Abstract
The mitochondrial sirtuin SIRT3 is a protein deacylase that regulates almost every major aspect of mitochondrial biology, including nutrient oxidation, ATP generation, reactive oxygen species detoxification, mitochondrial dynamics, and the mitochondrial unfolded protein response. Interestingly, mice lacking SIRT3 (SIRT3KO), either spontaneously or when crossed with mouse models of disease, develop several diseases of aging at an accelerated pace, such as cancer, metabolic syndrome, cardiovascular disease, and neurodegenerative diseases, and thus might be a valuable model of accelerated aging. In this review we discuss SIRT3 functions in pathways involved in diseases of aging, how lack of SIRT3 might accelerate the aging process, and suggest that further studies on SIRT3 might help uncover important new pathways driving the aging process.
Keywords: Mitochondria, Sirtuins, SIRT3, Aging, Disease
The role of sirtuins in aging and longevity
Aging is a complex process, marked by numerous cellular changes throughout an organism’s lifespan. A major goal in the study of aging is to identify the genetic determinants that regulate the aging process, influence progression of the diseases of aging, and ultimately govern longevity. The sirtuins are a conserved class of protein deacylases that were named after the founding member SIR2 (Sir2-ins) [1], and were among the first genes identified to extend lifespan in S. cerevisiae [1, 2]. An additional SIR2 allele can increase lifespan by 30%, whereas ablation of SIR2 decreased lifespan in yeast [2]. Consistent with this finding, the increased longevity that is seen with calorie restriction in yeast requires SIR2 activation [3]. These early studies set the stage for a nascent field to investigate the role of sirtuins in longevity and the biology of aging.
SIR2 has seven mammalian orthologs (Sirtuin 1-7, aka SIRT1-7), three of which (SIRT3, 4 and 5) are localized to the mitochondria. Given the importance of mitochondria in aging [4], the mitochondrial sirtuins are uniquely poised to regulate several aspects of the aging process. SIRT3 is a mitochondrial deacetylase [5], which is enriched in highly metabolic tissues such as the liver, heart, brain, and brown adipose tissue. SIRT3 has been shown to regulate almost every major aspect of mitochondrial biology, including reactive oxygen species (ROS) detoxification, ATP generation, mitochondrial dynamics, nutrient oxidation, and the mitochondrial unfolded protein response (UPR) [6-11]. SIRT3 regulates these processes by removing acetyl modifications from a growing list of mitochondrial proteins [5, 12]. SIRT3 has also been reported to remove other acyl modifications such as long-chain fatty acyl modifications and histone decrotonylation [13, 14], although understanding the biological relevance of these modifications requires further work.
A major challenge in the field is to disentangle the direct effects of SIRT3 from the indirect changes seen after SIRT3 manipulation, and ultimately to identify SIRT3 targets of deacetylation. Interestingly, an emerging sense in the field is that no single protein target can fully explain the biological and physiological effects of SIRT3. For example, SIRT3 does not likely regulate fatty acid oxidation (FAO) through deacetylation of a single enzyme [9]; instead, it likely imparts control over FAO at multiple enzymes in the pathways [15-17]. More work is needed to identify the primary targets and activities of SIRT3, and to understand the mechanism(s) by which SIRT3 regulates proteins and pathways, over a wide array of biology.
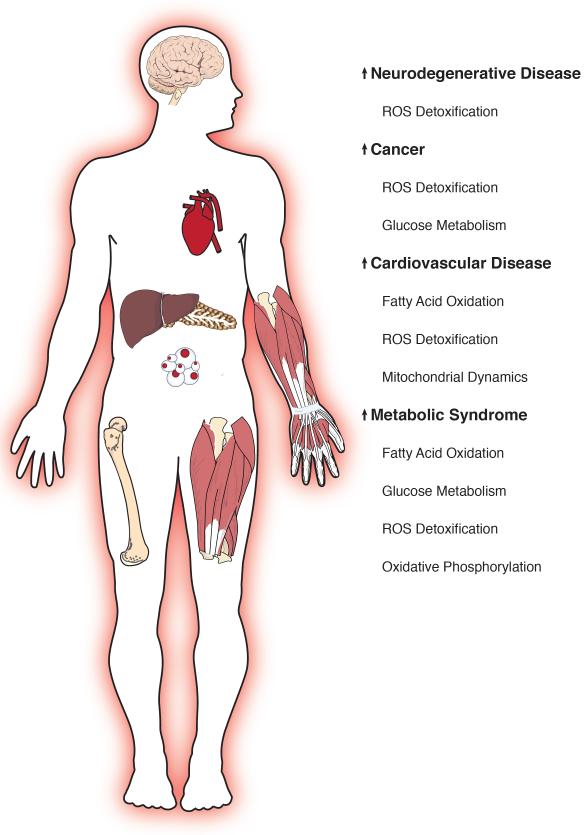
Despite these challenges, one theme that emerges from its continued study is that SIRT3 attenuation or ablation is associated with accelerated development of several diseases of aging. Indeed, SIRT3KO mice alone or when crossed with mouse models of disease, develop nearly every disease of aging at an accelerated pace (Fig. 1). Thus, the SIRT3KO mouse might be considered a model of accelerated aging. Below, we review the roles for SIRT3 in cancer, the metabolic syndrome, cardiovascular disease, and neurodegenerative diseases (Table 1), and discuss what the study of SIRT3 might reveal about the aging process.
Figure 1.
SIRT3 regulates pathways in the diseases of aging.
Loss of SIRT3 leads to deregulation of several mitochondrial pathways, which contributes to the accelerated development of the disease of aging. See text for details.
Table 1.
SIRT3 regulates pathways involved in the diseases of aging.
| Disease | Age-related Pathway |
SIRT3KO molecular phenotype |
[Patho]Physiology | Ref |
|---|---|---|---|---|
| Cancer | ROS detoxification |
 ROS ROS oxidative stress oxidative stress genomic instability genomic instability |
 tumorigenesis, tumorigenesis, cellular cellulartransformation.  SIRT3 expression in SIRT3 expression inseveral human cancers. |
[22, 26- 29, 34, 39, 40, 42] |
| Glucose metabolism |
 Glucose uptake Glucose uptake glycolysis, glycolysis,  biosynthetic biosyntheticintermediates –  “Warburg effect” “Warburg effect” |
|||
|
Metabolic
disease/insulin resistance |
Oxidative phosphorylation |
 activity of oxidative activity of oxidativephosphorylation machinery |
 Insulin resistance, Insulin resistance, nonalcoholic nonalcoholichepatosteatosis  basal metabolic rate basal metabolic rate glucose-stimulated glucose-stimulatedinsulin secretion |
[7, 9, 10, 52- 54, 56, 61] |
| Glucose metabolism |
 glucose uptake, glucose uptake, glucose oxidation glucose oxidation |
|||
| Fatty acid oxidation |
 decreased fat decreased fatmetabolism |
|||
| ROS detoxification |
 oxidative stress oxidative stress |
|||
|
Cardiovascular
Disease |
Fatty acid oxidation |
 ATP production ATP production |
 Cardiac hypertrophy Cardiac hypertrophy |
[7-9, 72, 73] |
| ROS detoxification |
 ROS in cardiomyocytes ROS in cardiomyocytes |
|||
| Mitochondrial dynamics |
 mitochondrial mitochondrialpermeability,  mitochondrial fusion mitochondrial fusion |
|||
|
Neurodegenerative
disease |
ROS detoxification |
 Oxidative damage Oxidative damage GSH levels GSH levelsElevated ROS leads to  SIRT3 expression SIRT3 expression |
 Hearing loss Hearing loss |
[79, 82, 83] |
SIRT3 and Diseases Of Aging
SIRT3 and Cancer
Cancer is a major disease of aging, where almost 90% of all cancer deaths occur in people over 50 years of age [18]. Cancer is thought to arise from an accumulation of mutations that occur slowly overtime; as such, cancer incidence increases dramatically with age. Further supporting this idea, individual tumor biopsies show multiple genetic lesions, indicative of the clonal evolution of cancer cells during tumorigenesis [19]. Environmental or genetic factors that increase the frequency of mutations or support cellular proliferation can increase the risk for and development of cancer. For example, a rise in oxidative stress can increase damage to DNA leading to genomic instability and increased mutagenesis and therefore the risk of oncogenic mutations [20]. Similarly, shifts in metabolism that promote anabolic growth may also support the growth of a tumor [21].
The role of SIRT3 in cancer was first shown by the discovery that SIRT3KO mouse embryonic fibroblasts (MEFs) could be transformed into cancer cells by the addition of a single oncogene (Myc or Ras) [22]. This single oncogene transformability in the setting of a gene knock-out is characteristic of a tumor suppressor gene, as wild-type primary MEFs require the expression of an oncogene in combination with either another oncogene (e.g. Myc or Ras) or the loss of a tumor suppressor gene (e.g. p53) in order to be transformed [23-25]. Seven out of 20 SIRT3KO mice spontaneously developed mammary tumors after two years of age, compared to none of the wild-type mice [22]. Furthermore, SIRT3 expression is decreased in several human cancers including breast, liver, and gastric cancers [22, 26-28], and low SIRT3 expression is associated with worse overall survival in hepatocellular carcinoma [29]. These studies collectively show the tumor suppressor potential of SIRT3.
Studies on the mechanisms linking SIRT3 ablation and accelerated cancer development have found increased oxidative stress and changes in cellular metabolism – both likely contributing to tumor initiation and/or progression [30]. Increased oxidative stress can promote genome instability and DNA damage, which may lead to increased mutation frequency [31]. ROS are also important signaling molecules that, when present at low levels, can alter gene transcription and promote cell proliferation [32]. Because mitochondria are the major source of ROS, SIRT3 may play a critical role in preventing tumor development by controlling ROS levels. Indeed, superoxide dismutase 2 (SOD2) is a target of SIRT3 and SIRT3KO MEFs have elevated levels of ROS [22, 33]. In support of this model, overexpression of SOD2 – a mitochondrial enzyme that detoxifies superoxide – in SIRT3KO MEFs slowed their growth and prevented their transformation in the presence of either Myc or Ras, indicating that mitochondrial ROS were required for the transformative potential of SIRT3KO cells [22]. SIRT3KO MEFs also had increased genomic instability when challenged with a genotoxic stress that was reduced by the overexpression of SOD2 [22]. Another study found that stable knockdown of SIRT3 in colon cancer cells led to larger tumor formation in mouse xenograft models, which could be prevented by the addition of the ROS scavenger N-acetyl cysteine (NAC) [34].
While the role of SIRT3 in controlling oxidative stress has been well described, SIRT3 may also affect metabolic reprogramming in tumor cells, an emerging hallmark of cancer [35]. Shifts in metabolism towards increased aerobic glycolysis have been well documented in cancer cells since Otto Warburg first reported that tumors metabolize more glucose through glycolysis and produce more lactate even in the presence of oxygen [36]. While Warburg’s initial hypothesis that mitochondrial defects appear to cause cancer in only a few isolated settings (i.e. fumarate hydratase, succinate dehydrogenase, and IDH2 mutations), altered mitochondrial metabolism seems to be a common characteristic of all cancers. Many cancer cells shift their metabolism towards anabolic processes, with reduced emphasis on ATP generation and increased generation of biosynthetic intermediates required to build new cells [21].
Cancer cell lines with SIRT3 knockdown, and MEFs from SIRT3KO mice have a shift in metabolism toward a Warburg-like phenotype [26], and have increased glucose uptake and glycolysis, increased levels of biosynthetic intermediates, increased ROS production, and proliferate faster. Multiple studies have shown that SIRT3 ablation leads to increased hypoxia inducible factor 1 alpha (HIF1α) protein stabilization and a concomitant increase in mRNA levels of numerous HIF1α target genes, including genes involved in glucose uptake and glycolysis [26, 34]. These shifts in metabolism are due, at least in part, to the increase in ROS, which is known to lead to increased HIF1α protein levels by inhibiting prolyl hydroxlases (PHDs) that target HIF1α for degradation [37, 38]. Addition of NAC, or knockdown of HIF1α prevented this shift in metabolism in cells lacking SIRT3 [26].
However, shifts in cancer cell metabolism in the absence of SIRT3 cannot be explained by ROS alone. Several studies have found that pyruvate dehydrogenase (PDH) is hyperacetylated and inhibited in the absence of SIRT3, leading to alterations in pyruvate metabolism [39, 40]. PDH is inhibited in many cancers by phosphorylation by pyruvate dehydrogenase kinases (PDKs), leading to decreased flux of glucose-derived carbon into the TCA cycle and increased lactate production [41]. Acetylation of PDH in the absence of SIRT3 also inhibits its activity and supports increased glycolysis and thus a Warburg-like shift in metabolism [39, 40]. Another recent study showed that the mitochondrial enzyme glutamate oxaloacetate aminotransferase (GOT2) is deacetylated by SIRT3; increased GOT2 acetylation promoted the growth of pancreatic tumor xenografts [42]. These examples represent a growing list of metabolic pathways that are dysregulated upon loss of SIRT3, leading to increased tumor growth.
Overall, loss of SIRT3 accelerates tumorigenesis in mouse models, and decreased SIRT3 expression is associated with several human cancers. Loss of SIRT3 leads to acetylation and deregulation of several mitochondrial proteins, which can cause increased oxidative stress and shifts in cellular metabolism – conditions that favor accelerated tumorigenesis.
SIRT3 and metabolic disease
Aging is recognized as a major risk factor for the development of diabetes [43]. The pervasiveness of diabetes and its related etiologies including obesity, nonalcoholic fatty liver disease (NAFLD), and the metabolic syndrome are on the rise. Recent estimates report that 8.3% of the global population (382 million people) currently have diabetes and this is expected to reach 10.1% of the global population (592 million people) by 2035 [43]. Interestingly, the current prevalence of diabetes is estimated to be greater in people aged between 60 and 79 (18.6% or 134.6 million people) [43, 44], suggesting a link in the development of metabolic disease and aging.
Accumulating evidence shows that mitochondrial dysfunction plays a role in the metabolic syndrome and diabetes [45]. For example, deregulation of several mitochondrial pathways has been implicated in the pathogenesis of metabolic disease including impaired FAO [46], oxidative damage from ROS [47], inflammation [48], and loss of metabolic flexibility [45, 49]. Interestingly, the decline in mitochondrial function during aging is concomitant with the development of hyperglycemia and hyperinsulinemia [50], suggesting a mechanistic link between mitochondria and age-induced metabolic dysfunction.
The first indication that SIRT3 played a role in fat metabolism was the discovery that SIRT3KO mouse livers accumulate long-chain acylcarnitine species and have impaired FAO [9]. Further studies demonstrated that hepatic SIRT3 expression and activity decreases in response to chronic high-fat diet (HFD) feeding, resulting in protein hyperacetylation, an impaired ability to protect against lipotoxic conditions, and hepatic steatosis [51-53]. SIRT3KO mice fed a HFD showed accelerated development of several hallmarks of the metabolic syndrome, including weight gain, impaired glucose tolerance, and insulin resistance (IR)[52]. Thus, SIRT3 plays a critical role in several key pathways involved in whole-body energy homeostasis.
SIRT3KO mice display peripheral insulin resistance due, in part, to decreased pyruvate dehydrogenase (PDH) activity [10] and hexokinase 2 (HK2) activity in skeletal muscle [54]. Together, these impairments lead to increased FAO and reduced glucose uptake [10, 54]. Furthermore, ROS is elevated in skeletal muscle of SIRT3KO mice, which is implicated in the development of insulin resistance in metabolically active tissues including the skeletal muscle, liver, and white adipose tissue [55-59]. Consistent with this idea, mice with heterozygous SOD2 deletion display impaired glucose tolerance on a low fat diet (LFD), and exacerbated β-cell dysfunction in the setting of a HFD feeding [55, 60].
β-cell failure is a critical event in the progression from insulin resistance to Type 2 Diabetes (T2D). Interestingly, pancreatic islets from human diabetic donors have lower SIRT3 levels [61]. Whole-body SIRT3KO mice fed a chronic HFD secrete less insulin in response to glucose, suggesting diet-induced pancreatic β-cell dysfunction in the absence of SIRT3, and supporting a model whereby SIRT3 serves to protect β-cells from nutritional stress [52]. Not surprisingly, transient knockdown of SIRT3 in β-cells in culture results in elevated ROS and impaired insulin secretion [61]. Further, adenoviral overexpression of SIRT3 in primary rat islets rescues lipotoxic impairment glucose-stimulated insulin secretion [62]. Thus, SIRT3 is emerging as a critical player in pancreatic β-cell survival and/or compensation in the presence of a hyperglycemic-hyperlipidemic insult, similar to its role in protecting against hepatic lipotoxicity. Taken together, SIRT3 plays a crucial role in whole-body metabolism, and its absence leads to accelerated metabolic deregulation and development of the metabolic syndrome.
SIRT3 and cardiac function
As humans age, cardiac function decreases, and understanding the underlying molecular mechanisms driving this dysfunction is crucial for the development of future therapies. Mitochondria in the heart are highly oxidative, and this capacity decreases with age in humans [63, 64]. In mice, decreased mitochondrial function is thought to contribute to a decline in heart health and to lead to accelerated cardiac hypertrophy. The mechanisms implicated in cardiac hypertrophy and subsequent progression to heart failure, include impaired FAO, reduced oxidative phosphorylation, increased ROS production, and dysregulated mitochondrial dynamics [64-66].
Not surprisingly, SIRT3 is proposed to play an important role in regulating cardiac function. Mice lacking SIRT3 have altered mitochondrial FAO in the heart [9, 67], and reduced oxidative phosphorylation (OXPHOS) complex activity and ATP production [7]. Because the heart relies on FAO for most of its energy production, several mouse models of deficient lipid oxidation develop cardiac hypertrophy, including mice deficient in SIRT3, AMP-activated protein kinase (AMPK) [68], peroxisome proliferator-activated receptor α (PPARα) [69], long-chain acyl-CoA dehydrogenase (LCAD) [70], and oxidative phosphorylation [71].
Consistent with these findings, increased ROS and subsequent oxidative damage can independently lead to cardiac hypertrophy [64]. Cardiomyocytes from SIRT3KO mice show increased ROS under basal conditions, as well as after phenylephrine treatment, supporting the idea that SIRT3 is required to prevent cellular ROS accumulation in the heart [72]. Further supporting the decline in antioxidant defense, Cyclophilin D is thought to be a structural component of the mitochondrial permeability transition pore and is a SIRT3 target. Cyclophilin D is hyperacetylated in SIRT3KO hearts, resulting in increased mitochondrial permeability, mitochondrial dysfunction, and altered energetics [73].
Impaired mitochondrial dynamics has also been associated with several cardiac diseases, including cardiac hypertrophy and heart failure [65]. Generally, fusion is a mitochondrial stress response that promotes mitochondrial function and is regulated by a family of fusion proteins that includes optic atrophy 1 (OPA1). OPA1 is a target of SIRT3, and loss of SIRT3 leads to OPA1 hyperacetylation and an impairment in mitochondrial fusion [8]. Stressors associated with heart failure, including cardiac hypertrophy and obesity, also result in hyperacetylation of OPA1 [8].
Taken together, these studies demonstrate that SIRT3 regulates several different mitochondrial pathways that are critical to maintaining cardiac health. In the absence of SIRT3, these pathways are dysregulated, which contributes to cardiac hypertrophy and accelerated heart failure.
SIRT3 and neurodegenerative disease
The onset of neurodegenerative diseases is a hallmark of aging, and susceptibility to Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis, and frontal temporal dementia, increases as humans age. Neurons are one of the highest energy consuming cell types, which make them particularly susceptible to metabolic and oxidative stressors. Indeed, loss of mitochondrial function plays a major role in neurodegeneration [74].
SIRT3 is expressed at high levels in the brain and other nervous system tissues [75-77]. Because SIRT3 regulates several aspects of metabolic homeostasis, it is predicted help protect against neurodegenerative diseases. SIRT3 can rescue neuronal loss in various neurodegenerative models. For example, primary cortical neurons grown in culture can be induced to undergo apoptosis in a manner similar to Alzheimer’s disease (AD) after treatment with beta-amyloid (Aβ In this model, co-treatment with pituitary adenylate cyclase-activating polypeptide (PACAP) protects primary cultured neurons from Aβ treatment by upregulating SIRT3 expression [78]. The PACAP mediated neuroprotective effect is lost if SIRT3 expression is knocked down by shRNA, in primary cultured neurons. Furthermore, SIRT3 overexpression confers resistance to oxidative stress and extends neuronal longevity in primary neuron cultures [79].
Similarly, amyotrophic lateral sclerosis (ALS) is associated with mitochondrial dysfunction [80], which can be modeled in cultured neurons by expressing the G93A mutant of superoxide dismutase 1 (SOD1) [81]. Mutant SOD1 induces mitochondrial dysfunction and apoptosis, which can be reduced by overexpression of either SIRT3 or peroxisome proliferator-activated receptor-gamma coactivator-1 alpha (PGC1α)— a transcription factor that regulates SIRT3 expression and other nuclear encoded mitochondrial genes [81].
Hearing declines with age and is associated with the progressive loss of hair cells in the ear, which convert acoustic vibrations into nerve impulses. Interestingly, calorie restriction can prevent age-related hearing loss, but only with SIRT3 present [82]. Overexpression of SIRT3 leads to activation of IDH2 and confers protection from oxidative stress. Similarly, noise-induced hearing loss can be reduced by nicotinamide riboside, an NAD+ precursor thought to boost SIRT3 activity [83].
Based on this early evidence, SIRT3 has a neuroprotective role and loss of SIRT3 accelerates the progression of neurodegenerative disease states. Even where a direct link between SIRT3 and a disease has not been established, growing evidence shows that mitochondrial dysfunction and oxidative stress play critical role in the pathogenesis of neurodegenerative diseases, highlighting a potentially broader role of SIRT3 in regulating neuronal health.
SIRT3 and acetylation
Much of the recent work on acetylation has been focused on using large-scale proteomic approaches to identify proteins that are hyperacetylated in the absence of SIRT3, many of which are in pathways with known roles in diseases of aging [84-88]. As the field moves towards more sophisticated proteomic technologies, an ever-increasing number of acetylated proteins will be identified. As this number grows, it is becoming clear that regulation of a single protein at a single site by SIRT3 cannot explain the role SIRT3 plays across a wide-array of biology and diseases of aging.
Supporting this idea are recent studies investigating the stoichiometry of acetylation sites. Early estimates of the stoichiometry of any given site of acetylation are thought to be low [89, 90]. If these studies hold, then again the acetylation status of a single protein at one site is unlikely to account for the changes in physiology or disease observed.
Instead, an emerging sense in the field is that multiple enzymes of a given pathway that are acetylated, even at low stoichiometry, could have a measurable effect on the activity of the entire pathway [89]. In this setting, SIRT3 might influence the pathway through sites of deacetylation. Thus, the overall picture emerging from these proteomic studies is that the sum of all acetyl modifications, or the sum of a set of SIRT3-regulated modifications in a given pathway, could be critical for the regulation of metabolism. Future proteomic and physiological studies will test this idea.
Does SIRT3 have a role in longevity?
Recently, nine hallmarks of aging were described that contribute to the aging process [4]. Current thinking on aging places time-dependent accumulation of cellular damage at the core of the aging process. Based on this model, current work should be focused on the sources of damage and the compensatory processes in response to damage. Along these lines, SIRT3’s regulation of ROS and ROS-induced damage could be an important mechanism by which SIRT3 influences aging and the diseases of aging. Furthermore, protein acetylation and acylation could represent a type of protein damage induced by “carbon stress” [91].
Early studies investigating a role for SIRT3 in longevity took a genomics approach by screening long-lived humans for possible variations in the SIRT3 gene. While early studies reported a possible correlation between a unique SIRT3 genotype and longevity [92] or a novel VNTR (variable number tandem repeat) enhancer within SIRT3 [93], a later study by the same group reported no association [94]. Furthermore, an independent large-scale investigation into the genetics of longevity found no role for SIRT3 [95]. Thus, a direct link between SIRT3 and longevity has not yet been established.
Additionally, no studies in model organisms have formally tested the role of SIRT3 in longevity. Part of this could be due to the fact that some model organisms often used to study longevity (i.e. S. cerevisiae, C. elegans, D. melanogaster) have sirtuin genes, but don’t have a SIRT3 ortholog. Furthermore, a SIRT3 transgenic mouse, which was made relatively recently, has been used to only study SIRT3 in the context of noise-induced hearing loss [83]. Thus, while some studies show that SIRT3 is a compelling regulator of aging [96], future studies should be focused on directly testing the role of SIRT3 in longevity.
Concluding Remarks
Even in the absence of understanding the complete molecular mechanisms of SIRT3 action (Box 1), it’s clear that SIRT3 ablation leads to accelerated aging in several models. SIRT3KO mice have decreased time to development and/or severity of age-related diseases. Thus, identifying the pathways that are affected when SIRT3 is ablated may aid in identifying pathways that become dysfunctional during the aging process. From this perspective, continued work on SIRT3 may uncover important new pathways of the aging process.
Box 1. Outstanding Questions.
What are the primary targets and/or pathways regulated by SIRT3?
Do low stoichiometry sites of acetylation influence protein function?
Does the reported in vitro deacylation activity of SIRT3, in addition to deacetylation, contribute its regulatory role in vivo?
Is the primary role of SIRT3-dependent deacetylation to restore activity of metabolic enzymes?
Which other disease states or diseases of aging does SIRT3 influence?
Does SIRT3 play a role in regulating longevity in model organisms or human populations?
Highlights.
SIRT3 regulates almost every major mitochondrial pathway
Mice lacking SIRT3 develop several diseases of aging at an accelerated rate
SIRT3KO mice might be considered a model of accelerated aging
Acknowledgements
We would like to thank the Hirschey lab for thoughtful feedback. We would also like to acknowledge funding support for the lab from the American Heart Association grants 12SDG8840004 and 12IRG9010008, Friedreich’s Ataxia Research Alliance, The Ellison Medical Foundation, the National Institutes of Health grants R01AA022146, R01AG045351, and R24DK085610-05; The Duke Pepper Older Americans Independence Center (OAIC) Program in Aging Research supported by the National Institute of Aging (P30AG028716-01); EM is supported by an NIH training grant to Duke University Cancer Biology Training Program (5T32-CA059365); BSP is supported by an NIH/NIGMS training grant to Duke University Pharmacological Sciences Training Program (5T32GM007105-40).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260(1):273–9. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 2.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13(19):2570–80. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and Cellular Biology. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs KM, et al. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int. J. Biol. Sci. 2008:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn B-H, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2008:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samant SA, et al. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Molecular and Cellular Biology. 2014:807–819. doi: 10.1128/MCB.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschey MD, et al. Nature. Nature Publishing Group; 2010. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation; pp. 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing E, et al. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62(10):3404–17. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papa L, Germain D. SirT3 regulates a novel arm of the mitochondrial unfolded protein response. Molecular and Cellular Biology. 2013 doi: 10.1128/MCB.00191-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onyango P, et al. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2002:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman JL, Baeza J, Denu JM. Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao X, et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014;3 doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peek CB, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342(6158):1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. SIRT3 and SIRT5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-CoA dehydrogenase. PLoS One. 2015;10(3):e0122297. doi: 10.1371/journal.pone.0122297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bharathi SS, et al. SIRT3 Regulates Long-chain Acyl-CoA Dehydrogenase by Deacetylating Conserved Lysines Near the Active Site. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altekruse S, et al. SEER Cancer Statistics Review. National Cancer Institute; Bethesda, MD: pp. 1975–2007. [Google Scholar]
- 19.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 20.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477(1-2):7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 21.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Kim H-S, et al. Cancer Cell. Elsevier Ltd; 2010. SIRT3 Is a Mitochondria-Localized Tumor Suppressor Required for Maintenance of Mitochondrial Integrity and Metabolism during Stress; pp. 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinds P, Finlay C, Levine AJ. Mutation is required to activate the p53 gene for cooperation with the ras oncogene and transformation. J Virol. 1989;63(2):739–46. doi: 10.1128/jvi.63.2.739-746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 25.Sun H, Taneja R. Analysis of transformation and tumorigenicity using mouse embryonic fibroblast cells. Methods Mol Biol. 2007;383:303–10. doi: 10.1007/978-1-59745-335-6_19. [DOI] [PubMed] [Google Scholar]
- 26.Finley LWS, et al. Cancer Cell. Elsevier Inc; 2011. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization; pp. 416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y-Y, Zhou L-M. Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochemical and Biophysical Research Communications. 2012:26–31. doi: 10.1016/j.bbrc.2012.05.053. [DOI] [PubMed] [Google Scholar]
- 28.Yang B, et al. Aberrant expression of SIRT3 is conversely correlated with the progression and prognosis of human gastric cancer. Biochem Biophys Res Commun. 2014;443(1):156–60. doi: 10.1016/j.bbrc.2013.11.068. [DOI] [PubMed] [Google Scholar]
- 29.Zhang CZ, et al. Low SIRT3 Expression Correlates with Poor Differentiation and Unfavorable Prognosis in Primary Hepatocellular Carcinoma. PLoS ONE. 2012:e51703. doi: 10.1371/journal.pone.0051703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haigis MC, et al. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. 2012;72(10):2468–72. doi: 10.1158/0008-5472.CAN-11-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tudek B, et al. Involvement of oxidatively damaged DNA and repair in cancer development and aging. Am J Transl Res. 2010;2(3):254–84. [PMC free article] [PubMed] [Google Scholar]
- 32.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014:R453–62. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X, et al. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metabolism. 2010:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Bell EL, et al. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–70. [PubMed] [Google Scholar]
- 37.Chandel NS. Reactive Oxygen Species Generated at Mitochondrial Complex III Stabilize Hypoxia-inducible Factor-1alpha during Hypoxia. A MECHANISM OF O2 SENSING. Journal of Biological Chemistry. 2000:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 38.Bell EL, et al. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177(6):1029–36. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozden O, et al. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radical Biology and Medicine. 2014:163–172. doi: 10.1016/j.freeradbiomed.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan J, et al. Molecular Cell. Elsevier Inc; 2014. Tyr Phosphorylation of PDP1 Toggles Recruitment between ACAT1 and SIRT3 to Regulate the Pyruvate Dehydrogenase Complex; pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McFate T, et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283(33):22700–8. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H, et al. SIRT3-dependent GOT2 acetylation status affects the malate aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 2015 doi: 10.15252/embj.201591041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.International Diabetes Federation . IDF Diabetes Atlas. 6th edn International Diabetes Federation; Brussels, Belgium: 2013. [Google Scholar]
- 44.Federation ID. IDF Diabetes Atlas. 6th Edition [Google Scholar]
- 45.Gao AW, Canto C, Houtkooper RH. Mitochondrial response to nutrient availability and its role in metabolic disease. EMBO Mol Med. 2014;6(5):580–9. doi: 10.1002/emmm.201303782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji H, Friedman MI. Reduced capacity for fatty acid oxidation in rats with inherited susceptibility to diet-induced obesity. Metabolism. 2007;56(8):1124–30. doi: 10.1016/j.metabol.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonomini F, Rodella LF, Rezzani R. Metabolic syndrome, aging and involvement of oxidative stress. Aging Dis. 2015;6(2):109–20. doi: 10.14336/AD.2014.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–7. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 49.Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;12(6):465–83. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen KF, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300(5622):1140–2. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao J, et al. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med. 2010;49(7):1230–7. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirschey MD, et al. Molecular Cell. Elsevier Inc; 2011. SIRT3 Deficiency and Mitochondrial Protein Hyperacetylation Accelerate the Development of the Metabolic Syndrome; pp. 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kendrick AA, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem. J. 2011:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lantier L, et al. SIRT3 is crucial for maintaining skeletal muscle insulin action and protects against severe insulin resistance in high fat fed mice. Diabetes. 2015 doi: 10.2337/db14-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoehn KL, et al. Insulin resistance is a cellular antioxidant defense mechanism. Proc Natl Acad Sci U S A. 2009;106(42):17787–92. doi: 10.1073/pnas.0902380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jing E, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci U S A. 2011;108(35):14608–13. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuzawa N, et al. Lipid-induced oxidative stress causes steatohepatitis in mice fed an atherogenic diet. Hepatology. 2007;46(5):1392–403. doi: 10.1002/hep.21874. [DOI] [PubMed] [Google Scholar]
- 58.Nakamura S, et al. Palmitate induces insulin resistance in H4IIEC3 hepatocytes through reactive oxygen species produced by mitochondria. J Biol Chem. 2009;284(22):14809–18. doi: 10.1074/jbc.M901488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paglialunga S, et al. In adipose tissue, increased mitochondrial emission of reactive oxygen species is important for short-term high-fat diet-induced insulin resistance in mice. Diabetologia. 2015 doi: 10.1007/s00125-015-3531-x. [DOI] [PubMed] [Google Scholar]
- 60.Kang L, et al. Heterozygous SOD2 deletion impairs glucose-stimulated insulin secretion, but not insulin action, in high-fat-fed mice. Diabetes. 2014;63(11):3699–710. doi: 10.2337/db13-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caton PW, et al. Sirtuin 3 regulates mouse pancreatic beta cell function and is suppressed in pancreatic islets isolated from human type 2 diabetic patients. Diabetologia. 2013;56(5):1068–77. doi: 10.1007/s00125-013-2851-y. [DOI] [PubMed] [Google Scholar]
- 62.Kim M, et al. SIRT3 Overexpression Attenuates Palmitate-Induced Pancreatic beta-Cell Dysfunction. PLoS One. 2015;10(4):e0124744. doi: 10.1371/journal.pone.0124744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jarreta D, et al. Mitochondrial function in heart muscle from patients with idiopathic dilated cardiomyopathy. Cardiovasc Res. 2000;45(4):860–5. doi: 10.1016/s0008-6363(99)00388-0. [DOI] [PubMed] [Google Scholar]
- 64.Dai DF, Rabinovitch PS, Ungvari Z. Mitochondria and cardiovascular aging. Circ Res. 2012;110(8):1109–24. doi: 10.1161/CIRCRESAHA.111.246140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikeda Y, et al. Molecular mechanisms mediating mitochondrial dynamics and mitophagy and their functional roles in the cardiovascular system. J Mol Cell Cardiol. 2015;78:116–22. doi: 10.1016/j.yjmcc.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyyti OM, et al. Aging impairs myocardial fatty acid and ketone oxidation and modifies cardiac functional and metabolic responses to insulin in mice. Am J Physiol Heart Circ Physiol. 2010;299(3):H868–75. doi: 10.1152/ajpheart.00931.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alrob OA, et al. Obesity-induced lysine acetylation increases cardiac fatty acid oxidation and impairs insulin signalling. Cardiovasc Res. 2014;103(4):485–97. doi: 10.1093/cvr/cvu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmad F, et al. Increased alpha2 subunit-associated AMPK activity and PRKAG2 cardiomyopathy. Circulation. 2005;112(20):3140–8. doi: 10.1161/CIRCULATIONAHA.105.550806. [DOI] [PubMed] [Google Scholar]
- 69.Smeets PJH, et al. Cardiac hypertrophy is enhanced in PPAR alpha−/− mice in response to chronic pressure overload. Cardiovasc Res. 2008;78(1):79–89. doi: 10.1093/cvr/cvn001. [DOI] [PubMed] [Google Scholar]
- 70.Cox K, et al. Cardiac hypertrophy in mice with long-chain acyl-CoA dehydrogenase or very long-chain acyl-CoA dehydrogenase deficiency. Lab Invest. 2009 doi: 10.1038/labinvest.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suenaga M, et al. Functional disorders of the oxidative phosphorylation system in the heart mitochondria of mice with juvenile visceral steatosis. Biol Pharm Bull. 2003;26(3):289–94. doi: 10.1248/bpb.26.289. [DOI] [PubMed] [Google Scholar]
- 72.Sundaresan NR, et al. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119(9):2758–71. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hafner AV, et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2010;2(12):914–23. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58(4):495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- 75.Ban N, et al. Light-dark condition regulates sirtuin mRNA levels in the retina. Exp Gerontol. 2013;48(11):1212–7. doi: 10.1016/j.exger.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 76.Sidorova-Darmos E, et al. Differential expression of sirtuin family members in the developing, adult, and aged rat brain. Front Aging Neurosci. 2014;6:333. doi: 10.3389/fnagi.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng L, et al. Age-related decrease in the mitochondrial sirtuin deacetylase Sirt3 expression associated with ROS accumulation in the auditory cortex of the mimetic aging rat model. PLoS One. 2014;9(2):e88019. doi: 10.1371/journal.pone.0088019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han P, et al. Pituitary adenylate cyclase-activating polypeptide protects against beta-amyloid toxicity. Neurobiol Aging. 2014;35(9):2064–71. doi: 10.1016/j.neurobiolaging.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 79.Weir HJ, et al. CNS SIRT3 expression is altered by reactive oxygen species and in Alzheimer’s disease. PLoS One. 2012;7(11):e48225. doi: 10.1371/journal.pone.0048225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manfredi G, Xu Z. Mitochondrial dysfunction and its role in motor neuron degeneration in ALS. Mitochondrion. 2005;5(2):77–87. doi: 10.1016/j.mito.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 81.Song W, et al. Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: neuroprotection by SIRT3 and PGC-1alpha. Neurobiol Dis. 2013;51:72–81. doi: 10.1016/j.nbd.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Someya S, et al. Sirt3 Mediates Reduction of Oxidative Damage and Prevention of Age-Related Hearing Loss under Caloric Restriction. Cell. 2010:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brown KD, et al. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014;20(6):1059–68. doi: 10.1016/j.cmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rardin MJ, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proceedings of the National Academy of Sciences. 2013:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hebert AS, et al. Molecular Cell. Elsevier Inc; 2013. Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome; pp. 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sol EM, et al. Proteomic Investigations of Lysine Acetylation Identify Diverse Substrates of Mitochondrial Deacetylase Sirt3. PLoS ONE. 2012:e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dittenhafer-Reed KE, et al. SIRT3 Mediates Multi-Tissue Coupling for Metabolic Fuel Switching. Cell Metab. 2015;21(4):637–46. doi: 10.1016/j.cmet.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Still AJ, et al. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem. 2013;288(36):26209–19. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baeza J, et al. Stoichiometry of site-specific lysine acetylation in an entire proteome. Journal of Biological Chemistry. 2014 doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weinert BT, et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wagner G, Hirschey MD. Non-enzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54(1):5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rose G, et al. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38(10):1065–70. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 93.Bellizzi D, et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85(2):258–63. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 94.De Rango F, et al. A novel sampling design to explore gene-longevity associations: the ECHA study. Eur J Hum Genet. 2008;16(2):236–42. doi: 10.1038/sj.ejhg.5201950. [DOI] [PubMed] [Google Scholar]
- 95.Lescai F, et al. Human longevity and 11p15.5: a study in 1321 centenarians. Eur J Hum Genet. 2009;17(11):1515–9. doi: 10.1038/ejhg.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benigni A, et al. Disruption of the Ang II type 1 receptor promotes longevity in mice. J Clin Invest. 2009 doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]