Abstract
Rationale
During embryogenesis, hematopoietic cells appear in the myocardium prior to the initiation of coronary formation. However, their role is unknown.
Objective
Here we investigate whether pre-existing hematopoietic cells are required for the formation of coronary vasculature.
Methods and Results
As a model of for hematopoietic cell deficient animals, we used Runx1 knockout embryos and Vav1-cre; R26-DTA embryos, latter of which genetically ablates 2/3 of CD45+ hematopoietic cells. Both Runx1 knockout embryos and Vav1-cre; R26-DTA embryos revealed disorganized, hypoplastic microvasculature of coronary vessels on section and whole-mount stainings. Furthermore, coronary explant experiments showed that the mouse heart explants from Runx1 and Vav1-cre; R26-DTA embryos exhibited impaired coronary formation ex vivo. Interestingly, in both models it appears that epicardial to mesenchymal transition is adversely affected in the absence of hematopoietic progenitors.
Conclusion
Hematopoietic cells are not merely passively transported via coronary vessel, but substantially involved in the induction of the coronary growth. Our findings suggest a novel mechanism of coronary growth.
Keywords: coronary formation, hematopoietic progenitors, epicardium, EMT, cardiac development
Introduction
Coronary artery disease (CAD) kills more than seven million people every year, making it the leading cause of deaths worldwide [1]. In the United States, CAD is the cause of death in one out of five people and bears an annual cost of 165 billion dollars [2]. Unlike many other organs, the heart has a very limited capacity for repair and regeneration after damage [3, 4]. Therefore, cardiac regeneration has been an increasingly important area of research in general, and development and regeneration of coronary arteries in particular. Despite remarkable advances in the study of the mechanisms of blood vessel development, little is known, and controversy still exists, on the cellular origins and developmental pathways that govern coronary artery development.
Initial findings suggest that coronary arteries originate from the aorta [5, 6]. However, only recent studies have shed light into the proepicardium, a transitory embryonic structure, as the origin of epicardium, which is the source of coronary artery building blocks [7, 8]. These experiments have paved the way to the current understanding that the epicardial cells, which undergo epithelial to mesenchymal transition (EMT) and migrate to the subepicardial space. Subsequently, these epicardially-derived cells (EPDCs) differentiate into coronary smooth muscle cells, perivascular fibroblasts and interstitial fibroblasts [9–13]. The origin of endothelial cells of coronary vessels is still an area of active research. Previous studies suggest that these endothelial cells originate from epicardium [14, 15], sinus venosus [16], or endocardium [17, 18].
Coronary plexus does not establish a connection with aortic root until E13.5 in mouse (Carenegie stage 18) [19]. Interestingly, previous studies demonstrate that isolated hematopoietic cells exist in the myocardium as early as E10.5, when coronary plexus has not yet established a connection with systemic circulation at aortic root [20, 21]. The appearance of isolated hematopoietic cells is followed by the formation of blood island-like structure. However, the role of pre-existing hematopoietic cells in the coronary formation is unknown. While hematopoietic cells are known to share their origins with endothelial cells, it is also proposed that hematopoietic cells in turn play an inductive role via paracrine factor(s) in the process of angiogenesis during development [22]. Disruption of Runx1/AML1 gene, a key regulator of definitive hematopoiesis [23], results in complete failure of definitive hematopoiesis of all cell lineages [24, 25], making them a model to study the role of angiogenesis up to E12.5 [24, 26]. Runx1 mutant mice lacking definitive hematopoiesis show abnormal vessels in many organs and the addition of hematopoietic cells rescues the phenotype ex vivo, suggesting that the hematopoietic cells are not just circulating through the vessels but are significant contributors to the vascular formation [22]. Here, using hematopoietic ablation mouse models, we show that development of coronary vessels are disrupted in the absence of definitive hematopoiesis. Furthermore, disruption of hematopoietic cells adversely affects EMT, a key process in coronary vessel formation. Together, our data suggest that hematopoietic cells pre-existing in the myocardium plays proangiogenic role during the formation of coronary vessels.
METHODS
Mice
Mice were maintained according to the Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (NIH Publication No. 85-23, revised 1996). Furthermore, housing, experiments, and protocols were performed according to the Institutional Approval for Appropriate Care and Use of Laboratory Animals by the UCLA Institutional Animal Care and Use Committee. All mouse strains were maintained in outbred backgrounds. Runx1+/− [24], vav1-cre [27, 28], Runx1-LacZ [26], and cre-inducible diphtheria toxin receptor mice [29] have been described previously.
Histological processing and immunostaining
At the appropriate calculated embryonic age, the pregnant females were sacrificed by cervical dislocation, and the embryos and hearts were carefully dissected followed by subsequent isolation in cold Phosphate Buffered Solution (PBS). The pericardial wall was removed and embryos were beheaded before fixation on ice in 4% paraformaldehyde/PBS or 2–4 hours depending on age. This was followed by washing embryos with PBS, and then cryoprotected in 30% sucrose/PBS solution overnight at 4°C. Next, tissues were placed in 1:1 30% sucrose/PBS and OCT (Sakura Torrance, CA) solution for 1 hour followed by 1 hour in 100% OCT compound at 4°C. Thereafter, the tissues were embedded in 100% OCT compounds, meticulously oriented in Peel-A-Way (Polysciences, Warrington, PA), followed by immediate freezing on dry ice with isopropanol and placed at −20°C. These blocks were cut to 8–10 µm thin sections with a Leica CM3050 S cryostat and collected on the glass slides sequentially to make serial sections. These sections were blocked with 10% normal goat serum; 0.1% TritonX-100. Primary antibody reactions were carried out in 5% normal goat serum for 1 hour at room temperature or at 4°C overnight. Secondary fluorescent conjugated antibody reactions were completed in 2% normal goat serum for 1 hour at room temperature. Primary antibodies used in this study were: rat anti-CD31 (BD Pharmingen, 1:200) and chicken anti-vimentin (Covance, 1:200). The following secondary antibodies were used: Biotinylated IgG antibodies (Vector Laboratories) for colorimetric staining, Alexa Fluor 488 (green), Alexa Fluor 594 (red)-conjugated secondary antibodies specific to the appropriate species were used (Invitrogen, 1:1000) for fluorescent staining. Next these slides were mounted with ProLong Gold DAPI media (Invitrogen, Carlsbad, CA) and analyzed by using AxioImager D1 (Carl Zeiss Microimaging, Inc). For non-fluorescent immunostaining, tissues were incubated with primary antibodies and biotinylated antibodies (Jackson ImmunoResearch Laboratories), and treated with Vectastain ABC Kit reagents (Vector Labs) followed by DAB substrate (Vector Labs). Standard protocols for Hematoxylin and Eosin (H&E) were used, and β-Galactosidase staining was carried out as previously described [30]. For the whole-mount images, CD31 positive areas were converted to 8-bit black and white images and analyzed as the ratio of the area covered by coronary vessels to three randomly selected mid-ventricular areas using ImageJ, for each heart analyzed (version 1.46r, Wayne Rasband, NIH, USA). The relative size of the major coronary vessels was expressed as the ratio of the coronary vessel length before it tapers down to capillary size to the length of the heart from the base to the apex of the heart.
Echocardiography
B- and M-mode ultrasound imaging was performed on E14.5 embryos after anesthesia with isoflurane of the pregnant mouse, using a high-resolution Vevo 2100 micro-ultrasound system with a 30 MHz transducer (Visual Sonics, Toronto, Ontario, Canada). The dimensions and functional parameters of left ventricle were measured from the short axis and mid-ventricular view with 2D oriented and M-mode imaging.
Heart explant culture
Hearts were dissected out at E11.5, atria separated and discarded from the ventricles, which were washed three times and then co-cultured on mouse OP9 stromal cells in 24 well plates for 7 days, in 500 µl of α-MEM (GIBCO/Invitrogen) containing 20% fetal bovine serum (Hyclone), 1% penicillin/streptomycin. The media was exchanged every 48 hours. For the rescue experiments, the Runx1 null hearts were co-cultured with CD45+ cells which were FACS-sorted from yolk sac of the wild-type littermate embryos. Approximately 5000 CD45+ cells were used for each Runx1 null heart, corresponding approximately to 2–3% of total number of yolk sac cells from each wild-type embryo. After 7 days the cultures were stained as described above, in immunostaining section. CD31 positive areas were analyzed as the ratio of the area covered by vessel sprouts to the entire microscope field area in 8-bit black and white images using ImageJ for each dissected heart (version 1.46r, Wayne Rasband, NIH, USA).
Fluorescent dye tracing of epicardial cells
Embryos were dissected at the appropriate embryonic age in PBS. Next, 10 µl of 5-(and 6-) carboxyfluorescein diacetate succinimidyl ester (CFSE) [31] was injected into the pericardial cavity (CFSE, Molecular Probes, C1157) as previously described. Expansion of the pericardium under pressure was observed. After 10 minutes at room temperature in the dark, hearts were removed and washed in PBS for 5 minutes. Next, hearts were randomly selected for fixation in 4% paraformaldehyde/PBS or for flow cytometry.
Flow cytometry for sorting cells of mouse tissues
After harvesting hearts from mouse embryos, they were washed three times and incubated at 37°C in a dissociation enzyme solution, pipetting every 10 minutes to a single-cell suspension. The enzyme solution contained 1% Penicillin/Streptomycin (Invitrogen, 15140-122), 10%Fetal Bovine Serum (Hyclone), collagenase 2mg/ml (Worthington, CLS-2), dispase 0.25mg/ml (Gibco, 17105-041), DNAase I (Invitrogen) in PBS. The cells were analyzed and sorted by a BD FACSAria with the rat anti-monoclonal antibody CD45APC (BD Pharmingen), CFSE, and 7-AAD (7-amino-actinomycinD, BD Biosciences), which was used to exclude non-viable cells.
Hematopoietic colony-forming assays
Sorted cells were cultured on OP9 stromal cells for 4 days in 48-well plates in 500 ml of a-MEM (Gibco/Invitrogen) containing 20% fetal bovine serum (Hyclone), 1% penicillin/ streptomycin and supplemented with stem cell factor (SCF, 50 ng/ml), inter- leukin-3 (IL-3, 5 ng/ml), IL-6 (5 ng/ml), thrombopoietin (5 ng/ml) and Flt-3 ligand (Flt-3L, 10 ng/ml) as previously described [32]. The cells were then dissociated mechanically from stroma by transfer pipette and filtered to remove the stromal cells (celltreck). The filtered cells were transferred into 1.5 ml methylcellulose with SCF, IL-6, IL-3 and EPO (MethoCult 3434, Stem Cell Technologies) supplemented with thrombopoietin (5 ng/ml) to determine multilineage potential. Colonies were scored 7–10 days later.
Gelatinase assay
Net matrix metalloproteinase activity (MMP) in heart tissue of embryonic E12.5 wild-type and Runx1 null hearts was determined using the EnzCheck Gelatinase Assay according to manufacturer’s instructions (Mocular Probes; Eugene, OR). MMP activity is reported as the rate of fluorescence increase over two hours normalized to the negative control containing DQ gelatin and reaction buffer with no extracted proteins from the heart tissue. Clostridium collagenase at a final concentration of 0.2 U/ml was used as a positive control.
Gene expression analysis by qRT-PCR
RNA was extracted from sorted cells using RNeasy Micro kit (Qiagen) according to manufacturer’s instructions. RNA was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (BioRad). qRT-PCR was performed using Lightcycler 480 SYBR Green I (Roche Applied Science). Forward and reverse primer sequences are summarized in Supplementary Table 1.
Statistics
Values are reported as mean ± SEM. P was calculated using the Student’s ttest. A P value < 0.05 was considered statistically significant. All calculations were performed using Prism 6 (GraphPad, La Jolla, CA).
RESULTS
Runx1 null embryos exhibit defective cardiac development and hypoplastic coronary vessels
Runx1 is a key regulator for definitive hematopoiesis expressed in and required for hemogenic endocardium. Recent report suggests that endocardial cells display hemogenic potential at around E8.5–10.5 [32]. Consistently, analyses of Runx1-lacZ embryos revealed that Runx1 is expressed in the endocardium, particularly in the cushion endocardium, where CD41+ hemogenic endocardial cells are enriched [32] (Supp Fig. 1). There are CD31+ cells in the endocardium, that also are Runx1 positive (Supp Fig. 1a, arrows). A subset of epicardial cells are also positive for lacZ (Supp Fig. 1). To examine whether endothelial cells in the epicardial/subepicardial layer display hemogenic activity, the epicardial cells of embryonic hearts at E11.5 were selectively labeled with CFSE injected into pericardial cavity (Supp Fig. 2A) and FACS-sorted for CFSE+/CD31+/CD41−/CD45− population. This endothelial population from epicardium does not include endocardial cells as CSFE dye labels only one or two cell layers of the epicardium (see also ref. [31] for sorting strategy; Supp Fig. 2B). Colony assay from this population showed no blood colony formation (Supp Fig. 2C), suggesting that, despite the fact that blood island-like structure develops in the epicardium at around E11.5, coronary endothelial cells are not hemogenic at least at this stage.
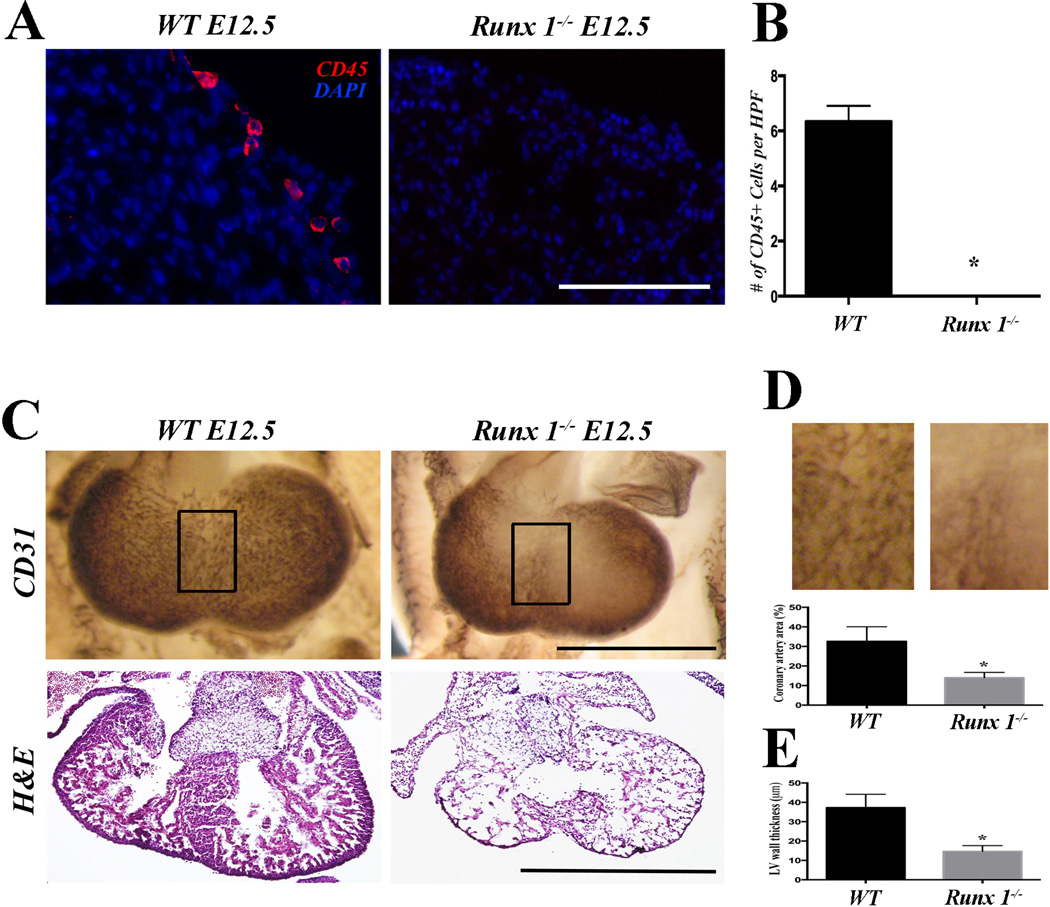
To examine the role of hematopoietic cells during coronary development, we first employed Runx1 mutant model. Runx1 null mice are embryonically lethal at around E12.5 with complete absence of definitive hematopoiesis [23, 24], and thus being used as a model for hematopoietic ablation [22]. Runx1 null embryos showed normal overall size and morphology, except for subcutaneous hemorrhages in the head at E11.5 as reported [22] (Supp Fig. 3A). Absence of definitive hematopoiesis in Runx1 null mice was confirmed by FACS analysis of the yolk sac for CD45 (Supp Fig. 3B). Although CD45+ hematopoietic cells are already identified in the wild-type heart at this stage [20, 21] (Figure 1A and B), Runx1 mutants showed no CD45 positive cells in the heart (Figures 1A and B). Interestingly, whole mount staining of E12.5 heart revealed that Runx1 null mice have an underdeveloped coronary plexus when compared to the wild-type (Figure 1C, first row, and 1D). Section staining confirmed that coronary vessels are smaller in the ventricular free wall (Figure 5B first row) and interventricular septum (Figure 5B second row) of Runx1-null embryos when compared to comparable regions in wild-type. In addition, morphological analysis by H&E staining of Runx1 null mice at E12.5 (Figure 1C, bottom row) revealed a thin compact myocardium and absence of ventricular septum when compared to wild-type (Figure 1E). These data suggest that Runx1 is required for the proper formation of coronary vessels during cardiogenesis, either cell-autonomously or non-autonomously.
Figure 1. Runx1 mutant embryos show defects in cardiac development and coronary formation.
(A) Representative CD45 staining of E12.5 Runx1-null embryos. No CD45+ cells are identified in the mutants. (B) Quantification of the number of CD45+ cells per high power field section (n=6). (C) Whole mount CD31 staining and H&E staining of E12.5 hearts. Coronary plexus formation is adversely affected in Runx1 null embryos (first row) when compared to wild-type embryos. H&E staining shows thin myocardium and ventricular septal defect (second row). (D) Quantification of the coronary plexus in the interventricular surface (boxed area in C; n=6). The density of the coronary plexus is reduced in Runx1 mutants. (E) Quantification of the thickness of ventricular free wall (n=6). Mutants show significantly thinner myocardium.
Scale bars: A, 0.1 mm. C, 1 mm. * p < 0.05.
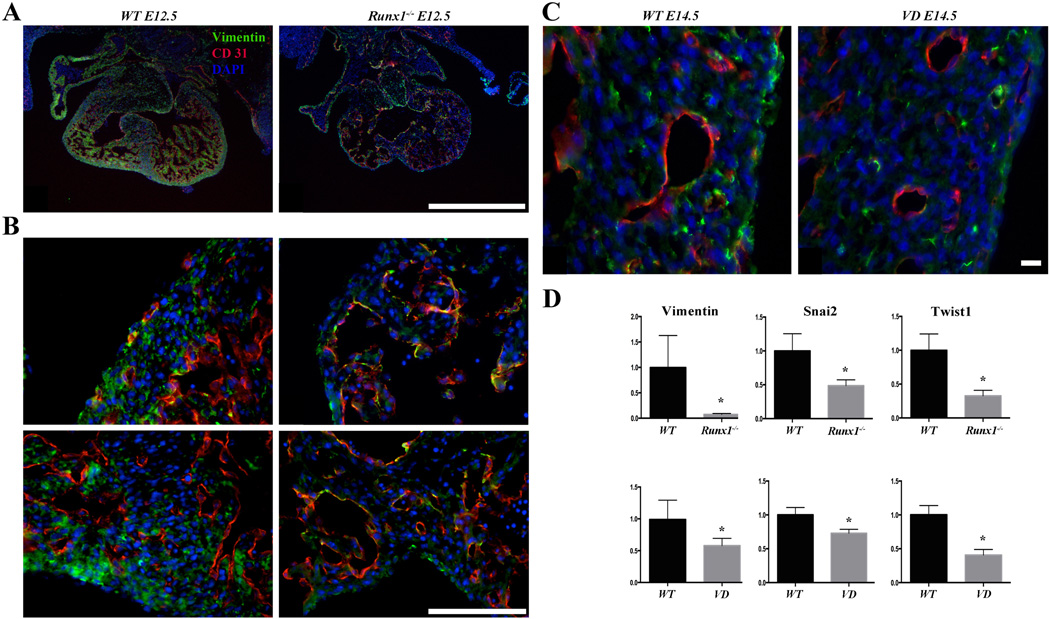
Figure 5. Hematopoietic deficient embryos show EMT defect.
(A) Representative vimentin (green) and CD31 (red) staining of E12.5 Runx1 mutant embryos. Markedly reduced vimentin staining is observed in the mutants (n=5). (B) High magnification of vimentin (green) and CD31 (red) staining of E12.5 Runx1 mutant embryos. The reduction of vimentin is observed throughout, including the lateral ventricular wall and interventricular septum as well (n=5). (C) Representative vimentin (green) and CD31 (red) staining of E14.5 VD embryos The reduction of vimentin is observed in VD hearts and it is associated with smaller size CD31+ vessels (n=4). (D) qRT-PCR of vimentin, snail2, and twist1. mRNA levels of vimentin, snail2, and twist1 are markedly downregulated in the epicardial cells of the E12.5 Runx1 mutant hearts (n=5) and E14.5 VD hearts (n=4).
Scale bars: two left columns, top row 1 mm, bottom rows and two right columns 0.1 mm. * p < 0.05.
Vav1-Cre; R26-DTA (VD) mice exhibit abnormal hypoplastic coronary vessels and absence of atrial septum
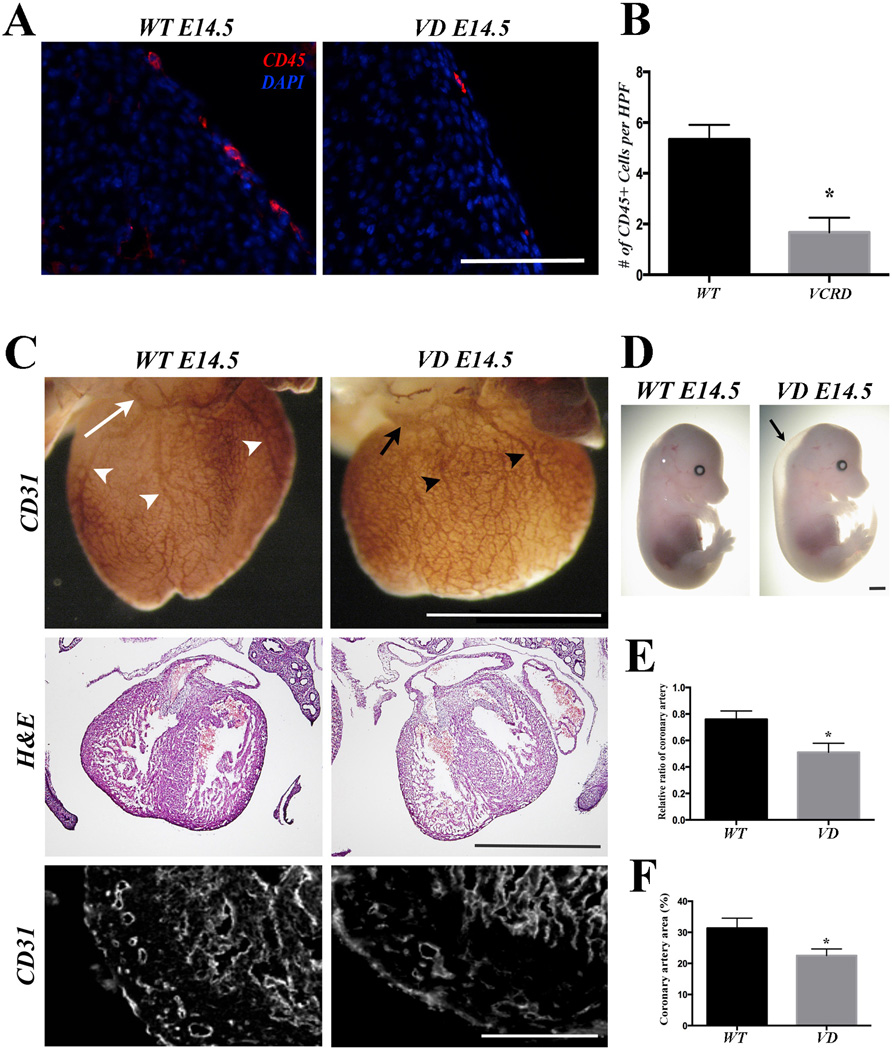
As Runx1 is expressed in the endocardial cells and a subset of epicardial cells, (Supp Fig. 1), Runx1 mutant data (Figure 1) do not distinguish whether Runx1 is required in cell autonomous fashion in the heart or non-autonomously in the existing hematopoietic cells via their interaction with coronary capillary. To test the latter possibility, we employed Vav1-Cretg; R26DTA/+ (VD) embryos in which Vav1+ hematopoietic cells were ablated by overexpression of Diphtheria toxin A [29]. Vav1 is a small GTPase that is specifically expressed in the hematopoietic cells [33]. Vav1-Cre labels 68% of hematopoietic cells at around E11.5 [28]. Importantly, analyses of Vav1-Cretg; R26YFP reporter/+ revealed that there is no YFP expression in the CD31 expressing cells in the endothelium or coronary vessels (Supp Fig. 4 arrowhead and arrow) [34], suggesting that CD31+ endocardial cells do not express Vav1 at E14.5 or any earlier stage. Therefore, ablating Vav1+ cells does not primarily affect CD31 positive cells in the endocardium or myocardium. VD mice are embryonically lethal at around E15.5, allowing the analyses of the role of hematopoietic cells on coronary development beyond E12.5 when Runx1 mutants die. At E14.5, VD embryos were normal in overall size, but exhibited subcutaneous edema (Figure 2D). As expected, VD heart showed significant reduction in the number of CD45+ cells by immunostaining (Figure 2A and B). H&E staining revealed a large atrial septal defect in VD (Supp Fig. 5, arrowhead) versus wild-type embryos (Supp Fig. 5, arrow), although ventricular septum and compact layer are well developed (Figure 2C second row). Interestingly, whole mount CD31 staining revealed that major coronary vessels of VD hearts are smaller and taper down half way to the length of the heart (Figure 2C first row, black arrowheads), when compared to wild-type hearts (Figure 2C first row, white arrowheads and E), the stem of the coronary artery appears to be comparable in both wild-type hearts and VD hearts (Figure 2C, first row, white and black arrows). In addition, coronary plexus was less dense in VD hearts (Figure 2C third row and F). These data suggest that ablation of hematopoietic progenitor cells in the VD model leads to aberrant coronary vessel formation.
Figure 2. VD embryos shows abnormal hypoplastic coronary vessels and structural heart defects.
(A) Representative CD45 staining of E14.5 VD embryos. Markedly reduced CD45+ cells are identified in the treated embryos. (B) Quantification of the number of the CD45+ cells per high power field section (n=5). (C) Whole mount CD31 staining, H&E staining, and section CD31 immunofluorescence staining of E14.5 hearts. Coronary plexus and major coronary vessels are adversely affected in the VD embryos (first row) when compared to wild-type embryos. H&E staining shows intact ventricular septum and myocardium (second row). Section CD31 immunostaining shows smaller coronary vessels (bottom row) in the VD embryos. (D) Representative E14.5 VD and wild-type embryos. VD embryos are of comparable size as wild-type embryos but exhibit subcutaneous edema (arrow). (E) Quantification of the relative length of the major coronary vessels in the wild-type hearts (white arrowheads in C) and VD hearts (black arrowheads in C) (n=5). (F) Quantification of the coronary plexus. The density of the coronary plexus is reduced in VD embryos (n=5).
Scale bars: A, 0.1 mm. C (first and second row), 2 mm. C (bottom row), 0.1 mm. * p < 0.05.
Vav1 mutant embryos have reduced cardiac systolic function
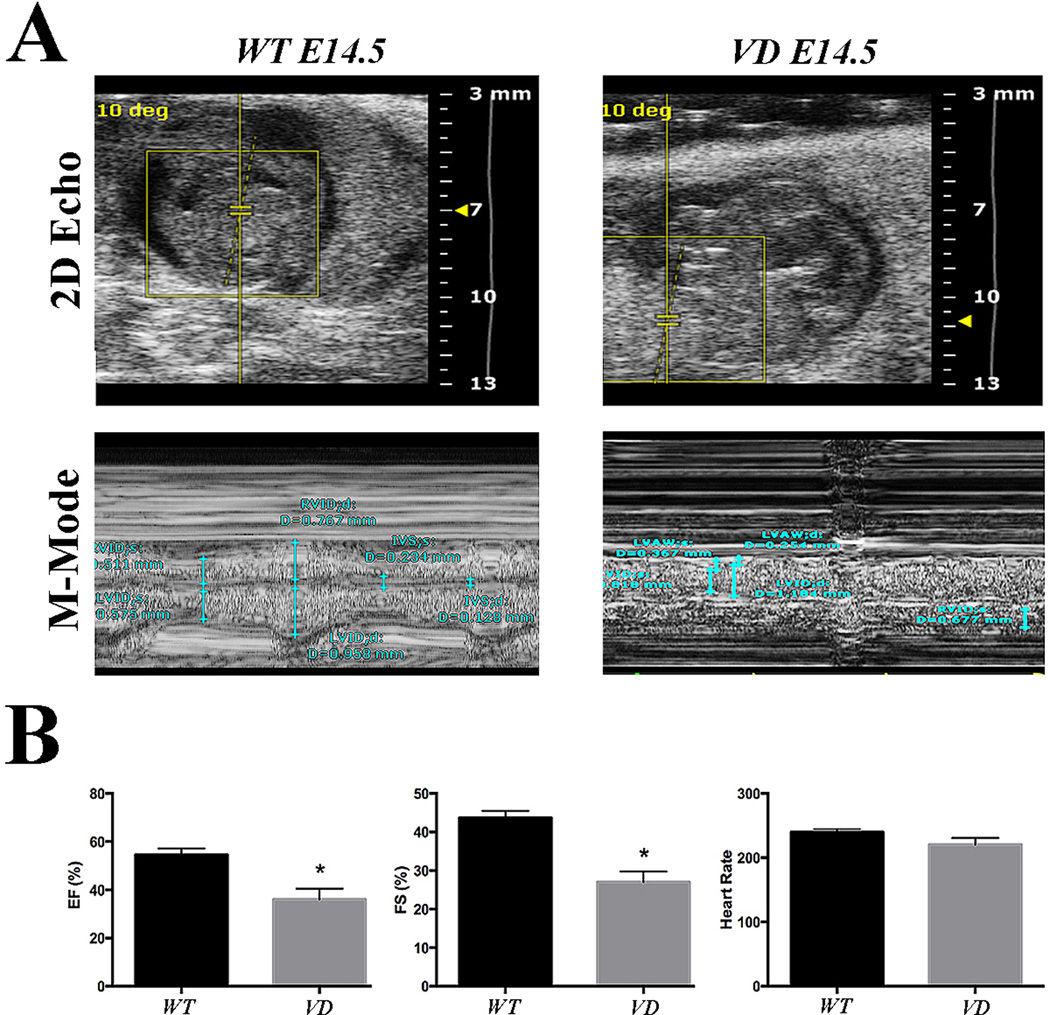
In order to evaluate the functional consequence of abnormal development of coronary vasculature, we performed echocardiographic studies on wild-type and VD embryos at E14.5. No arrhythmia or ventricular dilatation were detected during the procedure. However, left ventricular ejection fraction (EF) and fractional shortening (FS) were significantly reduced in VD (Figure 3A, B). These data suggest that VD embryos have reduced cardiac systolic function.
Figure 3. VD embryos reveal reduced cardiac function.
(A) Representative 2D images and M-mode analysis of E14.5 embryos. VD hearts show reduced systolic function (n=4). (B) Quantification of the ejection fraction (EF), fractional shortening (FS) and heart rate. The EF and FS are significantly reduced in the VD embryonic hearts (n=4). *p < 0.05
Ventricular explant cultures of Runx1 null and VD hearts exhibit reduced CD31 positive sprouting and endothelial networking
It is possible that the coronary phenotypes described above could be a result rather than a cause of potential hemodynamic abnormalities secondary to abnormal hematopoiesis. To exclude this possibility, we performed ex-vivo experiments. Ventricles were explanted from Runx1 null and VD embryos and control wild-type embryos at E11.5, and cultured for 7 days on OP9 feeder cells, which support the growth of both hematopoietic cells [35] and cardiomyocytes [36]. After 7 days, the cultures were stained for the endothelial marker CD31 (Figure 4A and C). The area covered by CD31 positive endothelial sprouting and networks were analyzed, showing a reduced network in Runx1 null (Figure 4B) and VD explants (Figure 4D). Furthermore, co-culture of Runx1 null explants with CD45+ cells leads to the rescue of the CD31 positive endothelial sprouting when compared to wild-type explants (Supplementary Figure 6). These data suggest that ablation of hematopoietic progenitors resulted in hypoplastic coronary vasculature independently from potential hemodynamic alteration.
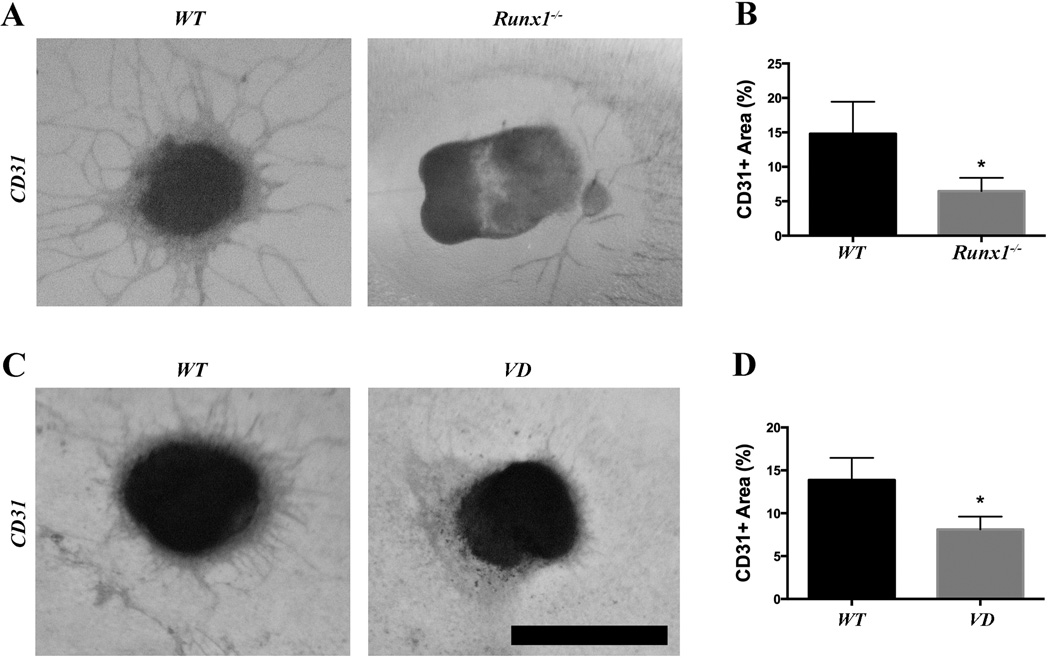
Figure 4. Runx1 and VD heart explants show reduced endothelial sprouting.
(A) Representative ventricular explants from Runx1 mutant embryos. Sprouting of CD31+ cells is reduced in the mutants. (B) Quantification of the CD31+ sprouting area from Runx1 mutants (n=6). (C) Representative ventricular explants from VD embryos. CD31+ sprouting is reduced in the VD explants. (D) Quantification of the CD31+ sprouting area from VD explants (n=5).
Scale bar: 1 mm. * p < 0.05.
Epicardial EMT is adversely affected in the absence of hematopoietic progenitor cells
Epicardial EMT is crucial for proper coronary development [9]. To explore how deficiency of definitive hematopoiesis impacts the coronary growth, we examined the marker expression in the two mouse models. First, immunofluorescence staining for vimentin, a marker for mesenchyme, revealed that its levels are reduced in Runx1 null mice as compared to wild-type (Figure 5A and B). The reduction in the number of mesenchymal cells suggests that EMT is adversely affected in Runx1 null mice. Reduced numbers of vimentin positive cells are also observed in the free left ventricular wall of the VD embryos at E14.5 (Figure 5C) when compared to wild-type. Therefore, we proceeded with FACS sorting of epicardial cells to analyze mRNA levels of EMT factors in E12.5 Runx1 mutant embryos and E14.5 VD embryos. qRT-PCR for Wt1, a marker of epicardial cells, shows that our FACS sorting successfully separates the epicardial cells (Supp Fig. 2B). Next, qRT-PCR of Runx1 and VD epicardial cells shows a marked decrease of Vimentin, Snail2, and Twist1 (Figure 5D). Snail1, a key transcription factor known to affect EMT, appears not to be affected (data not shown). This is consistent with prior studies showing that the EMT process in epicardial cells is independent of Snail1 [37]. E-cadherin (a marker for epithelial identity) or Notch signaling genes (Jagged1, Delta3, Delta4 and Hey1; involved in EMT and coronary development [38] [39]) showed no significant change in the mutant samples (data not shown). Expression level of Hif1a was not increased, suggesting that such abnormalities of coronary vessels are not secondary to hypoxia (data not shown). Furthermore, in order to determine if changes in MMP activity may affect the coronary growth in the Runx1 null heart, we analyzed the MMP activity of these hearts and compared them to wildtype hearts. The MMP activity in the Runx1 null heart is similar to the activity in wild-type hearts (Supplementary Figure 6). Together, absence of hematopoietic progenitor cells leads to a decrease of epicardially derived mesenchymal cells.
DISCUSSION
Circulatory system has evolved to support the growth and homeostasis of the organisms. The induction of cardiac, vascular and hematopoietic lineages is mutually linked during the formation of organs. The hematopoietic cells are not just passengers passively transferred through the vascular conduit, but substantially involved in the development and repair of the organs through paracrine factors [22, 40–43]. The process of coronary formation is no exception. Our study demonstrates a role of hematopoietic progenitor cells for proper epicardial EMT and subsequent coronary vessel development. In Runx1 null mice, we observed defective plexus formation and structural heart defects, which precede coronary connections to the aorta, suggesting that such abnormalities are unlikely to be secondary to any hemodynamic alterations. Furthermore, our explant cardiac culture data show reduced CD31+ sprouting in mutant explants, therefore such findings are not likely secondary to hemodynamic abnormalities.
Previous studies have shown that hematopoietic progenitors play an inductive role in angiogenic sprout of dorsal aorta [22, 39]. One possible mechanism is the induction of EMT by the presence of hematopoietic progenitor cells. A direct role of Runx1 in the upregulation of mesenchymal markers in the epicardial cells and facilitation of EMT cannot be ruled out from this study. Furthermore, it has been suggested that endocardial cells play a role in coronary development [17]. Our results suggest that a subset of endocardial cells express Runx1. As a result, we cannot exclude a non-hemogenic direct role of Runx1 in coronary development via the endocardium. However, aberrant epicardial EMT occurs when hematopoiesis is affected by ablating Vav1 expressing cells, suggesting that hematopoietic cells play an inductive role during the EMT process. This is most likely through the secreted factor(s), although the origin and the nature of such hematopoietic cells are also yet to be identified. Reassuringly, co-culturing of mutant hearts with CD45+ cells rescues the CD31+ sprouting in the mutant hearts. It is well established that epicardial cells give rise to coronary smooth muscle cells. Epicardial cells upregulate mesenchymal markers (such as vimentin), delaminate, and migrate in the subepicardial space [7, 10, 44]. These cells then differentiate into smooth muscle as well as adventitial cells of coronary arteries. Consistent with these events, we observed a reduction of vimentin in the hematopoietic progenitor cell deficient mice. In addition to defective EMT, our observations of the reduction of vimentin-positive cells could also be secondary to abnormal proliferative activity or cellular survival of vimentin-positive mesenchymal cells, although our data suggests that this process is not mediated by any alterations in MMP activity. In accordance with prior studies [37], Snail1, a transcription factor closely involved in EMT, does not appear to participate in epicardium to mesenchymal transition. In our study, Snail2 and Twist1 are downregulated in the absence of hematopoietic cells. The role of Snail2 in epicardial EMT is controversial [37, 45]. This could be due to the different embryonic time points and techniques used [37]. Several studies have suggested that Snail2 is an essential component during Twist1 mediated EMT [10, 46]. The addition of hematopoietic component will contribute to the deeper understanding of the coronary development.
Coronary collaterals to the jeopardized myocardium mitigate myocardial infarction and improve the survival. Whereas the anti-angiogenic therapies have potential promise for treatment of certain cancers and of age-related macular degeneration, the pro-angiogenic therapy for coronary and peripheral artery diseases have not so far proven successful, indicating the complexity of the regulation of the blood vessel growth. Numerous studies in animal models of myocardial infarction/heart failure and clinical trials has shown the therapeutic benefit of cytokines and growth factors [47] including colony-stimulating factors (G-CSF and GM-CSF) [48–54], VEGFA [55–57], FGF [58–60], stem cell factor (SCF) [61–63], etc. However, to date, randomized, controlled clinical trials have not reproduced the efficacy observed in pre-clinical and small-scale clinical investigations. Nevertheless, the list of promising cytokines continues to grow, with the precise mechanism of the biological effects of pleiotropic cytokines and growth factors left unknown. The fact that only a few of the cytokine receptors are expressed in the cardiomyocytes or coronary vessels raises the possibility that these cytokines may exert cardioprotective effects through hematopoietic cells. Further identification and characterization of the signaling pathways from cells originating from the hematopoietic progenitors can help us not only understand the tightly regulated process of coronary development but also engineer new avenues for the treatment of ischemic heart disease and anomalous coronary arteries, by charting new frontiers in the induction of new coronary vessels.
Supplementary Material
Highlights.
We studied the role of hematopoietic progenitor cells on the coronary development.
Mice lacking hematopoietic progenitor cells have hypoplastic coronary vessels.
These cells are not merely passengers, but substantially involved in coronary growth.
Acknowledgments
Founding sources: This work is funded by AHA (GRNT9420039) to A.N. NIH/ Child Health Research Center and the UCLA-CDI (Children’s Discovery and Innovation Institute) M.T. G.L. is supported by the UCLA STAR program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures and conflicts of interest: None
References
- 1.Mathers C, Fat DM, Boerma JT World Health Organization. The global burden of disease: 2004 update. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 2.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, et al. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:e21–e181. doi: 10.1161/CIRCULATIONAHA.108.191261. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aikawa E, Kawano J. Formation of coronary arteries sprouting from the primitive aortic sinus wall of the chick embryo. Experientia. 1982;38:816–818. doi: 10.1007/BF01972290. [DOI] [PubMed] [Google Scholar]
- 6.Conte G, Pellegrini A. On the development of the coronary arteries in human embryos, stages 14–19. Anatomy and embryology. 1984;169:209–218. doi: 10.1007/BF00303151. [DOI] [PubMed] [Google Scholar]
- 7.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Developmental biology. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. The International journal of developmental biology. 2002;46:1005–1013. [PubMed] [Google Scholar]
- 9.Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circulation research. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circulation research. 2012;110:1628–1645. doi: 10.1161/CIRCRESAHA.111.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sridurongrit S, Larsson J, Schwartz R, Ruiz-Lozano P, Kaartinen V. Signaling via the Tgf-beta type I receptor Alk5 in heart development. Developmental biology. 2008;322:208–218. doi: 10.1016/j.ydbio.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, et al. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes & development. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majesky MW. Development of coronary vessels. Current topics in developmental biology. 2004;62:225–259. doi: 10.1016/S0070-2153(04)62008-4. [DOI] [PubMed] [Google Scholar]
- 15.Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, et al. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Developmental cell. 2012;22:639–650. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151:1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, et al. Vessel formation. De novo formation of a distinct coronary vascular population in neonatal heart. Science. 2014;345:90–94. doi: 10.1126/science.1251487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchins GM, Kessler-Hanna A, Moore GW. Development of the coronary arteries in the embryonic human heart. Circulation. 1988;77:1250–1257. doi: 10.1161/01.cir.77.6.1250. [DOI] [PubMed] [Google Scholar]
- 20.Ratajska A, Czarnowska E, Kolodzinska A, Kluzek W, Lesniak W. Vasculogenesis of the embryonic heart: origin of blood island-like structures. The anatomical record Part A, Discoveries in molecular, cellular, and evolutionary biology. 2006;288:223–232. doi: 10.1002/ar.a.20311. [DOI] [PubMed] [Google Scholar]
- 21.Jankowska-Steifer E, Madej M, Niderla-Bielinska J, Ruminski S, Flaht-Zabost A, Czarnowska E, et al. Vasculogenic and hematopoietic cellular progenitors are scattered within the prenatal mouse heart. Histochemistry and cell biology. 2014 doi: 10.1007/s00418-014-1269-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takakura N, Watanabe T, Suenobu S, Yamada Y, Noda T, Ito Y, et al. A role for hematopoietic stem cells in promoting angiogenesis. Cell. 2000;102:199–209. doi: 10.1016/s0092-8674(00)00025-8. [DOI] [PubMed] [Google Scholar]
- 23.Okuda T, Nishimura M, Nakao M, Fujita Y. RUNX1/AML1: a central player in hematopoiesis. International journal of hematology. 2001;74:252–257. doi: 10.1007/BF02982057. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 26.North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 27.Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- 28.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nature methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 30.Nakano H, Williams E, Hoshijima M, Sasaki M, Minamisawa S, Chien KR, et al. Cardiac origin of smooth muscle cells in the inflow tract. Journal of molecular and cellular cardiology. 2011;50:337–345. doi: 10.1016/j.yjmcc.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano H, Liu X, Arshi A, Nakashima Y, van Handel B, Sasidharan R, et al. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nature communications. 2013;4:1564. doi: 10.1038/ncomms2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogilvy S, Metcalf D, Gibson L, Bath ML, Harris AW, Adams JM. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94:1855–1863. [PubMed] [Google Scholar]
- 34.Balmer GM, Bollini S, Dube KN, Martinez-Barbera JP, Williams O, Riley PR. Dynamic haematopoietic cell contribution to the developing and adult epicardium. Nature communications. 2014;5:4054. doi: 10.1038/ncomms5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakano T, Kodama H, Honjo T. In vitro development of primitive and definitive erythrocytes from different precursors. Science. 1996;272:722–724. doi: 10.1126/science.272.5262.722. [DOI] [PubMed] [Google Scholar]
- 36.Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, et al. IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature. 2008;454:345–349. doi: 10.1038/nature07027. [DOI] [PubMed] [Google Scholar]
- 37.Casanova JC, Travisano S, de la Pompa JL. Epithelial-to-mesenchymal transition in epicardium is independent of Snail1. Genesis. 2013;51:32–40. doi: 10.1002/dvg.22353. [DOI] [PubMed] [Google Scholar]
- 38.Grieskamp T, Rudat C, Ludtke TH, Norden J, Kispert A. Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circulation research. 2011;108:813–823. doi: 10.1161/CIRCRESAHA.110.228809. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez-Bergeron DL, Runge A, Adelman DM, Gohil M, Simon MC. HIF-dependent hematopoietic factors regulate the development of the embryonic vasculature. Developmental cell. 2006;11:81–92. doi: 10.1016/j.devcel.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David Dong ZM, Aplin AC, Nicosia RF. Regulation of angiogenesis by macrophages, dendritic cells, and circulating myelomonocytic cells. Current pharmaceutical design. 2009;15:365–379. doi: 10.2174/138161209787315783. [DOI] [PubMed] [Google Scholar]
- 41.Zhou P, Wirthlin L, McGee J, Annett G, Nolta J. Contribution of human hematopoietic stem cells to liver repair. Seminars in immunopathology. 2009;31:411–419. doi: 10.1007/s00281-009-0166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Zhao RC, Tredget EE. Concise review: bone marrow-derived stem/progenitor cells in cutaneous repair and regeneration. Stem cells. 2010;28:905–915. doi: 10.1002/stem.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stefater JA, 3rd, Lewkowich I, Rao S, Mariggi G, Carpenter AC, Burr AR, et al. Regulation of angiogenesis by a non-canonical Wnt-Flt1 pathway in myeloid cells. Nature. 2011;474:511–515. doi: 10.1038/nature10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Developmental biology. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 45.Takeichi M, Nimura K, Mori M, Nakagami H, Kaneda Y. The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bidirectional regulation of Slug in murine primary epicardial cells. PloS one. 2013;8:e57829. doi: 10.1371/journal.pone.0057829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer research. 2011;71:245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanganalmath SK, Abdel-Latif A, Bolli R, Xuan YT, Dawn B. Hematopoietic cytokines for cardiac repair: mobilization of bone marrow cells and beyond. Basic research in cardiology. 2011;106:709–733. doi: 10.1007/s00395-011-0183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ince H, Petzsch M, Kleine HD, Schmidt H, Rehders T, Korber T, et al. Preservation from left ventricular remodeling by front-integrated revascularization and stem cell liberation in evolving acute myocardial infarction by use of granulocyte-colony-stimulating factor (FIRSTLINE-AMI) Circulation. 2005;112:3097–3106. doi: 10.1161/CIRCULATIONAHA.105.541433. [DOI] [PubMed] [Google Scholar]
- 49.Valgimigli M, Rigolin GM, Cittanti C, Malagutti P, Curello S, Percoco G, et al. Use of granulocyte-colony stimulating factor during acute myocardial infarction to enhance bone marrow stem cell mobilization in humans: clinical and angiographic safety profile. European heart journal. 2005;26:1838–1845. doi: 10.1093/eurheartj/ehi289. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Tagil K, Ripa RS, Nilsson JC, Carstensen S, Jorgensen E, et al. Effect of mobilization of bone marrow stem cells by granulocyte colony stimulating factor on clinical symptoms, left ventricular perfusion and function in patients with severe chronic ischemic heart disease. International journal of cardiology. 2005;100:477–483. doi: 10.1016/j.ijcard.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Boyle AJ, Whitbourn R, Schlicht S, Krum H, Kocher A, Nandurkar H, et al. Intracoronary high-dose CD34+ stem cells in patients with chronic ischemic heart disease: a 12-month follow-up. International journal of cardiology. 2006;109:21–27. doi: 10.1016/j.ijcard.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 52.Engelmann MG, Theiss HD, Hennig-Theiss C, Huber A, Wintersperger BJ, Werle-Ruedinger AE, et al. Autologous bone marrow stem cell mobilization induced by granulocyte colony-stimulating factor after subacute ST-segment elevation myocardial infarction undergoing late revascularization: final results from the G-CSF-STEMI (Granulocyte Colony-Stimulating Factor ST-Segment Elevation Myocardial Infarction) trial. Journal of the American College of Cardiology. 2006;48:1712–1721. doi: 10.1016/j.jacc.2006.07.044. [DOI] [PubMed] [Google Scholar]
- 53.Ripa RS, Jorgensen E, Wang Y, Thune JJ, Nilsson JC, Sondergaard L, et al. Stem cell mobilization induced by subcutaneous granulocyte-colony stimulating factor to improve cardiac regeneration after acute ST-elevation myocardial infarction: result of the double-blind, randomized, placebo-controlled stem cells in myocardial infarction (STEMMI) trial. Circulation. 2006;113:1983–1992. doi: 10.1161/CIRCULATIONAHA.105.610469. [DOI] [PubMed] [Google Scholar]
- 54.Zohlnhofer D, Ott I, Mehilli J, Schomig K, Michalk F, Ibrahim T, et al. Stem cell mobilization by granulocyte colony-stimulating factor in patients with acute myocardial infarction: a randomized controlled trial. Jama. 2006;295:1003–1010. doi: 10.1001/jama.295.9.1003. [DOI] [PubMed] [Google Scholar]
- 55.Hedman M, Hartikainen J, Syvanne M, Stjernvall J, Hedman A, Kivela A, et al. Safety and feasibility of catheter-based local intracoronary vascular endothelial growth factor gene transfer in the prevention of postangioplasty and in-stent restenosis and in the treatment of chronic myocardial ischemia: phase II results of the Kuopio Angiogenesis Trial (KAT) Circulation. 2003;107:2677–2683. doi: 10.1161/01.CIR.0000070540.80780.92. [DOI] [PubMed] [Google Scholar]
- 56.Henry TD, Annex BH, McKendall GR, Azrin MA, Lopez JJ, Giordano FJ, et al. The VIVA trial: Vascular endothelial growth factor in Ischemia for Vascular Angiogenesis. Circulation. 2003;107:1359–1365. doi: 10.1161/01.cir.0000061911.47710.8a. [DOI] [PubMed] [Google Scholar]
- 57.Kastrup J, Jorgensen E, Ruck A, Tagil K, Glogar D, Ruzyllo W, et al. Direct intramyocardial plasmid vascular endothelial growth factor-A165 gene therapy in patients with stable severe angina pectoris A randomized double-blind placebo-controlled study: the Euroinject One trial. Journal of the American College of Cardiology. 2005;45:982–988. doi: 10.1016/j.jacc.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 58.Grines CL, Watkins MW, Helmer G, Penny W, Brinker J, Marmur JD, et al. Angiogenic Gene Therapy (AGENT) trial in patients with stable angina pectoris. Circulation. 2002;105:1291–1297. doi: 10.1161/hc1102.105595. [DOI] [PubMed] [Google Scholar]
- 59.Grines CL, Watkins MW, Mahmarian JJ, Iskandrian AE, Rade JJ, Marrott P, et al. A randomized, double-blind, placebo-controlled trial of Ad5FGF-4 gene therapy and its effect on myocardial perfusion in patients with stable angina. Journal of the American College of Cardiology. 2003;42:1339–1347. doi: 10.1016/s0735-1097(03)00988-4. [DOI] [PubMed] [Google Scholar]
- 60.Henry TD, Grines CL, Watkins MW, Dib N, Barbeau G, Moreadith R, et al. Effects of Ad5FGF-4 in patients with angina: an analysis of pooled data from the AGENT-3 and AGENT-4 trials. Journal of the American College of Cardiology. 2007;50:1038–1046. doi: 10.1016/j.jacc.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 61.Norol F, Merlet P, Isnard R, Sebillon P, Bonnet N, Cailliot C, et al. Influence of mobilized stem cells on myocardial infarct repair in a nonhuman primate model. Blood. 2003;102:4361–4368. doi: 10.1182/blood-2003-03-0685. [DOI] [PubMed] [Google Scholar]
- 62.Ohtsuka M, Takano H, Zou Y, Toko H, Akazawa H, Qin Y, et al. Cytokine therapy prevents left ventricular remodeling and dysfunction after myocardial infarction through neovascularization. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18:851–853. doi: 10.1096/fj.03-0637fje. [DOI] [PubMed] [Google Scholar]
- 63.Dawn B, Guo Y, Rezazadeh A, Huang Y, Stein AB, Hunt G, et al. Postinfarct cytokine therapy regenerates cardiac tissue and improves left ventricular function. Circulation research. 2006;98:1098–1105. doi: 10.1161/01.RES.0000218454.76784.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.