Abstract
Diffuse large B cell lymphoma (DLBCL) is a hematological cancer associated with an aggressive clinical course. The predominant subtypes of DLBCL display features of chronic or tonic B cell antigen receptor (BCR) signaling. However, it is not known if the spatial organization of the BCR contributes to regulation of pro-survival signaling pathways and cell growth. Here, we show that primary DLBCL tumors and patient-derived DLBCL cell lines contain high levels of phosphorylated Ezrin-Radixin-Moesin (ERM) proteins. The surface BCRs in both activated B cell and germinal B cell subtype DLBCL cells co-segregate with phosphoERM suggesting that the cytoskeletal network may support localized BCR signaling and contribute to pathogenesis. Indeed, ablation of membrane-cytoskeletal linkages by dominant negative mutants, pharmacological inhibition and knockdown of ERM proteins disrupted cell surface BCR organization, inhibited proximal and distal BCR signaling, and reduced the growth of DLBCL cell lines. In vivo administration of the ezrin inhibitor retarded the growth of DLBCL tumor xenografts, concomitant with reduction in intratumor phosphoERM levels, dampened pro-survival signaling and induction of apoptosis. Our results reveal a novel ERM-based spatial mechanism that is coopted by DLBCL cells to sustain tumor cell growth and survival.
Keywords: DLBCL, BCR, signaling, Ezrin-Radixin-Moesin, cytoskeleton
Introduction
DLBCL accounts for approximately 40% of all non-Hodgkin’s lymphomas.1 Gene expression profiling has revealed the existence of two germinal center (GC)-derived molecular subtypes of DLBCL, including germinal center B cell-like (GCB) and activated B cell-like (ABC). These subtypes differ in B cell differentiation stage, oncogenic aberrations, and responsiveness to standard therapy.2–6 Patients with ABC-DLBCL have inferior response to conventional chemotherapy with the B cell-depleting CD20 monoclonal antibody rituximab (i.e., R-CHOP) and poorer overall survival.7, 8 On the other hand, DLBCL patients with GCB subtype show an initial favorable response to R-CHOP but relapses are fatal,7, 8 necessitating the development of alternate/combinatorial treatment strategies, and a better understanding of mechanisms involved in the pathobiology of DLBCL. ABC-DLBCL pathogenesis is associated with activating mutations in CD79A and CD79B, which leads to chronic BCR signaling, or in CARD11 or TAK1 proteins, causing constitutive activation of NF-κB and increased B cell survival.9–12 GCB-DLBCLs exhibit activating mutations in Bcl2 and constitutive activation of the phosphatidylinositol 3-kinase (PI3K) pathway, which is a key element of tonic BCR signaling in B cells.3 Targeting of critical activating enzymes in BCR signaling10, 12–15 has demonstrated that this pathway may be a viable therapeutic target in DLBCL. Thus, identification of novel regulators of B cell activation in the germinal center should aid in expanding drug development efforts.
Ezrin-Radixin-Moesin (ERM) family proteins crosslink the plasma membrane and actin cytoskeleton, and facilitate several key cellular processes, including membrane dynamics, adhesion, cell survival, cell motility and morphogenesis.16 An N-terminal four-point-one, ezrin, radixin, moesin (FERM) domain anchors ERMs in the plasma membrane through direct or indirect interaction with transmembrane proteins, and a C-terminal domain interacts with cortical actin filaments. In the inactive/dormant state, actin and membrane-binding sites are masked by intramolecular interaction between the N- and C-termini.16 Functional activation of ERM proteins occurs upon phosphorylation of a conserved threonine residue at the C-terminus.17, 18 Phosphorylated ezrin and actin filaments collaborate to form the structural core of surface microvilli19, 20 where they can regulate diverse processes, including signal transduction.21 We have previously shown that in naïve B cells, ezrin exists in an open/active conformation defined by basal phosphorylation at Thr567. Ezrin and actin physically collaborate to control the diffusion of the BCR in the absence of antigen.22 BCR stimulation by antigen induces its transient dephosphorylation leading to uncoupling of the actin cytoskeleton from the plasma membrane.23, 24 Simultaneously, B cells spread to gather antigen and the BCR organizes itself into microclusters and recruits key signaling proteins.25–28 Local rephosphorylation of ERM proteins reinstates the membrane-cytoskeletal barriers, trapping BCR microclusters within compartments and facilitating signal transduction29. Together, these features of ezrin enable it to regulate antigen-induced BCR signal transduction and humoral immunity.30
Interestingly, ABC-DLBCL cells with mutations in the BCR exhibit spontaneous BCR microclustering, whereas in GCB-DLBCL cells the BCR is present in surface microvilli.9 In this study, we investigated the hypothesis that regulation of BCR organization by ERM proteins mediates DLBCL pathogenesis. We report that primary germinal center B cells, lymphoma biopsies from DLBCL patients and representative DLBCL cell lines contain high levels of ERM phosphorylation. Interference with ERM function decreases DLBCL cell growth and survival by disrupting BCR localization and impairing pro-survival signaling pathways. Therefore, we suggest that ERM proteins are novel molecular mediators of DLBCL pathogenesis whose activity may be targeted for intervention in the future.
Materials and Methods
Human tissues
Frozen DLBCL patient tumor biopsy samples were obtained according to guidelines approved by the Institutional Review Board of the Cleveland Clinic. DLBCL patients were identified as GCB or ABC based on application of the Hans immunochemistry algorithm to fresh-frozen paraffin-embedded tumor biopsies.31 B cells were purified from PBMCs of healthy individuals using the human B cell isolation kit (Miltenyi Biotec). Tonsils were obtained from healthy individuals undergoing routine tonsillectomies in accordance with ethical recommendations. After mincing, tonsillar mononuclear cells were isolated by Ficoll density centrifugation (GE Healthcare). GC B cells were enriched by magnetic cell separation with anti-CD77, MARM-4 (Abcam) and IgG1 microbeads (Miltenyi Biotec).
Cell lines, transfection and knockdown
All DLBCL cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, except for OCI-LY-10 cells, which were grown in IMDM and 10% human serum. DLBCL cell lines were transiently transfected using the Lonza nucleofector and nucleofection kit V. Briefly, three million cells and 3 µg DNA were resuspended in 100 µl solution V, and nucleofected using program A-020. The nucleofected cells were plated in 2 ml DLBCL growth medium until analyzed. The plasmid constructs used were wild type ezrin in plasmid pEYFP (Ez-WT),32 a truncation mutant of ezrin in plasmid pCB6 (Ez-DN, a.a. 1–310 containing the FERM domain but lacking the T567 phosphorylation site,33 phosphomimetic mutant T567D in pEYFP (Ez-TD)32 and a non-phosphorylatable mutant T567A in pEYFP (Ez-TA). For siRNA knockdown pools of untagged Accell siRNAs against human ezrin, radixin, moesin and scrambled siRNAs were purchased from Dharmacon RNAi Technologies. Briefly, DLBCL cells (1.5 million/ml) were incubated with the Accell delivery medium containing 1–2 µM of siRNAs at 37 °C according to the manufacturer's instructions (this procedure does not require electroporation). Following overnight incubation, 1 ml of fresh delivery medium was added to each well. Cells were counted every day up to six days using trypan blue to assess viability. Lysates collected at 96 hours were analyzed by immunoblotting with antibodies to ERM proteins, pThrERM, actin, pAkt, Akt, pIκB and IκB.
Lysis and immunoblotting
DLBCL cell lines were either treated with DMSO, NSC668394 (Calbiochem), or NSC305787 (obtained from the National Cancer Institute), or transfected with different ezrin constructs, followed by lysis as described.34 Freshly harvested tumor tissues from DMSO- and ezrin-inhibitor treated mice were homogenized in lysis buffer34 using a TissueRuptor (Qiagen), and lysates were analyzed by immunoblotting.
Cell growth, apoptosis and flow cytometry assays
DLBCL cell lines were either transfected with different mutant constructs of ezrin, or incubated with ezrin inhibitors, or siRNAs, or treated with the IκB kinase (IKK) inhibitor PS-1145 (Sigma-Aldrich) and viable cell counts were obtained using trypan blue staining. For tumor cell apoptosis assay, single-cell suspensions were obtained by placing tumors in 1 ml dissociation buffer (100 U/ml collagenase type I and 100 µg/ml DNase in RPMI+10% FBS) and incubating at 37°C for 30 min. Cell suspensions were passed through a 40 µ strainer to harvest mononuclear cells, and stained with Annexin V-phycoerythrin conjugate. Surface BCR levels were analyzed by fixing the DLBCL cells with 4% paraformaldehyde and labeling IgM or IgG with Rhodamine Red™-X-conjugated goat anti-human IgM (µ chain-specific) Fab fragment or AlexaFluor 488-conjugated goat anti-human IgG (H+L specific) Fab fragment (Jackson Immunoresearch) for 30 min at 4°C. The cells were subjected to flow cytometry on a FACSCalibur and data analyzed using FlowJo software (Treestar, Inc.).
Mouse xenograft assay
Ten million OCI-LY-10 or SU-DHL-6 cells were suspended in PBS and injected subcutaneously into the flanks of six week-old female NSG mice. After the tumor size reached ~200 mm3, half the mice were injected intraperitoneally with 5% DMSO and the other half with 13.5 mg/kg/day of NSC668394. Tumor volume was determined by digital calipering three times a week and calculated using the formula (smallest diameter2 X largest diameter)/2. Mice were periodically weighed and observed for discomfort and grooming behavior. Animal studies were conducted in accordance with guidelines approved by the Cleveland Clinic Institutional Animal Care and Use Committee.
Immunocytochemistry
DLBCL cells were fixed in 10% buffered formalin overnight. After washing with PBS, the cells were mixed with HistoGel (Richard-Allan Scientific), placed in cellblock cassettes, processed overnight using conventional histological techniques and embedded in paraffin. Immunocytochemistry was performed using an automated immunostainer (Discovery Ultra, Ventana Medical Systems, Tucson, AZ). After de-paraffinization and heat-induced epitope retrieval, slides were incubated with pThrERM antibody and OmniMap anti-rabbit HRP. Staining was developed using ChromoMap DAB kit (Ventana), and the cells counterstained with hematoxylin. The sections were dehydrated and mounted in Cytoseal XYL (Richard-Allan Scientific), and the slides viewed using a digital light microscope (Olympus) with a 10X objective.
TIRF microscopy
DLBCL cells were left untreated, or transfected with Ez-DN or incubated with Ez-Inh, following by fixation in 4% paraformaldehyde. For co-localization analysis of BCR and tyrosine phosphorylated proteins, the former were labeled with Rhodamine Red™-X-conjugated goat anti-human IgM or IgG Fab fragment (Invitrogen) for 30 min at 4°C, followed staining with a fluorescein isothiocyanate-conjugated mouse monoclonal antibody to pY (clone 4G10; EMD Millipore). For co-localization analysis of BCR and phosphorylated ERM proteins, the former were labeled with AlexaFluor 488 conjugated goat anti-human IgM or IgG Fab fragment (Invitrogen) for 30 min at 4°C, followed by fixation and staining with a rabbit polyclonal pThrERM antibody (Cell Signaling) and a rhodamine-conjugated goat anti-rabbit IgG antibody (Jackson Immunoresearch). The cells were imaged at the depth of 130–150 nm using a Leica-AM total internal reflection fluorescence (TIRF) microscope DMI6000 (Leica Microsystems) with an attached Hamamatsu EM-CCD camera and Leica acquisition software LAS AF Version 2.2.0. An HCX PL APO 100X oil objective (NA = 1.47) was used at an additional 1.6X magnification with appropriate filter cubes. The images were digitally deconvolved using the “iterative restoration” function, and analyzed for BCR cluster number and area, and microvilli number and length in Volocity 6.0. For co-localization analysis, the deconvolved images were thresholded and Pearson’s correlation coefficients calculated using Volocity 6.0.
Statistical Analysis
Statistical analysis was performed using Prism 4 (GraphPad Software, Inc.), and significance determined by employing Student t test. P value of < 0.05 was considered significant.
Results
Interference with ERM function inhibits DLBCL growth
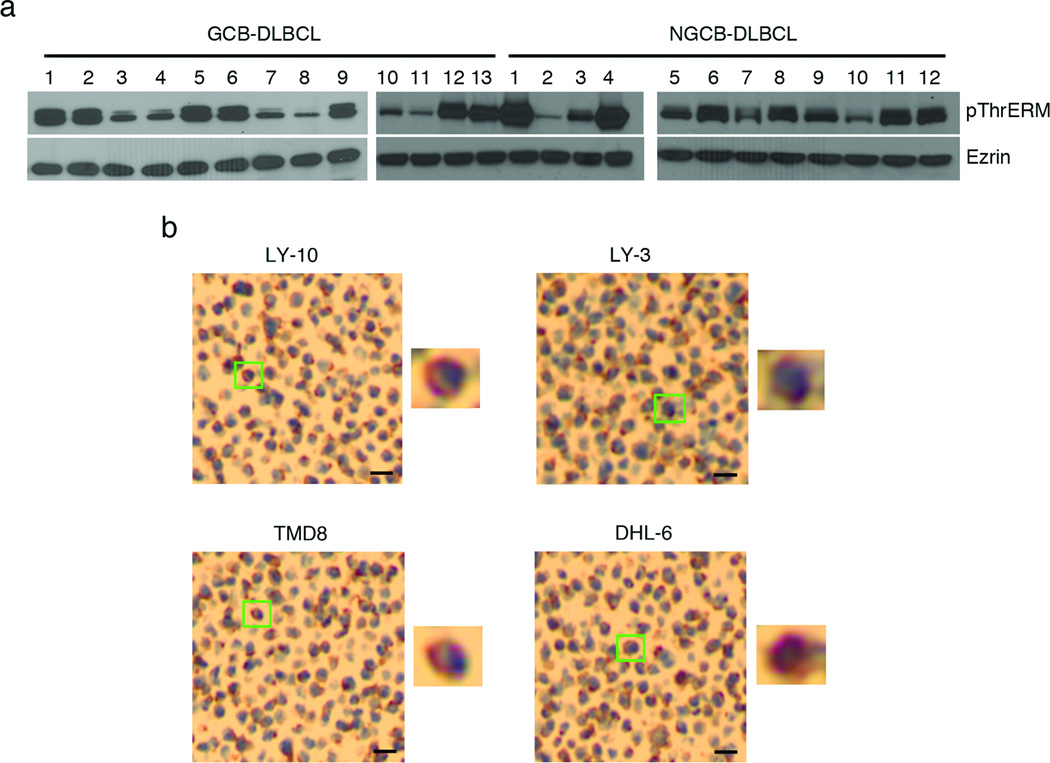
To examine if ERM proteins were phosphorylated at the C-terminal conserved threonine residue in DLBCL tumors and cell lines we employed an antibody to pThrERM, which binds to phosphorylated ezrin, moesin and radixin. Lysates prepared from lymphoma biopsy tissues from 12 ABC- and 13 GCB-DLBCL patients showed heterogeneous but high pThrERM levels (Figure 1a). Immunohistochemical analysis of four of the representative DLBCL cell lines OCI-LY-10, OCI-LY-3, TMD8 and SU-DHL-6 showed punctate pThrERM staining at the cell periphery (Figure 1b and zoomed-in panels). To test if high ERM phosphorylation in DLBCL tissues and cell lines was tumor-specific, we purified circulating B cells from blood and GC B cells from tonsils of three healthy individuals and compared their pThrERM levels. ERM phosphorylation was barely detectable in healthy peripheral B cells but primary GC B cells contained high pThrERM levels (Supplementary Figure 1a).
Figure 1. Phosphorylation of ERM proteins in DLBCL patient tissues.
(a) Lysates from GCB-DLBCL and non-GCB-DLBCL patient tumor biopsies were subjected to immunoblotting with pThrERM and ezrin antibodies. (b) Immunohistochemistry images of indicated DLBCL cell lines using antibody to pThrERM. Scale bar, 20 µm. Magnified images of individual cells indicated by green boxes are shown next to each panel. Blue color indicates DAPI-stained nuclei and brown/magenta color indicates pThrERM staining. Data are representative of two independent experiments.
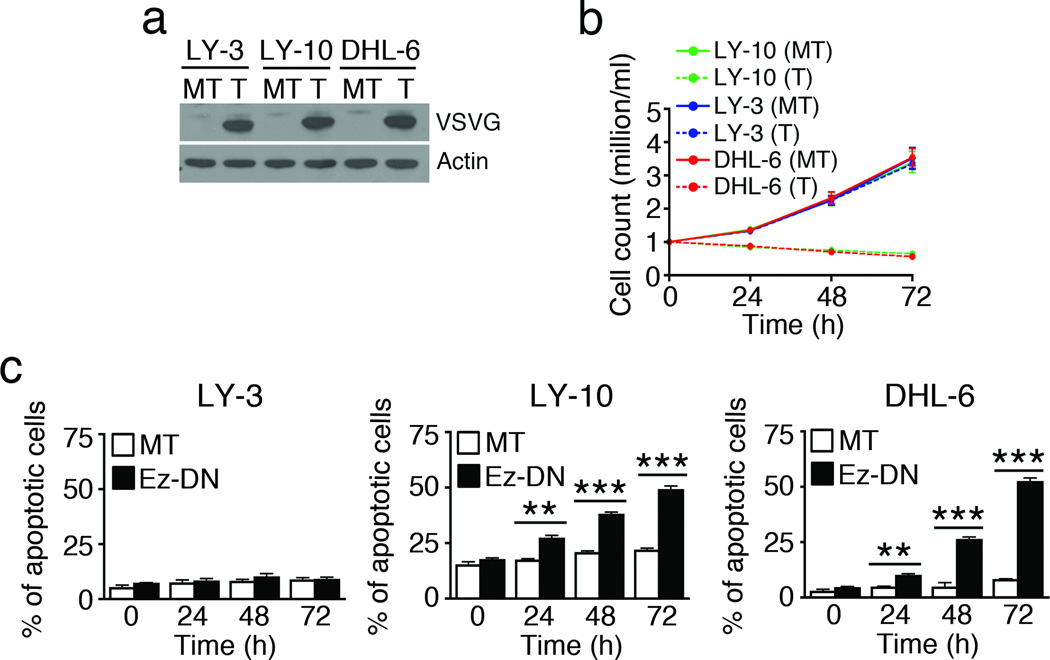
As phosphorylated ezrin regulates tumor cell growth and metastasis in several epithelial cell-derived cancers, we tested if interfering with the function of ERM proteins would affect the growth of DLBCL cells. ERM proteins do not possess intrinsic enzymatic activity; therefore, targeting their function has relied largely on ectopic expression of dominant negative mutants of ezrin or moesin, which contain the FERM domain but lack the conserved threonine phosphorylation site and thus compete with endogenous ERM proteins for binding to transmembrane proteins. This results in removal of endogenous ERM proteins from the cell surface and threonine dephosphorylation.35–37 We employed the dominant negative mutant of ezrin (Ez-DN; Supplementary Figure 1b) to inhibit ERM function. Expression of Ez-DN in OCI-LY-10 cells by transient transfection led to reduction in ERM phosphorylation within 24 h (Supplementary Figure 1c). OCI-LY-10 (CD79 mutant ABC-DLBCL), OCI-LY-3 (CARD11 mutant ABC-DLBCL) and SU-DHL-6 (GCB-DLBCL) cells were transiently transfected with the Ez-DN construct, and similar expression of VSVG-tagged Ez-DN was detected in all cell lines (Figure 2a). Interestingly, transient expression of Ez-DN led to loss in viable cell number in OCI-LY-10 and SU-DHL-6 but not in OCI-LY-3 cells (Figure 2b). As compared to mock-transfection, Ez-DN expression caused significant accumulation of Annexin-V+ apoptotic cells in OCI-LY-10 and SU-DHL-6, but not in OCI-LY-3 cells (Figure 2c). Apoptosis associated specifically with Ez-DN expression was calculated by subtracting the mock-transfected values from Ez-DN-transfected values. The results indicate that over 72 hours, up to 27% of OCI-LY-10, 44% of SU-DHL-6 and <1% of OCI-LY-3 cells underwent apoptosis upon expression of Ez-DN. The effect of wild type and other phosphorylation site mutants of ezrin on DLBCL cell growth was tested by transfecting OCI-LY-10 cells with pEYFP vector, YFP-fused wild type ezrin or YFP-fused mutants of ezrin Ez-TD (phosphomimetic mutant T567D) and Ez-TA (non-phosphorylatable mutant T567A)32, 33 (Supplementary Figure 1b). Lysates of OCI-LY-10 transfectants showed comparable expression of Ez-WT, Ez-TD and Ez-TA (Supplementary Figure 1d). As the Ez-DN construct is not fluorescently tagged, we employed the Ez-WT-YFP construct as a reporter of transfection efficiency by flow cytometry. Data in Supplementary Figure 1e show that 61.9% of OCI-LY-10, 68.2% of OCI-LY-3 and 74.2% of SU-DHL-6 cells express Ez-WT-YFP. Interestingly, Ez-WT boosted cell growth 72 h onwards, whereas despite its constitutive activation Ez-TD did not have an appreciable effect on cell growth (Supplementary Figure 1f). Ez-TA, which also acts as a dominant negative mutant like Ez-DN, resulted in progressive loss of cell viability (Supplementary Figure 1f).
Figure 2. Interference with ERM function inhibits DLBCL growth.
OCI-LY-10, SU-DHL-6 and OCI-LY-3 cells were mock-transfected (MT) or transfected with Ez-DN (T) for 24 h. (a) Cell lysates were probed with antibody to VSVG and actin. Viable cell counts were obtained by trypan blue exclusion (b), and induction of apoptosis quantified by Annexin V staining and flow cytometry (c). Mean ± SEM of three independent experiments is shown (**P < 0.01; ***P < 0.001).
Pharmacological inhibition of ERM phosphorylation impairs DLBCL cell growth
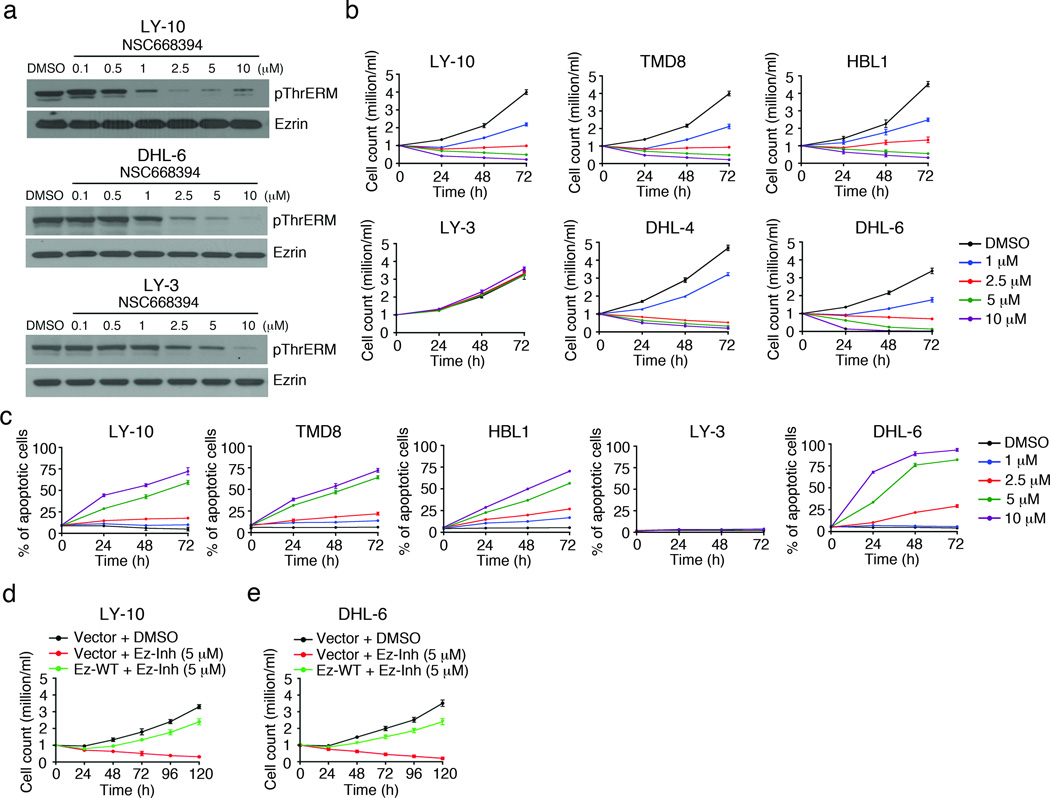
As an alternate strategy to inhibit ERM function, we employed novel compounds (NSC668394, NSC305787) identified in a recent small molecule screen in osteosarcoma cells that directly bind to ezrin, and abrogate its phosphorylation.38 To test the efficacy of these compounds on DLBCL growth, we treated OCI-LY-10 cells with varying doses of NSC305787 or NSC668394 for 1 h. Both compounds led to dephosphorylation of all three ERM proteins (Supplementary Figure 2a, b), and impaired cell growth (Supplementary Figure 2c, d) in a dose- and time-dependent manner. As the IC50 value for inhibition of cell growth by NSC668394 (5.8 µM) was slightly lower than that for NSC305787 (8.7 µM), we performed all subsequent inhibition experiments with NSC668394. DLBCL cell lines OCI-LY-10, SU-DHL-6 and OCI-LY-3 were treated with increasing concentrations of NSC668394 for 1 h, and dose-dependent dephosphorylation of the three ERM proteins was observed in all cell lines, albeit the effect was the strongest in OCI-LY-10 and weakest in OCI-LY-3 cells (Figure 3a). NSC668394 treatment led to a dose-dependent decrease in the growth of CD79 mutant ABC-DLBCL cell lines OCI-LY-10, TMD8, and HBL1, and the GCB-DLBCL cell lines SU-DHL-6 and SU-DHL-4, but not the CARD11 mutant OCI-LY-3 cell line (Figure 3b). As reported previously39 the growth of OCI-LY-3 cells was inhibited upon treatment with the IKK inhibitor PS-1145 (Supplementary Figure 3). Induction of apoptosis was observed in OCI-LY-10, TMD8, HBL1 and SU-DHL-6 but not OCI-LY-3 cells (Figure 3c). To ensure that NSC668394 specifically targets ERM proteins, a rescue experiment was performed in which we transiently transfected OCI-LY-10 and SU-DHL-6 cells with pEYFP vector or Ez-WT, followed by treatment with 5 µM of NSC668394 for five days. Expression of Ez-WT overcame the decrease in viability of OCI-LY-10 (Figure 3d) and SU-DHL-6 (Figure 3e) cells in the presence of NSC668394 by greater than 60%. To further confirm that ERM proteins contribute to DLBCL cell growth we knocked down their expression using siRNAs (siERM) in OCI-LY-10 and SU-DHL-6 cells. The expression and phosphorylation of ERM proteins was decreased by >80% in cells treated with siERM as compared to those treated with scrambled siRNAs (siControl) after 96 h (Supplementary Figure 4a). ERM knockdown did not affect the expression of actin (Supplementary Figure 4a). ERM knockdown also led to a time-dependent decrease in the growth of OCI-LY-10 and SU-DHL-6 cells (Supplementary Figure 4b).
Figure 3. Pharmacological inhibition of ERM proteins impairs growth of DLBCL cell lines.
(a) OCI-LY-10, OCI-LY-3 and SU-DHL-6 cells were treated with indicated concentrations of NSC668394 (Ez-Inh) and cell lysates probed with antibodies to pThrERM and ezrin. (b, c) DLBCL cell lines were treated with DMSO or indicated concentrations of NSC668394 for 72 h. Viable cell count was obtained by trypan blue exclusion (b), and induction of apoptosis quantified by Annexin V staining (c). Mean ± SEM of three independent experiments is shown (**P < 0.01; ***P < 0.001). OCI-LY-10 (d) and SU-DHL-6 (e) cells were transfected with Ez-WT for 24 h, followed by treatment with 5 µM of Ez-Inh for 120 h, and cell viability quantified as above. Data in d and e are representative of two independent experiments.
ERM proteins are co-localized with the BCRs in DLBCL
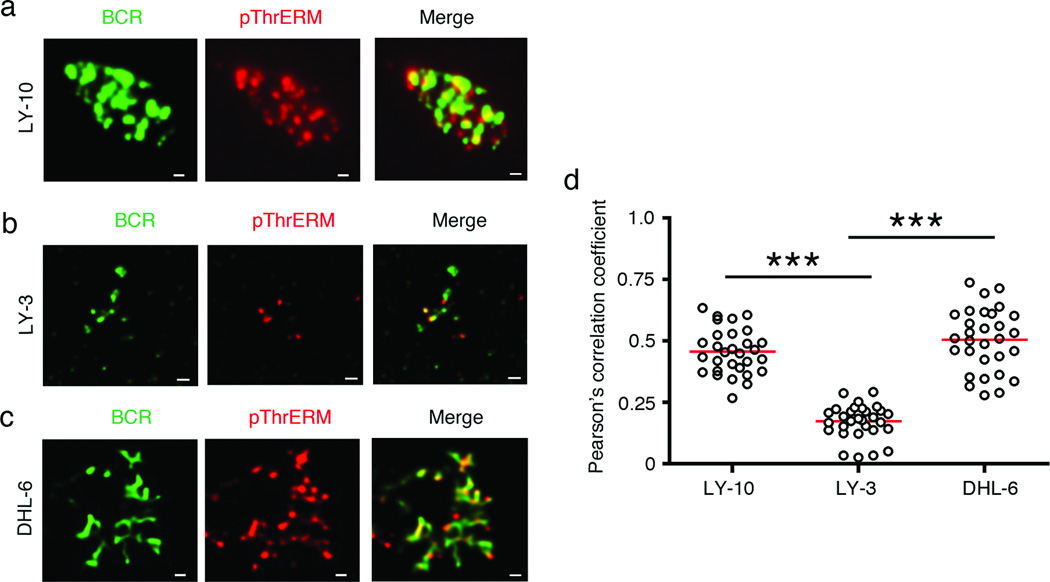
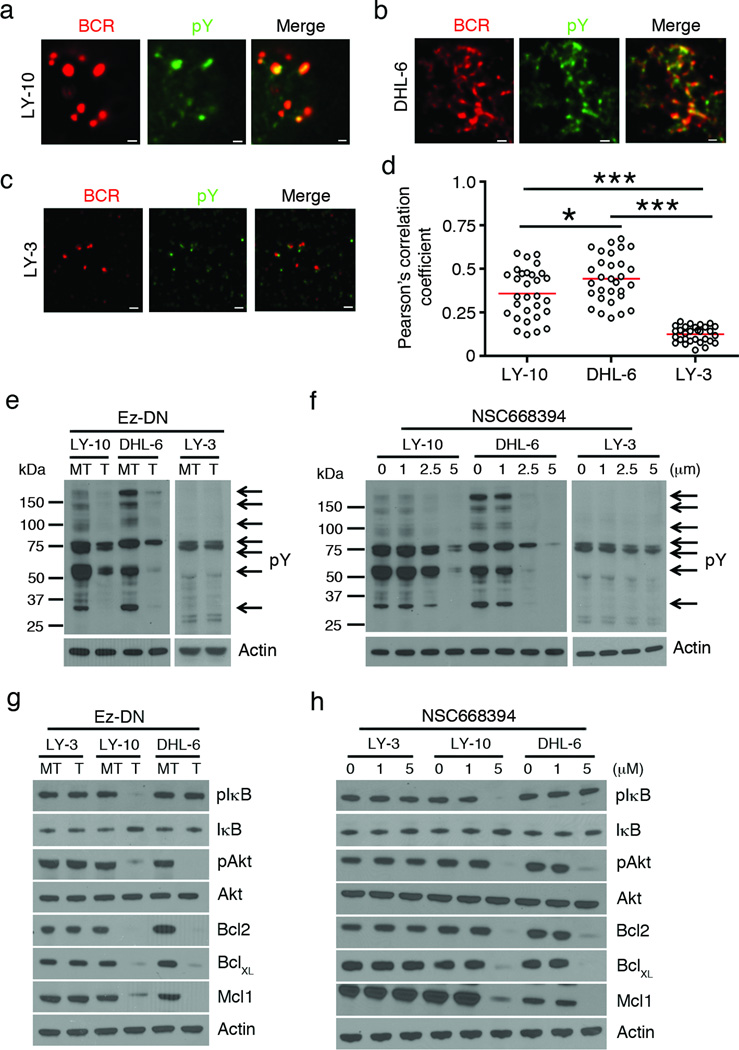
It is well established that dynamic phosphorylation-dephosphorylation of ERM proteins regulates the formation and maintenance of membrane microvilli and ruffles, as well as BCR clustering in antigen-stimulated mouse B cells.21, 22, 29, 30, 32 Therefore, we examined the spatial relationship of the BCR in DLBCL cells with phosphorylated ERM proteins by TIRF microscopy at a depth of 130–150 nm, a region that encompasses the plasma membrane and cell cortex. The CD79 mutant ABC-DLBCL cell lines (OCI-LY-10, TMD8 and HBL1) exhibited BCR arrangement in spontaneous clusters as previously reported9 (Supplementary Figure 5a). The CARD11 mutant OCI-LY-3 cell line also showed surface BCR clusters, but these were much smaller (Supplementary Figure 5a). The GCB-DLBCL cell lines (SU-DHL-4 and SU-DHL-6) showed a streaky BCR distribution (Supplementary Figure 5b), reminiscent of membrane microvilli9, 32 The BCRs showed co-segregation with threonine-phosphorylated ERM proteins in OCI-LY-10 cells (Figure 4a), OCI-LY-3 (Figure 4b) and SU-DHL-6 (Figure 4c) cells. Quantification by Pearson’s correlation analysis showed that the co-localization of the BCR with pThrERM was similar in OCI-LY-10 and SU-DHL-6 cells, but weaker in OCI-LY-3 cells (Figure 4d).
Figure 4. Co-localization of BCR with phosphorylated ERM proteins in DLBCL cells.
OCI-LY-10 (a), OCI-LY-3 (b) and SU-DHL-6 (c) cells were stained for the BCR (green) and pThrERM (red), and imaged by TIRF microscopy. Scale bar, 1 µm. Data are representative of two independent experiments with ~30 cells imaged per experiment. (d) Pearson’s co-localization coefficients are plotted for the indicated cell lines. Each dot represents an individual cell and horizontal lines indicate the mean. Mean ± SEM is shown (***P < 0.001) for the indicated comparisons.
ERM inhibition abrogates BCR clustering and localization in microvilli
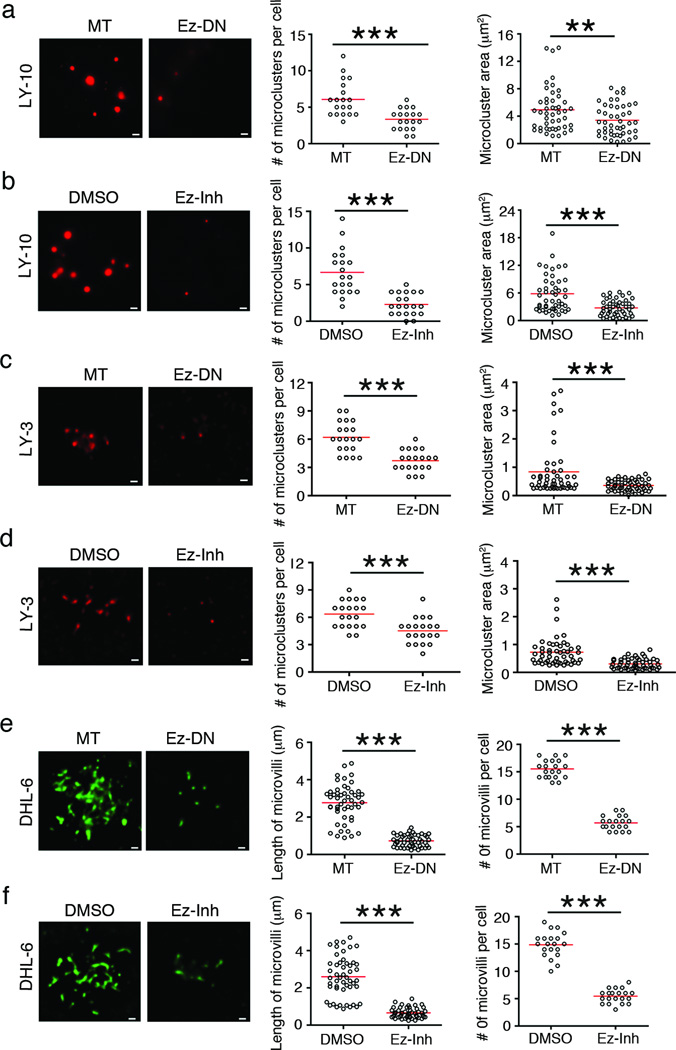
To investigate if ERM inhibition would affect BCR localization and distribution in DLBCL cells, we either transfected OCI-LY-10, OCI-LY-3 and SU-DHL-6 cells with Ez-DN or treated them with 5 µM of NSC668394. Mock-transfected OCI-LY-10 cells showed large BCR clusters ranging from 2–12 per cell with an average of 5 clusters per cell. Ez-DN-transfected cells showed a 50% reduction in the number of large clusters (Figure 5a), as well as a decrease in the area of the BCR clusters from 5 µm2 to 3 µm2 (Figure 5a). Treatment of OCI-LY-10 cells with NSC668394 led to a similar decrease in the number and size of spontaneous BCR clusters as compared to DMSO-treated cells (Figure 5b). Expression of wild type ezrin (Ez-WT) in OCI-LY-10 cells treated with 5 µM of Ez-Inh restored surface BCR clusters (Supplementary Figure 6a, b). While the number of BCR clusters in OCI-LY-3 cells was similar to that in OCI-LY-10 cells, the area of these clusters was only ~1 µm2 (Figure 5c, d). Both the number and size of BCR clusters in OCI-LY-3 cells was significantly reduced in the presence of Ez-DN (Figure 5c) and NSC668394 (Figure 5d). As the streaky BCR distribution in GCB-DLBCL cell lines resembled that of the microvillar ezrin localization reported by us in murine IgG+ B lymphoma cell line 2PK3,32 we quantified the length and number of the BCR-rich microvilli in the absence and presence of Ez-DN or NSC668394. SU-DHL-6 cells showed numerous BCR streaks, whose length ranged from 1 to 5 µm. Both, expression of Ez-DN (Figure 5e) and treatment with NSC668394 (Figure 5f) caused a significant reduction in the number and length of BCR-containing microvilli. Inhibition of ERM proteins by NSC668394 (left panels) or Ez-DN (right panels) did not alter the overall surface IgM expression in OCI-LY-10 (Supplementary Figure 6c) or OCI-LY-3 (Supplementary Figure 6d) cells. However, both inhibition methods reduced the surface IgG expression in SU-DHL-6 cells by 30–40% (Supplementary Figure 6e).
Figure 5. Inhibition of ERM function disrupts BCR organization.
OCI-LY-10 (a, b), OCI-LY-3 (c, d) and SU-DHL-6 (e, f) cells were transfected with Ez-DN (a, c, e), or treated with 5 µM NSC668394 (Ez-Inh) (b, d, f), and BCRs were imaged by TIRF microscopy. The number and area of BCR microclusters were quantified in a-d, and the number and length of microvilli in e and f. Scale bar 1 µm. Data are representative of two independent experiments with ~20 cells imaged per experiment (**P < 0.01; ***P < 0.001).
Inhibition of ERM function impairs proximal and distal signaling
As alterations in cell surface distribution of the BCRs regulates protein tyrosine phosphorylation and downstream activation signals in antigen-stimulated B cells,29, 30 we imaged the spatial interaction of surface BCRs with tyrosine phosphorylated (pY) proteins in DLBCL cells. The BCRs co-localized with pY-containing proteins in OCI-LY-10 (Figure 6a) and SU-DHL-6 (Figure 6b) cells but not in OCI-LY-3 cells (Figure 6c). The extent of co-localization between BCR and pY was higher in SU-DHL-6 cells than OCI-LY-10 cells, and it was the weakest in OCI-LY-3 cells (Figure 6d). Next, we tested if disruption of surface BCR organization by ERM inhibition would affect proximal BCR signaling. For this, OCI-LY-3, OCI-LY-10 and SU-DHL-6 cells were either transfected with Ez-DN (Figure 6e) or treated with varying doses of NSC668394 (Figure 6f). Immunoblots of control OCI-LY-10 and SU-DHL-6 cells showed several constitutive pY-containing protein bands (Figure 6e, lane marked MT and Figure 6f, lane marked 0). In contrast, OCI-LY-3 cells contained very few tyrosine-phosphorylated proteins (Figure 6e, MT lanes and Figure 6f, lane 0). Expression of Ez-DN (Figure 6e) or treatment with NSC668394 (Figure 6f) led to reduction in tyrosine phosphorylation in both OCI-LY-10 and SU-DHL-6 cells but the effect was minimal in OCI-LY-3 cells (Figure 6e, f). We further examined the effect of ERM inhibition on components of downstream NF-κB and PI3K activation, which are constitutively turned on in ABC-DLBCL and GCB-DLBCL.10, 40 Expression of Ez-DN resulted in a marked reduction in the phosphorylation of IκBα and Akt in OCI-LY-10 but not OCI-LY-3 cells (Figure 6g). SU-DHL-6 cells expressing Ez-DN showed reduced pAkt levels but no effect on pIκBα (Figure 6g). Expression of anti-apoptotic proteins Bcl2, BclXL and Mcl1, was significantly reduced in both OCI-LY-10 and SU-DHL-6 but not in OCI-LY-3 cells (Figure 6g). Like Ez-DN, NSC668394 also inhibited the phosphorylation of IκB and Akt, and lowered the expression of Bcl2, BclXL and Mcl1 in OCI-LY-10 but not in OCI-LY-3 cells (Figure 6h). Treatment of SU-DHL-6 cells with NSC668394 caused inactivation of Akt, and lower expression of Bcl2, BclXL and Mcl1 (Figure 6h). Ez-DN (Supplementary Figure 7a) and NSC668394 (Supplementary Figure 7b) also reduced the phosphorylation of IκB and Akt in TMD8 cells, but only that of Akt in SU-DHL-4 cells. NSC668394-induced reduction in tyrosine phosphorylation (Supplementary Figure 7c), impairment of NF-κB and PI3K signaling and Bcl2 expression (Supplementary Figure 7d) were all reversed by overexpression of Ez-WT. This effect could already be observed at 48 h of Ez-WT expression, but was maximal at 120 h (Supplementary Figure 7c, d). Furthermore, siRNA-induced knockdown of ERM expression reduced PI3K and NF-κB activation in OCI-LY-10, and only PI3K activation in SU-DHL-6 cells (Supplementary Figure 7e).
Figure 6. ERM inhibition impairs proximal and distal signaling in DLBCL cells.
BCRs (red) were stained on OCI-LY-10 (a) SU-DHL-6 (b) and OCI-LY-3 (c) cells, followed by staining of phosphotyrosine (pY; green), and imaging by TIRF microscopy. (d) Pearson’s co-localization coefficients are plotted for the indicated cell lines. Each dot represents an individual cell and horizontal lines indicate the mean. Mean ± SEM is shown (*P < 0.05; ***P < 0.001) for the indicated comparisons. Representative data from two independent experiments are shown, and ~25 cells were imaged per experiment. Indicated DLBCL cell lines were mock-transfected (MT) or transfected with Ez-DN (T) for 24 h (e), or treated with DMSO or indicated concentrations of NSC668394 for 24 h (f), and cell lysates were probed with antibodies to pY and actin. Indicated DLBCL cell lines were mock-transfected (MT) or transfected with Ez-DN (T) (g), or treated with DMSO or indicated concentrations of NSC668394 (h), and after 24 h the cell lysates were probed with antibodies to pIκB, IκB, pAkt, Akt, Bcl2, BclXL, Mcl1 and actin. Representative blots from three independent experiments are shown for e-h.
Inhibition of ERM function in vivo retards the growth of DLBCL xenografts
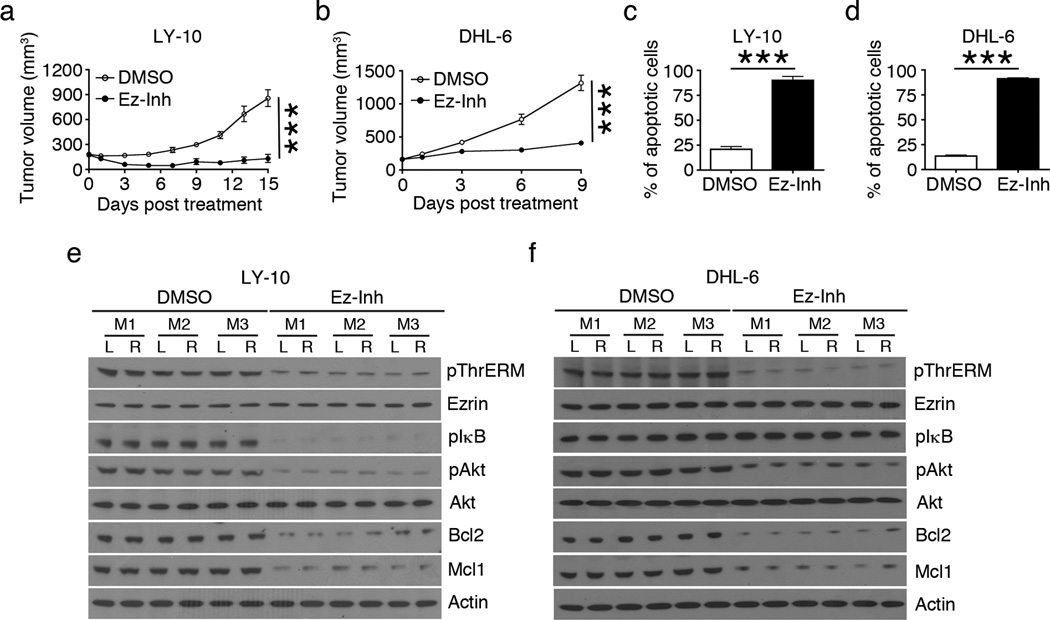
Next, we investigated the effect of ERM inhibition on lymphoma growth in vivo. For this, we employed DLBCL cell line transplantation into NOD-SCID-γ−/− (NSG) mice, which lack B, T and natural killer cells and cannot reject human lymphoma cell xenografts. We generated twelve OCI-LY-10 and twenty SU-DHL-6 tumor xenografts by injecting both flanks of six and ten NSG mice, respectively. After the tumor volume reached ~200 mm3, half of the mice in each cell line group received 5% DMSO and the other half received 13.5 mg/kg/day NSC668394. The growth of SU-DHL-6 xenografts was more robust than OCI-LY-10 xenografts (Figure 7a, b), and reached IACUC-approved limit for euthanasia sooner, thus limiting the duration of NSC668394 treatment to 9 days in SU-DHL-6 tumor-bearing mice. NSC668394-treated mice showed significant reduction in the growth of both OCI-LY-10 (Figure 7a) and SU-DHL-6 (Figure 7b) xenografts as compared those treated with DMSO. The mice tolerated the inhibitor well with no evidence of toxicity in terms of grooming behavior or weight loss (Supplementary Tables S1 and S2). Tumor cells from OCI-LY-10 (Figure 7c) and SU-DHL-6 (Figure 7d) xenografts harvested from NSC668394-treated mice exhibited a marked increase in the proportion of Annexin V+ apoptotic cells as compared to those from DMSO-treated mice. Furthermore, tumor tissue lysates showed that NSC668394 treatment reduced the pThrERM levels in both OCI-LY-10 (Figure 7e) and SU-DHL-6 (Figure 7f) xenografts as compared to DMSO treatment, demonstrating in vivo activity of the inhibitor towards ERM proteins. Reduced pThrERM levels were accompanied by reduction in levels of pIκB, pAkt, Bcl2 and Mcl1 in OCI-LY-10 xenografts (Figure 7e), and pAkt, Bcl2 and Mcl1 levels in SU-DHL-6 xenografts (Figure 7f).
Figure 7. ERM inhibition retards DLBCL growth in vivo.
Tumor xenograft growth in NSG mice injected with OCI-LY-10 (a) or SU-DHL-6 (b) cells and treated with DMSO or 13.5 mg/kg/day of NSC668394 (Ez-Inh) after tumor volume reached ~200 mm3 (n=3 mice per treatment group in a and n=5 mice per treatment group in b). Proportion of apoptotic cells per 105 cells from freshly harvested OCI-LY-10 (c) and SU-DHL-6 (d) tumor tissues was quantified by Annexin V staining. Mean ± SEM is shown (***P < 0.001) for data in a-d. Tissue lysates from 6 OCI-LY-10 (e) and 6 SU-DHL-6 (f) tumors were analyzed by western blotting using antibodies to pThrERM, ezrin, pIκB, pAkt, Akt, Bcl2, Mcl1 and actin (M,mouse; R,right flank; L,left flank).
Taken together, our data demonstrate that phosphorylated ERM proteins constitute an important pathogenic axis that promotes growth and survival of DLBCLs.
Discussion
In this study, we demonstrate that ERM proteins are key participants in DLBCL pathogenesis. We show that ERM proteins interact with spontaneously occurring cell surface BCR assemblies in the ABC and GCB subtypes of DLBCL. Abrogation of ERM function results in loss of BCR organization, blocks transmission of proximal BCR signals to NF-κB and PI3K pathways and retards DLBCL growth in vivo. As BCR organization is an important upstream step in the nucleation of B cell growth,27, 41–43 our data underscore the importance of spatially organized signaling in DLBCL cells, and suggest that pathogenic DLBCL cells co-opt the “normal” function of ERM proteins to promote their own growth and survival. Our findings further suggest that blocking ERM function may represent a novel strategy for controlling the growth of DLBCL tumors.
Cell lines representing the different subtypes of DLBCL demonstrated a graded response to ERM inhibition in our study; the GCB and CD79 mutant ABC-DLBCL cells showed similar sensitivity whereas the CARD11 mutant ABC-DLBCL cells were largely insensitive. We observed that instead of organizing into microclusters, the BCRs in GCB-DLBCL are localized in membrane microvilli, consistent with previous studies9 and the observation that mouse and human GC B cells bear dendritic structures during T cell-dependent immune response.44 The decrease in the number and length of microvilli upon ERM inhibition suggests that these membrane structures are supported by ERM proteins, which is in agreement with their known function.21, 45 The coincident reduction in surface BCR expression upon ERM inhibition in GCB-DLBCL suggests that the BCRs may be internalized upon microvillar resorption, and that this at least partially contributes towards quantitative reduction in the cell surface signaling apparatus. On the other hand ERM inhibition did not affect the surface BCR expression but resulted in smaller BCR clusters in ABC-DLBCL cell lines, suggesting that BCR single molecules or nanoclusters may be released from ERM network-based corrals. These smaller sized BCR assemblies are not detectable by the resolution of TIRF imaging employed in this study, but may be more readily visualized by super resolution imaging modalities such as photoactivated localization microscopy46, 47 or stochastic optical reconstruction microscopy30, 48, 49. In the CD79 mutant ABC-DLBCL, de-clustering of the BCRs was associated with decreased pro-survival signaling with impairment of cell growth. The CARD11 mutant ABC-DLBCL cell line OCI-LY-3 showed much lower basal tyrosine phosphorylation than OCI-LY-10 cells, and their BCRs were not co-localized with tyrosine-phosphorylated proteins, supporting the notion that proximal BCR signaling is not a remarkable feature of this mutant subtype.12 OCI-LY-3 cells were killed upon inhibition of IKK, an enzyme that functions downstream of CARD11 in the NF-κB activation pathway,50 further emphasizing the importance of NF-κB signaling as the main pathogenic mechanism in CARD11 mutant DLBCL cells.39 Our data also indicate ERM proteins promote DLBCL growth by acting upstream of CARD11, and that agents altering BCR organization may not be effective in patients whose lymphomas bear mutations in NF-κB signaling proteins.
A role for both chronic active BCR signaling and tonic BCR signaling in maintaining lymphoma survival has been appreciated in recent years10, 51, 52. Chronic active signaling was described for ABC-DLBCL with gain-of-function mutations in Igα and Igβ, whose sustained signaling at the surface maintains NF-κB activation. On the other hand, tonic signaling is largely associated with the surface BCR and PI3K pathway51, 52. Here, we use the term "pathogenic BCR signaling", which is conventionally associated with the ABC subtype of DLBCL, to also encompass the GCB subtype because of the following evidence. The BCRs co-localize with tyrosine-phosphorylated proteins in membrane microvilli in GCB-DLBCL, and lysates of these cells contain several prominent tyrosine phosphorylated protein bands. These data indicate BCR-associated basal tyrosine kinase activity in GCB-DLBCL, which may represent tonic signaling. ERM inhibition causes reduction in surface BCR levels in GCB-DLBCL (because of resorption of microvilli), and concomitant inhibition of PI3K signaling. As these effects of ERM inhibition are associated with loss of cell and tumor growth, we suggest that ERM-mediated subcellular BCR organization mediates “pathogenic” BCR signaling in the GCB subtype. Overall, our data suggest that ERM-mediated subcellular BCR organization in microclusters or microvilli supports pathogenic BCR signaling and DLBCL growth (Figure 8). PI3K is not only a component of GCB survival, but also a critical feature of tonic BCR signaling, as it can rescue mature B cell survival in the absence of the BCR.53 The p110δ catalytic subunit of PI3K is recruited to Igα and Igβ via the small Ras family GTPase TC21.53, 54 Alternatively, PI3K can be included within the BCR complex through direct binding to CD19, a co-receptor that is known to promote BCR signaling.49, 55, 56 As ERM inhibition disrupts BCR assembly in the microvilli in GCB-DLBCL cells, associated with loss of tyrosine phosphorylation, PI3K signaling and cell growth, we propose that ERM proteins may support tonic BCR signaling in microvilli in GCB-DLBCL by enabling BCR-CD19/TC21-PI3K assembly.
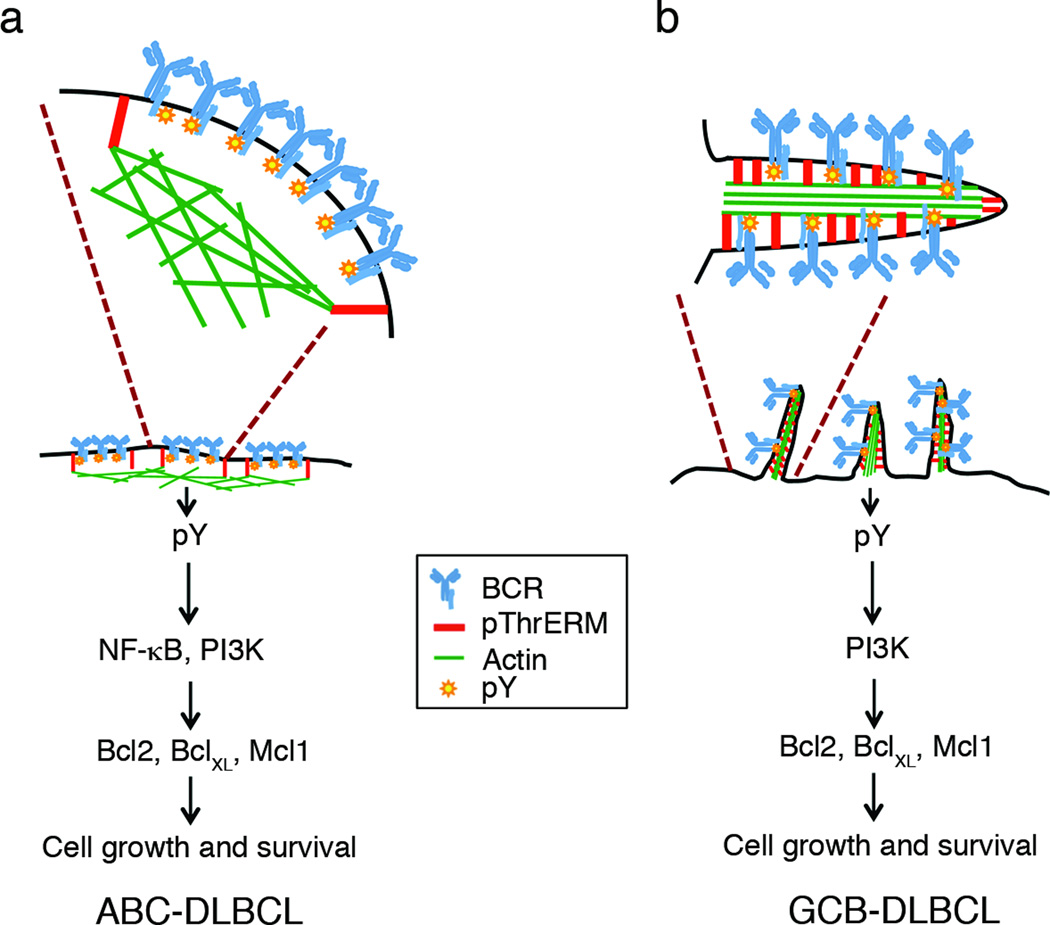
Figure 8. Model for regulation of BCR signaling by ERM proteins in DLBCL.
(A) In ABC-DLBCL with activating CD79 mutations, the pre-clustered BCRs are trapped within compartments supported by phosphorylated ERM proteins through linkage of plasma membrane and actin filaments. These BCR clusters continuously transduce NF-κB and PI3K pathway activation signals to promote cell growth. (B) In GCB-DLBCL, the BCRs are present in surface microvilli, whose backbone consists of phosphorylated ERM proteins and actin filaments. The BCRs transmit tonic signals through the PI3K pathway in this location. ERM inhibition disrupts both types of organization by interfering with plasma membrane-actin cytoskeletal linkage.
Phosphorylation at the conserved threonine site in the C-terminal F-actin-binding domain is believed to be a necessary step in the conformational activation of ERM proteins,18 and also responsible for the functional redundancy between them45. The two non-phosphorylatable mutants Ez-DN and Ez-TA interfered with DLBCL growth but the constitutively active mutant Ez-TD, which is irreversibly trapped in the open conformation, did not. On the other hand, overexpression of Ez-WT significantly enhanced the growth of OCI-LY-10 cells, suggesting that dynamic and reversible regulation of ERM phosphorylation is necessary for supporting cell growth in DLBCL. We have previously reported that genetic deletion of ezrin amplifies BCR signaling in naïve mouse B cells,30 whereas in this study interfering with ERM function disrupted BCR signaling in DLBCL cell lines. The data obtained in DLBCL cell lines are in agreement with those of Treanor et al29 who reported that expression of dominant negative mutants of ezrin or knocking down expression of ezrin and moesin disrupts the integrity of BCR clusters and reduces the strength of BCR signaling in mouse lymphoma B cell lines.29 The opposite effects of ERM inhibition in naïve and lymphoma B cells suggest that the impact of the cytoskeletal network on B cell activation is context-dependent. The differences may stem from the type of stimulation experienced by naïve and GC origin B cells. Naïve B cells respond to BCR crosslinking acutely by dephosphorylating ERM proteins, and rephosphorylating them upon continuous stimulation. On the other hand, continual survey of antigens in the germinal center by the BCR is a property of GC B cells and must be necessary for selection and survival.57, 58 We observed that like DLBCL patient tissue and cell lines, primary GC B cells also contain high levels of phosphorylated ERM proteins. It is possible that high pThrERM levels in GC B cells serve to tether/corral BCRs at the cell surface for efficient antigen recognition, selection and differentiation. In DLBCL cells, a “self antigen” may drive continuous stimulation of the BCRs and aid in lymphoma cell survival.51, 59 Thus, DLBCLs likely represent transformed germinal center B cells that are undergoing chronic active or tonic BCR signaling supported by the ERM network. The outcome of ERM inhibition in naïve mouse B cells and GC-derived lymphomas may be additionally influenced by other microenvironmental stimuli. Indeed, a detailed investigation of the role for the cytoskeletal network in primary naïve versus GC B cells is warranted, and should provide mechanistic insights into spatial and molecular control of stage-specific B cell activation, selection and differentiation.
Hyperphosphorylation of ezrin is frequently associated with growth, metastasis and poor prognosis in a variety of clinically aggressive human cancers, including hepatocellular carcinoma, breast cancer and osteosarcomas.35, 60, 61 Knockdown of ezrin expression abrogates invasion, early metastatic survival as well as lung metastases in osteosarcoma,35 and breast carcinoma.36 Likewise, our data support an important role for phosphorylated ERM proteins in promoting DLBCL cell survival, albeit through regulation of BCR organization. As ERM proteins also facilitate communication between cells and their microenvironment, and response to chemokines and cytokines,32, 62 it will be interesting to explore additional roles for ERM proteins in DLBCL cell dissemination and interaction with the tumor microenvironment. Finally, a variety of novel kinase inhibitors have been developed to target various proteins associated with lymphoma cell survival, including Syk, PI3K, Btk and PKCβ and JAK2/STAT3.10, 14, 15, 63–65 Combining ERM inhibitors with these and other novel therapies may suppress multiple critical cell survival pathways simultaneously and offer therapeutic synergy.
Supplementary Material
Acknowledgements
This study was funded by grants to N.G. (AI081743 from NIH and Cancer Research Institute Investigator Award). We thank Yvonne Parker for assistance with the animal experiments.
Footnotes
Conflict-of-interest statement: The authors declare no conflict of interest.
Supplementary information is available at Leukemia’s website.
References
- 1.Coiffier B. Current strategies for the treatment of diffuse large B cell lymphoma. Curr Opin Hematol. 2005 Jul;12(4):259–265. doi: 10.1097/01.moh.0000168519.98990.94. [DOI] [PubMed] [Google Scholar]
- 2.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2003 Aug 19;100(17):9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2008 Sep 9;105(36):13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. The Journal of experimental medicine. 2003 Sep 15;198(6):851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider C, Pasqualucci L, Dalla-Favera R. Molecular pathogenesis of diffuse large B-cell lymphoma. Seminars in diagnostic pathology. 2011 May;28(2):167–177. doi: 10.1053/j.semdp.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annual review of immunology. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sehn LH, Connors JM. Oncology. 4 Suppl 1. Vol. 19. Williston Park: 2005. Apr, Treatment of aggressive non-Hodgkin's lymphoma: a north American perspective; pp. 26–34. [PubMed] [Google Scholar]
- 8.Sehn LH, Connors JM. Treatment of diffuse large B-cell lymphoma: a risk-based approach. Clinical lymphoma & myeloma. 2006 Oct;(7 Suppl 1):S14–S19. doi: 10.3816/clm.2006.s.003. [DOI] [PubMed] [Google Scholar]
- 9.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010 Jan 7;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013 Mar;12(3):229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansell SM, Hodge LS, Secreto FJ, Manske M, Braggio E, Price-Troska T, et al. Activation of TAK1 by MYD88 L265P drives malignant B-cell Growth in non-Hodgkin lymphoma. Blood cancer journal. 2014;4:e183. doi: 10.1038/bcj.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008 Mar 21;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 13.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009 Jun 4;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrutinib approved for mantle cell lymphoma. Cancer discovery. 2014 Jan;4(1):OF1. doi: 10.1158/2159-8290.CD-NB2013-172. [DOI] [PubMed] [Google Scholar]
- 15.Cheng S, Ma J, Guo A, Lu P, Leonard JP, Coleman M, et al. BTK inhibition targets in vivo CLL proliferation through its effects on B-cell receptor signaling activity. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK; 2013. Nov 25, [DOI] [PubMed] [Google Scholar]
- 16.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nature reviews Molecular cell biology. 2010 Apr;11(4):276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yonemura S, Matsui T, Tsukita S. Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. Journal of cell science. 2002 Jun 15;115(Pt 12):2569–2580. doi: 10.1242/jcs.115.12.2569. [DOI] [PubMed] [Google Scholar]
- 18.Fievet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, et al. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. The Journal of cell biology. 2004 Mar 1;164(5):653–659. doi: 10.1083/jcb.200307032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bretscher A, Chambers D, Nguyen R, Reczek D. ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annual review of cell and developmental biology. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- 20.Yonemura S, Tsukita S, Tsukita S. Direct involvement of ezrin/radixin/moesin (ERM)-binding membrane proteins in the organization of microvilli in collaboration with activated ERM proteins. The Journal of cell biology. 1999 Jun 28;145(7):1497–1509. doi: 10.1083/jcb.145.7.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viswanatha R, Bretscher A, Garbett D. Dynamics of ezrin and EBP50 in regulating microvilli on the apical aspect of epithelial cells. Biochem Soc Trans. 2014 Feb;42(1):189–194. doi: 10.1042/BST20130263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treanor B, Depoil D, Gonzalez-Granja A, Barral P, Weber M, Dushek O, et al. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity. 2010 Feb 26;32(2):187–199. doi: 10.1016/j.immuni.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta N, Wollscheid B, Watts JD, Scheer B, Aebersold R, DeFranco AL. Quantitative proteomic analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid raft dynamics. Nature immunology. 2006 Jun;7(6):625–633. doi: 10.1038/ni1337. [DOI] [PubMed] [Google Scholar]
- 24.Viola A, Gupta N. Tether and trap: regulation of membrane-raft dynamics by actin-binding proteins. Nature reviews Immunology. 2007 Nov;7(11):889–896. doi: 10.1038/nri2193. [DOI] [PubMed] [Google Scholar]
- 25.Batista FD, Treanor B, Harwood NE. Visualizing a role for the actin cytoskeleton in the regulation of B-cell activation. Immunological reviews. 2010 Sep;237(1):191–204. doi: 10.1111/j.1600-065X.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Meckel T, Tolar P, Sohn HW, Pierce SK. Intrinsic properties of immunoglobulin IgG1 isotype-switched B cell receptors promote microclustering and the initiation of signaling. Immunity. 2010 Jun 25;32(6):778–789. doi: 10.1016/j.immuni.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W, Meckel T, Tolar P, Sohn HW, Pierce SK. Antigen affinity discrimination is an intrinsic function of the B cell receptor. The Journal of experimental medicine. 2010 May 10;207(5):1095–1111. doi: 10.1084/jem.20092123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klasener K, Maity PC, Hobeika E, Yang J, Reth M. B cell activation involves nanoscale receptor reorganizations and inside-out signaling by Syk. eLife. 2014;3:e02069. doi: 10.7554/eLife.02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treanor B, Depoil D, Bruckbauer A, Batista FD. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. The Journal of experimental medicine. 2011 May 9;208(5):1055–1068. doi: 10.1084/jem.20101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pore D, Parameswaran N, Matsui K, Stone MB, Saotome I, McClatchey AI, et al. Ezrin Tunes the Magnitude of Humoral Immunity. J Immunol. 2013 Oct 15;191(8):4048–4058. doi: 10.4049/jimmunol.1301315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004 Jan 1;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 32.Parameswaran N, Matsui K, Gupta N. Conformational switching in ezrin regulates morphological and cytoskeletal changes required for B cell chemotaxis. J Immunol. 2011 Apr 1;186(7):4088–4097. doi: 10.4049/jimmunol.1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crepaldi T, Gautreau A, Comoglio PM, Louvard D, Arpin M. Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. The Journal of cell biology. 1997 Jul 28;138(2):423–434. doi: 10.1083/jcb.138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parameswaran N, Enyindah-Asonye G, Bagheri N, Shah NB, Gupta N. Spatial coupling of JNK activation to the B cell antigen receptor by tyrosine-phosphorylated ezrin. J Immunol. 2013 Mar 1;190(5):2017–2026. doi: 10.4049/jimmunol.1201292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004 Feb;10(2):182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Wu M, Wang H, Xu G, Zhu T, Zhang Y, et al. Ezrin silencing by small hairpin RNA reverses metastatic behaviors of human breast cancer cells. Cancer Lett. 2008 Mar 8;261(1):55–63. doi: 10.1016/j.canlet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 37.Monni R, Haddaoui L, Naba A, Gallais I, Arpin M, Mayeux P, et al. Ezrin is a target for oncogenic Kit mutants in murine erythroleukemia. Blood. 2008 Mar 15;111(6):3163–3172. doi: 10.1182/blood-2007-09-110510. [DOI] [PubMed] [Google Scholar]
- 38.Bulut G, Hong SH, Chen K, Beauchamp EM, Rahim S, Kosturko GW, et al. Small molecule inhibitors of ezrin inhibit the invasive phenotype of osteosarcoma cells. Oncogene. 2012 Jan 19;31(3):269–281. doi: 10.1038/onc.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam LT, Davis RE, Pierce J, Hepperle M, Xu Y, Hottelet M, et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005 Jan 1;11(1):28–40. [PubMed] [Google Scholar]
- 40.Kloo B, Nagel D, Pfeifer M, Grau M, Duwel M, Vincendeau M, et al. Critical role of PI3K signaling for NF-kappaB-dependent survival in a subset of activated B-cell-like diffuse large B-cell lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2011 Jan 4;108(1):272–277. doi: 10.1073/pnas.1008969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce SK, Liu W. The tipping points in the initiation of B cell signalling: how small changes make big differences. Nature reviews Immunology. 2010 Nov;10(11):767–777. doi: 10.1038/nri2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolar P, Hanna J, Krueger PD, Pierce SK. The constant region of the membrane immunoglobulin mediates B cell-receptor clustering and signaling in response to membrane antigens. Immunity. 2009 Jan 16;30(1):44–55. doi: 10.1016/j.immuni.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolar P, Meckel T. Imaging B-cell receptor signaling by single-molecule techniques. Methods Mol Biol. 2009;571:437–453. doi: 10.1007/978-1-60761-198-1_29. [DOI] [PubMed] [Google Scholar]
- 44.Yu D, Cook MC, Shin DM, Silva DG, Marshall J, Toellner KM, et al. Axon growth and guidance genes identify T-dependent germinal centre B cells. Immunology and cell biology. 2008 Jan;86(1):3–14. doi: 10.1038/sj.icb.7100123. [DOI] [PubMed] [Google Scholar]
- 45.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nature reviews Molecular cell biology. 2002 Aug;3(8):586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 46.Fornasiero EF, Opazo F. Super-resolution imaging for cell biologists: Concepts, applications, current challenges and developments. BioEssays : news and reviews in molecular, cellular and developmental biology. 2015 Jan 12; doi: 10.1002/bies.201400170. [DOI] [PubMed] [Google Scholar]
- 47.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nature immunology. 2010 Jan;11(1):90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shelby SA, Holowka D, Baird B, Veatch SL. Distinct stages of stimulated FcepsilonRI receptor clustering and immobilization are identified through superresolution imaging. Biophysical journal. 2013 Nov 19;105(10):2343–2354. doi: 10.1016/j.bpj.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, et al. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013 Mar 21;38(3):461–474. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 50.Blonska M, Lin X. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunological reviews. 2009 Mar;228(1):199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nature reviews Immunology. 2013 Aug;13(8):578–591. doi: 10.1038/nri3487. [DOI] [PubMed] [Google Scholar]
- 52.Baracho GV, Miletic AV, Omori SA, Cato MH, Rickert RC. Emergence of the PI3-kinase pathway as a central modulator of normal and aberrant B cell differentiation. Current opinion in immunology. 2011 Apr;23(2):178–183. doi: 10.1016/j.coi.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delgado P, Cubelos B, Calleja E, Martinez-Martin N, Cipres A, Merida I, et al. Essential function for the GTPase TC21 in homeostatic antigen receptor signaling. Nature immunology. 2009 Aug;10(8):880–888. doi: 10.1038/ni.1749. [DOI] [PubMed] [Google Scholar]
- 54.Clark MR, Campbell KS, Kazlauskas A, Johnson SA, Hertz M, Potter TA, et al. The B cell antigen receptor complex: association of Ig-alpha and Ig-beta with distinct cytoplasmic effectors. Science. 1992 Oct 2;258(5079):123–126. doi: 10.1126/science.1439759. [DOI] [PubMed] [Google Scholar]
- 55.Cherukuri A, Cheng PC, Sohn HW, Pierce SK. The CD19/CD21 complex functions to prolong B cell antigen receptor signaling from lipid rafts. Immunity. 2001 Feb;14(2):169–179. doi: 10.1016/s1074-7613(01)00098-x. [DOI] [PubMed] [Google Scholar]
- 56.Depoil D, Fleire S, Treanor BL, Weber M, Harwood NE, Marchbank KL, et al. CD19 is essential for B cell activation by promoting B cell receptor-antigen microcluster formation in response to membrane-bound ligand. Nature immunology. 2008 Jan;9(1):63–72. doi: 10.1038/ni1547. [DOI] [PubMed] [Google Scholar]
- 57.Zotos D, Tarlinton DM. Determining germinal centre B cell fate. Trends in immunology. 2012 Jun;33(6):281–288. doi: 10.1016/j.it.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007 Jan 26;315(5811):528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 59.Ohnishi K, Melchers F. The nonimmunoglobulin portion of lambda5 mediates cell-autonomous pre-B cell receptor signaling. Nature immunology. 2003 Sep;4(9):849–856. doi: 10.1038/ni959. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Wang D, Guo Z, Zhao J, Wu B, Deng H, et al. Rho kinase phosphorylation promotes ezrin-mediated metastasis in hepatocellular carcinoma. Cancer Res. 2011 Mar 1;71(5):1721–1729. doi: 10.1158/0008-5472.CAN-09-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elliott BE, Meens JA, SenGupta SK, Louvard D, Arpin M. The membrane cytoskeletal crosslinker ezrin is required for metastasis of breast carcinoma cells. Breast Cancer Res. 2005;7(3):R365–R373. doi: 10.1186/bcr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legg JW, Lewis CA, Parsons M, Ng T, Isacke CM. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nature cell biology. 2002 Jun;4(6):399–407. doi: 10.1038/ncb797. [DOI] [PubMed] [Google Scholar]
- 63.Chen L, Monti S, Juszczynski P, Daley J, Chen W, Witzig TE, et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008 Feb 15;111(4):2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Idelalisib approved for trio of blood cancers. Cancer discovery. 2014 Oct;4(10):OF6. doi: 10.1158/2159-8290.CD-NB2014-122. [DOI] [PubMed] [Google Scholar]
- 65.Naylor TL, Tang H, Ratsch BA, Enns A, Loo A, Chen L, et al. Protein kinase C inhibitor sotrastaurin selectively inhibits the growth of CD79 mutant diffuse large B-cell lymphomas. Cancer Res. 2011 Apr 1;71(7):2643–2653. doi: 10.1158/0008-5472.CAN-10-2525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.