Abstract
Low versus high glycemic load (GL) diet patterns are inversely associated with obesity and chronic diseases such as cancer and cardiovascular disease. These associations persist beyond the protection afforded by increased fiber alone, representing an important gap in our understanding of the metabolic effects of GL. We conducted a randomized, controlled, crossover feeding trial of two 28-day diet periods of high and low GL. Using LC-MS, targeted metabolomics analysis of 155 metabolites was performed on plasma samples from 19 healthy adults aged 18-45 years. Fourteen metabolites differed significantly between diets (P<0.05), with kynurenate remaining significant after Bonferroni correction (P<4×10-4). Metabolites with the largest difference in abundance were kynurenate and trimethylamine-N-oxide (TMAO), both significantly higher after consumption of the low GL diet. Partial least squares-discriminant analysis showed clear separation between the two diets; however no specific pathway was identified in pathway analyses. We found significant differences in 14 plasma metabolites suggesting a differing metabolic response to low and high GL diets. Kynurenate is associated with reduced inflammation, and may be one mechanism through which protective effects of a low GL diet are manifested and warrants further evaluation. This trial was registered at clinicaltrials.gov as NCT00622661.
Keywords: dietary intervention, fiber, glycemic load, kynurenine, metabolomics
Introduction
Obesity and chronic diseases such as cancer and cardiovascular disease (CVD), collectively account for roughly half of the deaths in the United States 1-3. Numerous epidemiologic studies find that low glycemic load (LGL) diets, characterized by consumption of foods that result in a relatively small postprandial increase in blood glucose4, are associated with reduced risk for obesity5, some types of cancer5, 6, diabetes5, CVD5, and coronary heart disease (CHD)5, 7. In contrast, high glycemic load (HGL) diets are associated with an increased postprandial glucose response, post-prandial hyperglycemia, and oxidative stress, which are thought to contribute to the negative health outcomes linked to chronic consumption of HGL diets6, 8.
Several explanations have been proposed to explain the protective effects of LGL diets. In addition to averting the hyperglycemic and hyperinsulinemic oxidative stress of HGL diets, higher fiber content is a suggested means by which LGL mitigates chronic disease risk. While LGL diets are inherently higher in dietary fiber, human studies have shown that increased fiber alone does not always confer the same degree of risk reduction for chronic disease as does lowering the GL of the diet6. Other factors, such as type of fiber (e.g., soluble vs insoluble), structural form of foods (e.g, whole grain vs milled), or the type of foods selected (high- vs low-phytochemical) may also contribute to response to a LGL diet. The protective effects of LGL beyond increased fiber represent an important gap in our understanding of the metabolic effects of differing GL.
Metabolomics is becoming a commonly used approach to identify metabolic differences resulting from various conditions, such as a diseased or non-diseased state, or exposures, e.g., before and after a drug or dietary treatment9. Metabolomics captures the current status of an organism's biochemistry and can illuminate consequences of environmental exposures (e.g., food), gene expression, and their combined implications10. In nutritional research, metabolomics has been used to identify biomarkers of dietary intake11, validate food frequency questionnaires (FFQs)12, and in dietary intervention studies to elucidate dietary effects on metabolic pathways13. A few studies have used metabolomics to identify biomarkers associated with glycemic index (GI) or exposure of dietary fiber intake14, 15. For example, after 77 overweight adults were randomized to high or low glycemic index diet for 6 months, authors found urinary formate discriminated between the two dietary patterns 14. In another study, plasma concentrations of 2,6-dihyroxybenzoic acid and 2-aminophenol sulfate were increased after 5 weeks of consuming a high fiber diet compared to a low fiber diet15. However, no controlled feeding study has investigated metabolic changes in the plasma metabolome based on dietary GL in a crossover design.
In the present study we take an agnostic approach to evaluate the extent to which HGL and LGL diets modify the plasma metabolome using a targeted metabolomics approach on specimens collected from a controlled feeding trial. We hypothesize that LGL diets influence metabolism differently than HGL diets and that these effects will result in differences in metabolite abundances between the two diets. Results from this study may provide insight on the protective benefits of consuming low glycemic load diets.
Materials and Methods
The metabolomics analysis was a secondary analysis using samples derived from the “Carbohydrate and Related Biomarkers” study (CARB) conducted between June 2006 and July 2009 at the Fred Hutchinson Cancer Research Center (FHCRC) in Seattle, Washington. This trial was registered at clinicaltrials.gov as NCT00622661. CARB was a crossover dietary intervention with two 4-week feeding periods of HGL and LGL diets given in a computer-generated, randomly assigned order16. Metabolomics analysis was conducted on plasma collected from a subset of 20 participants at the end of each intervention period, from which samples from 19 pairs could be analyzed. The study protocol was approved by the FHCRC Institutional Review Board and all participants gave written informed consent.
Participants and Study Design
Details on recruitment and study design have been published elsewhere 16. In brief, non-smoking, healthy individuals between the ages of 18-45 years were recruited from the Seattle area. Exclusion criteria included impaired fasting glucose (fasting blood glucose ≥ 5.6 mmol/L), any physician-diagnosed condition requiring a restricted diet, food allergies, regular use of hormones or anti-inflammatory medication, pregnancy or lactation, or heavy use of alcohol (>2 drinks per day). A total of 82 individuals completed all study activities. The study population was 50% male and was evenly distributed between normal weight (BMI >18.5 ≤ 25.0 kg/m2) and overweight/obese (BMI ≥ 28.0 ≤ 40.0 kg/m2)17. Diet order was randomly assigned, with half of the participants receiving the HGL diet first. Each intervention was four weeks long, with a four week washout period in between study periods16. A subset of 20 individuals was selected for this pilot metabolomics analysis. The group was selected to represent the larger study population: 50% male, evenly distributed between normal and overweight, with half having received the HGL diet first. Blood was drawn at the end of each intervention period for a total of 40 samples; however, one sample was compromised during sample preparation. Thus both time-points for that individual were excluded and final analysis was conducted on the remaining 19 pairs.
Study Diets
The study diets were based on a repeating seven-day menu, with identical distribution of macronutrients for both the HGL and LGL diets (see Table 1 for sample menu). The diets were designed to differ largely in GL, with the LGL diet being half the GL of the HGL intervention (125 and 250 GL/d respectively), and differed notably in fiber (55 and 28 g/d for the LGL and HGL, respectively). Total GL for each mixed meal was calculated by multiplying the grams of carbohydrate in each food by that food's GI value. The diets were isocaloric for each individual, although minor adjustments were made as necessary to keep participants' weight stable during the intervention (Table 2). Further details of the study diet have been published previously16. All food was provided by the FHCRC during the intervention, with weekday dinners consumed under supervision at the Center and the next day's breakfast and lunch taken home for consumption. On Fridays, participants received all weekend meals. Any unconsumed food was returned to the study center, weighed and recorded. During the washout period individuals returned to their habitual diet patterns.
Table 1.
Sample daily meal plans for high glycemic load (HGL) and low glycemic load (LGL) intervention diets in the Carbohydrate and Related Biomarkers study.1
| HGL Breakfast | LGL Breakfast | ||
|---|---|---|---|
| Grape-nut cereal | Dates | All Bran | Strawberries/blueberries |
| 2% milk | Dried cranberries | 2% milk | Nut crunch |
| Sweetener | Sweetener | Tomato juice | |
|
| |||
| HGL Lunch | LGL Lunch | ||
|
| |||
| White bread | Cauliflower | Pumpernickel bread | Carrots |
| Roast beef | Potato salad | Roast beef | Tabouli |
| Mayo | Ranch dressing | Mayo | Hummus |
| Mustard | Mustard | ||
| Tomatoes | Dessert | Tomato | Dessert |
| Lettuce | Fruit roll ups | Lettuce | M&Ms |
| Onions | Jellybeans | Onions | Pears |
| Pickles | Apricots | Pickles | |
|
| |||
| HGL Dinner | LGL Dinner | ||
|
| |||
| Chicken breast | Sour cream | Chicken breast | Sour cream |
| Green pepper | White rice | Green pepper | Tortilla |
| Red pepper | Taco shell | Red pepper | |
| Onions | Dessert | Onions | Dessert |
| Mexican sauce | Rice pudding | Mexican sauce | Chocolate mousse |
| Salsa | Cranberry juice | Salsa | Apple juice |
| HGL Snack | LGL Snack | ||
|
| |||
| Energy bar | Dried apple | Chocolate power bar | |
Menuswere designed to contain similar foods with specific items substituted to alter glycemic load (GL) of overall meal so as to minimize change to the diet outside of GL
Table 2.
Summary of average daily intake macronutrient and glycemic load (GL)in high glycemic load (HGL) and low glycemic load (LGL) intervention diets.1
| Daily intake | HGL diet | LGL diet | ||
|---|---|---|---|---|
| Energy, kcal | 2610 (476) | 2719 (563) | ||
| Carbohydrate, g; % | 364 (69) | 67% | 386 (84) | 68% |
| Protein, g; % | 95 (18) | 17% | 98 (22) | 17% |
| Fat, g; % | 86 (16) | 16% | 87 (20) | 15% |
| GL | 259 (50) | 125 (27) | ||
| Fiber, g | 28 (5) | 56 (12) | ||
Mean (SD)
Sample Collection And Analysis
At baseline, height and weight measurements were recorded for assignment to the overweight + obese (BMI > 28 kg/m2) or normal weight (BMI < 25 kg/m2) category. Those with BMI 25-27.9 kg/m2 were excluded to ensure sufficient contrast between the normal weight and overweight/obese groups. Whole-body dual-energy X-ray absorptiometry (DEXA) scans (GE Lunar DPX-Pro) were also completed for each participant to accurately assess body fat percent. Weight measurements were taken regularly throughout the study period, with adjustments made to the study food provided as necessary to keep participants weight stable16. Blood was taken from each participant on Day 28 of both treatment periods after a 12-hour overnight fast, and processed and stored at -80°C using a standard protocol.
Metabolomics analysis
Metabolite profiling of plasma was completed at the University of Washington's Northwest Metabolomics Research Center. Targeted metabolomics analysis on the plasma of the subset of 19 participants was carried out using a liquid chromatography tandem mass spectrometry (LC-MS/MS) platform in both positive and negative ion modes against 155 standard metabolites from 25 metabolic pathways, (e.g., Glycolysis, TCA cycle, Amino Acid Metabolism, etc.) of potential significance to monitor diet effects (see Supplemental Table)18-20.
All plasma samples were prepared at the same time. A standard protocol was used, where 25 μL plasma and 150 μL high performance liquid chromatography (HPLC) grade methanol were combined in an Eppendorf vial and vortexed for 2 min. After 20 min storage at -20 °C the samples were centrifuged at 18,000 g for 10 min. A fixed volume of 150 μL supernatant was collected and placed in a new Eppendorf vial. The protein pellets were mixed with another 300 μL HPLC grade methanol, then vortexed for 10 min and centrifuged for 10 min at 18,000 g. 250 μL was collected and combined with the previous 150 μL sample. Samples were then dried at 30 °C in a Speed Vac for 3 h.
The samples were analyzed in two sequential batches, each containing two quality control (QC) samples so that batch variation could be assessed. Prior to each LC run, samples were reconstituted with 100 μL 5 mM ammonium acetate in 95% water/5% acetonitrile + 0.5% acetic acid, and filtered through 0.45 μm PVDF filters (Phenomenex, Torrance, CA) prior to analysis on an AB Sciex QTrap 5500 LC-MS/MS system (AB Sciex, Toronto, ON, Canada)21-23. The LC system was composed of two Agilent 1260 binary pumps, an Agilent 1260 auto-sampler and Agilent 1290 column compartment containing a column-switching valve (Agilent Technologies, Santa Clara, CA). Each sample was injected twice, 10 μL for analysis using negative ionization mode and 2 μL for analysis using positive ionization mode. Both chromatographic separations were performed in reverse phase (RP) on Thermo Accucore PFP columns (150 × 2.1 mm, 2.6 μm particle size, Thermo Fisher Scientific Inc., Waltham, MA). The flow rate was 0.250 mL/min, auto-sampler temperature was kept at 4 °C, and the column compartment was set at 40 °C. The mobile phase was composed of Solvents A (5 mM ammonium acetate in H2O + 0.5% acetic acid + 0.5% acetonitrile) and B (acetonitrile + 0.5% acetic acid + 0.5% water). After chromatographic separation, MS ionization and data acquisition was performed using AB Sciex QTrap 5500 mass spectrometer (AB Sciex, Toronto, ON, Canada) with electrospray ionization (ESI) source. The collision gas was 99.99% pure nitrogen. The data gathered through the multiple reaction monitoring were integrated using MultiQuant 2.1 software (AB Sciex, Toronto, ON, Canada).20 Intra-assay CVs were 9.4% and 6.3 for negative and positive ion mode, respectively and 7.4% across all samples.
Statistical Analysis
Of the 155 metabolites measured, 125 had detectable signal in all study samples and were retained for analysis. Plasma data showed between-batch variability, and were therefore normalized using the mean signal among all study samples for a given batch as the constant by which to standardize the values for that metabolite within-batch. Paired t-tests were conducted on log-transformed metabolite values to identify statistically significant (P<0.05) within-person differences in plasma metabolite abundances, comparing Day 28 measurements from the LGL versus the HGL diet. Bonferroni and Benjamini-Hochberg (false discovery rate, FDR) methods were used to correct for multiple comparisons (Stata v13.1, College Station, TX)24. To compare the relative abundance of metabolites (within-person) between diet interventions, geometric mean ratios of detected metabolite concentrations after the LGL diet versus the HGL diet were calculated by exponentiating the mean within-person differences in log-transformed metabolite signals.
While the study protocol attempted to achieve weight maintenance, most participants experienced a minor change in weight (∼1%). Up to 3% change was permitted, consistent with many dietary intervention studies as it represents minor fluctuations in fluid balance and normal day to day variation. To ensure that these minor fluctuations did not impact the findings, a linear mixed regression model was used to test the effects of weight change and diet on metabolite concentrations, modeled as a fixed effect, and individual baseline metabolite concentrations as a random effect (R statistical package v3.01, with package lme4 1.0). Models were also carried out to evaluate metabolites adjusted for fat distribution as determined by android to gynoid body fat % ratio, and body fat % using data derived from DEXA scans. Finally, to ensure that there were no carry over effects, diet sequence and period were evaluated. As these covariates did not have any effect on point estimates, data are presented without adjustment.
To consider differences occurring beyond the level of individual metabolites, pathway level analyses were conducted using the global test for changes among groups of metabolites described by Goeman, et al.25 (Globaltest R statistical package v3.02). A priori pathways assessed were Krebs cycle, Gluconeogenesis and Glycolysis. Among inflammation-related pathways, the Tryptophan pathway was the only one with a sufficient number of metabolites to allow for meaningful analysis. We evaluated each pathway using 4 to 7 metabolites from the respective pathways, as identified in the Human Metabolome Database (HMDB)26 and Kyoto Encyclopedia of Genes and Genomes (KEGG)27 databases.
To determine whether the intervention diets led to holistic differences in metabolic responses, partial least squares discriminant analysis (PLS-DA) plots were generated via class separation in MetaboAnalyst 2.0 (http://www.metaboanalyst.ca/) with input of all metabolites, using the method described by Westerhuis, et. al., which considers the paired data structure.28 Leave one out cross-validation method was used to test the predictive performance of the model, and R2 and Q2 values were calculated. The variable importance in projection (VIP) score was used to estimate the influence of each metabolite to the first two components of the PLS model. Variables with VIP scores >1 are generally considered to have an important contribution to the model.29
Results
Characteristics for the 19 participants stratified by sex are given in Table 3. There are approximately an equal number of men and women evenly distributed between normal and overweight/obese.
Table 3. Baseline characteristics of study participants (n=19).
| Demographics | Men | Women |
|---|---|---|
| N | 9 | 10 |
| Age, y1 | 31.3 (9.2) | 31.9 (9.5) |
| BMI2 group (N) | ||
| Normal (BMI < 25) | 4 | 4 |
| Overweight/obese (BMI > 28) | 5 | 6 |
| Mean body fat %3 | ||
| BMI < 25 | 20.57 (4.4) | 28.74 (5.6) |
| BMI > 28 | 36.19 (6.0) | 45.58 (4.6) |
| Mean waist/hip ratio4 | ||
| BMI < 25 | 0.86 (0.0) | 1.08 (0.2) |
| BMI > 28 | (0.11) | 1.21 (0.1) |
| Fasting blood glucose, mmol/L1 | ||
| BMI < 25 | 4.9 (0.3) | 4.9 (0.4) |
| BMI > 28 | 4.8 (0.8) | 4.8 (0.7) |
Mean (SD)
BMI, body mass index= weight in kg/m2
Body fat % (SD), measured by DEXA
Waist to hip ratio (SD), calculated using android body fat % divided by gynoid body fat % from DEXA scan data
Of the 125 plasma metabolites detected, a total of 14 metabolites differed significantly between the LGL and HGL interventions (P<0.05; Table 4). After Bonferonni correction for multiple comparisons (P<0.05/125=4×10-4), one metabolite (kynurenate) remained significant, while two additional metabolites, cystamine and methyl succinate, satisfied the less stringent threshold of FDR q<0.20. Geometric mean changes for metabolites in LGL compared to HGL ranged from 0.77-1.37 with a near equal number of analytes increasing as decreasing after the LGL diet relative to the HGL diet. Metabolites with the greatest fold change between diets were kynurenate and trimethylamine N-oxide (TMAO). Adjusting for weight change, body fat %, and fat distribution did not alter these findings (data not shown). Pathway analyses for Krebs cycle, Glycolysis and Gluconeogenesis, and Tryptophan metabolism did not yield significant findings (data not shown).
Table 4. Plasma metabolites significantly different at Day 28 between LGL/HGL dietary intervention (N=19).
| Plasma Metabolite | Pathway | Function | Ion mode | Ratio1 | P-value2 | q-value3 | |
|---|---|---|---|---|---|---|---|
| Kynurenate | Tryptophan metabolism | NMDA receptor antagonist; modulates inflammation | - | 1.40 | 0.0002* | 0.02 | p < 0.01 |
| Methyl succinate | Dicarboxylic acids and derivatives | Energy production | - | 1.14 | 0.004 | 0.18 | |
| Cystamine | Taurine and hypotaurine metabolism | Amino acid metabolite | + | 0.77 | 0.004 | 0.18 | |
|
| |||||||
| Proline | Arginine and proline metabolism | Amino acid | + | 1.11 | 0.010 | 0.29 | p > 0.01 and p < 0.05 |
| Acetylcholine | Glycerophospholipid metabolism | Neurotransmitter; in the periphery it activates skeletal muscles; inhibits cardiac muscles to slow heart rate | + | 0.86 | 0.014 | 0.29 | |
| Hydroxyproline | Arginine and proline metabolism | Component of collagen | + | 0.83 | 0.016 | 0.29 | |
| Creatine | Glycine, serine, threonine metabolism; Arginine and proline metabolism |
Energy production | + | 0.83 | 0.016 | 0.29 | |
| Trimethylamine N-oxide | Gut bacterial metabolite | Osmolyte | + | 1.37 | 0.025 | 0.39 | |
| Carnitine | Fatty acid metabolism | Energy production via beta oxidation | + | 1.11 | 0.030 | 0.42 | |
| Homovanillate | Tyrosine metabolism | Catecholamine/dopamine metabolite | - | 1.18 | 0.036 | 0.42 | |
| Lysine | Biotin metabolism;Carnitine synthesis | Amino acid | + | 1.05 | 0.039 | 0.42 | |
| Nitrotyrosine | Product of reactive nitrogen species | Marker of oxidative stress | - | 1.12 | 0.042 | 0.42 | |
| Niacinamide | Vitamin B3 | Coenzyme in oxidation-reduction reactions | + | 0.85 | 0.046 | 0.42 | |
| Dimethylguanosine | Nucleoside | RNA/DNA synthesis | + | 1.10 | 0.047 | 0.42 | |
Geometric mean within-person ratio of plasma metabolite concentrations comparing LGL diet versus HGL diet at Day 28; a ratio >1 means that the metabolite was higher in the LGL diet relative to the HGL diet, whereas a ratio <1 means that the metabolite was lower
Paired t-test P value
False discovery rate (Benjamini-Hochberg);
Significant after Bonferroni correction (P<4×10-4)
HGL, high glycemic load; LGL, low glycemic load; NMDA, N-methyl-D-aspartate
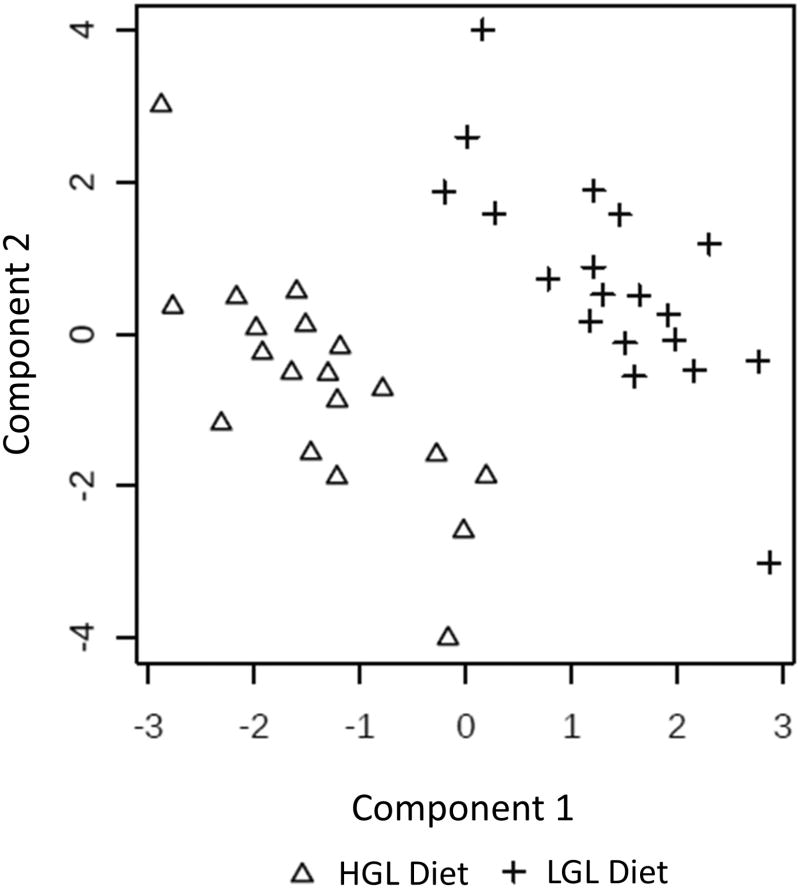
PLS-DA using all 125 detected metabolites showed good separation between the diets by the primary and secondary components (Figure 1). Together, these two components accounted for ∼23% of the variability (11.7% and 10.7% for the first and second components, respectively). The R2 and Q2 values were 0.72 and 0.45, and 0.91 and 0.75, for components 1 and 2 respectively. The metabolites with the greatest contribution in distinguishing the diets in the first component were kynurenate, cystathionine, glycocholate, glycochenodeoxycholate, adenylosuccinate, glyceraldehyde 3-phosphate, and biotin, all with VIP scores >2. The metabolites with the greatest contribution in the second component based on VIP scores of >2, were kynurenate, cystathionine, glycochenodeoxycholate, hippuric acid, glycerol-3-phosphate, and biotin.
Figure 1.
PLS-DA score plot of plasma metabolites after 28-d consumption of high glycemic load (HGL, Δ) and low glycemic load (LGL, +) diets in a randomized crossover-feeding study. The within individual variation and class-separated score plot between the selected components showed clear separation. The first component accounted for 11.7% and the second for 10.7% of the variance. R2 and Q2 values were 0.0.72 and 0.45, and 0.91 and 0.75 for components 1 and 2, respectively.
Discussion
In this randomized crossover feeding trial, concentrations of 14 plasma metabolites differed significantly in abundance between the HGL and LGL intervention diets, with a near-equal number increasing as decreasing after the LGL. Differentially-abundant metabolites could be classified into several different metabolic pathways, but we were not able to unequivocally identify pathways that would be uniquely related to GL. Further analyses, focused on energy metabolism and the Tryptophan pathway using all retained metabolites, did not reveal statistically significant pathway perturbations between the diets. Differences in some metabolites between diets were small (5-37%), yet all but one were larger than the overall intra-assay variation (7%). Although modest, such differences compounded over decades may, in part, account for observed differences in disease risk associated with differing habitual dietary patterns. Further, these differences may increase over time.
Despite the lack of significant pathway differences, the PLS-DA plot showed good separation between the diets with several metabolites, including kynurenate contributing considerably (VIP scores >2) to the model. Kynurenate was also identified in univariate analyses as significantly different between interventions, satisfying the stringent Bonferroni significance threshold (P<0.05/125=4×10-4). While plasma kynurenate was ∼40% higher after the LGL diet compared with the HGL diet, its precursor, tryptophan, was not appreciably altered.
Tryptophan is converted to kynurenine, and then further degraded to quinolinate or kynurenate, which agonize and antagonize N-methyl-D-aspartate (NMDA) receptors, respectively30. While the effect of tryptophan metabolites is most pronounced in cerebrospinal fluid, NMDA receptors are also found in peripheral tissue, where they elicit the same excitatory and inhibitory responses31. Among other functions (e.g., cognition and memory), these receptors modulate inflammation pathways31, 32. Whereas quinolinate activates NMDA receptors, stimulating immune activation, kynurenate inhibits these effects. Modulatory effects on inflammation have been noted apart from the effects these metabolites have on receptor activity. For example, quinolinate is also produced by macrophages in the periphery in response to cytokine activation which, in turn upregulates other inflammatory mediators, such as MCP-1, a potent chemoattractant33, as well as enzymes involved in the conversion of tryptophan to quinolinate31. Therefore, quinolinate not only stimulates pro-inflammatory pathways, but also upregulates the enzymes which lead to its further production, in a forward-feed cycle. Further, quinolinate, through interaction with hydroxyl kynurenine, an intermediate in the quinolinate pathway, is a pro-oxidant34. Consequently, a shift towards the kynurenate, or inhibitory side of the pathway, as observed after the LGL diet, suggests potential anti-inflammatory effects of the intervention35, 36. This finding is consistent with our previous report of reduced C-reactive protein concentrations after the LGL diet 16.
When considering directionality of all plasma metabolites in the Tryptophan pathway measured in our metabolite panel, regardless of significance (e.g., tryptophan, L-kynurinine, kynuranate, 3-hydroxykynurenine, xanthurinate, and quinolinate), a consistent shift away from quinolinate, and toward the kynurenate side of the pathway was observed; however, there was no significant difference in quinolinate concentrations between the diets. Although we did not determine absolute concentrations in plasma, studies suggest that the ratio of kynurenate to quinolinate is what determines its protective effects. Indeed, a higher ratio of quinolinate to kynurenate has been implicated in many neurodegenerative diseases thought to have an inflammatory basis, e.g., Alzheimer's disease, Parkinson's disease and Huntington's 34, 37. Thus, increases in kynurenate relative to quinolinate, as observed after the LGL diet in the present study, is hypothesized to be beneficial35, 36, 38, and may be one mechanism by which LGL diets contribute to reduced chronic disease risk.
TMAO was 37% higher at the end of the LGL compared to the HGL intervention. Generation of TMAO occurs via oxidation of gut bacterially-derived trimethylamine (TMA) in the liver. Therate of TMA to TMAO conversion is determined by a combination of genetics and substrate availability39. Given the crossover nature of this study, differences are likely due to substrate availability, although diet itself may alter the gut microbial community, in turn affecting TMAO production40. Precursors for TMA include carnitine, choline and betaine41, 42. These substrates are absorbed by a combination of active and passive transport in the small intestine, only reaching the microbiota responsible for their conversion to TMA in the large intestine when doses are high enough to saturate these transport mechanisms43-45. The LGL diet had slightly less protein from animal sources, a rich source of carnitine, than the HGL diet (daily average of 47% of total animal protein compared to 59%) to accommodate other foods associated with lower GL (e.g., beans and whole grains). As these lower GL foods are higher in protein, meat content was reduced to keep macronutrient ratios constant between the diets. Carnitine was therefore lower in the LGL diet (Table 4). In contrast, the inclusion of more whole grains and leafy greens on the LGL diet resulted in ∼80% higher betaine, based on the nutrient data available, although it should be noted that betaine content of diet is not as well-documented as that of other micronutrients. Dietary intake of choline appeared to be similar between the diets. Thus, only one of the TMAO precursors —betaine, was higher in the LGL diet. It is possible that consuming a LGL diet, inherently high in fiber, results in changes to the gut microbiome community structure that enhance production of TMAO, however studies done on individuals who consume vegan or vegetarian diets, or other diets characteristically high in fiber, would suggest otherwise46, 47.
TMAO is hypothesized to be associated with CVD risk due to its interference with cholesterol clearance, and the association between red meat intake and CVD is attributed, in part, to carnitine-derived TMAO 42, 48. Therefore, the increase in TMAO after the LGL diet is contrary to what might be expected. Increased intakes of whole grains and leafy greens are widely accepted as healthy dietary behaviors49, with betaine specifically identified as contributing beneficially as a methyl group donor to homocysteine50. A recent review evaluating dietary betaine and choline intakes not only found no association between increased intake and CVD risk, but reported reduced inflammation and other CVD risk factors51. Solanky et al. reported similar findings of increased TMAO after a dietary intervention with soy52, and TMAO is found in high quantities in fish53, both foods to which health benefits have been ascribed.
To our knowledge, this is the first study to use metabolomics to interrogate metabolic differences in specific pathways as a result of high and low GL dietary patterns in a crossover study design. While not specifically evaluated in other studies of GL, targeted metabolomics has been applied in the context of glycemic index, fiber and whole grain intake, mainly for dietary biomarker discovery14, 15, 54. An additional strength is the crossover design where individuals serve as their own control. The 4-week duration of the controlled diet intervention, where all foods were provided, was of sufficient length to allow us to capture changes in metabolism16. The participants largely maintained their weight on the standardized diets where macronutrient levels, with the exception of fiber, were held constant, differing only in GL. Adherence to the diets was excellent, with 97% of the participants consuming > 90% of the study provided food, with no difference in compliance between the two study diets. Targeted metabolomics, unlike global metabolomics, also enabled us to be more certain about the identities and relative abundances of the metabolites, although our targeted profile was limited to 125 mostly aqueous metabolites.
This study was limited in that LC-MS was used, and some compounds, such as some volatile compounds, are not well detected with this method. Additional analysis with GC-MS and NMR, as well as lipidomics could provide more comprehensive insight into metabolic changes between the two GL diets. Fitness and activity level impact the metabolome55 and while participants were asked to keep physical activity constant, detailed data on daily physical activity were not collected, and thus no adjustments were made. Although we were able to adjust for body fat and weight change, these variables account for only a small proportion of total metabolome variation. Finally, although fiber is a fundamental determinant of GL, it is difficult to ascertain whether the results of consuming a LGL diet pattern are due to differences in amount of fiber, type of fiber, structure of the foods, or other dietary constituents associated with low GI.
Conclusion
In summary, we found significant differences in plasma metabolites between a high and low GL diet which appear to suggest a metabolic response to differing GL; however, there were no statistically significant discernable shifts in the specific pathways examined that explain reported associations between LGL diet and reduced chronic disease risk. While the first two components in the PLS-DA analysis accounted for ∼22% of the variance, and only 14 metabolites were significantly altered between the diets, our ability to distinguish between the diets in this discriminate analysis suggests the GL of a diet affects plasma metabolites broadly. The metabolite with the largest difference in abundance was kynurenate, which increased after the LGL interventionm was identified as one of the small number of metabolites driving the separation between diets seen in the PLS-DA. Kynurenate, the only metabolite remaining statistically significant after Bonferonni correction, is associated with reduced inflammation and may be one mechanism through which protective effects of a LGL dietary pattern are manifested.
Supplementary Material
Acknowledgments
The authors would like to thank Andrew McDavid, postdoctoral fellow of the University of Washington for his statistical support with the linear regression models, and the University of Washington Nutrition and Obesity Research Center (P30 DK035816) for their support of the metabolomics analysis. This work was supported by NIH grant U54CA116847 and Fred Hutchinson Cancer Research Center. S.L.N. and M.F.B. were supported by grant T32CA09168. D.R. holds equity and an executive position at Matrix-Bio, Inc. S. B., S.L.N., M.F.B., Y.S., H.G., D.J., M.N., M.K., and J.W.L. declare no conflict of interest.
Abbreviations
- GI
glycemic index
- GL
glycemic load
- LGL
low glycemic load
- HGL
high glycemic load
- TMAO
Trimethylamine-N-oxide
- TMA
Trimethylamine
- CVD
cardiovascular disease
- CHD
coronary heart disease
- FFQ
food frequency questionnaire
- CARB
Carbohydrate and Related Biomarkers
- FHCRC
Fred Hutchinson Cancer Research Center
- DEXA
dual-energy X-ray absorptiometry
- LC-MS
liquid chromatography mass spectrometry
- GC-MS
gas chromatography mass spectrometry
- NMR
nuclear magnetic resonance
- HPLC
high performance liquid chromatography, QC, quality control
- FDR
false discovery rate
- PLS-DA
partial least squares discriminant analysis
- VIP
variable importance in projection
References
- 1.D. o. V. Statistics, editor. N C f H Statistics. Centers for Disease Control and Prevention; Hyattsville, MD: 2010. [Google Scholar]
- 2.U. S. C. S. W. Group, editor. D o H a H Services. Centers for Disease Control and Prevention and National Cancer Institute; Atlanta, U.S: 2013. [Google Scholar]
- 3.Kochanek KD, Murphy SL, Xu J, Arias E. NCHS Data Brief. 2014:1–8. [PubMed] [Google Scholar]
- 4.Jenkins DJ, Kendall CW, Augustin LS, Franceschi S, Hamidi M, Marchie A, Jenkins AL, Axelsen M. Am J Clin Nutr. 2002;76:266S–273S. doi: 10.1093/ajcn/76/1.266S. [DOI] [PubMed] [Google Scholar]
- 5.Augustin LS, Franceschi S, Jenkins DJ, Kendall CW, La Vecchia C. Eur J Clin Nutr. 2002;56:1049–1071. doi: 10.1038/sj.ejcn.1601454. [DOI] [PubMed] [Google Scholar]
- 6.Barclay AW, Petocz P, McMillan-Price J, Flood VM, Prvan T, Mitchell P, Brand-Miller JC. Am J Clin Nutr. 2008;87:627–637. doi: 10.1093/ajcn/87.3.627. [DOI] [PubMed] [Google Scholar]
- 7.Frost G, Leeds AA, Dore CJ, Madeiros S, Brading S, Dornhorst A. Lancet. 1999;353:1045–1048. doi: 10.1016/s0140-6736(98)07164-5. [DOI] [PubMed] [Google Scholar]
- 8.Brand-Miller JC. Nutrition Rev. 2003;61:S49–S55. doi: 10.1301/nr.2003.may.S49-S55. [DOI] [PubMed] [Google Scholar]
- 9.Putri SP, Nakayama Y, Matsuda F, Uchikata T, Kobayashi S, Matsubara A, Fukusaki E. J Biosci Bioeng. 2013;115:579–589. doi: 10.1016/j.jbiosc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Malkaram SA, Hassan YI, Zempleni J. Adv Nutr. 2012;3:654–665. doi: 10.3945/an.112.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menni C, Zhai GJ, MacGregor A, Prehn C, Romisch-Margl W, Suhre K, Adamski J, Cassidy A, Illig T, Spector TD, Valdes AM. Metabolomics. 2013;9:506–514. doi: 10.1007/s11306-012-0469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Sullivan A, Gibney MJ, Brennan L. Am J Clin Nutr. 2011;93:314–321. doi: 10.3945/ajcn.110.000950. [DOI] [PubMed] [Google Scholar]
- 13.Brennan L. Biochem Soc Transac. 2013;41:670–673. doi: 10.1042/BST20120350. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen LG, Winning H, Savorani F, Ritz C, Engelsen SB, Astrup A, Larsen TM, Dragsted LO. Genes Nutr. 2012;7:281–293. doi: 10.1007/s12263-011-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson-Persson A, Barri T, Ulmius M, Onning G, Dragsted LO. Anal Bioanal Chem. 2013;405:4799–4809. doi: 10.1007/s00216-013-6874-5. [DOI] [PubMed] [Google Scholar]
- 16.Neuhouser ML, Schwarz Y, Wang C, Breymeyer K, Coronado G, Wang CY, Noar K, Song X, Lampe JW. J Nutr. 2012;142:369–374. doi: 10.3945/jn.111.149807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Runchey SS, Pollak MN, Valsta LM, Coronado GD, Schwarz Y, Breymeyer KL, Wang C, Wang CY, Lampe JW, Neuhouser ML. Eur J Clin Nutr. 2012;66:1146–1152. doi: 10.1038/ejcn.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei R, Li G, Seymour AB. Anal Chem. 2010;82:5527–5533. doi: 10.1021/ac100331b. [DOI] [PubMed] [Google Scholar]
- 19.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D. J Proteome Res. 2014;13:4120–4130. doi: 10.1021/pr500494u. [DOI] [PubMed] [Google Scholar]
- 21.Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, Beyer RP, MacCoss MJ, Rabinovitch PS. Aging Cell. 2014;13:529–539. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu H, Du J, Carnevale Neto F, Carroll PA, Turner SJ, Chiorean EG, Eisenman RN, Raftery D. Analyst. 2015;140:2726–2734. doi: 10.1039/c4an02386b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll PA, Diolaiti D, McFerrin L, Gu H, Djukovic D, Du J, Cheng PF, Anderson S, Ulrich M, Hurley JB, Raftery D, Ayer DE, Eisenman RN. Cancer Cell. 2015;27:271–285. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. J Royal Statist Soc Serial B. 1995;57:289–300. [Google Scholar]
- 25.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 26.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westerhuis JA, van Velzen EJ, Hoefsloot HC, Smilde AK. Metabolomics. 2010;6:119–128. doi: 10.1007/s11306-009-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehmood T, L KH, Snipen L, Saebo S. Chemometr Intell Lab. 2012;118:62–69. [Google Scholar]
- 30.Stone TW. Pharmacol Rev. 1993;45:309–379. [PubMed] [Google Scholar]
- 31.Guillemin GJ. FEBS J. 2012;279:1356–1365. doi: 10.1111/j.1742-4658.2012.08485.x. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lugo-Huitron R, Ugalde Muniz P, Pineda B, Pedraza-Chaverri J, Rios C, Perez-de la Cruz V. Oxid Med Cell Longev. 2013;2013:104024. doi: 10.1155/2013/104024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell BM, Charych E, Lee AW, Moller T. Front Neurosci. 2014;8:12. doi: 10.3389/fnins.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyes MP, Saito K, Crowley JS, Davis LE, Demitrack MA, Der M, Dilling LA, Elia J, Kruesi MJ, Lackner A, et al. Brain. 1992;115(Pt 5):1249–1273. doi: 10.1093/brain/115.5.1249. [DOI] [PubMed] [Google Scholar]
- 36.Grohmann U, Fallarino F, Puccetti P. Trends Immunol. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 37.Gong CY, Li Z, Wang HM, Liu J, Chen L, Zhang HW, Wang X, Yang J. Med Hypotheses. 2011;77:383–385. doi: 10.1016/j.mehy.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Wu HQ, Guidetti P, Goodman JH, Varasi M, Ceresoli-Borroni G, Speciale C, Scharfman HE, Schwarcz R. Neuroscience. 2000;97:243–251. doi: 10.1016/s0306-4522(00)00030-0. [DOI] [PubMed] [Google Scholar]
- 39.Bennett BJ, d A Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craig SAS. Am J Clin Nutr. 2004;80:539–549. doi: 10.1093/ajcn/80.3.539. [DOI] [PubMed] [Google Scholar]
- 44.Zeisel SH, Dacosta KA, Youssef M, Hensey S. J Nutr. 1989;119:800–804. doi: 10.1093/jn/119.5.800. [DOI] [PubMed] [Google Scholar]
- 45.Rebouche CJ. Ann N Y Acad Sci. 2004;1033:30–41. doi: 10.1196/annals.1320.003. [DOI] [PubMed] [Google Scholar]
- 46.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lappi J, Salojarvi J, Kolehmainen M, Mykkanen H, Poutanen K, de Vos WM, Salonen A. J Nutr. 2013;143:648–655. doi: 10.3945/jn.112.172668. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. Nature. 2011;472:57–U82. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson JW, Hanna TJ. Am J Clin Nutr. 1999;70:307–308. doi: 10.1093/ajcn/70.3.307. [DOI] [PubMed] [Google Scholar]
- 50.Obeid R. Nutrients. 2013;5:3481–3495. doi: 10.3390/nu5093481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajaie S, Esmaillzadeh A. ARYA Atheroscler. 2011;7:78–86. [PMC free article] [PubMed] [Google Scholar]
- 52.Solanky KS, Bailey NJ, Beckwith-Hall BM, Bingham S, Davis A, Holmes E, Nicholson JK, Cassidy A. J Nutr Biochem. 2005;16:236–244. doi: 10.1016/j.jnutbio.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Ussher JR, Lopaschuk GD, Arduini A. Atherosclerosis. 2013;231:456–461. doi: 10.1016/j.atherosclerosis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 54.Moazzami AA, Zhang JX, Kamal-Eldin A, Aman P, Hallmans G, Johansson JE, Andersson SO. J Nutr. 2011;141:2126–2132. doi: 10.3945/jn.111.148239. [DOI] [PubMed] [Google Scholar]
- 55.Morris C, Grada CO, Ryan M, Roche HM, De Vito G, Gibney MJ, Gibney ER, Brennan L. Mol Nutr Food Res. 2013;57:1246–1254. doi: 10.1002/mnfr.201200629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.