Abstract
Identification of essential genes is critical to understand the physiology of a species, propose novel drug targets and uncover minimal gene sets required for life. Although essential gene sets of several organisms have been determined using large-scale mutagenesis techniques, systematic studies addressing their conservation, genomic context and functions remain scant. Here we integrate 17 essential gene sets from genome-wide in vitro screenings and three gene collections required for growth in vivo, encompassing 15 Bacteria and one Archaea. We refine and generalize important theories proposed using Escherichia coli. Essential genes are typically monogenic and more conserved than nonessential genes. Genes required in vivo are less conserved than those essential in vitro, suggesting that more divergent strategies are deployed when the organism is stressed by the host immune system and unstable nutrient availability. We identified essential analogous pathways that would probably be missed by orthology-based essentiality prediction strategies. For example, Streptococcus sanguinis carries horizontally-transferred isoprenoid biosynthesis genes that are widespread in Archaea. Genes specifically essential in Mycobacterium tuberculosis and Burkholderia pseudomallei are reported as potential drug targets. Moreover, essential genes are not only preferentially located in operons, but also occupy the first position therein, supporting the influence of their regulatory regions in driving transcription of whole operons. Finally, these important genomic features are shared between Bacteria and at least one Archaea, suggesting that high order properties of gene essentiality and genome architecture were probably present in the last universal common ancestor or evolved independently in the prokaryotic domains.
Keywords: essential genes, operons, genome organization, prokaryotes, transposon mutagenesis, genome evolution
Graphical Abstract
INTRODUCTION
Bacteria and Archaea are widely diversified prokaryotic domains [1], adapted to a wide range of niches [2]. Prokaryotes evolved over billions of years and divergence of the major groups of Bacteria and of Archaea occurred between 2.5-3.2 and 3.1-4.1 billion years, respectively [3]. Prokaryotic genomes have been carved by selection pressures, population size bottlenecks, mutation and recombination rates and mobile genetic elements [4], resulting in highly variable genome sizes and contents in different phylogenetic groups [5, 6]. Genome sequencing efforts over the past two decades fueled the search for a universal set of genes that would represent the minimal genome. However, it has been demonstrated that as phylogenetic distance increases, the number of universal genes is reduced to a level that is unlikely to support cellular life [7, 8]. Single-gene deletion techniques and modern approaches such as large-scale transposon mutagenesis followed by high-throughput sequencing allowed simultaneous screening of as many as 1 million mutants with high-resolution [9-11]. Determination of bacterial essential genes using these approaches improved gene annotations [9], mapping of metabolic pathways [10, 12], identification of genotype to phenotype associations [12] and helped to define genome-wide essential gene sets in vitro [13] and during infection and colonization [14, 15]. As gene essentiality is condition-dependent, in vivo screenings have gained attention [15-17] because of their potential to uncover genes involved in pathogenesis, which are of particular interest in the case of resistant bacteria [9, 13]. Moreover, identification of essential genes is critical for the development of engineered cells for compound production [18].
Integrative analyses of computationally and experimentally-determined essential gene sets uncovered important features that constitute the basis of essentiality prediction in bacteria, such as their lower substitution rates when compared to nonessential genes [19] and the correlation of essentiality with high gene expression [20] and conservation [21]. Functional analysis using Clusters of Orthologous Groups (COGs) [22] revealed that Information storage and processing genes are overrepresented among essential genes in most species, whereas species specificity was found in more peripheral metabolic pathways [23]. Moreover, only 34% and 61% of the Bacillus subtilis and Escherichia coli essential genes are universally conserved in their phyla (Firmicutes and Gamma-proteobacteria, respectively) [21], favoring essentiality prediction based on persistence, according to which persistent genes are those shared by most genomes [21]. Persistence analysis allowed the identification of truly essential genes that are frequently missing in essential gene sets (e.g. DNA repair genes) [21, 24]. Further, essential and persistent nonessential genes share common characteristics such as: high sequence conservation and expression rates (predicted by the codon adaptation index); preferential localization at the chromosomal leading strand, minimizing the risk of head-on collisions between DNA and RNA polymerases [20], and tendency to be in operons [21]. Approximately 60% of the bacterial genes are co-transcribed in polycistronic RNAs derived from operons [25]. The most accepted theory of operon formation is the co-regulation hypothesis [26, 27], which postulates that operons are formed by rearrangements that place two or more genes together, with subsequent maintenance of such structure by selection for concerted transcriptional regulation and translation of functionally related proteins. Further, according to the co-regulation hypothesis, essential genes would be preferentially located in operons, as observed in E. coli [26, 27].
Although important discoveries have been reported by means of comparative genomics and experimental data on gene essentiality in E. coli, the progress brought upon by next-generation sequencing and mutagenesis methods allowed the evaluation of distantly related species with high resolution. Here we integrate data from 20 genome-wide screenings in 16 organisms, encompassing 17 saturated in vitro and 3 in vivo datasets. Unlike previous studies, we took advantage of a recently published archaeal essential gene set [28], extending our analyses to the two prokaryotic domains of life. Important trends discovered in E. coli are also present in all major bacterial groups, such as the extensive conservation of essential genes and their propensity to be in operons. Moreover, essential gene sets in vitro are very different from those in vivo, probably because of the recruitment of a less conserved gene set to survive under stress conditions and limited nutrient sources. We have also demonstrated that essential genes do not only tend to be in operons, but also occupy the first position therein. Finally, many genomic features of the bacterial essential genes are also present in the only archaeal species for which a large-scale essentiality screening is available, suggesting that a high level genomic organization could have been either present in the last universal common ancestor (LUCA) or evolved independently in the two prokaryotic domains of life.
RESULTS AND DISCUSSION
The number of essential genes is not correlated to genome size in Bacteria
We carefully selected a compendium of large-scale studies of gene essentiality across a wide range of phylogenetic groups and conditions (Table 1). Our curation process (see methods for details) resulted in 17 in vitro experiments for 16 organisms from 6 distinct phyla (Table 1). The datasets used here have important technical and biological differences. The three organisms (Streptococcus sanguinis, E. coli and B. subtilis) showing the lowest numbers of essential genes (Figure 1; Table 1) were evaluated by single-gene knockouts, which is considered the gold standard approach. On the other hand, Methanococcus maripaludis (an Archaea) and Mycobacterium tuberculosis showed the largest essential gene datasets (Figure 1; Table 1). Both species grow in the presence of CO2 and have complex nutritional requirements and more genes are required for growth due to suboptimal growth media. However, as shown throughout the manuscript, these datasets are unbiased and can be used in systematic analyses like that reported here.
Table 1.
Experimentally-determined essential gene sets used in the present study.
| Phylum | Species | Total of essential Genes | Number of CDSs | % of essential genes | Medium | Approach | Ref |
|---|---|---|---|---|---|---|---|
| Actinobacteria | Mycobacterium tuberculosis H37Rv | 774 | 4018 | 19.20% | Minimal | High-density transposon mutagenesis + Illumina sequencing | [85] |
| Bacteroides | Bacteroides fragilis 638R | 550 | 4290 | 12.80% | Rich | Transposon delivery vetor + Illumina sequencing | [86] |
| Bacteroides | Porphyromonas gingivalis ATCC 33277 | 463 | 2090 | 22.10% | Rich | Global transposon mutagenesis + Illumina sequencing (TnSeq) | [87] |
| Firmicutes | Bacillus subtilis 168 | 271 | 4176 | 6.40% | Rich | Gene-by-gene inactivation | [77] |
| Firmicutes | Streptococcus sanguinis SK36 | 218 | 2270 | 9.60% | Rich | Systematic gene replacement | [10] |
| Methanococci | Methanococcus maripaludis S2 | 526 | 1722 | 30.50% | Rich | Saturation mutagenesis technique + Illumina sequencing (TnSeq) | [28] |
| Methanococci | Methanococcus maripaludis S2 | 664 | 1722 | 38.50% | Minimal | Saturation mutagenesis technique + Illumina sequencing (TnSeq) | [28] |
| Proteobacteria (alpha) | Caulobacter crescentus NA1000 | 480 | 3877 | 12.40% | Rich | Hyper-saturated transposon mutagenesis + Illumina sequencing | [88] |
| Proteobacteria (beta) | Burkholderia pseudomallei K96243 | 505 | 5727 | 8.80% | Rich | Transposon directed insertion sequencing site (TRADIS) | [9] |
| Proteobacteria (beta) | Burkholderia thailandensis E264 | 406 | 5632 | 7.20% | Rich | Saturation level transposon mutagenesis + Illumina sequencing (TnSeq) | [89] |
| Proteobacteria (gamma) | Acinetobacter baylyi ADP1 | 499 | 3307 | 15.10% | Minimal | Single-gene-deletion | [90] |
| Proteobacteria (gamma) | Escherichia coli K12 | 303 | 4145 | 7.30% | Rich | In-frame single gene deletions | [91] |
| Proteobacteria (gamma) | Francisella tularensis novicida U112 | 396 | 1719 | 23.00% | Rich | Sequence-defined transposon mutant library + Sanger sequencing | [92] |
| Proteobacteria (gamma) | Salmonella enterica typhi Ty2 | 356 | 4370 | 8.10% | Rich | Transposon directed insertion sequencing site (TRADIS) | [76] |
| Proteobacteria (gamma) | Salmonella enterica typhimurium SL1344 | 353 | 4446 | 7.90% | Rich | Transposon directed insertion sequencing site (TRADIS) | [76] |
| Tenericutes | Mycoplasma genitalium G37 | 382 | 475 | 80.40% | Rich | Global transposon mutagenesis + Sanger sequencing | [93] |
| Tenericutes | Mycoplasma pulmonis CT | 310 | 782 | 39.60% | Rich | Global transposon mutagenesis + Sanger sequencing | [94] |
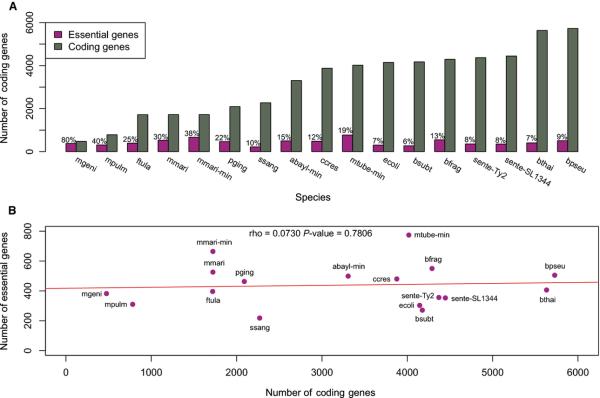
Figure 1. Essential genes obtained from 17 dispensability experiments and their correlation to the total gene complement.
A) Percentage of essential protein-coding genes; B) Correlation between essential gene set size and genome size. Abbreviations: abayl-min (Acinetobacter baylyi ADP1, minimal medium); bfrag (Bacteroides fragilis 638R); bpseu (Burkholderia pseudomallei K96243); bsubt (Bacillus subtilis 168); bthai (Burkholderia thailandensis E264); ccres (Caulobacter crescentus NA1000); ecoli (Escherichia coli K-12); ftula (Francisella tularensis novicida U112); mgeni (Mycoplasma genitalium G37); mmari (Methanococcus maripaludis S2, rich medium); mmarimin (Methanococcus maripaludis S2, minimal medium); mpulm (Mycoplasma pulmonis CT); mtube-min (Mycobacterium tuberculosis H37Rv, minimal medium); pging (Porphyromonas gingivalis ATCC 33277); sente-SL1344 (Salmonella enterica typhimurium SL1344); sente-Ty2 (Salmonella enterica typhi Ty2); ssang (Streptococcus sanguinis SK36, minimal medium).
We found that while genome sizes vary from 475 (Mycoplasma genitalium) up to 5,727 (Burkholderia pseudomallei) protein-coding genes, the number of essential genes varies from 218 (S. sanguinis) to 774 (M. tuberculosis) (Figure 1; Table 1), indicating a lack of correlation between the number of essential genes and genome size in Bacteria (Figure 1; Table 1). Essential gene sets are apparently more constrained in prokaryotes than in eukaryotes, in which the fraction of essential genes is more proportional to genome size. Saccharomyces cerevisiae and Schizosaccharomyces pombe have 1,100-1,300 genes that are essential for growth [29, 30]. Even though the gene complements of these fungi have sizes comparable to larger bacterial genomes, their essential gene sets are more than ~1.4 larger than the largest prokaryotic essential gene set studied here (i.e. Mycobacterium tuberculosis, with 774 essential genes). Although systematic screenings are yet unavailable for multicellular eukaryotes, ~3,000 genes have been demonstrably essential for viable development in mouse [31]. We will be able to have a better picture of this phenomenon when more systematic surveys become available for Archaea and eukaryotes.
Genes involved in cellular proliferation are enriched in essential genes
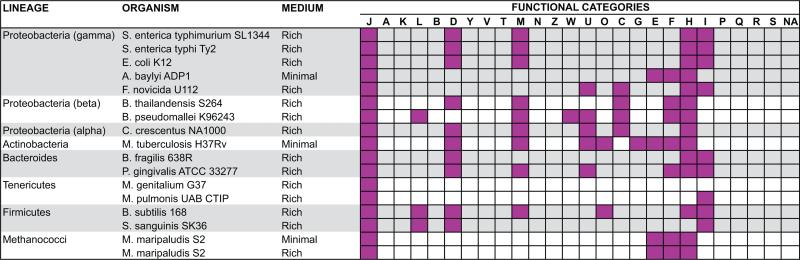
Most studies report genes indispensable for bacteria in rich medium, allowing growth without several biosynthetic pathways. Because these experiments are stress-free, essential genes mainly comprise the basic cellular machinery (e.g. DNA replication and protein translation genes). This observation is supported by the over-representation of essential genes in the Translation, ribosomal structure and biogenesis category (J) in all in vitro screenings (Figure 2). Our results also show other over-representation patterns in these gene sets (Figure 2). Because experimental determination is largely independent of evolutionary concepts and computational predictions, pathways conserved in few species can also be detected as functionally enriched. For example, Cell wall, membrane and envelope biogenesis (M) genes were enriched in many datasets (Figure 2) in spite of the distinct cell wall composition and biosynthetic pathways encoded in gram-positive and gram-negative bacterial genomes [32].
Figure 2. Functional categories enriched in essential gene datasets.
Squares in magenta represent functional categories enriched in the respective essential gene set (Fisher's exact test; P < 0.05).
We also analyzed individual essential genes, pathways and their phyletic patterns. Lipid transport and metabolism (I) holds relevant differences between Bacteria and Archaea. Isoprenoids (or terpenoids) are important elements of prokaryotic membrane and cell wall [33]. Phospholipids are other critical components of membranes and their biosynthesis is widely conserved in bacteria [34]. Phosphatidate cytidylyltransferase (EC 2.7.7.41) and CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase (EC 2.7.8.5), essential enzymes involved in glycerophospholipid metabolism (KEGG: ec00564), were conserved in nearly all bacteria (Table S1). On the other hand, the role of fatty acids in Archaea remains controversial, as the archaeal membrane depends on isoprenoids synthesized by the mevalonate pathway (KEGG: M00095) [35]. The mevalonate pathway comprises the first steps (from acetyl-CoA to isopentenyl-PP) in terpenoid backbone biosynthesis (KEGG: ec00900) in Archaea, fungi and metazoans, while Bacteria and Apicomplexa perform these steps up to isopentenyl-PP through the methylerythritol 4-phosphate pathway (KEGG: M00096) [36]. Enzymes from the mevalonate pathway (i.e. MMP1212, MMP1211, MMP0087 and MMP1335) are essential in M. maripaludis, as well as those from the methylerythritol 4-phosphate pathway are essential in Bacteria. Mevalonate pathway genes are essential in S. sanguinis (SSA_0338, SSA_0337, SSA_0333, SSA_0335, SSA_0334), as previously shown for Streptococcus pneumoniae [37]. Unlike most bacteria, S. sanguinis uses the mevalonate pathway, which was horizontally transferred from archaeal or eukaryotic cells [33, 37] and probably replaced the methylerythritol 4-phosphate pathway. Finally, the undecaprenyl pyrophosphate synthase (EC 2.5.1.31), a lipid carrier for peptidoglycan synthesis in bacteria (probably a glycosyl carrier in Archaea) and involved in the final step of terpenoid backbone biosynthesis (ec00900), is essential in archaea and most bacteria, except for mycoplasmas, which do not have cell wall.
Cell cycle control, cell division, chromosome partitioning (D) and Coenzyme transport and metabolism (H) were enriched in 58% and 82% of experiments, respectively. Cell division is directly related to cellular mass increase, cytoplasm and DNA partitioning between daughter cells and membrane remodeling. Defective cell division genes may result in asymmetrically-sized daughters [38] or impaired cell division [39]. Many studies on bacterial cell division focus on FtsZ [39], a protein conserved in most bacteria and in Euryarchaeota [40], a phylum that includes M. maripaludis and presents a bacterial-type division mechanism [41]. FtsZ is important for septum formation [42] and is essential in all species except Porphyromonas gingivalis and M. maripaludis (Table S1). The P. gingivalis FtsZ gene had 2 insertions in both technical replicates and was not considered essential (Brian Klein, personal communication). M. maripaludis has two FtsZ genes (MMP1436 and MMP1500) with identical domain architectures, probably providing a genetic backup to each other.
Coenzyme transport and metabolism (H) comprises pathways whose products are critical for various other pathways. Genes required for the synthesis of coenzyme A (CoA) and biotin (coenzyme R/vitamin H), critical coenzymes in fatty acid oxidation and other metabolic pathways, are required in most species (Table S1). Further, genes involved in the production of nicotinamide (vitamin B3), riboflavin (vitamin B2), folate (vitamin B9) and S-adenosylmethionine are also essential. Nevertheless, genes that are poorly characterized or have no COG annotation account for 28-54% of the essential genes sets. Hence, other functional trends are likely to emerge as genome annotations improve.
Conservation, gene families and the composition of essential gene complements
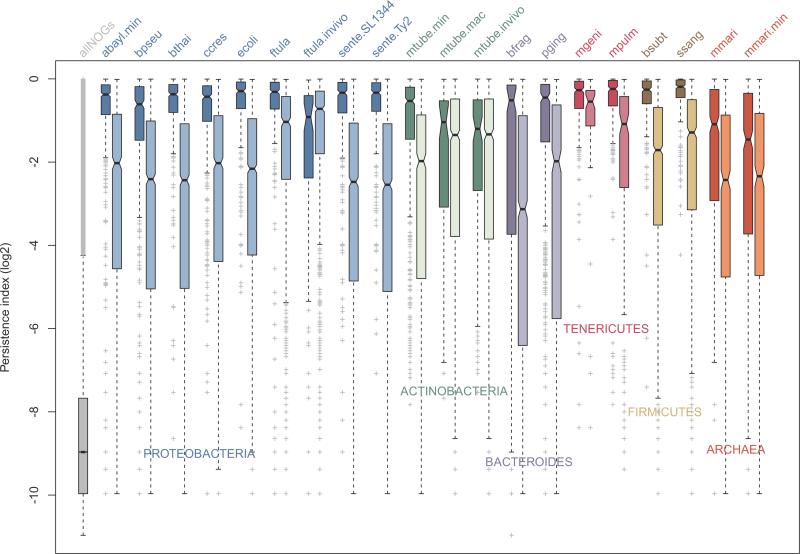
To investigate the conservation of the essential gene sets, we mapped all protein-coding genes from the 16 prokaryotes studied here to the eggNOG database [43]. Genes assigned to the same non-supervised orthologous group (NOG) were considered homologs. Conservation across 2,031 genomes available in eggNOG was assessed using the Persistence Index [44] (see methods for details). To avoid biases from the phylogenetic composition of the database, essential gene properties were compared with their non-essential counterparts in the same genome. Strikingly, all essential datasets determined in vitro comprise genes that are far more conserved than the non-essential genes (Figure 3). These results support the existence of a highly-conserved core of genes responsible for growth in a common condition (i.e. rich medium), in spite of the wide evolutionary range and other phenomena that affect gene retention/loss (e.g. non-orthologous gene displacement [45]). These observations generalize concepts developed using E. coli and B. subtilis [21, 44] to the other major bacterial groups and Archaea, implying that higher conservation of essential genes is a common feature in prokaryotes. Interestingly, this trend is apparently attenuated in vivo (Figure 3), likely as a consequence of divergent survival strategies to grow under unstable nutrient offer and attacks from the immune system. It is important to bear in mind that in vivo screenings typically do not reach saturation and have in vitro steps before inoculation, resulting in a potentially underappreciated gene set. However, due to their medical relevance, the lower conservation of genes required in vivo deserves further investigation, as discussed below.
Figure 3. Conservation of essential and nonessential gene sets across thousands of species.
Boxplot representation of essential gene sets across thousands of species available in the eggNOG database (see methods for details). Unless indicated otherwise, rich media were used in the screenings. For abbreviations of in vitro experiments refer to Figure 1. For in vivo experiments: ftula-invivo (F. tularensis novicida U112, in vivo); mtube-invivo (Mycobacterium tuberculosis H37Rv, in vivo); and mtube-mac (Mycobacterium tuberculosis H37Rv, macrophages). Essential and nonessential gene sets for each condition are side-by-side, in dark and light colors. Proteobacteria, Actinobacteria, Bacteroides, Tenericutes, Firmicutes and Archaea are represented in blue, green, purple, magenta, brown and red, respectively.
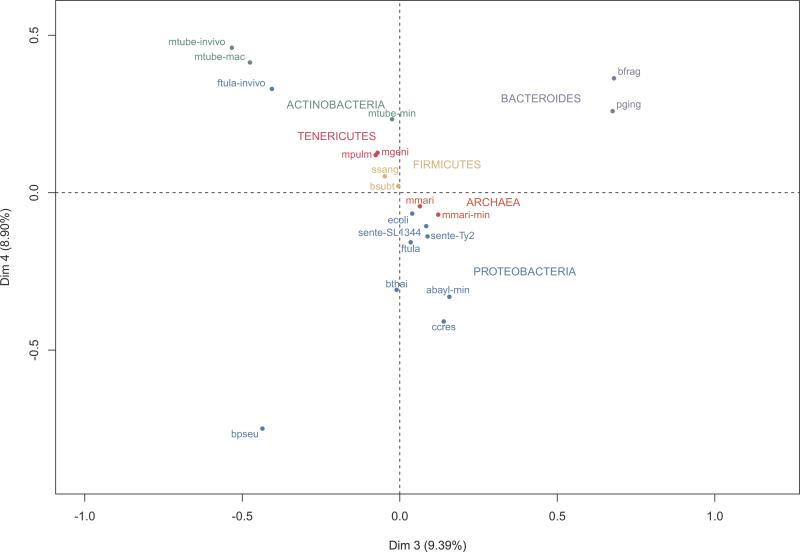
In order to evaluate the diversity of the essential gene repertories, we performed a Multiple Correspondence Analysis (MCA) [46], a multivariate method to reduce data dimension and identify systematic patterns of variations in categorical data. We defined the categorical data as the presence/absence of essential genes from each gene set in each NOG. Besides clustering closely-related organisms, MCA allowed us to capture an influence of the environment, reflected by departures from the phylogeny-driven clustering (Figure 4). The three in vivo required gene complements from the distantly related M. tuberculosis and F. tularensis novicida were closely positioned to each other, suggesting that common strategies might be employed when in contact with the immune system (Figure 4). Although there are limitations in these in vivo datasets (discussed above), we investigated this important trend in further detail. NOGs that are exclusively essential in M. tuberculosis in vivo conditions (i.e. those in which there is no other essential gene in any of the other datasets) were retrieved, aiming to find genes related to infection and pathogenesis (Table S2). This set comprises several lipid metabolism genes, which is coherent with previous reports emphasizing the energetic roles of lipids in M. tuberculosis in vivo [15, 47, 48]. Transporters from the MFS and ABC superfamilies and the tetR helix-turn-helix transcriptional repressor Rv3050c were also found as essential in this gene set. Since members of the tetR family have been related to multiple biological processes related with stress [49], Rv3050c might be a critical regulator during M. tuberculosis infection. It is also clear that Burkholderia pseudomallei, the causative agent of melioidosis, has important peculiarities concerning its essential gene complement (Figure 4) and we analyzed its exclusively essential NOGs, as explained above (Table S2). Strikingly, there are 105 NOGs from which members are essential only in B. pseudomallei, including many ABC transporters and metabolic enzymes, supporting its complex metabolic landscape. This gene set also contains transcriptional regulators from the GntR, LysR and AraC families [50, 51], which might be related to the extraordinary antibiotic resistance of B. pseudomallei and its capacity to occupy a wide range of niches, from soil to intracellular environments [52]. Importantly, these results cannot be solely explained by B. pseudomallei genome size or plasticity [9, 53]. Burkholderia thailandensis, which diverged from B. pseudomallei around 47 million years ago [9] and also harbors a large and highly plastic genome, has its essential gene set closely grouped to other proteobacteria (Figure 4). Given the scarce treatment options and the classification of B. pseudomallei as a potential bioterrorism threat by the U.S. Centers for Disease Control and Prevention, this gene set constitute a valuable source of candidates for downstream experiments to validate their potential as drug or vaccine candidates.
Figure 4. Multiple Correspondence Analysis (MCA) of the presence/absence of essential genes in NOGs.
MCA analysis of essential gene sets based on the presence/absence profiles of each mapped NOG. The first two dimensions obtained in MCA were dominated by one or two samples and therefore, are not very useful for separation purposes. Dimensions 3 and 4 allowed an evolutionarily coherent clustering, while still accounting for a significant amount of variance. For abbreviations of in vitro experiments refer to Figure 1. For in vivo experiments: ftula-invivo (F. tularensis novicida U112, in vivo); mtube-invivo (Mycobacterium tuberculosis H37Rv, in vivo); and mtube-mac (Mycobacterium tuberculosis H37Rv, macrophages). For color codes, refer to Figure 3.
Next, we compared essential genes from the archaea M. maripaludis to COG/NOG annotations from the 15 bacterial genomes with in vitro essential datasets available. Essential archaeal COG/NOGs with no orthologs in bacteria were considered M. maripaludis exclusive essential gene groups, while COG/NOGs with at least one bacterial ortholog were considered shared gene groups. Out of the 520 genes essential in rich medium, 10 were assigned to two COG/NOGs and not considered for further analysis; 194 genes were archaeal exclusive (including 26 genes with no COG/NOG annotation) and 316 genes had bacterial homologs. The eggNOG categories Replication, recombination and repair (L) and Poorly characterized (S and R) prevailed among exclusive essential genes (Figure S1). Interestingly, three large and almost entirely essential operons (6-8 genes) involved in methane metabolism were identified. Tungstein-containing formylmethanofuran dehydrogenase (DOOR operonID 95836, Table S3) catalyzes the dehydrogenation of formylmethanofuran to methanofuran and CO (EC: 1.2.99.5). This enzymatic complex, present in methanogenic and sulfate-reducing archaea, is related to the first steps in methanogenesis, responsible for CO2 reduction to methane and autotrophic CO2 fixation, being crucial for archaeal metabolism [54]. Tetrahydromethanopterin S-methyltransferase and Methyl-coenzyme M reductase operons (DOOR operonIDs 95906 and 95905, respectively) are related to Coenzyme transport and metabolism. The former catalyzes the formation of methyl-coenzyme M and tetrahydromethanopterin from coenzyme M and methyltetrahydromethanopterin (EC: 2.1.1.86) [55], whereas the latter plays a role in the final step of methane biosynthesis, reducing methyl-coenzyme M and coenzyme B to methane (EC: 2.8.4.1) [56]. Large-scale methanogen production is of great biotechnological interest and understanding which genes and operons are involved in this process is critical. Since methanogenic Archaea are extremely important in anaerobic decomposition of sewage, optimized methane-producing cells could be used in industrial waste management and as a methane-renewable energy source.
We also sought to identify universal essential genes, as this set may help us to understand some features of the LUCA. We found 19 of such genes in all 16 organisms tested in vitro (Table 2), mostly belonging to Information storage and processing (J). This gene set comprises 6 aminoacyl-tRNA synthetases, 8 ribosomal proteins and the alpha subunit of DNA polymerase. Other major proteins are secY, the main transmembrane subunit of type II secretion system (Intracellular trafficking, secretion, and vesicular transport, U), and prs, which converts ribose 5-phosphate into phosphoribosyl pyrophosphate (EC: 2.7.6.1), which are essential for purine metabolism. Taken together, these observations are in agreement with the status of protein translation and DNA replication as central biological processes in all organisms. Nevertheless, some groups have long been reported non-orthologous or distantly related genes performing the same functions even in the core machinery between Bacteria and Archaea [45, 57, 58], explaining the small number of universal genes, especially when only experimentally-determined essential genes are considered. Our findings demonstrate the existence of an important core of experimentally determined essential genes shared by Bacteria and an Archaea, which could have been essential in the LUCA as well. Availability of other archaeal essential gene sets will certainly help to evaluate this hypothesis.
Table 2. Universally conserved essential COG/NOGs.
Only essential genes experimentally determined were considered.
| INFORMATION STORAGE AND PROCESSING | ||
|---|---|---|
| Translation, ribosomal structure and biogenesis (J) | COG0018 | Arginyl-tRNA synthetase |
| COG0008 | Glutamyl- and glutaminyl-tRNA synthetases | |
| COG0124 | Histidyl-tRNA synthetase | |
| COG0495 | Leucyl-tRNA synthetase | |
| COG0442 | Prolyl-tRNA synthetase | |
| COG0172 | Seryl-tRNA synthetase | |
| COG0090 | Ribosomal protein L2 | |
| COG0087 | Ribosomal protein L3 | |
| COG0088 | Ribosomal protein L4 | |
| COG0097 | Ribosomal protein L6P/L9E | |
| COG0102 | Ribosomal protein L13 | |
| COG0092 | Ribosomal protein S3 | |
| COG0522 | Ribosomal protein S4 and related proteins | |
| COG0098 | Ribosomal protein S5 | |
| Transcription (K) | COG0202 | DNA-directed RNA polymerase, alpha subunit/40 kD subunit |
| Replication, recombination and repair (L) | COG0592 | DNA polymerase III sliding clamp (beta) subunit, PCNA homolog |
| CELLULAR PROCESSES AND SIGNALING | ||
|---|---|---|
| Cell cycle control, cell division, chromosome partitioning (D) | COG0037 | Predicted ATPase of the PP-loop superfamily implicated in cell cycle control |
| Intracellular trafficking, secretion, and vesicular transport (U) | COG0201 | Preprotein translocase subunit SecY |
| COG0552 | Signal recognition particle GTPase (protein FtsY) ** |
| METABOLISM | ||
|---|---|---|
| Nucleotide transport and metabolism (F) | COG0462 | Phosphoribosylpyrophosphate synthetase |
* When considering only the Methanococcus maripaludis minimal medium;
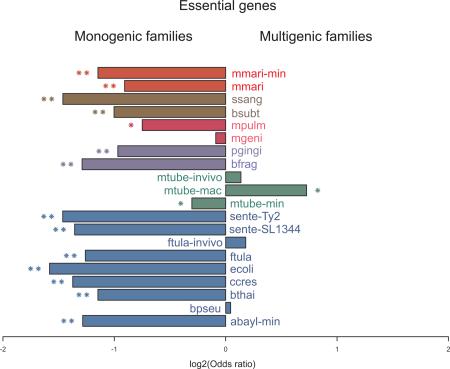
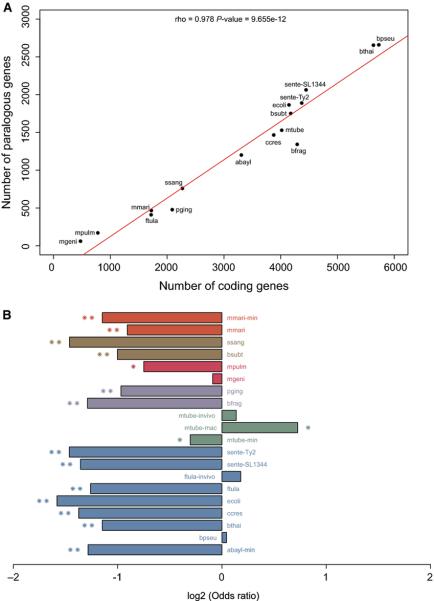
Multigene families may be of great adaptive value in the evolution of novel functions and providing biochemical backups [59-61]. We found a strong correlation between the number of genes from multigene families and number of CDSs in the species analyzed here (see methods for details) (Figure 5A; Table S4); together with the lack of correlation between the number of essential genes and CDSs (Figure 1), this result suggests that homologs often compensate single gene loss. We tested this hypothesis and found a strong negative association between the presence of homologs in the genome and essentiality in vitro (13/17 with P < 10−5; Figure 5B). In other words, under controlled conditions essential genes tend to come from monogenic families. Surprisingly, when we performed the same analysis on conditions closer to the organism lifestyle, such as growth in vivo or in macrophages, there is an apparent reversion of this trend with the recruitment of genes from multigene families to essential roles (Figure 5B). A clearer picture is likely to emerge when saturated in vivo screenings become available. One may argue that the enrichment of essential genes in monogenic families derive from a technical limitation in single-gene deletion/disruption screening when a homologous backup is available. Nevertheless, the theoretical foundations of gene essentiality lie on a single-gene framework, which is biologically relevant and has proven extremely successful over the past two decades; therefore, our observations have implications for bacterial evolution, as discussed below.
Figure 5. Association between the number of coding genes and gene essentiality with the presence of homologs.
A) Total number of coding genes versus genes in multigene families: genes with same COG/NOG assignment in a genome were considered part of multigene families. B) Gene essentiality versus presence of a homolog in the genome: Fisher's exact tests were performed to assess the enrichment of essential genes in multigene families. Bars with one and two asterisks represent P ≤ 10−2 and P ≤ 10−5, respectively. For abbreviations of in vitro experiments refer to Figure 1. For in vivo experiments: ftula-invivo (F. tularensis novicida U112, in vivo); mtubeinvivo (Mycobacterium tuberculosis H37Rv, in vivo); and mtube-mac (Mycobacterium tuberculosis H37Rv, macrophages). For color codes, refer to Figure 3.
Finally, we have also tested the prominence of horizontal gene transfers (HGTs) in shaping prokaryotic essential gene sets by performing all the analyses described in this section after excluding all genes predicted to have high probability of an HGT event (see methods for details). Very low numbers of essential HGT genes (less than 2%) were found in all species (Table S5), as expected from previous reports [27, 62]; the removal of HGT genes did not affect the statistical significance of our results (data not shown).
In vitro and in vivo essential gene sets are extremely dissimilar
The identification of genes required in vivo is of great interest, not only in biomedical research, but also from an evolutionary perspective, as these genes are likely to be required in nature. In vivo screenings are mostly based on mutant fitness -- when a mutant fails to grow or shows reduced counting in the output pool, it is inferred that the disrupted gene is important for the infection and survival of the pathogen inside the host [15, 17, 47]. Among the selected studies, unique gene sets are found to be required in vivo and essential in vitro (Figure S2). As discussed above, this overlap might be underappreciated because the mutants are passed in vitro before inoculation and many important genes in vitro are likely to be critical in vivo as well. Nevertheless, genes required in vivo but not in vitro are functionally diverse (Figure S2), being important candidates for drug intervention.
Over disease progression, mutants must colonize, disseminate and persist under unstable nutrient supply and continuous attacks from the immune system. Accordingly, many genes required in vivo are related to metabolism categories (Figure S2). Interestingly, the most predominant categories within the in vivo required gene sets are poorly characterized or have no COG assignment (Figure S2). As these genes may play important roles in pathogenesis, we analyzed their protein domain architectures. Some genes required for F. tularensis novicida during mice infection are related to coenzyme pyrrolo-quinoline-quinone (PQQ) biosynthesis (FTN_0933, Pfam: PF05402), variable adherence-associated antigen adhesins (FTN_1133, Pfam: PF01540) and DNA repair (FTN_1196, Pfam: PF02575) (Table S6). Further, genes involved in fatty acid synthesis (Rv0100, Pfam: PF00550) and processome of tRNAs or rRNAs (Rv0207c, Pfam: PF01936) are essential in M. tuberculosis in vivo conditions and categorized as poorly characterized (Table S6). These results illustrate an open field to phenotypic/functional genomic studies that could shed light on the roles of those genes in complex host-pathogen interactions.
Essential genes are not uniformly distributed inside operons
It has been demonstrated that essential genes are enriched in operons in E. coli [26] and we tested whether this is a general feature in other Bacteria and in Archaea. There is a clear trend for essential genes to occupy operons across all 16 prokaryotic genomes (P ≤ 0.05; Fisher's exact test) (Table S7). Further, the statistical significance is very high in 13 of these conditions (P < 1.6×10−4; Fisher's exact test) (Table S7). Importantly, these results were largely supported even without considering genes encoding ribosomal proteins, which are clustered in large, widely-conserved operons (Table S8). The tendency of essential genes to be in operons was further corroborated using simulated datasets (data not shown). Thus, the prevalence of essential genes in operons is an ancient, high-level prokaryotic feature, probably present in the LUCA. Alternatively, the adaptive value of arranging essential genes in operons is so high that it might have evolved independently in Bacteria and Archaea. Further, we analyzed the association between gene order and essentiality. Gene order is generally preserved in closely related organisms but rapidly decreases with phylogenetic distance [6], except for a few widely-conserved operons [63-65]. However, even such extremely conserved operons are found in distinct arrangements across bacterial and archaeal genomes [63, 66]. Remarkably, we observed that essential genes preferentially occupy the first position in operons containing at least one essential gene (Table 3). Two- and three-sized operons account for the majority (52.3-74.9%) of operons [25, 67, 68], regardless of the presence of essential genes in their structures. Thus, we performed a chi-square test in these 2- and 3-sized operons and confirmed the enrichment of essential genes in the first operon positions in most species, including M. maripaludis (P < 0.01; Table S9). This scenario is in agreement with a previous observation that essential genes are biased towards the 5′-end half of operons, while pseudogenes tend to be in the 3′-end half of Mycobacterium leprae operons [69].
Table 3.
Relationship between essential genes and operon position.
| Species | Medium | Position in the operon | Ess-opa | Ess-genesb | Operonsc | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 13 | 14 | |||||
| A. baylyi ADP1 | Minimal | 120 (65.2%) | 50 | 7 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 184 | 348 | 643 |
| B. subtilis 168 | Rich | 59 (58.4%) | 33 | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 101 | 206 | 818 |
| B. fragilis 638R | Rich | 155 (69.1%) | 42 | 19 | 4 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 224 | 407 | 956 |
| B. pseudomallei K96243 | Rich | 126 (55.2%) | 58 | 21 | 13 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 228 | 403 | 1146 |
| B. thailandensis S264 | Rich | 97 (57.4%) | 47 | 13 | 8 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 169 | 310 | 1155 |
| C. crescentus NA1000 | Rich | 123 (67.2%) | 38 | 14 | 3 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 183 | 338 | 844 |
| E. coli K-12 | Rich | 67 (52.8%) | 38 | 7 | 8 | 5 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 127 | 232 | 851 |
| F. tularensis novicida U112 | Rich | 94 (59.9%) | 42 | 12 | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 157 | 321 | 373 |
| M. maripaludis S2 | Rich | 102 (62.2%) | 44 | 13 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 164 | 351 | 362 |
| M. maripaludis S2 | Minimal | 128 (62.7%) | 56 | 12 | 5 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 204 | 423 | 362 |
| M. tuberculosis H37Rv | Minimal | 193 (59.2%) | 92 | 24 | 10 | 4 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 326 | 582 | 895 |
| M. genitalium G37 | Rich | 69 (82.1%) | 12 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 84 | 343 | 89 |
| M. pulmonis UAB | Rich | 81 (74.3%) | 21 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 109 | 247 | 175 |
| P. gingivalis ATCC | Rich | 144 (83.7%) | 16 | 6 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 172 | 385 | 455 |
| S. enterica typhimurium SL1344 | Rich | 75 (57.7%) | 37 | 10 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 130 | 238 | 881 |
| S. enterica typhi Ty2 | Rich | 76 (54.7%) | 44 | 7 | 8 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 139 | 264 | 838 |
| S. sanguinis SK36 | Rich | 50 (57.5%) | 27 | 6 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 87 | 159 | 489 |
Number of operons with at least one essential gene
Number of polycistronic essential genes
Total number of operons.
Prokaryotes have ~50% of their genes present in operons [70] and we found that essential genes are enriched in operons in several species (Table S7). Probably due to their higher expression [20], essential genes are more conserved than nonessential genes in terms of phyletic patterns (Figure 3) and sequence similarity [19]. Moreover, essential genes tend to be hubs and form cliques (complete sub-graphs) with each other in protein interaction networks [71, 72]. These features may contribute to the propensity of essential genes to become coordinately expressed with other genes (essential or otherwise) required under similar conditions. Operons reduce the amount of regulatory information needed for optimized transcription of co-regulated genes [27] and under complex regulatory requirements, operons are more likely to evolve than independent promoters in distinct genes [27]. This observation is supported by the more complex regulatory regions of operons when compared to monocistronic genes [27].
Genes from an operon are typically expressed according to their position and a strong correlation between operon length, order and expression has been proposed [73, 74]. Further, these genes generally display decaying expression in a staircase-like manner, with proximal genes (5′) being more expressed than 3′ genes [73, 74]. Based on their codon adaptation index and microarray data [75], essential genes are known to be highly expressed [20, 75]. Thus, the presence of essential genes in the first position in operons (Table 3; Table S9) has direct implications in their higher expression levels [73]. Further, the presence of upstream essential genes in operons increases their chances of being expressed if a mutation hampers the transcription of downstream genes. Taken together the results presented here and elsewhere, we hypothesize that the regulatory regions of 5’-essential genes may drive the regulation and, ultimately, the evolution of whole operons.
Here we reported a systematic analysis of experimentally determined essential genes. We found that essential genes are typically monogenic and more conserved than their nonessential counterparts across thousands of genomes. Extreme gene retention rates are at the foundations of gene persistence, which has been related to gene essentiality and genome organization [44]. Persistence was also extremely useful in the identification of truly essential genes that are not detected in controlled stress-free conditions (e.g. DNA repair genes) [44]. Nevertheless, we showed that many genes essential for growth in vivo are not widely conserved and often recruited from multigenic families, suggesting the existence of different survival strategies that co-evolved with the respective bacterial hosts. We propose novel targets for therapeutic or vaccine intervention by exploiting the phyletic patterns of such genes. Moreover, we showed that essential genes are not only preferentially located in operons, but tend to occupy the first position therein, supporting the importance of their regulatory regions in driving the expression of operons. Importantly, many features of gene essentiality in Bacteria are also present in the extremophile archaea M. maripaludis, suggesting that there are high order prokaryotic features that could have been either present in the LUCA or evolved independently in the two prokaryotic domains. We believe that the development of synthetic bacterial genomes for biotechnological applications may seriously benefit from an integrated computational and experimental approach based on the many features of essential and persistent genes reported here and elsewhere [27, 44], including not only the essential gene content, but also their specific positioning in operons.
MATERIALS AND METHODS
Data sources
Gene sets were selected after careful assessment of their original publications, using the following criteria for in vitro studies: 1) Essentiality must be supported by experimental evidence; 2) the experimental approach must have been systematic, covering at least 80% of the genome when single-gene deletions were used; 3) when transposon mutagenesis was employed, the screening should have reached saturation or near saturation. Only protein-coding genes were analyzed. The only studies that did not have strictly met the criteria above was with Salmonella enterica typhimurium SL1344 [76], which was based on cell fitness reduction when a gene was disrupted by a transposon and; Bacillus subtilis [77], for which essential genes were identified with a single crossover recombination technique complemented by a predicted set of essential genes. The former study was included because 95% of required gene set overlaps the essential gene set from a previous study [11], whereas the latter was considered because only 4% (185/4100) of the genes were used for essentiality prediction. A total of 16 organisms (1 Archaea and 15 Bacteria) and 17 in vitro screenings were selected (Table 1). Genomes were retrieved from Genbank [78], along with their gene identifiers, coordinates, gene names, strand information and protein sequence, which were extracted from Genbank files. Operon predictions were downloaded from DOOR2 [79], which was reported as one of the most accurate operon prediction repositories [80]. Simulations and data processing were conducted using in-house Perl, R (www.r-project.org) and shell scripts (available upon request).
Homology analysis
Homology data were obtained from the eggNOG database v4.0 [43]. Protein sequences were mapped to eggNOG (2,031 core-periphery species) using BLAST [81]. NOGs are an extension of the manually curated COGs [22, 43]. Since the species considered here are already part of the eggNOG database or have close relatives therein, we used strict BLAST criteria, e-value ≤ 10−10 and S ≥ 60%, where S is the coverage of the shortest sequence (either query or hit). The persistence index of a NOG was computed as previously described [44] as the fraction of the species with at least one homolog in that NOG. The presence/absence patterns of essential genes from a given species in a NOG in each condition were used to create a Boolean matrix that was used to perform a MCA using the FactoMineR package [46]. Functional category enrichment analyses were calculated using the Fisher's exact test (P < 0.05). Two genes from the same species were considered part of a multigene family if they share the same NOG. Genes associated with potential HGT events for all species (except S. enterica typhimurium SL1344 and B. fragilis 638R) were identified with the DarkHorse database [82], which is based on a statistical analysis of archaeal and bacterial genomes for the identification of phylogenetically atypical proteins [82]. Domain architectures were computed using HMMer v3 (E-value ≤ 0.01) [83] and the Pfam 27.0 database [84].
Genome organization
Presence of essential genes in operons
For each dataset, all protein-coding genes were used to build a 2×2 contingency table with the following categories: polycistronic essential genes; polycistronic nonessential genes; monocistronic essential genes and; monocistronic nonessential genes. Statistical associations were evaluated using the Fisher's exact test (P ≤ 0.05). In addition, the same analysis was performed in 10,000 simulated genomes with the same number of essential genes, operons and genes in each operon. Statistical analyses were performed in R.
Position of essential genes in operons
Only operons with at least one essential gene were considered. As most operons have 2 or 3 genes, we analyzed those for the preferential location of essential genes at the first position using chi-squared tests (P ≤ 0.01).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the following institutions for funding our research: Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (E-26/110.236/2011 and E-26/102.259/2013), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Universidade Estadual do Norte Fluminense Darcy Ribeiro (UENF) (A.L.G PhD fellowship). N.M.V. postdoctoral fellowship is funded by a partnership between CNPq and National Institutes of Health (NIH). T.M.V. is a recipient of an established investigator fellowship award from CNPq. This research was supported in part by the Intramural Research Program of the National Library of Medicine, NIH. We would also like to thank Dr. Robson Francisco de Souza for sharing the tjoin script and Dr. Fenglou Mao for helping with DOOR2.
Abbreviations
- LUCA
Last Universal Common Ancestor
- NOG
non-supervised orthologous group
- MCA
Multiple Correspondence Analysis
- COG
Clusters of Orthologous Group
Footnotes
AUTHOR CONTRIBUTIONS
Conceived the study: ALG, NMV, TMV; planned the analyses: ALG, NMV, TMV; performed the analyses: ALG, NMV, TMV; Wrote the paper: ALG, NMV, TMV.
REFERENCES
- 1.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nature reviews Microbiology. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 2.Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT. Microbial biogeography: putting microorganisms on the map. Nature reviews Microbiology. 2006;4:102–12. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 3.Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC evolutionary biology. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin EV. Evolution of genome architecture. The international journal of biochemistry & cell biology. 2009;41:298–306. doi: 10.1016/j.biocel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omelchenko MV, Makarova KS, Wolf YI, Rogozin IB, Koonin EV. Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome biology. 2003;4:R55. doi: 10.1186/gb-2003-4-9-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamames J. Evolution of gene order conservation in prokaryotes. Genome biology. 2001;2:RESEARCH0020. doi: 10.1186/gb-2001-2-6-research0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV. How many genes can make a cell: the minimal-gene-set concept. Annual review of genomics and human genetics. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10268–73. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moule MG, Hemsley CM, Seet Q, Guerra-Assuncao JA, Lim J, Sarkar-Tyson M, Clark TG, Tan PB, Titball RW, Cuccui J, Wren BW. Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio. 2014;5:e00926–13. doi: 10.1128/mBio.00926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu P, Ge X, Chen L, Wang X, Dou Y, Xu JZ, Patel JR, Stone V, Trinh M, Evans K, Kitten T, Bonchev D, Buck GA. Genome-wide essential gene identification in Streptococcus sanguinis. Scientific reports. 2011;1:125. doi: 10.1038/srep00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome research. 2009;19:2308–16. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome research. 2012;22:2541–51. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juhas M, Eberl L, Glass JI. Essence of life: essential genes of minimal genomes. Trends in cell biology. 2011;21:562–8. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell host & microbe. 2009;6:279–89. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12989–94. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS genetics. 2013;9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, Brittnacher MJ, Manoil C, Skerett SJ, Salama NR. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infection and immunity. 2009;77:232–44. doi: 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidl SR, Sheth RU, Wu A, Tabor JJ. Refactoring and optimization of light-switchable escherichia coli two-component systems. ACS synthetic biology. 2014;3:820–31. doi: 10.1021/sb500273n. [DOI] [PubMed] [Google Scholar]
- 19.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome research. 2002;12:962–8. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocha EP, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Molecular biology and evolution. 2004;21:108–16. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- 21.Fang G, Rocha E, Danchin A. How essential are nonessential genes? Molecular biology and evolution. 2005;22:2147–56. doi: 10.1093/molbev/msi211. [DOI] [PubMed] [Google Scholar]
- 22.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 23.Zhang CT, Zhang R. Gene essentiality analysis based on DEG, a database of essential genes. Methods in molecular biology. 2008;416:391–400. doi: 10.1007/978-1-59745-321-9_27. [DOI] [PubMed] [Google Scholar]
- 24.Acevedo-Rocha CG, Fang G, Schmidt M, Ussery DW, Danchin A. From essential to persistent genes: a functional approach to constructing synthetic life. Trends in genetics : TIG. 2013;29:273–9. doi: 10.1016/j.tig.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway T, Creecy JP, Maddox SM, Grissom JE, Conkle TL, Shadid TM, Teramoto J, San Miguel P, Shimada T, Ishihama A, Mori H, Wanner BL. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. mBio. 2014;5:e01442–14. doi: 10.1128/mBio.01442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pal C, Hurst LD. Evidence against the selfish operon theory, Trends in genetics. TIG. 2004;20:232–4. doi: 10.1016/j.tig.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Price MN, Huang KH, Arkin AP, Alm EJ. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome research. 2005;15:809–19. doi: 10.1101/gr.3368805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarmiento F, Mrazek J, Whitman WB. Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4726–31. doi: 10.1073/pnas.1220225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 30.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR, Jang YJ, Chung KS, Choi SJ, Park JY, Park Y, Kim HM, Park SK, Park HJ, Kang EJ, Kim HB, Kang HS, Park HM, Kim K, Song K, Song KB, Nurse P, Hoe KL. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nature biotechnology. 2010;28:617–23. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT, Mouse Genome Database G. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic acids research. 2011;39:D842–8. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swoboda JG, Campbell J, Meredith TC, Walker S. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem : a European journal of chemical biology. 2010;11:35–45. doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boucher Y, Kamekura M, Doolittle WF. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Molecular microbiology. 2004;52:515–27. doi: 10.1111/j.1365-2958.2004.03992.x. [DOI] [PubMed] [Google Scholar]
- 34.Koga Y, Kyuragi T, Nishihara M, Sone N. Did archaeal and bacterial cells arise independently from noncellular Precursors? A hypothesis stating that the advent of membrane phospholipid with enantiomeric glycerophosphate backbones caused the separation of the two lines of descent. Journal of molecular evolution. 1998;47:631. doi: 10.1007/pl00006419. [DOI] [PubMed] [Google Scholar]
- 35.Dibrova DV, Galperin MY, Mulkidjanian AY. Phylogenomic reconstruction of archaeal fatty acid metabolism. Environmental microbiology. 2014;16:907–18. doi: 10.1111/1462-2920.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Gil J, Rodriguez-Concepcion M. Metabolic plasticity for isoprenoid biosynthesis in bacteria. The Biochemical journal. 2013;452:19–25. doi: 10.1042/BJ20121899. [DOI] [PubMed] [Google Scholar]
- 37.Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. Journal of bacteriology. 2000;182:4319–27. doi: 10.1128/jb.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS computational biology. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivastava A, Taneja A, Collins I, Errington J, Czaplewski LG. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321:1673–5. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 40.Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. Journal of molecular evolution. 2004;58:19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 41.Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nature reviews Microbiology. 2010;8:731–41. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss DS. Bacterial cell division and the septal ring. Molecular microbiology. 2004;54:588–97. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- 43.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldon T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic acids research. 2014;42:D231–9. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang G, Rocha EP, Danchin A. Persistence drives gene clustering in bacterial genomes. BMC genomics. 2008;9:4. doi: 10.1186/1471-2164-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends in genetics : TIG. 1996;12:334–6. [PubMed] [Google Scholar]
- 46.Le S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J Stat Softw. 2008;25:1–18. [Google Scholar]
- 47.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8327–32. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–8. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 49.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiology and molecular biology reviews : MMBR. 2005;69:326–56. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiology and molecular biology reviews : MMBR. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–23. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 52.Thibault FM, Hernandez E, Vidal DR, Girardet M, Cavallo JD. Antibiotic susceptibility of 65 isolates of Burkholderia pseudomallei and Burkholderia mallei to 35 antimicrobial agents. The Journal of antimicrobial chemotherapy. 2004;54:1134–8. doi: 10.1093/jac/dkh471. [DOI] [PubMed] [Google Scholar]
- 53.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14240–5. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vorholt JA, Thauer RK. The active species of 'CO2' utilized by formylmethanofuran dehydrogenase from methanogenic Archaea. European journal of biochemistry / FEBS. 1997;248:919–24. doi: 10.1111/j.1432-1033.1997.00919.x. [DOI] [PubMed] [Google Scholar]
- 55.Gartner P, Ecker A, Fischer R, Linder D, Fuchs G, Thauer RK. Purification and properties of N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. European journal of biochemistry / FEBS. 1993;213:537–45. doi: 10.1111/j.1432-1033.1993.tb17792.x. [DOI] [PubMed] [Google Scholar]
- 56.Ermler U. On the mechanism of methyl-coenzyme M reductase. Dalton transactions. 2005:3451–8. doi: 10.1039/b506697b. [DOI] [PubMed] [Google Scholar]
- 57.Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic acids research. 2002;30:5382–90. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic acids research. 1999;27:3389–401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gossani C, Bellieny-Rabelo D, Venancio TM. Evolutionary analysis of multidrug resistance genes in fungi - impact of gene duplication and family conservation. The FEBS journal. 2014;281:4967–77. doi: 10.1111/febs.13046. [DOI] [PubMed] [Google Scholar]
- 60.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–6. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 61.Mendonca AG, Alves RJ, Pereira-Leal JB. Loss of genetic redundancy in reductive genome evolution. PLoS computational biology. 2011;7:e1001082. doi: 10.1371/journal.pcbi.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS biology. 2003;1:E19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome research. 2001;11:356–72. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 64.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends in biochemical sciences. 1998;23:324–8. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 65.Lathe WC, 3rd, Snel B, Bork P. Gene context conservation of a higher order than operons. Trends in biochemical sciences. 2000;25:474–9. doi: 10.1016/s0968-0004(00)01663-7. [DOI] [PubMed] [Google Scholar]
- 66.Coenye T, Vandamme P. Organisation of the S10, spc and alpha ribosomal protein gene clusters in prokaryotic genomes. FEMS microbiology letters. 2005;242:117–26. doi: 10.1016/j.femsle.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 67.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6652–7. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng Y, Szustakowski JD, Fortnow L, Roberts RJ, Kasif S. Computational identification of operons in microbial genomes. Genome research. 2002;12:1221–30. doi: 10.1101/gr.200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muro EM, Mah N, Moreno-Hagelsieb G, Andrade-Navarro MA. The pseudogenes of Mycobacterium leprae reveal the functional relevance of gene order within operons. Nucleic acids research. 2011;39:1732–8. doi: 10.1093/nar/gkq1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price MN, Arkin AP, Alm EJ. The life-cycle of operons. PLoS genetics. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ning K, Ng HK, Srihari S, Leong HW, Nesvizhskii AI. Examination of the relationship between essential genes in PPI network and hub proteins in reverse nearest neighbor topology. BMC bioinformatics. 2010;11:505. doi: 10.1186/1471-2105-11-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin CC, Juan HF, Hsiang JT, Hwang YC, Mori H, Huang HC. Essential core of protein-protein interaction network in Escherichia coli. Journal of proteome research. 2009;8:1925–31. doi: 10.1021/pr8008786. [DOI] [PubMed] [Google Scholar]
- 73.Lim HN, Lee Y, Hussein R. Fundamental relationship between operon organization and gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10626–31. doi: 10.1073/pnas.1105692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, Rode M, Suyama M, Schmidt S, Gavin AC, Bork P, Serrano L. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–71. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 75.Dotsch A, Klawonn F, Jarek M, Scharfe M, Blocker H, Haussler S. Evolutionary conservation of essential and highly expressed genes in Pseudomonas aeruginosa. BMC genomics. 2010;11:234. doi: 10.1186/1471-2164-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barquist L, Langridge GC, Turner DJ, Phan MD, Turner AK, Bateman A, Parkhill J, Wain J, Gardner PP. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic acids research. 2013;41:4549–64. doi: 10.1093/nar/gkt148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4678–83. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic acids research. 2014;42:D32–7. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mao X, Ma Q, Zhou C, Chen X, Zhang H, Yang J, Mao F, Lai W, Xu Y. DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic acids research. 2014;42:D654–9. doi: 10.1093/nar/gkt1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brouwer RW, Kuipers OP, van Hijum SA. The relative value of operon predictions. Briefings in bioinformatics. 2008;9:367–75. doi: 10.1093/bib/bbn019. [DOI] [PubMed] [Google Scholar]
- 81.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Podell S, Gaasterland T, Allen EE. A database of phylogenetically atypical genes in archaeal and bacterial genomes, identified using the DarkHorse algorithm. BMC bioinformatics. 2008;9:419. doi: 10.1186/1471-2105-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome informatics International Conference on Genome Informatics. 2009;23:205–11. [PubMed] [Google Scholar]
- 84.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS pathogens. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Veeranagouda Y, Husain F, Tenorio EL, Wexler HM. Identification of genes required for the survival of B. fragilis using massive parallel sequencing of a saturated transposon mutant library. BMC genomics. 2014;15:429. doi: 10.1186/1471-2164-15-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. The essential genome of a bacterium. Molecular systems biology. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baugh L, Gallagher LA, Patrapuvich R, Clifton MC, Gardberg AS, Edwards TE, Armour B, Begley DW, Dieterich SH, Dranow DM, Abendroth J, Fairman JW, Fox D, 3rd, Staker BL, Phan I, Gillespie A, Choi R, Nakazawa-Hewitt S, Nguyen MT, Napuli A, Barrett L, Buchko GW, Stacy R, Myler PJ, Stewart LJ, Manoil C, Van Voorhis WC. Combining functional and structural genomics to sample the essential Burkholderia structome. PloS one. 2013;8:e53851. doi: 10.1371/journal.pone.0053851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, Durot M, Kreimeyer A, Le Fevre F, Schachter V, Pezo V, Doring V, Scarpelli C, Medigue C, Cohen GN, Marliere P, Salanoubat M, Weissenbach J. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Molecular systems biology. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baba T, Huan HC, Datsenko K, Wanner BL, Mori H. The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12. Methods in molecular biology. 2008;416:183–94. doi: 10.1007/978-1-59745-321-9_12. [DOI] [PubMed] [Google Scholar]
- 92.Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1009–14. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, 3rd, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:425–30. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Molecular microbiology. 2008;69:67–76. doi: 10.1111/j.1365-2958.2008.06262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.