Abstract
The human (h) ZIP4 transporter is a plasma membrane protein which functions to increase the cytosolic concentration of zinc. hZIP4 transports zinc into intestinal cells and therefore has a central role in the absorption of dietary zinc. hZIP4 has eight transmembrane domains and encodes a large intracellular loop between transmembrane domains III and IV, M3M4. Previously, it has been postulated that this domain regulates hZIP4 levels in the plasma membrane in a zinc-dependent manner. The objective of this research was to examine the zinc binding properties of the large intracellular loop of hZIP4. Therefore, we have recombineantly expressed and purified M3M4 and showed that this domain binds two zinc ions. Using a combination of site-directed mutagenesis, metal binding affinity assays, and X-ray absorption spectroscopy, we demonstrated that the two Zn2+ ions bind sequentially, with the first Zn2+ binding to a CysHis3 site with a nanomolar binding affinity, and the second Zn2+ binding to a His4 site with a weaker affinity. Circular dichroism spectroscopy revealed that the M3M4 domain is intrinsically disordered, with only a small structural change induced upon Zn2+ coordination. Our data supports a model in which the intracellular M3M4 domain senses high cytosolic Zn2+ concentrations and regulates the plasma membrane levels of the hZIP4 transporter in response to Zn2+ binding.
Introduction
Zinc is the second most abundant transition metal in cells. Zinc has catalytic, structural and regulatory roles in vivo.1 This is demonstrated by the presence of more than 3000 Zn2+-containing proteins encoded in the human genome.1 Cellular Zn2+ deficiency impairs protein synthesis, cell growth and metabolism, whereas excessive Zn2+ levels cause protein misfolding and aggregation, leading to toxic effects in the cell.1 Consequently, intracellular Zn2+ concentrations are tightly regulated. The ZIP (SLC39) family of transporters function to increase cytoplasmic Zn2+ levels by importing Zn2+ from the extracellular environment or by exporting Zn2+ from organelles into the cytoplasm.2 The ZIP family, which stands for Zrt-, Irt-like proteins, derives its name from the first members of this family to be identified: the zinc transporters ZRT1 and ZRT2 in Saccharomyces cerevisiae3 and the iron transporter IRT1 in Arabidopsis thaliana.4 Eukaryotic ZIP transporters are predicted to have eight transmembrane domains with extracytoplasmic N- and C-termini and a large cytosolic loop between transmembrane domains III and IV.5
In humans, 14 ZIP proteins have been identified and classified into four sub-families, ZIPI, ZIPII, LIV-1 and gufA.6 Amongst these proteins, hZIP4, a member of the LIV-1 subfamily, plays an important role in Zn2+ homeostasis. hZIP4 was first identified due to its involvement in the lethal, childhood Zn2+ deficiency disease acrodermatitis enteropathica (AE).7,8 hZIP4 is the primary Zn2+ transporter expressed in the stomach, small intestine, colon and kidney.9 AE is an autosomal recessive genetic disorder whose symptoms include skin lesions, diarrhea, growth retardation, neurological disorders, severe infections and, if left untreated, death.10 The symptoms of AE can be reversed with increased dietary Zn2+ supplementation.11 In addition to its normal tissue distribution, the surface expression of hZIP4 is increased in pancreatic,12,13 liver14 and brain cancer cells,15 where hZIP4 surface expression has been correlated with metastatic stage and survival time. In cancer cells, hZIP4 overexpression was shown to increase the expression of growth factors and matrix metalloproteinases14 and to activate the interleukin 6 (IL-6) and signal transducer and activator of transcription 3 (STAT3) pathways that are implicated in cancer cell proliferation.14
Studies with hZIP4 and the mouse homologue have shown that surface expression of the transporter is regulated by the cytosolic concentration of Zn2+.5 At high cytosolic Zn2+ concentrations, ZIP4 undergoes Zn2+-dependent endocytosis, thereby reducing ZIP4 levels in the plasma membrane.16 At even higher Zn2+ concentrations, hZIP4 is ubiquitinated, presumably at a highly conserved lysine residue within a large intracellular loop between transmembrane domains III and IV and is further subjected to proteasomal degradation.17 The Zn2+-dependent ubiquitination and degradation required the presence of a histidine-rich domain located on the large cytosolic loop, leading to the hypothesis that the intracellular domain acts as a Zn2+ sensor that is involved in regulating hZIP4 levels in the plasma membrane.17
Recently, intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs) have become recognized as having important biological functions in cell signaling, regulation and control, although they lack secondary and tertiary structural elements.18 IDPRs are estimated to be present in over 35% of human proteins.19 In the case of membrane proteins, a survey of the Protein Data Bank identified disordered regions, as determined by missing electron density in crystal structures, in more than half of deposited membrane protein structures.20 Moreover, an analysis of human plasma membrane proteins found that over 40% contained disordered regions of more than 30 amino acids in length, and these disordered regions were three times more likely to occur on the cytoplasmic side than on the extracellular side of the membrane.21 Inside cells, IDPs and IDPRs participate in molecular recognition functions by binding to target molecules such as nucleic acid, proteins or small ligands.22 Among their molecular recognition functions, IDPs and IDPRs act as scavengers for ions or small molecules and provide display sites for post-translational modifications such as phosphorylation and ubiquitination.22
Here, we describe the metal binding properties of the single significant intracellular loop located between transmembrane domains III and IV (M3M4) of hZIP4. We provide the first direct evidence that this domain coordinates two Zn2+ ions. Moreover, Zn2+ coordination occurs in a sequential manner with the first Zn2+ binding with nanomolar affinity to a CysHis3 site and the second Zn2+ binding with a lower affinity to a His4 site. Finally, we show that the intracellular M3M4 loop is an intrinsically disordered region that serves as a protein-specific regulatory domain.
Results
The intracellular loop of hZIP4 is disordered
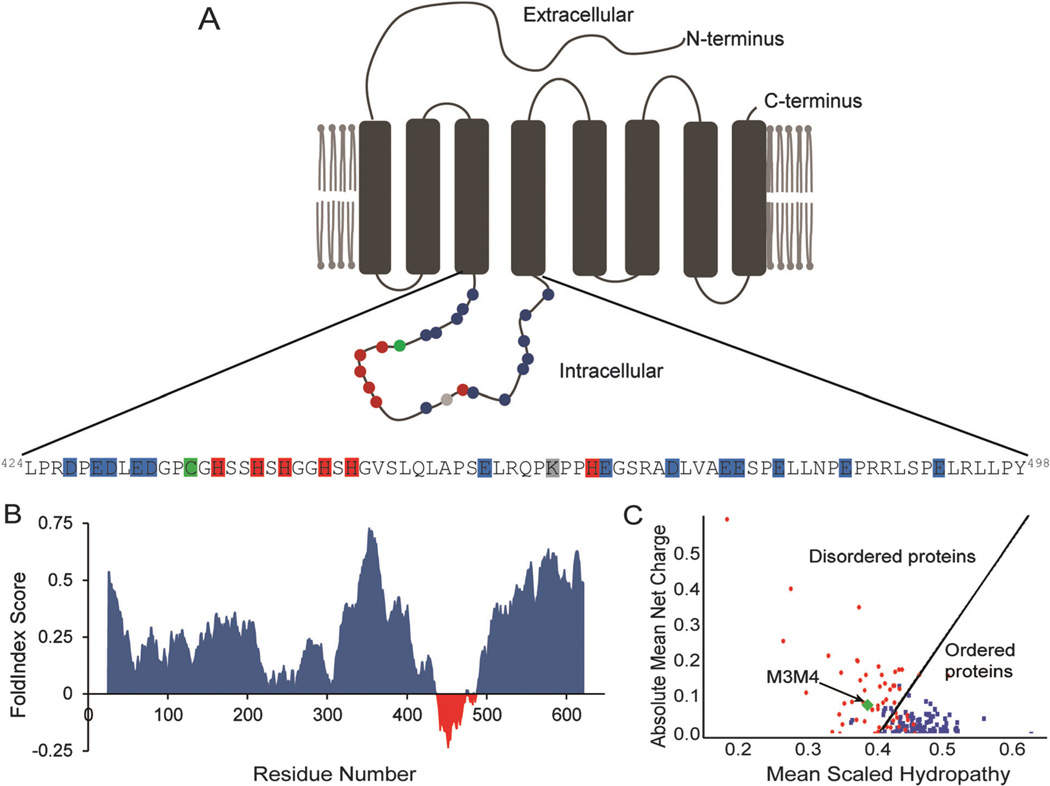
The hZIP4 protein (Fig. 1A) was analyzed using the disorder predictors FoldIndex23 and PONDR-Fit (Prediction of Natural Disordered Regions).24 The FoldIndex algorithm predicted that the majority of the hZIP4 protein is folded with the exception of the intracellular M3M4 loop (Fig. 1B). Similarly, the PONDR-Fit output predicted that the M3M4 loop is primarily disordered (data not shown). Further, the intracellular M3M4 domain was calculated to have a low overall hydrophobicity and a high mean net charge characteristic of disordered proteins (Fig. 1C).25 In support of the predictions, the amino acid sequence of the intracellular M3M4 domain contains a low proportion (17%) of order-promoting amino acids (W, C, F, I, Y, V, L, N) and a high proportion (64%) of disorder-promoting residues (A, R, G, Q, S, P, E, K).26
Fig. 1.
The hZIP4 domain structure and predicted regions of disorder. (A) Schematic of the hZIP4 transporter with the sequence of the large cytosolic loop M3M4 shown. Histidine residues are colored red, cysteine is colored green, acidic residues are colored blue, and the lysine residue is gray. (B) FoldIndex prediction of disordered regions within the hZIP4 protein. Amino acid residues with a negative FoldIndex score are considered as disordered while those with positive scores are considered to be in ordered regions. (C) Mean charge versus hydropathy analysis of the M3M4 domain indicates it lies in the disordered region. The dividing line represents the division between intrinsically disordered and folded proteins.
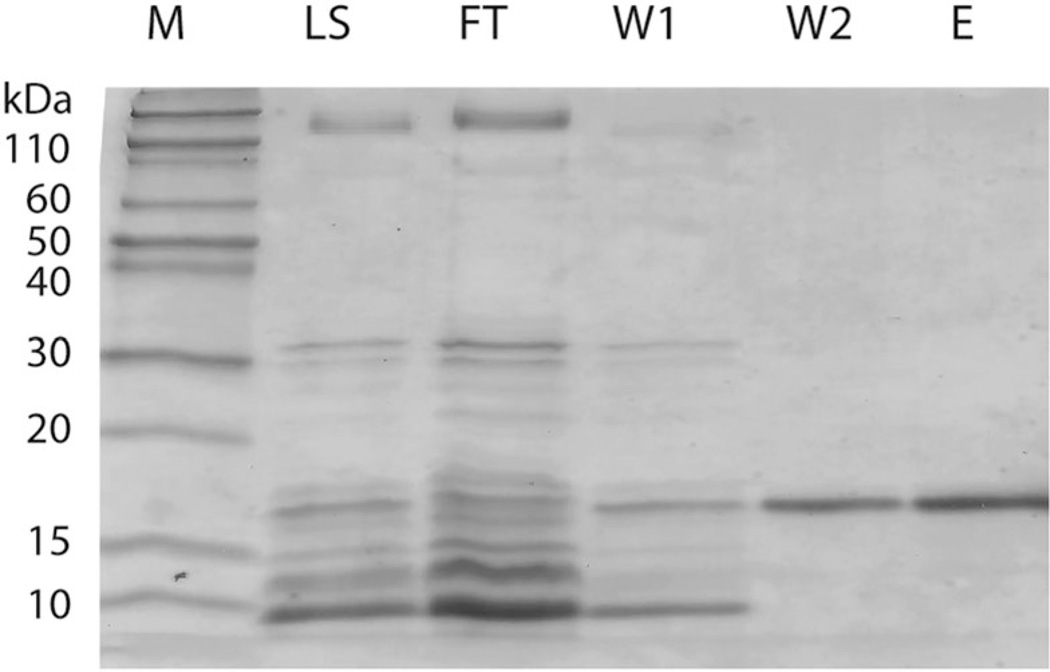
To investigate the structural and functional properties of the large intracellular loop of hZIP4, we expressed the intracellular domain (residues 424–498 of the full-length hZIP4) fused to a Strep-tag in Escherichia coli and purified the protein using a heat-cooling extraction method shown to improve the yield and purity for intrinsically disordered proteins.27,28 Following affinity purification, yields of 1 mg protein L−1 culture were obtained. N-terminal protein sequencing confirmed the identity of the purified protein as the M3M4 domain (data not shown). The purified protein had a slower than predicted mobility on SDS-PAGE, migrating at an apparent molecular mass of 16 kDa (Fig. 2), which is 1.4 times higher than the molecular mass calculated from the amino acid sequence (11.4 kDa). Slower mobilities in SDS-PAGE have been observed for IDPs, which bind less SDS than globular proteins due to their unique amino acid compositions.26
Fig. 2.
Purification of the recombinantly expressed M3M4 domain using the heat-cooling extraction method. Coomassie blue stained SDS-PAGE gel of the purification fractions with molecular weight markers (M) indicated. After cell lysis using the heat-cooling extraction method, the cell lysate supernatant (LS) was applied to a Strep-Tactin column and the flow-through (FT) was collected. The column was washed with buffer (20 mM Hepes pH 8, 150 mM NaCl (W1) and 20 mM Hepes pH 8, 20% glycerol (W2)), and the protein was eluted in buffer containing desthiobiotin (E).
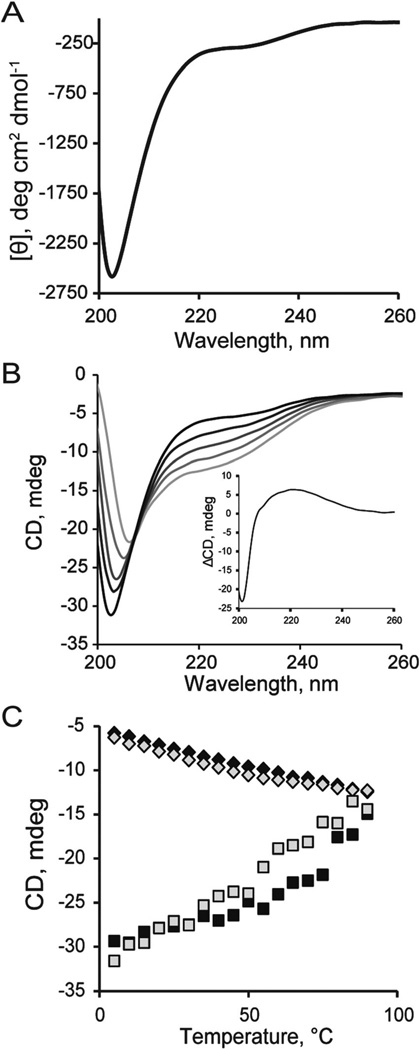
The disordered nature of the M3M4 domain was supported by far-UV circular dichroism (CD) spectroscopy. The CD spectrum (Fig. 3A) of the M3M4 domain showed a negative minimum at 203 nm, indicative of a random coil structure, and a weak negative shoulder at 220 nm, which may indicate a small degree of α-helical structure. The ellipticity values at 203 nm and 220 nm suggest that the M3M4 loop is coil-like, rather than pre-molten globule.29 Upon increasing the temperature from 5 °C to 85 °C, the CD spectra showed a reversible, linear increase in ellipticity at 203 nm along with a reversible, linear decrease in ellipticity at 220 nm and an isodichroic point at 208 nm (Fig. 3B and C). The isodichroic point indicates that the M3M4 protein undergoes a temperature-induced conformational change. The difference CD spectrum between 5 °C and 85 °C showed a peak at 220 nm (Fig. 3B inset) characteristic of a poly-l-proline type II (PPII) structure, and the temperature-induced changes in the CD spectra may reflect melting of the PPII helix.29,30 Overall, the CD data support the structural predictions that the intracellular M3M4 loop of hZIP4 is an intrinsically disordered domain.
Fig. 3.
CD spectra of the purified M3M4 domain in 20 mM MOPS, pH 7, 20% glycerol, 1 mM TCEP at (A) 5 °C and (B) at increasing temperatures from 5 °C (black) to 85 °C (light gray). Representative curves at 5 °C, 25 °C, 45 °C, 65 °C and 85 °C are shown. The inset shows the CD difference spectrum (5–85 °C) indicating the presence of polyproline type II helices in the M3M4 domain. (C) Temperature scans at 222 nm (diamonds) and 204 nm (squares) indicate a reversible, linear change in CD signal. The CD spectra were collected at increasing (black) and decreasing (gray) temperatures.
The intracellular M3M4 domain binds Zn2+ with nanomolar affinity
The number of Zn2+ ions that bind to the M3M4 domain was quantified by atomic absorption spectroscopy (AAS). For the AAS analysis, purified M3M4 protein was incubated with a fourfold molar excess of Zn2+ and loosely bound Zn2+ was removed by washing with buffer. As measured by AAS, the M3M4 domain binds Zn2+ with a stoichiometry of 2.2 ± 0.2 Zn2+ ions per protein molecule, demonstrating the presence of two binding sites for Zn2+ within the intracellular loop.
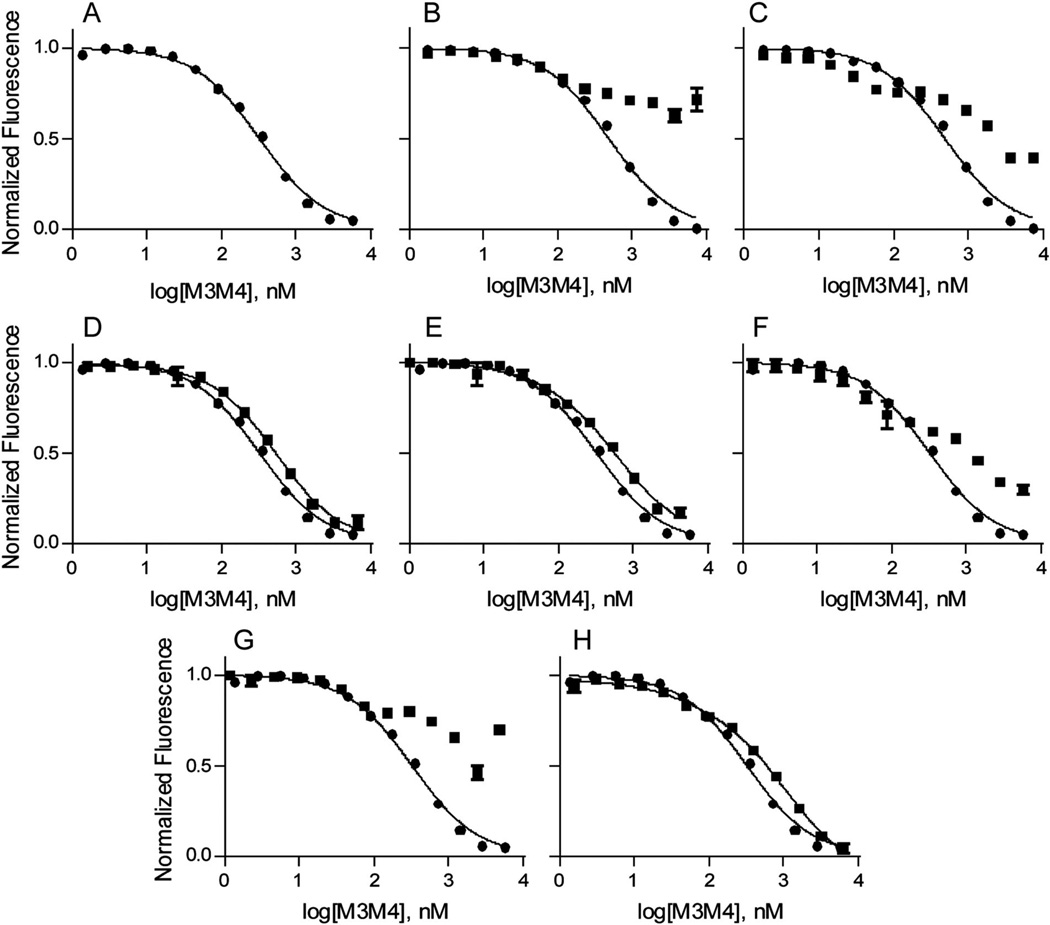
To investigate the binding affinity of the M3M4 domain for Zn2+, FluoZin-3 was used as a Zn2+ chelator in competition assays with the protein.31 FluoZin-3 shows an increase in fluorescence upon binding Zn2+ with a 1:1 stoichiometry. The dissociation constant of FluoZin-3 (Kdfluozin) for Zn2+ was determined to be 21 ± 5 nM under our experimental conditions. The dissociation constant of FluoZin-3 for Zn2+ was determined individually for each experimental data set, and this value was used to fit the binding data within the same set of competition experiments. The M3M4 dissociation constant for Zn2+ was measured using a competition experiment with FluoZin-3 as described previously.31 In the competition experiments, equimolar amounts of FluoZin-3 and Zn2+ were titrated with purified M3M4 protein. The resulting competition data for the wild-type M3M4 domain (Fig. 4A) were fit using one- and two-site binding models. Error analysis of the two models indicated that the one-site model provided the best fit for the data despite the measured 2:1 Zn2+:protein stoichiometry observed for the M3M4 protein domain. This suggests either that the two Zn2+ ions bind to the M3M4 domain with similar affinities, which cannot be distinguished using the competition assay, or that the second Zn2+ ion binds to the protein with a much weaker binding affinity, which cannot be measured using the current assay method. Unfortunately, our attempts to measure a weaker Zn2+ binding affinity using a fluorescent indicator (Newport Green) with a micromolar dissociation constant for Zn2+ were unsuccessful due to the tendency of the protein to aggregate at high Zn2+ concentrations as determined by dynamic light scattering experiments (data not shown). Thus, the intracellular M3M4 domain of the hZIP4 transporter binds at least one Zn2+ ion according to a one-site binding model with a macroscopic dissociation constant of 6 ± 1 nM (Fig. 4A).
Fig. 4.
Normalized competitive binding curves for the determination of Zn2+ dissociation constants to the wild-type, labeled and mutant M3M4 proteins. The fluorescence emitted in the presence of various concentrations of M3M4 was normalized to Fmax (the fluorescence of 1 µM Zn2+ and 1µM Fluozin-3) and dissociation constants were determined using a one-site binding model (eqn (1)). (A) Fluorescence inhibition curve for wild-type M3M4 protein. Representative fluorescence inhibition curves comparing wild-type (circles) and (B) DEPC treated wild-type protein (squares), (C) NEM treated wild-type protein (squares), (D) H438A mutant protein (squares), (E) H441A mutant protein (squares), (F) C436A (squares), (G) C436A/H438A/H441A triple mutant protein (squares), and (H) H443A/H446A/H448A triple mutant protein (squares). Data are the average of the assay done in triplicate for one protein preparation. Three independent protein preparations gave equivalent results. Error bars represent ± one standard deviation.
Histidine and cysteine residues are involved in Zn2+ binding in the M3M4 domain
Cysteine, histidine, and the acidic residues aspartate and glutamate most commonly coordinate Zn2+.1 The M3M4 domain contains one cysteine, six histidine and 13 acidic residues (Fig. 1A). In order to determine the contributions of these residues in Zn2+ binding, the M3M4 protein was treated with N-ethylmaleimide (NEM), diethyl pyrocarbonate (DEPC), and N,N′-dicyclohexyl carbodiimide (DCCD), which selectively labels cysteine, histidine, and aspartate and glutamate residues, respectively. Following labeling, the Zn2+ binding stoichiometry and dissociation constants were measured. The results (Table 1) indicate that labeling the acidic residues with DCCD did not affect either the Zn2+:protein binding stoichiometry or the dissociation constant compared to the wild-type protein. However, labeling the protein with DEPC resulted in a complete loss of Zn2+ binding to the M3M4 protein as measured by AAS (Table 1). Consistent with this result, the dissociation constant for the DEPC-labeled protein could not be measured using the FluoZin-3 competition assay (Fig. 4B). A slight decrease in fluorescence signal was observed in the competition assay at high concentrations of the DEPC-treated M3M4 protein, which we postulate is the result of nonspecific interactions between the free Zn2+ and the protein. Finally, labeling the single cysteine residue in the M3M4 domain by treatment with NEM lowered the protein’s binding affinity for Zn2+ such that the dissociation constant could not be measured using the competition assay (Fig. 4C), although the NEM-treated protein was still able to bind 1.7 ± 0.3 Zn2+ per protein molecule when treated with an excess of Zn2+ (Table 1). Based on the labeling data, we conclude that the histidine and cysteine residues, but not aspartate or glutamate residues, coordinate Zn2+ in the intracellular M3M4 loop of hZIP4.
Table 1.
Zn2+ binding stoichiometry and dissociation constants for M3M4 proteinsa
| Protein | Zinc:protein stoichiometry | Kd (nM) |
|---|---|---|
| Labeling reagent | ||
| Unlabeled | 2.2 ± 0.2 | 5 ± 1 (n = 3) |
| DCCD | 2.3 ± 0.2 | 6 ± 1 (n = 3) |
| DEPC | 0.3 ± 0.1 | No binding (n = 3) |
| N-Ethyl maleimide | 1.7 ± 0.3 | N.D. (n = 3) |
| Mutation | ||
| Wild-type | 2.2 ± 0.2 | 6 ± 1 (n = 5) |
| C436A | 2.2 ± 0.2 | N.D. (n = 3) |
| H438A | 2.1 ± 0.1 | 8 ± 1 (n = 3) |
| H441A | 2.0 ± 0.3 | 9 ± 2 (n = 3) |
| H443A | 2.4 ± 0.2 | 7 ± 1 (n = 3) |
| H446A | 2.4 ± 0.2 | 6 ± 2 (n = 3) |
| H448A | 2.4 ± 0.4 | 6 ± 2 (n = 3) |
| H466A | 2.7 ± 0.2 | 5.6 ± 0.2 (n= 3) |
| C436A/H438A/H441A | 1.3 ± 0.2 | N.D. (n = 3) |
| H443A/H446A/H448A | 1.1 ± 0.2 | 12 ± 2 (n = 3) |
N.D. – weaker binding. The dissociation constant could not be determined using the competition assay. Bold-faced are significantly different (p < 0.05) from wild-type. Kd values are average and S.E.M. from three measurements of three independent protein preparations. AAS values are average and S.E.M. of three independent protein preparations.
To further investigate the role of the histidine and cysteine residues in Zn2+ coordination by M3M4, we individually mutated the six histidine and one cysteine residues to alanine. All single mutants retained the ability to bind two Zn2+ ions per molecule protein (Table 1). Interestingly, the H466A mutant protein was able to bind significantly higher amounts of Zn2+. Competition assays with FluoZin-3 were performed. As with the wild-type protein, the binding data for the single histidine mutations were best fit to a one-site binding model, and the dissociation constants were calculated (Table 1). The single mutants H443A, H446A, H448A and H466A showed Zn2+ binding affinities that were comparable to the wild-type protein, whereas the single mutants H438A and H441A displayed binding affinities that were statistically different (p values < 0.05) from the wild-type protein (Table 1, Fig. 4D and E). Although the H438A and H441A mutant proteins exhibited weaker binding affinities compared to the wild-type, the change in dissociation constants was not as dramatic as observed for the DEPC-labeled protein. The inability of any single histidine mutation to produce a marked change in Zn2+ binding affinity is likely due to stabilization of the Zn2+ ion by the remaining histidines and neighboring residues. Ligand substitution by neighboring residues has also been observed in a Zn2+ finger protein.32 In contrast to the histidine mutants, mutation of the single cysteine residue to alanine (C436A) resulted in a protein with a substantially weaker Zn2+ binding affinity that could not be measured using the FluoZin-3 competition assay (Fig. 4F). Taken together, the single histidine and cysteine mutations in M3M4 indicated that C436, H438, and H441 are important residues contributing to Zn2+ binding.
Based on the results of the single mutants, we designed two triple mutants, C436A/H438A/H441A and H443A/H446A/H448A, and analyzed these mutants for Zn2+ binding. As expected, both triple mutant proteins bound only one Zn2+ ion per molecule protein (Table 1). Also, the triple mutants showed significantly weaker Zn2+ binding affinities when compared to the wild-type domain (Table 1, Fig. 4G and H). In the case of the H443A/H446A/H448A triple mutant, the measured dissociation constant was two-fold higher than that of the wild-type protein, whereas the C436A/H438A/H441A triple mutant yielded a substantially weaker dissociation constant that could not be measured using the FluoZin-3 competition assay. These data suggest that C436/ H438/H441 and H443/H446/H448 likely form the two coordination sites for Zn2+ binding to the intracellular M3M4 domain, with the C436/H438/H441 having a tighter binding affinity for Zn2+. The binding affinity data suggest that Zn2+ binds first to the higher affinity C436/H438/H441 site, followed by binding of the second Zn2+ to the H443/H446/H448 site.
EXAFS reveals the coordination geometry of Zn2+ bound to M3M4
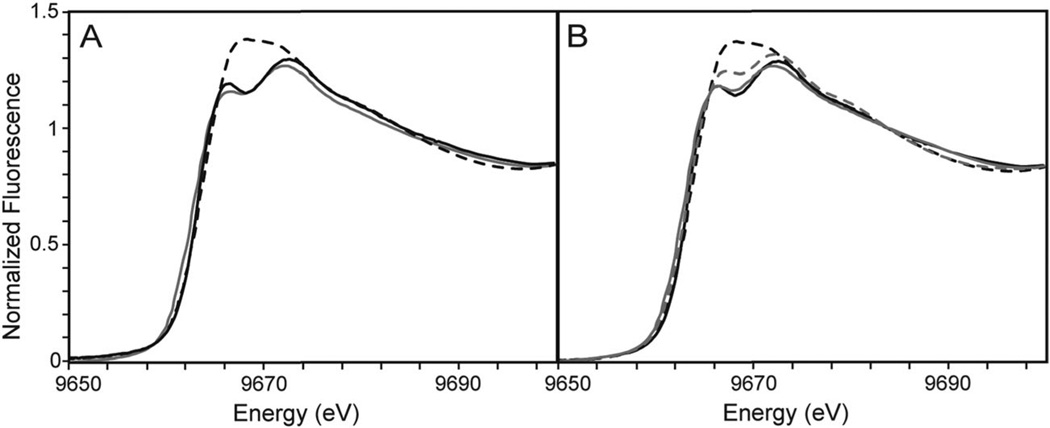
In order to further elucidate the Zn2+-binding properties of the M3M4 domain, the Zn2+-bound protein was analyzed by X-ray absorption spectroscopy (XAS). The X-ray absorption near edge structure (XANES) portion of an XAS spectrum provides qualitative details regarding metal site structure with ligand speciation and can be used to compare differences in metal binding sites on related protein samples. XANES spectra of M3M4 prepared with various stoichiometric amounts of Zn2+ revealed that the M3M4 protein with 0.5 and 1 molar equivalent of Zn2+ yielded similar XANES spectra, whereas the two Zn2+-bound protein produced a distinctly different XANES edge (Fig. 5A). These data are consistent with a model in which the two Zn2+ ions bind sequentially to the M3M4 domain.
Fig. 5.
Normalized XANES spectra for wild-type and triple mutant Zn2+-loaded M3M4 samples. (A) XANES spectra of wild-type M3M4 in the presence of 0.5 (gray line), 1 (black line) and 2 (dashed line) equivalents of Zn2+. (B) XANES spectra of C436A/H438A/H441A triple mutant (gray dashed line) and H443A/H446A/H448A triple mutant (gray solid line) loaded with 1 equivalent Zn2+ compared to wild-type M3M4 loaded with 1 (black solid line) or 2 (black dashed line) equivalents of Zn2+.
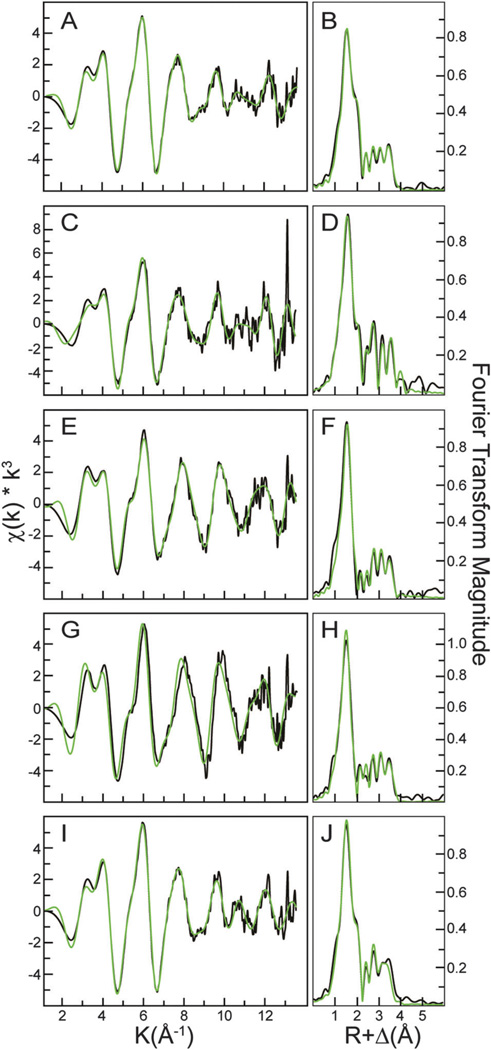
Further, simulations of the extended X-ray absorption fine structure (EXAFS) region of the XAS spectrum provide metrical details regarding the metal–ligand coordination environments for a metal in a metalloprotein at extremely high resolution (±0.02 Å).33 EXAFS spectra for the Zn2+-bound M3M4 proteins, along with the Fourier transforms of the EXAFS data, are given in Fig. 6. In each spectrum, the EXAFS at ca. k = 4 Å−1 shows a bead pattern characteristic of imidazole scattering from a histidine residue coordinated to the protein-bound metal.34 All spectra could be fit with nearest neighbor scattering constructed predominately with O/N ligands, and, in the case of M3M4 with 0.5 and 1 molar equivalent Zn2+, with an additional sulfur scattering ligand (Table 2). No sulfur ligation was observed in the M3M4 protein with two Zn2+ bound, presumably since the sulfur scattering is a low component of the overall ligand scattering signal. Within error of the technique (±0.5), all Zn2+ coordination numbers from the simulations are consistent with Zn2+ being tetra-coordinated. Based on the EXAFS analysis, we conclude that the first Zn2+ ion binds to a CysHis3 site and the second Zn2+ ion binds to a site comprised solely of histidines. Long range scattering (R > 2.8 Å) is observed in the Fourier transforms of the samples, as expected given the suggested presence of imidazole coordination. Long-range scattering interactions could be easily simulated for Zn2+ −C/N interactions above R = 2.8 Å using single scattering models, however coordination numbers for the fits were variable and consistent with an overlap between single and multiple scattering contributions at each of the long range bond lengths. A summary of the complete simulation parameters that led to the simulated spectra in Fig. 6 are provided in the ESI.† Attempts to fit this data with a multiple scattering theoretical model compound for a Zn2+ −imidazole interaction were unsuccessful.
Fig. 6.
EXAFS and Fourier transform of the EXAFS data for wild-type and mutant Zn2+-loaded M3M4 proteins. Comparison of raw data (black) and simulations (green) for both the EXAFS (left) and Fourier transform (right). Wild-type M3M4 with 0.5 equivalents of Zn2+ (A, B), wild-type with 1 equivalent of Zn2+ (C, D), wild-type with 2 equivalents of Zn2+ (E, F), C436A/H438A/H441A triple mutant (G, H), and H443A/H446A/H448A triple mutant (I, J).
Table 2.
Summary of Zn2+ EXAFS fitting analysis of wild-type and mutant M3M4 samples. Data fit over a k range of 1 to 13.5 Å−1, however fitting analysis was limited to only nearest neighbor ligand environment fits. Complete spectral fits parameters, including long-range simulation components, are provided in the ESI. Best fit simulation parameters for each sample are shown in bold
| Nearest-neighbor ligand environmenta |
||||||
|---|---|---|---|---|---|---|
| Sample | Fitb | Atomc | Rd (Å) | C.N.e | σ2f | F′g |
| Wild-type + 0.5Zn2+ | 1 | O/N | 2.01 | 2.5 | 5.4 | 1.55 |
| 2 | O/N | 1.99 | 2.5 | 5.1 | 0.52 | |
| S | 2.28 | 1.5 | 5.1 | |||
| Wild-type + 1Zn2+ | 1 | O/N | 2.01 | 3.0 | 4.6 | 1.90 |
| 2 | O/N | 1.99 | 3.0 | 4.0 | 1.41 | |
| S | 2.28 | 1.0 | 4.3 | |||
| Wild-type + 2Zn2+ | 1 | O/N | 1.99 | 3.5 | 4.8 | 0.54 |
| C436A/H438A/H441A | 1 | O/N | 1.98 | 4.0 | 4.6 | 0.84 |
| H443A/H446A/H448A | 1 | O/N | 2.01 | 3.0 | 5.3 | 1.62 |
| 2 | O/N | 1.99 | 3.0 | 4.8 | 0.71 | |
| S | 2.28 | 1.5 | 5.0 | |||
Independent metal–ligand scattering environments at R < 3.0 Å.
Iterative simulation fit number.
Scattering atoms: S (sulfur), C (carbon), O (oxygen), N (nitrogen).
Average metal–ligand bond length.
Average metal–ligand coordination number.
Average Debye–Waller factor in Å2 × 103.
Number of degrees of freedom weighted mean square deviation between data and fit.
XAS was also used to evaluate Zn2+ coordination in the triple mutants (C436A/H438A/H441A and H443A/H446A/H448A). XANES spectra of the H443A/H446A/H448A mutant resembled the one Zn2+-bound wild-type protein (Fig. 5B), and EXAFS data (Fig. 6G, Table 2) showed the same CysHis3 coordination geometry as the wild-type protein with a single Zn2+ ion bound. The C436A/H438A/ H441A mutant protein showed an XANES spectrum intermediate between the one and two Zn2+ bound wild-type (Fig. 5B), and the EXAFS analysis revealed a ligand environment consisting of four histidine residues (Fig. 6I, Table 2). Interestingly, addition of a second Zn2+ to the wild-type M3M4 protein distorts the average metal binding site away from a simple linear combination of the single loaded independent sites. Taken together, the XAS results are consistent with the model derived from mutagenesis analysis, in which the two Zn2+ ions bind sequentially to two distinct sites within the M3M4 domain, with the CysHis3 site comprising the first site and a histidine only site comprising the second site.
Structural changes to the M3M4 domain upon Zn2+ binding
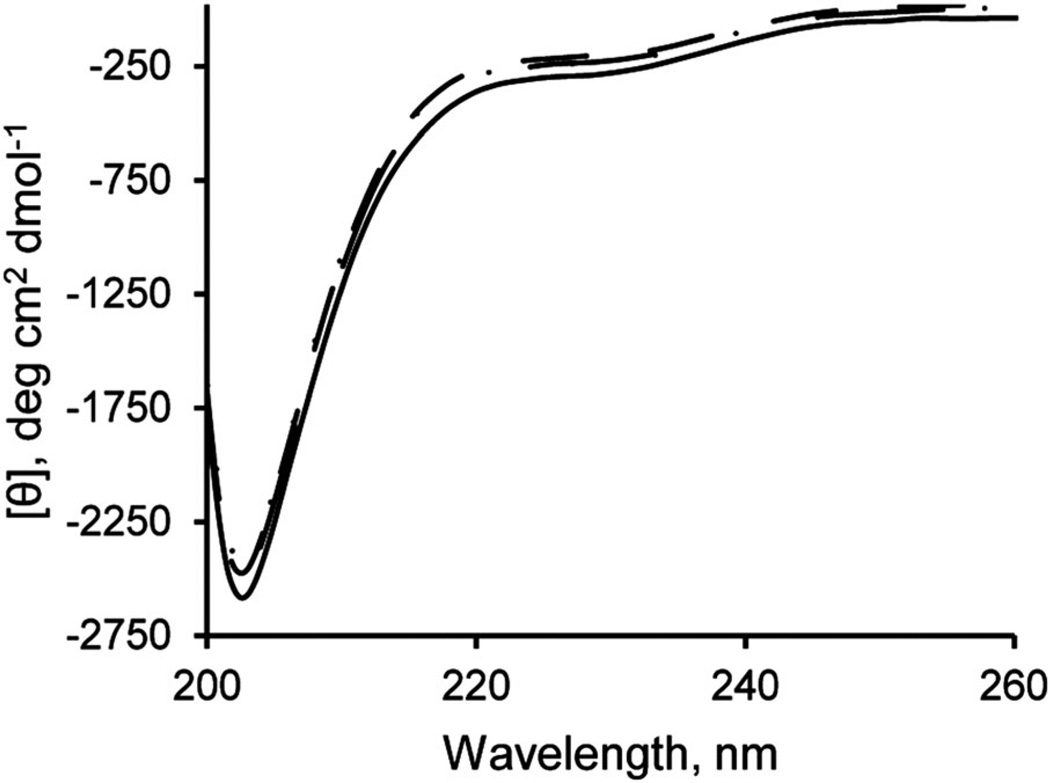
Structural changes to the intracellular domain upon Zn2+ binding were monitored by CD spectroscopy. Upon sequential addition of Zn2+ to M3M4, only minor changes in the CD spectra were observed (Fig. 7). Interestingly, an isodichroic point at 213 nm is present when the one Zn2+-bound and two Zn2+-bound CD spectra are compared (Fig. 7), indicating that the M3M4 protein undergoes a small structural change upon binding the second Zn2+. However, the presence of the large negative peak at 203 nm in all the spectra indicates that the M3M4 protein domain remains largely disordered even in the Zn2+-bound state.
Fig. 7.
CD spectra of the M3M4 domain in the absence (solid line) and presence of 0.5 (broken dashed line), 1 (dashed line) and 2 (dotted line) molar equivalents of Zn2+.
Discussion
As the primary Zn2+ importer in the gastrointestinal tract, hZIP4 plays a key role in dietary Zn2+ absorption. Moreover, the presence of hZIP4 in tissues other than gastrointestinal tract organs such as the eye, lung, heart, skin, prostate, ovary, skin and mammary glands indicates that the role of hZIP4 is not limited to dietary Zn2+ absorption but extends to maintaining cellular Zn2+ homeostasis.35 However, the mechanism for metal transport remains unclear due to the absence of structural data not only for hZIP4 but for any ZIP family member. Previously, in situ hZIP4 functional studies indicated that hZIP4 mediated metal transport is not limited to Zn2+ but extends to other metal ions such as Cu2+ and Ni2+.36 Additionally, the large histidine-rich cytoplasmic M3M4 loop has been hypothesized to undergo conformational changes upon Zn2+ binding that lead to post-translational modifications modulating hZIP4 levels in the plasma membrane.17 hZIP4 transporter was shown to undergo Zn2+-stimulated ubiquitination and this required the presence of the histidine-rich domain in the cytoplasmic M3M4 loop.17 Overexpression of hZIP4 has been associated with pancreatic,12,13 liver,14 and brain15 cancers where it has been observed that hZIP4 expression is vital for balancing the expression of pro-metastatic and pro-apoptotic genes.37 Thus, obtaining structural and functional data for the cytoplasmic M3M4 domain is important to understand not only how hZIP4 functions to maintain Zn2+ homeostasis in normal tissues but also how the plasma membrane expression of this transporter is regulated in cancer cells.
Our data provide the first direct evidence that the histidinerich domain within the cytoplasmic M3M4 loop of hZIP4 binds two Zn2+ ions (Table 1). Moreover, we have shown by using XAS (Fig. 5 and 6) that Zn2+ binding occurs sequentially with the first Zn2+ binding to a site involving the single cysteine residue and histidine residues (CysHis3) followed by a second Zn2+ binding event to a site consisting solely of histidine residues (His4). Based on the results of our mutagenesis analysis (Table 1), our data is consistent with the observation that single alanine replacement of histidine residues within the M3M4 domain does not eliminate zinc-dependent degradation of hZIP4.17 Equally, from the analysis of this data, we conclude that the CysHis3 site is comprised of C436, H438 and H441, and the second site (His4) is likely composed of H443, H446 and H448 and a fourth ligand, which may be H466. Although mutation of H466 to alanine did not affect the binding affinity for Zn2+, the H466A mutant protein consistently bound higher amounts of Zn2+ than the wild-type or any of the other single mutants. We, therefore, speculate that mutation of H466, increases the structural flexibility of the region C-terminal to the histidine-rich domain, which may allow more adventitious Zn2+ binding to the M3M4 domain (Fig. 1A). The fourth histidine ligand for the CysHis3 site is likely a bridging ligand shared by the two sites. As Zn2+ ions are typically found in tetra-coordinated environments,38 binding of two Zn2+ ions would require a minimum of seven protein ligands with one ligand serving as a bridge. Bridging ligands have been observed in other Zn2+ binding proteins; for example, cysteine is a bridging ligand in metallothionein39 and histidine serves as a bridging ligand in Cu, Zn superoxide dismutase.40 Based on our data, we speculate that one of the histidines in the loop acts as a bridging ligand, but we cannot predict which of the histidines functions as the bridging ligand. Additionally, the observation that a combination of residue mutations interferes with zinc coordination suggests a high degree of cooperativity between both sites to accomplish metal binding.
Using the FluoZin-3 competition assay, we measured a single apparent dissociation constant of 6 ± 1 nM for Zn2+ binding to the isolated M3M4 intracellular domain (Table 1, Fig. 4A). The measured one-site binding reflects Zn2+ binding to the CysHis3 site as the H443A/H446A/H448A triple mutant displayed a binding affinity in the low nanomolar range, whereas a significantly weaker binding affinity that could not be measured using the Fluozin-3 indicator was observed for the C436A/H438A/H441A triple mutant. Thus, the first Zn2+ binds to the CysHis3 site with a low nanomolar binding affinity followed by binding of the second Zn2+ to the His4 site with a weaker, likely micromolar or higher, binding affinity. It should be noted that the dissociation constant measured for the purified M3M4 protein may not necessarily reflect the in vivo binding affinity of Zn2+ for this domain as interactions of the cytoplasmic loop with other regions of the hZIP4 transporter or with the lipid environment may modulate its affinity for Zn2+. Nevertheless, the low nanomolar Zn2+ binding affinity of the CysHis3 site within the M3M4 domain is comparable to other Zn2+-binding proteins whose measured Zn2+ dissociation constants range from nanomolar to low picomolar or less.41 The cytoplasmic concentration of free Zn2+ in eukaryotic cells is estimated to be in the picomolar to low nanomolar range,41 which suggests that the CysHis3 site may be occupied with Zn2+ under normal physiological conditions. The second, His4, site would be unoccupied at normal cytosolic free Zn2+ concentrations. The His4 site would become occupied with Zn2+ as the local Zn2+ concentration near the M3M4 domain is expected to be higher as hZIP4 functions to transport Zn2+ across the membrane. Thus, we propose that the His4 site likely acts as a sensor to detect high cytosolic Zn2+ concentrations and control the level of hZIP4 in the plasma membrane accordingly. Only when both sites are occupied will hZIP4 be subjected to zincstimulated ubiquitination and degradation.17 A caveat to this analysis is that it is possible that the M3M4 domain may become more structured within the holo-hZIP4 transporter. However, considering that M3M4 is predicted to be disordered using multiple prediction tools as well as the observation that zinc coordination to this domain is congruent with the cellular zinc concentration, we consider this possibility unlikely.
To investigate the structural basis for Zn2+ sensing by the M3M4 loop, a combination of protein structure prediction algorithms and CD spectroscopy were used. The cytoplasmic M3M4 loop of hZIP4 was predicted (Fig. 1B) and shown to be intrinsically disordered (Fig. 3). As IDPs and IDPRs tend to undergo disorder-to-order transitions upon binding partner molecules, we sought to determine if the M3M4 cytosolic loop becomes ordered upon Zn2+ binding using CD spectroscopy to assess secondary structure. The M3M4 protein domain remains largely disordered in the Zn2+-bound state (Fig. 7). Recently, the occurrence of disorder in the bound state has been found to be a common feature of IDPs.22,42 More than 40 IDPs have been shown to form “fuzzy” complexes upon interaction with their partners.42 “Fuzziness” is believed to be functionally advantageous, allowing IDPs or IDPRs the flexibility to interact with multiple partners or to undergo a variety of post-translational modifications, such as phosphorylation and ubiquitination.22,42 A number of short linear motifs, which are located within disordered protein domains and modulate protein function,43 were predicted in the cytoplasmic M3M4 loop of hZIP4 using the eukaryotic linear motif (ELM) server (Table 3). Among these short linear motifs in M3M4 are targeting sites for membrane protein assembly, endocytosis and a number of phosphorylation sites. Phosphorylation of S490 in hZIP4 was identified in an analysis of the phosphoproteome of human embryonic stem cells.44 In addition to phosphorylation, hZIP4 was predicted using the UbPred server to undergo ubiquitination at K463 within the intracellular M3M4 loop.45 hZIP4 was shown to be ubiquitinated at high cytosolic Zn2+ concentrations and to undergo subsequent proteasomal degradation of the transporter.17 Taken together with our data on Zn2+ binding to the purified cytosolic domain, we propose a model whereby, at high cytosolic Zn2+ concentrations, both zinc sites are filled. This induces structural changes in the M3M4 intracellular loop, which may alter the post-translational modification status of hZIP4. Degradation in the proteasome is enhanced by the presence of a disordered protein region downstream of the ubiquitination site.17,46 Interestingly and correlated to this model is the observation that H438 and H441 are not conserved within the otherwise homologous mouse ZIP4. Therefore, it will be important to examine the metal binding properties of homologous loops and how these properties integrate with the results presented within this manuscript.
Table 3.
Predicted short linear motifs in the M3M4 sequence
| Position | Sequence | Short linear motif | Cell compartment |
|---|---|---|---|
| 487–489 | RRL | Metalloendopeptidase N-Arg dibasic convertase cleavage site | Cell surface |
| 487–491 | RRLSP | Cyclin docking site recognition sequence that increases phosphorylation levels | Cytosol |
| 494–498 | RLLPY | Cyclin docking site recognition sequence that increases phosphorylation levels | Cytosol |
| 487–495 | RRLSPELRL | Docking site for MAPK serine/threonine kinases | Cytosol |
| 476–481 | AEESPE | PinI docking site that controls dephosphorylation, degradation and targeting | Cytosol |
| 487–492 | RRLSPE | PinI docking site that controls dephosphorylation, degradation and targeting | Cytosol |
| 459–465 | LRQPKPP | Binding site for SH3 domains that require PPII helix | Plasma membrane, cytosol |
| 439–445 | SSHSHGG | CK1 phosphorylation site | Cytosol |
| 444–451 | GGHSHGVS | GSK3 phosphorylation recognition site | Cytosol |
| 475–481 | AEESPEL | Proline-directed kinase (e.g. MAPK) phosphorylation site | Cytosol |
| 487–493 | RRLSPEL | Proline-directed kinase (e.g. MAPK) phosphorylation site | Cytosol |
| 486–488 | PRR | di-Arg endoplasmic reticulum retention motif for proper assembly of multimeric membrane proteins, phosphorylation-dependent |
Cytosol, endoplasmic reticulum membrane |
| 487–489 | RRL | di-Arg endoplasmic reticulum retention motif for proper assembly of multimeric membrane proteins, phosphorylation-dependent |
Cytosol, endoplasmic reticulum membrane |
| 477–482 | ESPELL | Sorting and internalization signal in cytoplasmic domains of membrane proteins that is involved in endocytosis |
Cytosol, endocytic vesicle |
The cytosolic loop of hZIP4 likely fulfills a number of roles, including proper processing and recycling of the transporter and possibly modulating Zn2+ transport through changes in post-translational modifications, all of which rely on its disordered state. As disordered regions tend not to be under strong evolutionary conservation,47 it is not surprising that the large cytosolic loop between transmembrane domains III and IV, which is characteristic of the LIV-I subfamily of ZIP transporters, is nonconserved. Among the human LIV-I subfamily members, the amino acid sequence and the number and arrangement of potential Zn2+ binding ligands varies. Thus, the cytosolic M3M4 domain likely functions as a protein-specific regulatory domain.
Experimental
Materials
d-Desthiobiotin and Strep-Tactin Superflow resin were purchased from IBA Life Sciences. Zinc chloride, isopropyl-β-d-thiogalacto-pyranoside (IPTG), ethylenediaminetetraacetic acid (EDTA), ethylene glycol tetraacetic acid (EGTA) and N-ethylmaleimide (NEM) were purchased from Sigma-Aldrich. Glycerol, tris(2-chloro-ethyl) phosphate (TCEP), 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), diethyl pyrocarbonate (DEPC), and N,N′-dicyclohexyl carbodiimide (DCCD) were purchased from Amresco. 4-Morpholinepropanesulfonic acid (MOPS) was purchased from BDH Chemicals.
Bioinformatics
Disorder predictions were performed using FoldIndex (http://bip.weizmann.ac.il/fldbin/findex)23 and PONDR-FIT (http://www.disprot.org/metapredictor.php).24 The mean charge versus hydropathy of the M3M4 domain was also analyzed (http://pondr.com/cgi-bin/PONDR/pondr.cgi).25 The M3M4 sequence was analyzed for predicted short linear motifs using the Eukaryotic Linear Motif server (http://elm.eu.org/)43 and for ubiquitination sites using the UbPred server (http://www.ubpred.org/).45
Molecular cloning
The gene sequence corresponding to the predicted intracellular domain (M3M4) between residues 424 and 498 of the hZIP4 protein was amplified by PCR from the hZIP4 gene. The resulting PCR product was cloned into the overexpression vector pPR-IBA1 (IBA Life Sciences) to generate pPRIBA-M3M4 by using the unique restrictions sites KpnI and NcoI. The pPR-IBA1 vector introduced a C-terminal Strep tag for affinity purification. Site-directed mutagenesis to generate single cysteine or histidine mutant proteins was performed according to the manufacturer’s instructions (Agilent Technologies). Gene synthesis (GenScript) was used to generate triple mutants. All plasmid constructs were verified by DNA sequencing.
Protein expression, purification and labeling
The pPRIBA-M3M4 plasmid was transformed into E. coli BL21(DE3) pLysS cells carrying the pSJS1240 plasmid coding for rare tRNAs.48 The transformed E. coli cells were grown at 37 °C in TB medium containing 100 µg mL−1 ampicillin, 34 µg mL−1 chloramphenicol and 50 µg mL−1 spectinomycin. Protein expression was induced by adding 100 µM IPTG to a culture of OD600 0.6–0.8, and expression was carried out at 18 °C for 20 hours. The cells were harvested and washed with wash buffer (20 mM HEPES, 150 mM NaCl, pH 8). The protein was purified using a boiling lysis method previously reported for the purification of IDPs.27,28 The cell pellet from a 1 L culture was resuspended in 20 mL wash buffer containing 2 mM EDTA and 1 mM dithiothreitol. The cells were lysed by boiling for 20 minutes, followed by incubation in an ice-salt bath for 5 min. All remaining purification steps were performed at 4 °C. The lysed cells were sonicated for 30 s to shear the DNA and the cell lysate was clarified by ultracentrifugation for 30 minutes at 100 000 × g. The resulting soluble fraction was loaded onto a Strep-Tact in Superflow (IBA Life Sciences) gravity column. The column was washed first with wash buffer containing 0.2 mM EDTA and 0.1 mM TCEP, followed by buffer exchange with chelex-100 treated elution buffer (20 mM HEPES, 150 mM NaCl, 20% (v/v) glycerol, 0.1 mM TCEP, pH 7). The protein was eluted in chelex-100 treated elution buffer containing 2.5 mM d-desthiobiotin. Protein was quantified using the Reducing Agent Compatible Pierce™ Microplate BCA Protein Assay Kit (Thermo Scientific). For labeling, the purified protein was incubated with 0.5 mM DCCD, 10 mM DEPC, or 1 mM NEM for 30 minutes at 4 °C followed by extensive dialysis against chelex-100 treated elution buffer.
Circular dichroism spectroscopy
All CD spectra were recorded using a Jasco J-1500 CD spectrometer in 1 mm quartz cuvettes. Protein samples were prepared in 20 mM MOPS (pH 7), 20% (v/v) glycerol and 1 mM TCEP. Sample and baseline spectra were acquired at 5 °C using 20 consecutive scans collected in 0.5 nm increments with a 1 nm bandwidth, a scanning speed of 50 nm min−1, and a 4 s data integration time. The temperature dependence of the CD signal was measured between 5 and 90 °C at 5 °C intervals. Samples were heated at a rate of 2 °C min−1 before three scans were recorded at each temperature. The spectra were averaged, baseline-corrected and smoothed using a Savitzky–Golay filter in the Spectra Manager software (Jasco).
Binding affinity (Kd) determination of M3M4
The Zn2+ stock solution was prepared in chelex-100 treated elution buffer and the concentration of Zn2+ was quantified using a terpyridine–Zn2+ titration.49 The dissociation constant (Kd) for Zn2+ to FluoZin-3 was calculated as per the manufacturer’s instructions (Invitrogen). Briefly, 1.1 mM EGTA and 1.1 mM Zn2+ solutions were mixed to yield free Zn2+ concentrations from 0 to 100 nM, and these Zn2+ solutions were incubated with 1 µM FluoZin-3 in black 96-well plates (Thermo Scientific). Fluorescence was recorded on a Perkin Elmer VICTOR 1420 multilabel counter using a 485/14 excitation filter and a 535/25 emission filter. For competition assays with the purified M3M4 proteins, 1 µM FluoZin-3 and 1 µM Zn2+ were incubated with varying concentrations of M3M4 protein, and the fluorescence was measured. The Zn2+ dissociation constant for M3M4 was determined by fitting the curve to eqn (1) using GraphPad.
| (1) |
where IC50 is the concentration of M3M4 required to reduce the maximum fluorescence by 50%, [FluoZin-3] is the concentration of FluoZin-3, and Kd(FluoZin-3) is the experimentally determined Zn2+ dissociation constant for FluoZin-3. To fit the binding curves using GraphPad, the fluorescence was normalized, and the curves were fit by constraining the minimum and maximum normalized fluorescence values. The minimum fluorescence was obtained in the absence of zinc and maximum fluorescence was obtained in the absence of protein.
Atomic absorption spectroscopy
The purified M3M4 protein in elution buffer with an additional 1 mM TCEP was incubated with 4 molar equivalents of Zn2+ overnight at 4 °C. Excess Zn2+ was removed by washing with chelex-100 treated 20 mM Hepes, 20% (v/v) glycerol, 1 mM TCEP, pH 7 using 3 kDa molecular weight cut-off centricons (Millipore). The protein samples were diluted in 10% (v/v) nitric acid (trace metal-free) for AAS analysis. Zn2+ was quantified by graphite furnace atomic absorption spectroscopy (Perkin Elmer PiNNacle 900Z). Metal contents reported are the averages of at least three independent experiments. The metal content of proteins before Zn2+ addition was less than 0.1 moles Zn2+ per mole protein.
X-ray absorption spectroscopy
M3M4 samples were prepared at final concentrations of 1 mM Zn2+ in 20 mM HEPES and 30% glycerol at pH 7.0. Samples were loaded into Lucite XAS sample cells wrapped with Kapton tape. After loading, samples were flash frozen and stored in liquid N2 until data collection. XAS data were collected at the National Synchrotron Light Source (NSLS), on beamline X3-b. Beamline X3-b utilized a Si[111] single crystal monochromator equipped with a Ni plated harmonic rejection/focusing mirror. During data collection, samples were maintained at 24 K using a He Displex Cryostat. Protein fluorescence excitation spectra were collected using a 31-element Ge solid-state detector, with a 0.6 mM Cu fluorescence filter placed between the cryostat and detector. XAS spectra were measured in 5 eV increments in the pre-edge region (9600–9660 eV), 0.25 eV increments in the edge region (9660–9740 eV) and 0.05 Å−1 increments in the extended X-ray absorption fine structure (EXAFS) region (to k = 14 Å−1), integrating from 1 s to 25 s in a k3 weighted manner for a total scan length of approximately 50 minutes. X-ray energies were individually calibrated by collecting Zn-foil absorption spectra simultaneously with the protein data. The first inflection point of the Zn-foil spectrum was assigned to 9668 eV. Each fluorescence channel of each scan was examined for spectral anomalies, and data represent the average of 11 to 14 scans for each sample.
XAS data were processed using the Macintosh OS X version of the EXAFSPAK program suite50 integrated with the Feff v8 software package for theoretical model generation.51 Data reduction utilized a Gaussian function in the pre-edge region and a threeregion cubic spline throughout the EXAFS region. Data were converted to k-space using a Zn E0 value of 9668 eV. The k cubed weighted EXAFS was truncated at 1.0 and 13.5 Å−1 for filtering purposes. This k range corresponds to a spectral resolution of ca. 0.115 Å for all zinc-ligand interactions; therefore only independent scattering environments outside 0.115 Å were considered resolvable in the EXAFS fitting analysis.52 EXAFS fitting analysis was performed on raw/unfiltered data. EXAFS data were fit using both single and multiple scattering amplitude and phase functions calculated with the program Feff v8. Single scattering theoretical models were calculated for carbon, nitrogen, oxygen, and sulfur coordination to simulate zinc nearest-neighbor ligand environments. Scale factors (Sc) and E0 values used during the simulations were calibrated by fitting crystallographically characterized Zn models; specific values include a Scale Factor of 0.9, and E0 values for O, N, C and S of −15.54 eV were used in these simulations.9 Criteria for judging the best-fit simulation utilized both the lowest mean square deviation between data and fit (F′), corrected for the number of degrees of freedom and a reasonable Debye–Waller factor.33,53
Conclusions
Analysis of our experiments clearly indicates that the large intracellular loop of hZIP4 binds two Zn2+ ions sequentially, with the first Zn2+ ion binding with nanomolar affinity to a CysHis3 site and the second Zn2+ ion binding with weaker affinity to a His4 site. Further, we have shown that the M3M4 loop is disordered and contains a number of post-translational modification sites. Our findings underscore the importance of the large intracellular loop as a regulatory domain that responds to cytosolic zinc levels.
Supplementary Material
Acknowledgements
We thank Kumari Alka and Fei Huang for performing initial experiments. This work was supported by the NIH (R01 GM105964) to R.E.D. S.P.D and T.L.S. are supported by the NIH (DK101230 and DK068139, respectively). Portions of this research were carried out at the National Synchrotron Light Source (NSLS). NSLS, located at Brookhaven National Laboratory, is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, under Contract No. DE-AC02-98CH10886.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mt00066a
References
- 1.Maret W, Li Y. Chem. Rev. 2009;109:4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- 2.Kambe T, Suzuki T, Nagao M, Yamaguchi-Iwai Y. Genomics, Proteomics Bioinf. 2006;4:1–9. doi: 10.1016/S1672-0229(06)60010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao H, Eide D. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eide D, Broderius M, Fett J, Guerinot ML. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasana S, Din J, Maret W. J. Trace Elem. Med. Biol. 2015;29:47–62. doi: 10.1016/j.jtemb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Dempski RE. Curr. Top. Membr. 2012;69:221–245. doi: 10.1016/B978-0-12-394390-3.00009-4. [DOI] [PubMed] [Google Scholar]
- 7.Küry S, Dréno B, Bézieau S, Giraudet S, Kharfi M, Kamoun R, Moisan J-P. Nat. Genet. 2002;31:239–240. doi: 10.1038/ng913. [DOI] [PubMed] [Google Scholar]
- 8.Wang K, Zhou B, Kuo Y-M, Zemansky J, Gitschier J. Am. J. Hum. Genet. 2002;71:66–73. doi: 10.1086/341125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufner-Beattie J, Wang F, Kuo Y-M, Gitschier J, Eide D, Andrews GK. J. Biol. Chem. 2003;278:33474–33481. doi: 10.1074/jbc.M305000200. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt S, Küry S, Giraud M, Dréno B, Kharfi M, Bézieau S. Hum. Mutat. 2009;30:926–933. doi: 10.1002/humu.20988. [DOI] [PubMed] [Google Scholar]
- 11.Van Wouwe J. Eur. J. Pediatr. 1989;149:2–8. doi: 10.1007/BF02024322. [DOI] [PubMed] [Google Scholar]
- 12.Donahue T, Hines OJ. Cancer Biol. Ther. 2010;9:243–245. doi: 10.4161/cbt.9.3.11064. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H, Wang X, Zhang S, Liuzzi JP, Chang S-M, Cousins RJ. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18636–18641. doi: 10.1073/pnas.0709307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weaver BP, Zhang Y, Hiscox S, Guo GL, Apte U, Taylor KM, Sheline CT, Wang L, Andrews GK. PLoS One. 2010;5:e13158. doi: 10.1371/journal.pone.0013158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Chen Y, Wang Y, Yang J, Zhu VF, Liu Y, Cui X, Chen L, Yan W, Jiang T, Hergenroeder GW, Fletcher SA, Levine JM, Kim DH, Tandon N, Zhu J-j, Li M. Neuro Oncol. 2013;15:1008–1016. doi: 10.1093/neuonc/not042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim B-E, Wang F, Dufner-Beattie J, Andrews GK, Eide DJ, Petris MJ. J. Biol. Chem. 2004;279:4523–4530. doi: 10.1074/jbc.M310799200. [DOI] [PubMed] [Google Scholar]
- 17.Mao X, Kim B-E, Wang F, Eide DJ, Petris MJ. J. Biol. Chem. 2007;282:6992–7000. doi: 10.1074/jbc.M610552200. [DOI] [PubMed] [Google Scholar]
- 18.Uversky VN. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flock T, Weatheritt RJ, Latysheva NS, Babu MM. Curr. Opin. Struct. Biol. 2014;26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Xue B, Li L, Meroueh SO, Uversky VN, Dunker AK. Mol. BioSyst. 2009;5:1688–1702. doi: 10.1039/B905913J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minezaki Y, Homma K, Nishikawa K. J. Mol. Biol. 2007;368:902–913. doi: 10.1016/j.jmb.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 22.Habchi J, Tompa P, Longhi S, Uversky VN. Chem. Rev. 2014;114:6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- 23.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 24.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 26.Tompa P. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 27.Yi S, Brickenden A, Choy W-Y. Protein Expression Purif. 2008;57:1–8. doi: 10.1016/j.pep.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Campos F, Guillén G, Reyes JL, Covarrubias AA. Protein Expression Purif. 2011;80:47–51. doi: 10.1016/j.pep.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Uversky VN. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chemes LB, Alonso LG, Noval MG, de Prat-Gay G. Methods Mol. Biol. 2012;895:387–404. doi: 10.1007/978-1-61779-927-3_22. [DOI] [PubMed] [Google Scholar]
- 31.De León-Rodríguez LM, Lubag AJ, López JA, Andreu-de-Riquer G, Alvarado-Monzón JC, Sherry AD. MedChemComm. 2012;3:480–483. doi: 10.1039/C2MD00301E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blasie CA, Berg JM. Inorg. Chem. 2000;39:348–351. doi: 10.1021/ic990913y. [DOI] [PubMed] [Google Scholar]
- 33.Bencze KZ, Kondapalli KC, Stemmler TL. Applications of Physical Methods to Inorganic and Bioinorganic Chemistry. 2007:513–528. [Google Scholar]
- 34.Stemmler TL, Sossong TM, Goldstein JI, Ash DE, Elgren TE, Kurtz DM, Penner-Hahn JE. Biochemistry. 1997;36:9847–9858. doi: 10.1021/bi9702795. [DOI] [PubMed] [Google Scholar]
- 35.Colvin RA, Holmes WR, Fontaine CP, Maret W. Metallomics. 2010;2:306–317. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- 36.Antala S, Dempski RE. Biochemistry. 2012;51:963–973. doi: 10.1021/bi201553p. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Guo H-J, Xie H-Y, Li J, Zhuang R-Z, Ling Q, Zhou L, Wei X-Y, Liu Z-K, Ding S-M, Chen K-J, Xu Z-Y, Zheng S-S. Int. J. Biol. Sci. 2014;10:245–256. doi: 10.7150/ijbs.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laitaoja M, Valjakka J, Jänis J. Inorg. Chem. 2013;52:10983–10991. doi: 10.1021/ic401072d. [DOI] [PubMed] [Google Scholar]
- 39.Meloni G, Zovo K, Kazantseva J, Palumaa P, Vašák M. J. Biol. Chem. 2006;281:14588–14595. doi: 10.1074/jbc.M601724200. [DOI] [PubMed] [Google Scholar]
- 40.Parge HE, Hallewell RA, Tainer JA. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kochańczyk T, Drozd A, Krężel A. Metallomics. 2015;7:244–257. doi: 10.1039/c4mt00094c. [DOI] [PubMed] [Google Scholar]
- 42.Fuxreiter M, Tompa P. Adv. Exp. Med. Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 43.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Krüger D, Grebnev G, Kubań M, Strumillo M, Uyar B, Budd A, Altenberg B, Seiler M, Chemes LB, Glavina J, Sánchez IE, Diella F, Gibson TJ. Nucleic Acids Res. 2013:1–8. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brill LM, Xiong W, Lee K-B, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Proteins: Struct., Funct. Bioinf. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 47.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Curr. Opin. Struct. Biol. 2011;21:441–446. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim R, Sandler SJ, Goldman S, Yokota H, Clark AJ, Kim S-H. Biotechnol. Lett. 1998;20:207–210. [Google Scholar]
- 49.Burdette SC, Walkup GK, Spingler B, Tsien RY, Lippard SJ. J. Am. Chem. Soc. 2001;123:7831–7841. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 50.George G, Pickering I. EXAFSPAK: A Suite of Computer Programs for Analysis of X-ray Absorption Spectra. Stanford, CA: Stanford Synchrotron Radiation Laboratory; 2000. [Google Scholar]
- 51.Rehr J, Ankudinov A. J. Synchrotron Radiat. 2001;8:61–65. doi: 10.1107/s0909049500016423. [DOI] [PubMed] [Google Scholar]
- 52.Riggs-Gelasco PJ, Stemmler TL, Penner-Hahn JE. Coord. Chem. Rev. 1995;144:245–286. [Google Scholar]
- 53.Cotelesage JJ, Pushie MJ, Grochulski P, Pickering IJ, George GN. J. Inorg. Biochem. 2012;115:127–137. doi: 10.1016/j.jinorgbio.2012.06.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.