annotated transcripts and is designed to detect partially degraded mRNAs such as typically found in FFPE tissue specimens.8 Briefly, mRNA isolated from melanoma specimens was reverse transcribed and amplified by PCR using universal primers. PCR products were denatured and hybridized to the Illumina array, washed and scanned to obtain gene expression intensity data. A single intensity (expression) value for each Illumina probe on the DASL array was obtained using Illumina GenomeStudio software with standard settings and no background correction (the dataset is available at NCBI’s Gene Expression Omnibus under the GEO Series accession number GSE65041). Based on examining the histograms of the expression values for each sample, which were generally bimodal, a probe was considered to be Present in a given sample if its corresponding expression value was at or above 1024 (210), and no normalization was performed. The expression values for all probes and samples were log (base 2) transformed before performing statistical analysis. Analysis for differential expression was carried out for each Illumina microarray probe. Lists of genes passing specified distinguishing criteria were examined for significant enrichment in gene annotation categories, and in functionally related categories including KEGG pathways, using the DAVID web tool (http://david.abcc.ncifcrf.gov/).9.10 Additional details are provided in Supplementary Methods.
Multiplex qRT-PCR
Total RNA from each melanoma specimen was reverse-transcribed, pre-amplified, and added to TaqMan Array Micro Fluidic cards per protocol (Applied Biosystems, Foster City, CA) (see Supplementary Methods). These cards were custom-designed with 64 gene-specific primers/probes in triplicate, including internal controls (Supplementary Table S1). PCRs were run using a 7900 HT Fast Real Time PCR system, and data analysis and display were performed using the manufacturer’s software (Applied Biosystems). Analysis was based on tumor PD-L1 expression status (positive or negative by IHC), using the manufacture’s software.
Analysis of lymphocyte activation gene 3 (LAG-3) expression in tissue sections
LAG-3 protein expression was analyzed in 5-um-thick FFPE tissue sections by IHC. Antigen retrieval was performed at 120°C for 10 min in citrate buffer, pH 6.0. Primary anti-LAG-3 mAb (murine anti-human clone 17B4, LS Bio, Seattle, WA) was used at a concentration of 1.0 ug/mL and incubated for 2 hours at room temperature. An anti-mouse Ig HRP polymer detection kit was used for visualization (ImmPRESS, Vector Laboratories). Cases where ≥5% of TILs expressed LAG-3 were considered positive.
LAG3 mRNA expression was detected by amplified ISH, performed by an automated stainer (Ventana Discovery Ultra, Ventana Medical Systems, Tucson, AZ) using the RNAscope kit (Advanced Cell Diagnostics Inc., Hayward, CA) according to the manufacturer’s instructions. In brief, 5-µm-thick FFPE tissue sections were deparaffinized and rehydrated before pretreatment with heat and protease. They were then hybridized with LAG3-specific probes, followed by the application of the preamplifier, amplifier, and horseradish peroxidase-labeled probes (Advanced Cell Diagnostics). Color development was performed with diaminobenzidine. Probes for products of the bacterial gene dapB and the housekeeping gene PPIB (peptidylprolyl isomerase B, cyclophilin B) were used as negative and positive controls, respectively, for mRNA expression. Brown, punctate dots visualized in the cytoplasm by light microscopy were considered positive signals.
Cell cultures
Human monocyte and T cell cultures were generated from leukapheresis specimens from consenting donors under IRB-approved protocols. Peripheral blood mononuclear cells (PBMCs) were isolated following density gradient centrifugation (Ficoll-Hypaque, GE Healthcare, Uppsala, Sweden) and cryopreserved. Short-term monocyte cultures were established from thawed PBMCs by plastic adherence for 2 hours at 37° C, in RPMI 1640 medium with 10% heat-inactivated AB serum (Life Technologies, Carlsbad, CA). Non-adherent cells were removed by washing to enrich for adherent monocytes. CD3+ T cells were isolated from PBMCs using the MACS Pan T Cell Isolation Kit II (Miltenyi Biotec, Auburn, CA) and cultured in RPMI 1640 medium with 10% heat-inactivated AB serum and cytokines as described below. The melanoma cell lines 537-mel, 1363-mel, and 1558-mel, established from metastatic melanoma specimens as previously described,11 were cultured in RPMI 1640 medium with 10% heat-inactivated FBS (Life Technologies; or Sigma-Aldrich, St. Louis, MO). All cultures were maintained in a 37°C, 5% CO2 incubator.
Recombinant cytokines and chemokines
Commercially available recombinant human cytokines and chemokines were added to cell cultures at final concentrations of 100 or 250 ng/ml, except interferon-gamma (IFN-g) which was used at 100 or 250 IU/ml. These concentrations were selected as biologically active based on published literature. IL-10, CCL5 (RANTES), and CXCL1 were purchased from Peprotech (Rocky Hill, NJ); IL-18, IL-32-a, and IL-32-g from R&D Systems (Minneapolis, MN); and IL-21 from GIBCO (Frederick, MD). IFN-g was obtained from Biogen (Cambridge, MA).
Effects of cytokines and chemokines on PD-L1 expression by cultured melanomas
Cultured melanoma cells were seeded into 24-well plates at 50% confluence and allowed to adhere for at least 24 hours. Then they were cultured under 3 conditions: without cytokines, with individual cytokines or chemokines, or with a combination of IFN-g plus each cytokine or chemokine. Cells were harvested after 1, 2, or 3 days and co-stained with mAbs specific for PD-L1 (clone MIH1) and HLA-DR (clone L243), or isotype-matched negative control antibodies. Treatment with IFN-g alone provided a positive control for promoting PD-L1 and HLA-DR expression by melanoma cells.
Effects of cytokines and chemokines on PD-1/PD-L1 expression by human PBMCs (T cells and monocytes)
Unseparated PBMCs were cultured in 24-well plates (1.5–2.0e6 per well) in the presence or absence of a cytokine or chemokine, under each of the following conditions: 1) no T-cell stimulation; 2) anti-CD3 alone (plate-bound OKT3, 0.2 ug/ml); and 3) anti-CD3 plus anti-CD28 (soluble clone CD28.2, 0.2 ug/ml). Cells cultured for 1–3 days were co-stained for CD3 and the following markers: CD8, PD-1 (clone MIH4 or EH12.1), PD-L1, HLA-DR and CD69. Monocytes in these cultures were evaluated by gating on CD3(−)FSChiSSChi events.
To determine the effects of IL-32-g on T cells in the absence of monocytes, CD3+ cells were isolated (>98% purity) and cultured with or without anti-CD3/CD28 stimulation in the presence or absence of IL-32-g (100 ng/ml) for 1 or 3 days. Cells were then co-stained for CD8 or CD4, and the following markers: PD-1, PD-L1, and CD69.
To investigate the effects of IL-10 and IL-32-g on monocytes in the absence of lymphocytes, monocytes were enriched by plastic adherence and cultured under the following conditions: 1) no cytokines; 2) IFN-g alone (100 or 250 IU/ml); 3) IL-10 alone or IL-32-g alone (100 ng/ml); and 4) IFN-g plus either IL-10 or IL-32-g. After two days, cells were collected and co-stained for CD14 and the following markers: PD-1, PD-L1, PD-L2 (clone MIH18), CD86, and HLA-DR.
Flow cytometric analysis
Non-specific mouse IgG (Life Technologies) was used to block Fc receptors on PBMCs and enriched monocytes. All fluorochrome-conjugated specific mAbs and their isotype-matched controls were purchased from BD Biosciences (San Jose, CA) or eBioscience (San Diego, CA). Samples were acquired on the BD FACSCalibur and data were analyzed with FlowJo Software (TreeStar, Ashland, OR).
RESULTS
Whole genome microarray and functional clustering analysis
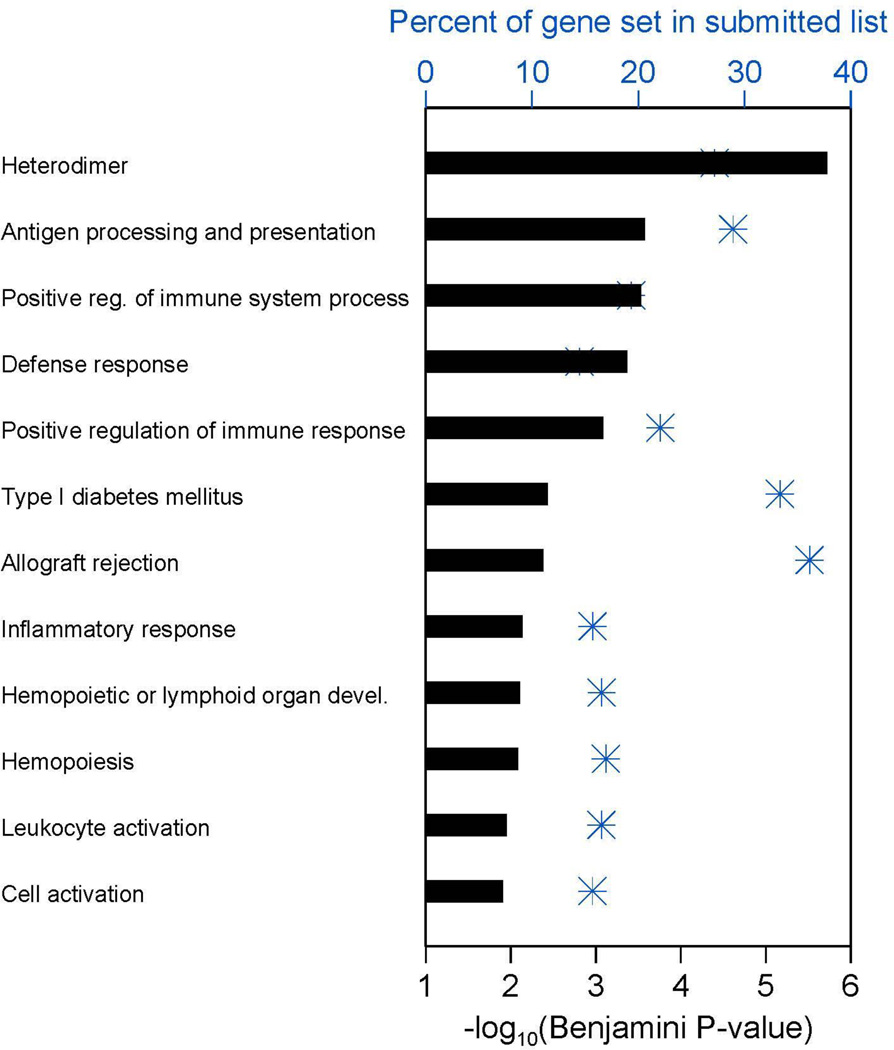
To identify genes differentially expressed between PD-L1+ and (−) melanomas, LCM was used to precisely capture boundary areas containing tumor cells and TILs (“immune fronts”) from 5 PD-L1+ melanoma specimens and 6 PD-L1(−) melanomas. While there is a general association between the presence of TILs and PD-L1 expression, a significant number of TIL-infiltrated tumors do not express PD-L1;5 this subset of tumors represented the PD-L1(−) subset. Cases with PD-L1 expression had moderate to severe TILs (grade 2–3), while PD-L1(−) cases had more modest immune infiltrates (grade 1). High-throughput whole genome microarray analysis was performed. The list of 1660 Illumina probes up-regulated at least 2-fold in PD-L1+ specimens (Supplementary Table S2) was submitted to the NIH DAVID database for functional annotation clustering. Twelve resulting categories containing ≥10 distinct genes from this list and having a Benjamini-Hochberg adjusted p-value (FDR) ≤0.015 were obtained.12 As shown in Figure 1 and Supplementary Tables S3 and S4, genes over-expressed in PD-L1+ melanomas were functionally related in pathways involving immune cell activation, inflammation, and antigen processing and presentation, among others. These results demonstrate an enhanced immune-reactive microenvironment in PD-L1+ compared to PD-L1(−) melanomas.
Figure 1. Whole genome microarray reveals functional gene categories that are differentially expressed in PD-L1+ vs. negative melanomas.
In PD-L1+ melanomas, TIL intensity was moderate to severe, while in PD-L1(−) melanomas it was mild. Blue asterisks show the percent of each gene set that occurred in the list submitted to DAVID analysis, using the x-axis scale at the top of the plot. The horizontal bar for each gene set shows –log10 of the corresponding Benjamini-Hochberg multiple comparison adjusted p-value as calculated by DAVID, using the x-axis scale at the bottom of the plot. Functional categories with a gene count ≥10 and a Benjamini p-value ≤0.015 are shown. Additional information, including the specific number of genes in the submitted list present in each DAVID functional category appearing in the plot, is provided in Supplementary Tables S3 and S4.
Gene expression profiling with multiplex qRT-PCR
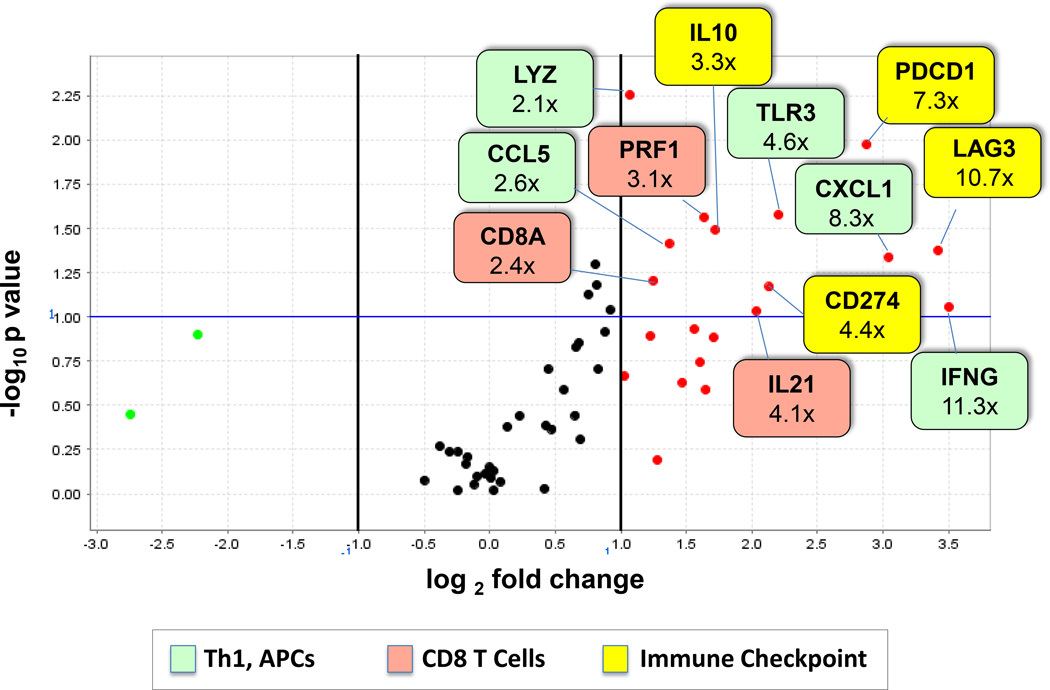
A custom qRT-PCR array was designed to validate differential expression of genes revealed by whole genome microarray analysis, and to test additional candidate genes potentially associated with tumor PD-L1 expression. A new cohort of 11 melanoma specimens [6 PD-L1+ and 5 PD-L1(−)] was subjected to LCM and RNA isolation. Sixty genes were analyzed including 45 candidate genes and 15 genes selected from the microarray analysis based on their degree of up-regulation in PD-L1(+) melanomas and functional relevance to immunoregulatory pathways (Supplementary Table S1). Figure 2 and Supplementary Table S5 show genes significantly up-regulated ≥ 2-fold in PD-L1+ specimens when normalized to expression of PTPRC (CD45, pan immune cell marker) or GUSB (beta-glucuronidase). They include functionally related genes whose expression is characteristic of activated CD8+ T cells and APCs, as well as several immune checkpoint molecules. In specimens that expressed PD-L1 protein by IHC, CD274 (PD-L1) mRNA expression was also up-regulated. IFNG expression was up-regulated in PD-L1+ vs. (−) melanomas, consistent with previous results.5,6 Also consistent with previous studies, PDCD1 (PD-1) was over-expressed in the PD-L1+ TME.13 Other immunosuppressive molecules coordinately expressed with CD274 included IL10 and LAG3.
Figure 2. Functionally related genes are over-expressed in PD-L1+ melanomas.
RNA isolated from 6 PD-L1+ and 5 PD-L1(−) melanomas was assessed for expression of immune-related genes in a multiplex qRT-PCR assay. Results were normalized to PTPRC (CD45, a pan immune cell marker). Red and green dots represent genes over- and under-expressed by at least 2-fold, respectively, in PD-L1+ tumors. The horizontal blue line represents a p-value = 0.10. Numbers in colored boxes denote fold-change in gene expression. Over-expressed genes are associated with immunosuppressive molecules, activated CD8+ T cells, and antigen presenting cells. Additional information is provided in Supplementary Table S5. APC, antigen presenting cell; Th1, T helper 1.
LAG-3 over-expression in the PD-L1+ tumor immune microenvironment
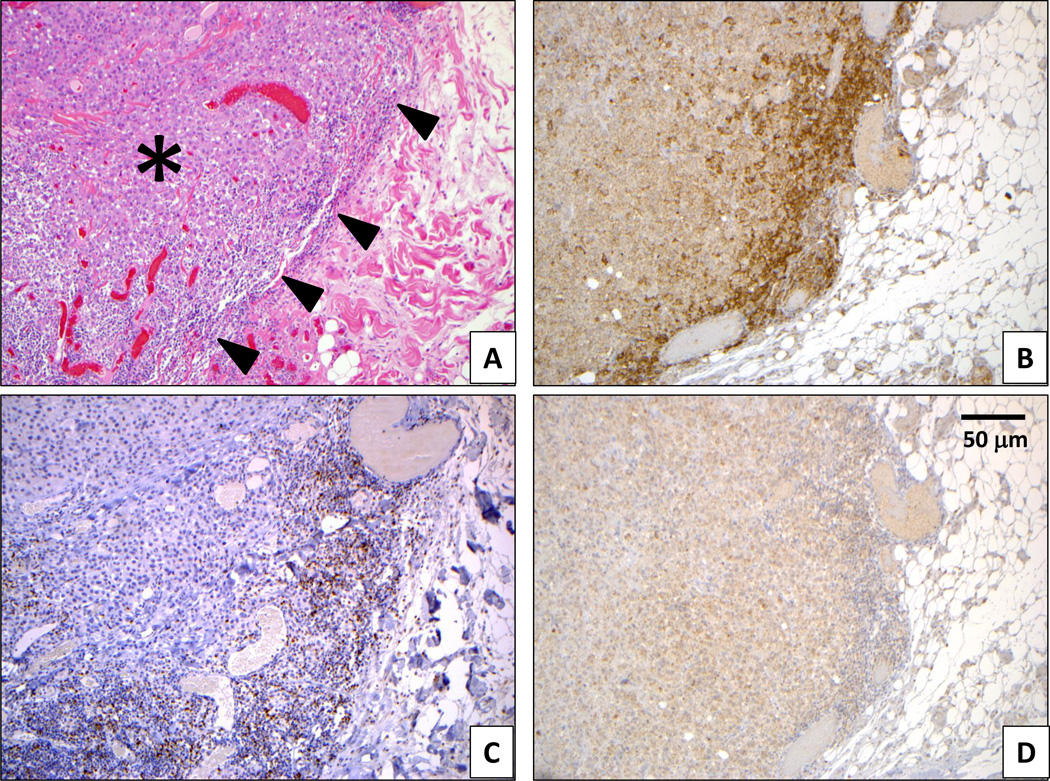
The coordinate expression of the LAG3 immune checkpoint with CD274, detected on mRNA analysis, was further explored on a protein level with IHC in a cohort of 25 melanoma lymph node metastases. Eleven of 12 cases (92%) demonstrating tumor cell PD-L1 protein expression at the interface between tumor and lymphocytes also showed LAG-3 protein expression by at least 5% of those lymphocytes (Figure 3, Supplementary Figure S1). In contrast, only 2 of 13 (15%) cases that were PD-L1(−) expressed LAG-3 (p=0.0002, Fisher’s Exact Test). Thus, LAG-3+ TILs significantly co-localized with PD-L1+ melanoma cells.
Figure 3. PD-L1 and LAG-3 are geographically associated at the interface of a metastatic melanoma deposit and immune cell infiltrates.
A subcutaneous deposit of metastatic melanoma is surrounded by a lymphohistiocytic host response (panel A, H&E stain). The tumor is denoted by an asterisk, and the host immune response is denoted by arrowheads. By IHC, both the immune infiltrates and adjacent tumor cells demonstrate PD-L1 expression (B), which co-localizes with LAG-3+ lymphocytes (C, brown stain). Cytoplasmic melanin granules in melanoma cells are evident with isotype control staining (D). Original magnification 200X.
LAG3 mRNA expression in melanoma tissue sections was also detected with amplified ISH and was compared to protein expression on IHC. In all 8 melanoma lymph node metastases examined, the density and location of lymphocytes expressing LAG-3 by IHC correlated with ISH results (Supplementary Figure S2).
Impact of cytokines and chemokines on melanoma cell expression of PD-L1
Several cytokines and chemokines that were over-expressed in the PD-L1+ melanoma microenvironment were tested in vitro for their potential effects on PD-L1 expression by three melanoma cell lines. These recombinant proteins included CCL5 (RANTES), CXCL1, IL-10, IL-18, and IL-21 (Supplementary Table S5). In addition, the alpha and gamma isoforms of IL-32 were assessed in vitro. IL32 mRNA, detected by one of two probes in the whole genome microarray recognizing a shared sequence in all IL32 isoforms, was up-regulated 15-fold in PD-L1+ melanomas (p=0.014) (Supplementary Table S2); although this was not validated by qRT-PCR, the available commercial probe detected a different region of IL32. The melanomas 537-mel, 1363-mel and 1558-mel, known to up-regulate cell surface PD-L1 and HLA-DR protein expression in response to IFN-g exposure in vitro,5 were incubated with each recombinant protein at 100 or 250 ng/ml, with or without IFN-g at 100 IU/ml. Notably, none of these individual factors or combinations affected the intensity of melanoma cell surface PD-L1 or HLA-DR expression after 1, 2 or 3 days of culture (not shown). Additionally, at 6 days, no effects were observed on melanoma proliferation. Therefore, these factors did not appear to directly affect melanoma cell expression of PD-L1 in vitro.
Impact of cytokines and chemokines on immune cell expression of PD-1 and its ligands
We next investigated the potential effects of factors over-expressed in PD-L1+ melanomas, on immune cell expression of PD-1 and its ligands. PBMCs were cultured for 1–3 days in the absence or presence of recombinant CCL5, CXCL1, IL-10, IL-18, IL-21, IL-32-a, or IL-32-g. Cultures were conducted with or without T cell stimulation by anti-CD3 alone (suboptimal) or in combination with anti-CD28 (optimal). Changes in cell surface expression of PD-1, PD-L1, and the activation markers HLA-DR and CD69 on CD3+ cells, or on CD3(−)FSChiSSChi cells (monocyte population), were examined.
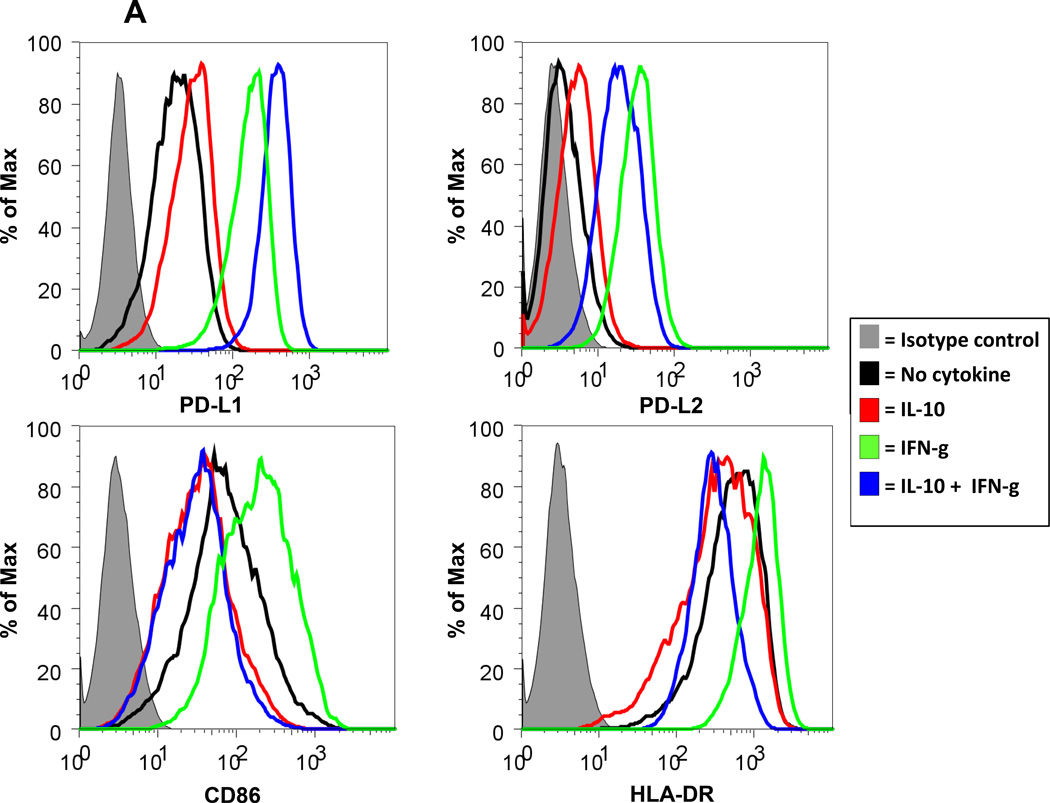
When IL-10 was present during T cell stimulation with anti-CD3 or anti-CD3/CD28, decreased activation was observed, evidenced by decreased expression of PD-1, PD-L1, CD69, and HLA-DR on CD3+ cells in 2 of 2 donors tested (not shown). At the same time, increased expression of PD-L1 and decreased expression of HLA-DR on CD3(−) FSChiSSChi cells (monocyte population) were seen. To determine if IL-10 directly affected monocytes in the absence of T cells, monocytes were enriched from PBMCs by plastic adherence and were exposed to IL-10 for 48 hours in the presence or absence of IFN-g. IL-10 alone selectively increased expression of the co-inhibitory ligands PD-L1 and PD-L2 on CD14+ cells, while decreasing the co-stimulatory molecules CD86 and HLA-DR. As expected, IFN-g alone up-regulated expression of all four molecules. Combining IL-10 with IFN-g further increased PD-L1 expression, compared to either cytokine alone. However, this cytokine combination reduced PD-L2, CD86 and HLA-DR expression below levels achieved with IFN-g alone, and in some cases below baseline levels in the absence of cytokines (Figure 4A). Thus, IL-10 appeared to dampen T cell activation while shifting the balance of monocyte expression of co-regulatory molecules towards an immunosuppressive profile.
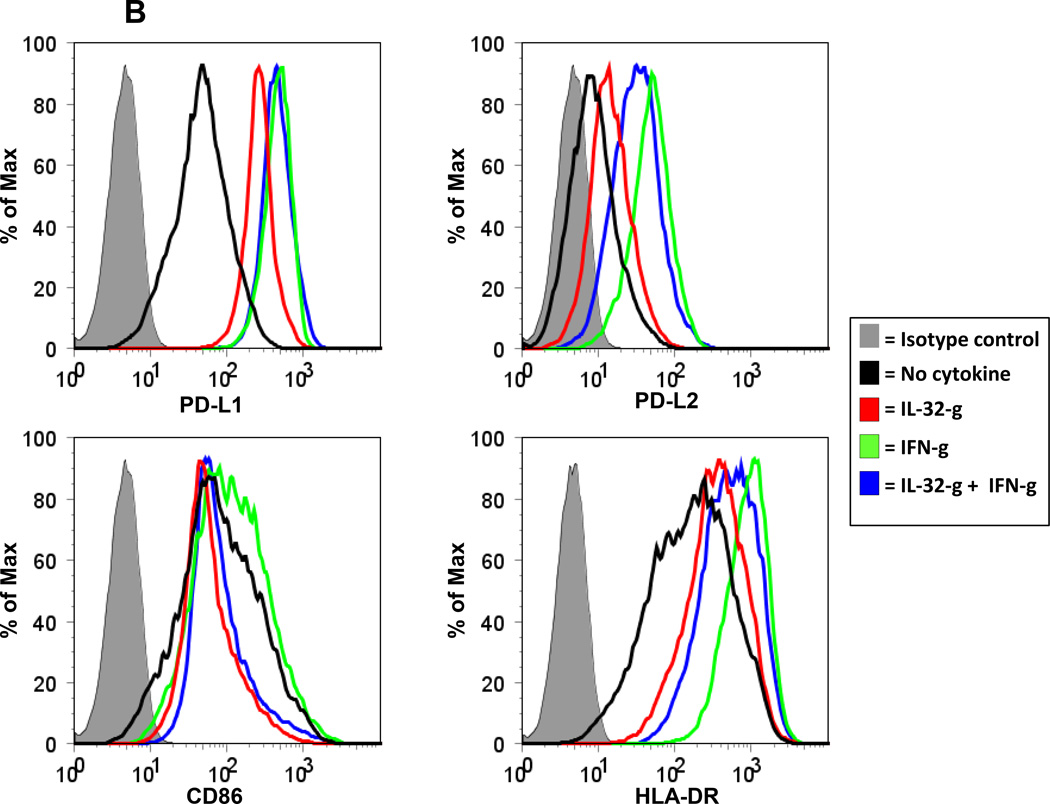
Figure 4. IL-10 and IL-32-g promote PD-L1 expression on monocytes.
Human monocytes enriched by plastic adherence were cultured in the presence IL-10 (A) or IL-32-g (B) at 100 ng/ml. Cells were also cultured with IFN-g alone (250 or 100 ng/ml, respectively, for A and B), or IFN-g plus IL-10 or 1L-32-g. After 48 hours, CD14+ gated events were analyzed for expression of PD-L1, PD-L2, CD86 (B7.2) and HLA-DR by flow cytometry. Panels in (A) are a composite of data from 3 donors and are representative of the majority of four donors tested for each marker. Results in (B) are derived from cells from a single donor and are representative of three donors tested.
Among the other factors tested, IL-32-g had a reproducible effect on the expression of PD-1 ligands by cultured PBMCs. When unseparated PBMCs were cultured for 1 to 3 days in the presence of IL-32-g, both the proportion and intensity of PD-L1 and CD69 expression on CD3+ T cells [CD8+ and CD8(−) ] increased (Supplementary Figure S3). No effects were observed on anti-CD3/CD28 stimulated cells, which abundantly expressed PD-L1 and CD69. Interestingly, purified CD3+ T cells were not affected by IL-32-g (3 of 3 donors), implying that effects on T cells in unseparated PBMCs were mediated by factors produced by non-T cells. Because PD-L1 expression was also increased on CD3(−) FSChiSSChi cells in unseparated PBMCs cultured with IL-32-g, we next exposed enriched monocyte cultures to IL-32-g. Similar to the effects observed with IL-10, IL-32-g alone was found to increase the expression of PD-L1 and PD-L2 on CD14+ cells, and in combination with IFN-g it mitigated IFN-induced enhancement of PD-L2, CD86 and HLA-DR expression (Figure 4B). These effects were specific to IL-32-g and were not observed with IL-32-a. Furthermore, when supernatants from IL-32-g-exposed monocytes were added to purified CD3+ T cells, a modest increase in PD-L1 expression was observed on both CD8+ and CD4+ T cells, suggesting complex mechanisms by which IL-32-g may modulate the expression of immune regulatory molecules intratumorally.
DISCUSSION
Many human cancers contain tumor and/or stromal cells expressing the immunosuppressive ligand PD-L1. Tumor cell PD-L1 expression may result from dysregulated signaling pathways or somatic gene alterations including amplifications and translocations.14 However, its expression in melanoma is characteristically associated with intratumoral immune infiltrates, a phenomenon termed “adaptive immune resistance”.5 IFN-g, a dominant product of human melanoma-specific TILs, is a major inducer of PD-L1 expression on tumor cells in vitro15 and is selectively over-expressed in the milieu of PD-L1+ solid tumors including melanomas.5,6 The current study confirms an association of IFN-g with PD-L1+ melanomas and goes beyond this to explore other factors coordinately expressed in the PD-L1+ melanoma microenvironment. It reveals a complex landscape of interacting receptors, ligands and soluble factors which may be exploited to therapeutic advantage.
Several of the molecules that were found to be significantly over-expressed in PD-L1+ vs. PD-L1(−) melanomas are associated with activated CD8+ T cells (CD8A, PRF1, IL18, IL21). The observed co-expression of mRNAs encoding CD274 and CD8A is consistent with published reports detecting the co-localization of PD-L1+ melanoma cells with infiltrating CD8+ T cells by IHC.16,17 The presence of CD8+ TILs has recently been proposed as a biomarker of response to PD-1 blockade in melanoma.16 In addition to markers of T cell activation, we also found evidence for over-expression of molecules associated with activated pro-inflammatory APCs, including CXCL1, TLR3, and LYZ. This constellation of factors characterizes an immune-reactive microenvironment poised to eliminate cancer cells, if not for dominant inhibition exerted by the PD-L1 checkpoint.
Importantly, additional checkpoints were found to be over-expressed in the PD-L1+ melanoma microenvironment, including PDCD1, LAG3, and IL10. While LAG-3 and IL-10 appear to be sub-dominant in the hierarchy of intratumoral immunosuppression, they may nevertheless provide bypass mechanisms for melanoma to evade anti-PD-1/PD-L1 therapies. These findings suggest opportunities for co-targeted combination treatment regimens. For instance, co-expression of PD-1 and LAG-3, a distinct inhibitory receptor expressed on activated T cells, has been demonstrated in murine and human TILs.18,4 Blocking LAG-3 is marginally effective as monotherapy but synergizes with anti-PD-1 in murine tumor models, providing a rationale for an ongoing clinical trial of anti-LAG-3 plus anti-PD-1 in patients with advanced solid tumors (NCT01968109). IL-10 is an anti-inflammatory cytokine produced by T helper cells, T regulatory cells, monocytes, and some human cancers.19 IL-10 secretion by T cells has been shown to be stimulated by PD-L1.20 In the current study, IL-10 exposure inhibited human T cell activation and promoted monocyte PD-L1 expression in vitro, consistent with published reports,21 supporting a checkpoint role for this cytokine. The potential for IL-10 to mediate non-antigen driven LAG-3 expression on human tumor-specific T cells has also been reported,22 further expanding the immunosuppressive profile of this cytokine. Finally, our study also revealed over-expression of the proinflammatory cytokine IL-32 in the PD-L1+ melanoma microenvironment, although the cellular source of this cytokine was not determined. IL-32 has been associated with progression and metastasis in some human cancers.23 Its most active isoform, IL-32-g, has been shown to activate monocytes and promote their secretion of IL-6 and TNF-a.24, 25 However, the specific effects of IL-32-g on lymphocyte and monocyte PD-L1 expression demonstrated here have not been reported previously to our knowledge, and suggest a complex functional profile for this cytokine which may contribute to local tumor immunosuppression. Notably, neither IL-10 nor IL32-g affected PD-L1 expression by melanoma cells, either with or without IFN-g, which could reflect lack of expression of the corresponding cytokine receptors by tumor cells or differences in downstream signaling between melanoma cells and monocytes.
In summary, while IFN-g appears to be a dominant factor mediating adaptive immune resistance in melanoma and other human tumors, the current study reveals additional interacting factors contributing to this phenomenon. Some of these factors may selectively enhance PD-L1 expression on tumor infiltrating myeloid cells rather than tumor cells. It is only by identifying and targeting such coordinated immunosuppressive molecular networks that the impact of anti-PD-1/PD-L1 immunotherapies can be fully realized.
Supplementary Material
Statement of translational relevance.
Although drugs blocking the PD-1/PD-L1 pathway have shown efficacy in some patients with advanced cancers, a deeper knowledge of coordinated immunosuppression in the PD-L1+ tumor microenvironment is needed to improve upon these therapeutic results. Here we show that PD-L1+ melanomas over-express PD-1, LAG-3, IL-10, and IL-32, which may contribute to local immunosuppression and therefore are candidates for co-targeting in combination treatment regimens. This study further reveals factors that selectively induce PD-L1 on myeloid cells but not tumor cells, and thus begins to elucidate novel mechanisms for PD-L1 up-regulation in the tumor microenvironment.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Lieping Chen (Yale University) for providing the anti-PD-L1 monoclonal antibody 5H1; Dr. Chris Umbricht and Dr. Mariana Brait (Johns Hopkins University) for technical advice; Dr. Maria Ascierto (Johns Hopkins University) for helpful discussions; and Jung H. Kim for technical assistance. This study was supported by the Dermatology Foundation (JMT), the National Cancer Institute NIH (R01 CA142779; JMT, DMP, SLT), the Melanoma Research Alliance (JMT, DMP, SLT), the Barney Family Foundation (JMT, SLT), the Laverna Hahn Charitable Trust (JMT, SLT), the Commonwealth Foundation (JMT, DMP), and Moving for Melanoma of Delaware (JMT, DMP, SLT). JMT, DMP and SLT were also supported by a Stand Up To Cancer—Cancer Research Institute Cancer Immunology Translational Cancer Research Grant (SU2C-AACR-DT1012). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
The following authors have declared relevant financial relationships: JMT, research support from Bristol-Myers Squibb, and consulting for Bristol-Myers Squibb. DMP, research grants from Bristol-Myers Squibb and Potenza Therapeutics; consulting for Amgen, Five Prime Therapeutics, GlaxoSmithKline, Jounce Therapeutics, MedImmune, Merck, Pfizer, Potenza Therapeutics, and Sanofi; stock options in Jounce and Potenza; and patent royalties through his institution, from Bristol-Myers Squibb and Potenza. SLT, research grants from Bristol-Myers Squibb, and consulting for Five Prime Therapeutics, GlaxoSmithKline, and Jounce Therapeutics.
Footnotes
The remaining authors have declared no financial relationships.
REFERENCES
- 1.Pardoll DM, Drake CG. Immunotherapy earns its spot in the ranks of cancer therapy. J Exp Med. 2012;209:201–209. doi: 10.1084/jem.20112275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2014 Nov 16; doi: 10.1056/NEJMoa1412082. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Llosa NJ, Cruise M, Tam A, Wick EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Co-localization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Science Transl Med. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyford-Pike S, Pen S, Young GD, Taube JM, Westra WH, Akpeng B, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73:1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikova M, Talantov D, Chudin E, Yeakley JM, Chen J, Doucet D, et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol. 2004;165:1799–1807. doi: 10.1016/S0002-9440(10)63435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 10.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topalian SL, Rivoltini L, Mancini M, Markus NR, Robbins PF, Kawakami Y, et al. Human CD4+ T cells specifically recognize a shared melanoma-associated antigen encoded by the tyrosinase gene. Proc Natl Acad Sci USA. 1994;91:9461–9465. doi: 10.1073/pnas.91.20.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 13.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2014 Dec 6; doi: 10.1056/NEJMoa1411087. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong H1, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 16.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spranger S1, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012;72:917–927. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennisa KL, Blatnera NR, Gounarib F, Khazaiea K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr Opin Oncol. 2013;25:637–645. doi: 10.1097/CCO.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong H1, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 21.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci USA. 2010;107:7875–7880. doi: 10.1073/pnas.1003345107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai CY, Wang CS, Tsai MM, Chi HC, Cheng WL, Tseng YH, et al. Interleukin-32 increases human gastric cancer cell invasion associated with tumor progression and metastasis. Clin Cancer Res. 2014;20:2276–2288. doi: 10.1158/1078-0432.CCR-13-1221. [DOI] [PubMed] [Google Scholar]
- 24.Choi J, Bae S, Hong J, Azam T, Dinarello CA, Her E, et al. Identification of the most active interleukin-32 isoform. Immunology. 2008;126:535–542. doi: 10.1111/j.1365-2567.2008.02917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu WD, Zhang M, Feng CC, Yang XK, Pan HF, Ye DQ. IL-32 with potential insights into rheumatoid arthritis. Clin Immunol. 2013;147:89–94. doi: 10.1016/j.clim.2013.02.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.