Abstract
Genome engineering relies on DNA modifying enzymes that are able to locate a DNA sequence of interest and initiate a desired genome rearrangement. Currently, the field predominantly utilizes site-specific DNA nucleases that depend on the host DNA repair machinery to complete a genome modification task. We show here that genome engineering approaches that employ target-specific variants of self-sufficient, versatile site-specific DNA recombinase Flp can be developed into promising alternatives. We demonstrate that the Flp variant evolved to recombine an FRT-like sequence FL-IL10A, which is located upstream of the human interleukin-10 gene, can target this sequence in the model setting of hamster CHO and human HEK293 cells. This target-specific Flp variant is able to perform the integration reaction and, when paired with another recombinase, the dual RMCE reaction. The efficiency of the integration reaction in human cells can be enhanced by ‘humanizing’ the Flp variant gene and by adding the NLS sequence to the recombinase.
Keywords: Flp, genome engineering, RMCE, site-specific recombination, gene targeting
Graphical abstract
Introduction
The ability to manipulate genomes at a desired location opens an opportunity to develop model cells or even model organisms or to correct genetic disorders. Recent advances in the developing field of genome engineering are primarily attributed to site-specific DNA nucleases: mainly zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and nucleases that are based on the clustered regularly interspaced short palindromic repeats/Cas9 systems (CRISPR/Cas9) [1–9]. The burst of the nuclease-based genome engineering applications is triggered by two main factors: the efficiency of cleavage of genomic targets by these composite nucleases and the relative ease with which their target recognition units can be assembled, especially for the CRISPR/Cas9-based systems. Although site-specific nucleases can locate and cleave their targets, these enzymes are not able to complete the genome modification task: the host DNA repair machinery is required to seal the introduced DNA breaks either by the non-homologous end joining (NHEJ) or by the homologous DNA repair mechanisms [2].
The alternative, ‘old-fashioned’ genome engineering systems are based on the versatile, self-sufficient site-specific DNA recombinases. These enzymes do not generate double strand breaks in DNA and can accomplish the genome modification task without any assistance from the host DNA repair machineries. Popular site-specific recombination systems employed for genome engineering applications include Cre/lox from coliphage P1, Flp/FRT from 2 micron plasmid of S. cerevisiae, φC31/att from Streptomyces phage φC31, as well as others [10–20]. Depending on the location and relative orientation of their targets, these systems can mediate insertions, deletions, replacements and translocations of DNA fragments. Moreover, site-specific recombinases are able to perform genetic rearrangements in all cell types tested: from bacteria to plant to human.
To be useful in advanced genome engineering applications, these handy recombinases have to be modified to recognize target-like sequences in the genome locale of interest. The quest to generate variants of tyrosine recombinases with desired target specificity began when several mutant and chimeric λ integrase variants were shown to recombine target sites for HK022 integrase [21, 22]. The research was boosted by the advances in two molecular evolution techniques: error-prone PCR and DNA shuffling which allow for the fast generation of recombinase variant libraries [23–25]. The solved structures of the Cre/loxP and Flp/FRT complexes [26, 27] provided another boost: they helped utilize another molecular evolution technique: site-directed mutagenesis of amino acids that can affect DNA binding.
The first success in applying directed protein evolution approaches to change the target specificity of the tyrosine recombinases was achieved when variants of Cre were evolved to recombine a loxP-like sequence [28]. Later, two other groups also showed that target specificity of Cre can be indeed intentionally modified [29, 30]. The major success was achieved when Cre variants were evolved to delete, in the model setting, a proviral DNA of HIV-1 pseudotype utilizing loxP-like sequences located in the long terminal repeats (LTRs) as targets [31].
Another popular tyrosine recombinase, Flp, has also been shown to be amenable to the modification of its target specificity: a number of Flp variants that can recombine FRT derivatives with point mutations and a combination of point mutations were evolved [32–34]. The technology developed in these experiments was applied to evolve Flp variants to be specific for an FRT-like sequence located upstream of the human interleukin-10 gene, dubbed FL-IL10A [35]. Then, a set of Flp variants, each specific for a unique FRT-like sequence located in the human beta-globin and interleukin-10 genes and in the human and mouse ROSA26 loci, was generated [36]. The knowledge acquired from these experiments was applied to develop algorithms to identify and rank genomic FRT-like sequences [35, 36].
In this report we show that Flp variants evolved to recombine genomic FRT-like sequence FL-IL10A can utilize this sequence as a target in the model setting of hamster CHO and human HEK293 cells to perform the integration and dual recombinase-mediated cassette exchange reactions. We also show that the efficiency of the integration reaction in the hamster cells is higher than in the human cells and that the efficiency in the human cells can be increased by ‘humanizing’ the codons of the Flp variant gene and by the addition of the NLS sequence to the N- or C-terminal ends of the variant. Under optimal conditions, the efficiency of the integration reaction in the model human cells can reach about 0.3% of the transfected cells.
Results
Target-Specific Flp Variant Recombines FRT-like Sequence Pre-integrated into CHO Genome
To test whether target-specific Flp variants can recombine their cognate DNA sequences in mammalian genome, we chose well-characterized Flp variant FV7 [35] that recognizes FRT-like sequence FL-IL10A (Fig. 1) located upstream of the human interleukin-10 gene. For these experiments, we generated a CHO cell line with FL-IL10A singly integrated into genome.
Fig. 1.
Recombination targets used in the study. (A) FRT, shown as a reference, consists of two inverted 13 bp Flp binding elements flanking the 8-bp strand exchange region (spacer). (B) FRT-like sequence FL-IL10A. (C) The hybrid FRT/FL-IL10A sequence is composed of the left recombinase binding element of FRT and right recombinase binding element of FL-IL10A separated by the spacer from FL-IL10A. The positions within the recombinase binding elements that differ from the corresponding positions in FRT are indicated by lower case red letters. (D) loxP consists of two inverted 14 bp Cre binding elements flanking the 6-bp strand exchange region. Points of single-stranded DNA exchange in FRT and loxP are indicated by vertical arrows.
A schematic of the assay to test the integration activity of FV7 is shown in Fig. 2A. The FL-IL10A site in the pre-integrated platform reporter pCMV-IL10A-DsRed is located between the CMV promoter and the DsRed gene. The incoming reporter pIL10A-EGFP bears the hybrid FRT/FL-IL10A site (Fig. 1C) followed by the promoterless EGFP gene. Upon integration of the incoming reporter into the FL-IL10A site, the EGFP gene is transferred under the control of the CMV promoter thus preventing the DsRed gene from being expressed. This integration event can be detected by the appearance of green, non-red cells.
Fig. 2.
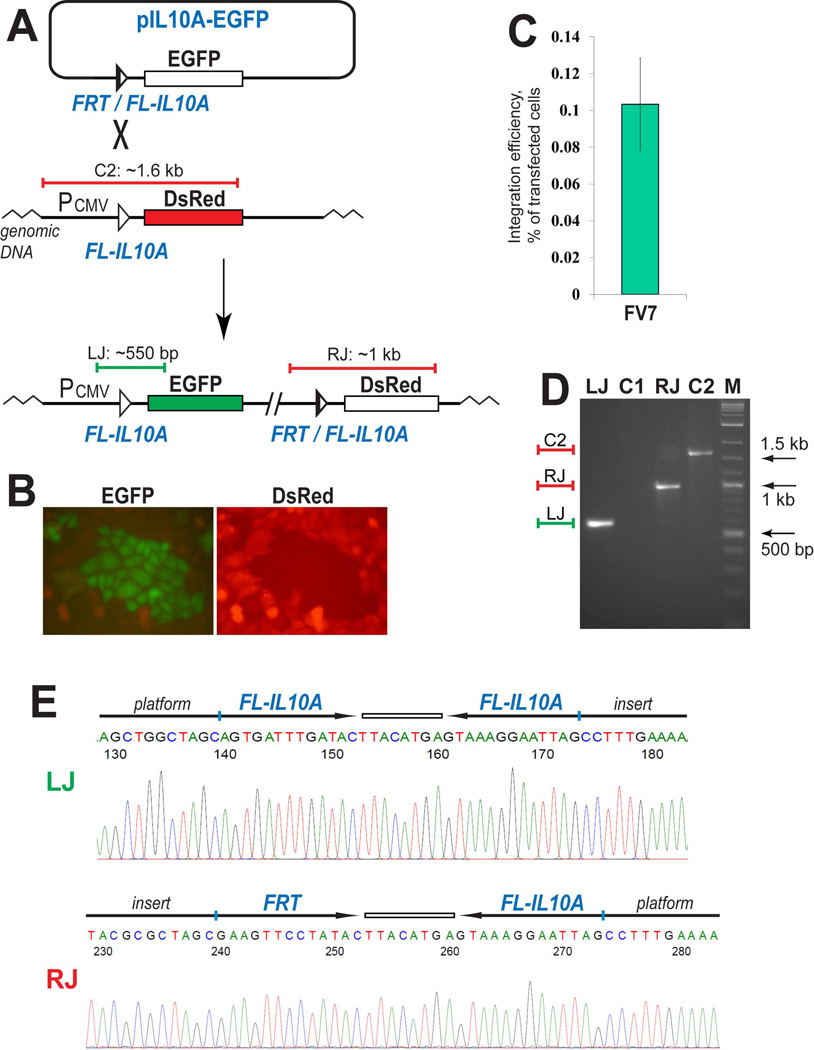
FV7-mediated integration of a reporter into FL-IL10A pre-integrated into the CHO genome. (A) Schematic outline of the integration assay. The horizontal blue and red bars in panels (A) and (D) schematically represent the PCR products at the left junction [LJ] and the right junction [RJ] of the integration product and the control PCR product of the platform reporter [C2]. (B) Green colonies are formed after the transfected cells are split and expanded. (C) The efficiency of the FV7-dependent integration is represented by the green bar that shows the mean value of three experiments; the error bar indicates standard deviation. (D) PCR analyses of a typical expanded green colony. LJ and C1 lanes show the results of the PCR analysis of the green cells and the original platform cells, respectively, using the same primers that anneal on the CMV promoter and the EGFP gene. RJ and C2, the PCR analysis of the green cells and the control platform cells using the primers that anneal on the vector backbone and the DsRed gene. M, DNA marker. (E) Representative sequencing snapshots of the recombination site regions in the diagnostic PCR products LJ and RJ (panel D). The schematics of the recombination sites and the surrounding sequences are shown above the sequencing snapshots.
The integration activity of FV7 was tested by transfecting the reporter cells with the incoming reporter and the FV7 expression vector. The experiments were performed in 6-well plates. 48 hours after transfection, the cells were expanded and screened for the groups of green cells (Fig. 2B). Only these groups and not individual green cells were counted as productive integration events. Based on the number of groups of green cells and the efficiency of the control transfection of an EGFP-expressing reporter similar in size to pIL10A-EGFP, we estimated the efficiency of the FV7-mediated integration into FL-IL10A as a fraction of the transfected cells. On average, in the one-sixth of the expanded transfected cells we observed 17 integration-positive green, non-red colonies (Fig. 2C). This number corresponded to about 0.1% of the transfected cells (Fig. 2C). In the expanded cells, in addition to the green, non-red cells, we also noticed rare green/red colonies: on average, 1 colony per plate of the expanded cells; these colonies were not analyzed. In the control experiments performed with the incoming reporter alone, no green or green/red colonies were observed.
We expanded and analyzed 10 green, non-red colonies obtained as described above. The integration events in all these colonies were confirmed by PCR (Fig. 2D) and sequencing of the integration-specific PCR products (Fig. 2E).
FV7 Supports Dual RMCE in CHO Cells
Recombinase-mediated cassette exchange, or RMCE, is a powerful method of unidirectional delivery of DNA fragments into genome using site-specific recombinases [37, 38]. In its original form, the cassette exchange is performed by one recombinase that acts on a pair of recombination targets that cannot cross-react. In the variation of RMCE, dubbed dual RMCE, the cassette exchange is performed by two recombinases that act on their respective cognate targets [16, 39]. Dual RMCE can be quite efficient: the Flp/Cre pair can perform cassette exchange in about 45% of the transfected cells, while the Flp/HK022 Int pair can replace the cassettes in 12% of the transfected cells [40, 41].
To test if a target-specific Flp variant can be combined with another site-specific recombinase to perform dual RMCE in mammalian cells, we paired FV7 with Cre in a model setting similar to that we used to analyze the replacement activity of the Flp/HK022 Int recombinase pair [41]. For this, we introduced the respective cognate targets for FV7 and Cre in the incoming and platform reporters of the replacement system (Fig. 3) and the platform reporter was then integrated into the genome of CHO cells.
Fig. 3.
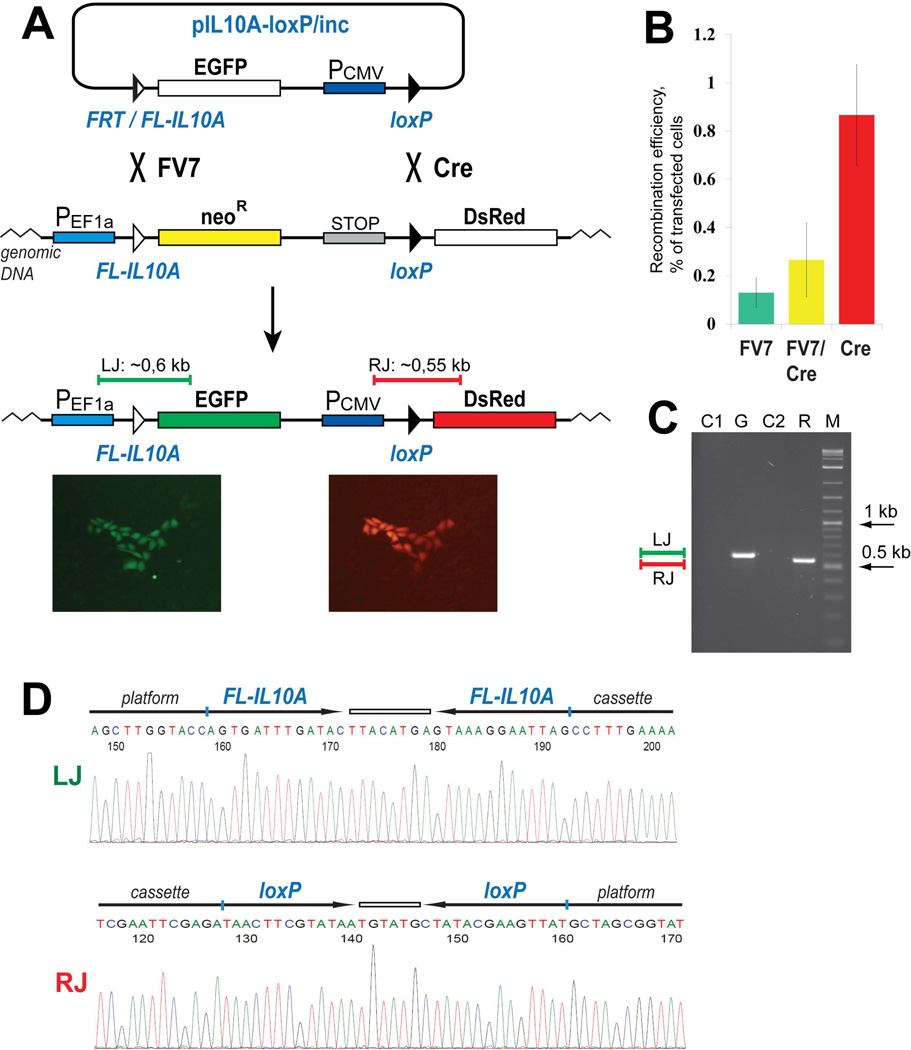
FV7 partnered with Cre can mediate dual RMCE in CHO cells. (A) Schematic outline of the replacement assay. An example of a green/red colony generated in the assay is shown below the scheme. (B) The efficiency of the FV7 catalyzed integration, FV7/Cre catalyzed cassette replacement, and the Cre catalyzed integration is represented by the green, yellow, and red bars, respectively. The bars show the mean value of three experiments; the error bars indicate standard deviation. (C) PCR analyses of a typical expanded green/red colony. The green and red bars in panels (A) and (C) represent the PCR products at the left and right junctions of the replacement construct that are expected if the replacement reaction is successful. G and R, PCR analysis of the expanded green/red cells with the primers that anneal on the EF1α promoter and the EGFP gene and on the CMV promoter and the DsRed gene, respectively. C1 and C2, control PCR analysis of the replacement platform reporter cells using the same set of primers as in lanes ‘G’ and ‘R’, respectively. M, DNA marker. (D) Representative sequencing snapshots of the recombination site regions in the diagnostic PCR products LJ and RJ (panel C). The schematics of the recombination sites and the surrounding sequences are shown above the sequencing snapshots.
A schematic of the assay to test the replacement activity of the FV7/Cre recombinase pair is shown in Fig. 3A. In the integrated platform reporter, FL-IL10A is located between the EF1α promoter and the neoR gene while loxP is located upstream of the promoterless DsRed gene. The incoming reporter bears the hybrid FRT/FL-IL10A site upstream of the promoterless EGFP gene whereas loxP is positioned downstream of the CMV promoter. If the FV7/Cre recombinase pair is active in the dual RMCE reaction, the neoR-STOP cassette of the platform reporter gets replaced with the EGFP-CMV cassette of the incoming reporter; this recombination event can be detected by the activation of the expression of both EGFP and DsRed genes in the same cell. The expression of just DsRed will indicate the Cre-dependent integration of the incoming reporter into the platform reporter at the loxP site while the expression of just EGFP will signify the FV7-dependent integration of the incoming reporter into the platform reporter at the FL-IL10A site.
To perform the replacement, we co-transfected the platform reporter cells with the incoming reporter and the vectors that express FV7 and Cre. The experiments were performed in 6-well plates. 48 hours post-transfection, the cells were expanded and examined for cell groups that express either EGFP or DsRed, or both (Fig. 3A). These cell groups and not individual cells that express EGFP and/or DsRed were considered true indicators of recombination events. The efficiency of the respective recombination events was assessed as a fraction of the transfected cells. On average, in the one-sixth of the expanded transfected cells we observed 19 just green colonies, 133 just red colonies and 46 green/red colonies. In the control experiments performed with the incoming reporter alone, no green, red or green/red colonies were observed. Based on the number of the cell groups that expressed both EGFP and DsRed and the efficiency of the control transfection of an EGFP-expressing plasmid similar in size to the incoming reporter, we estimated that the FV7/Cre-dependent cassette replacement occurred in about 0.3% of the transfected cells (Fig. 3B). The number of the cell groups that expressed just EGFP or just DsRed revealed that the reporter integration occurred in ~0.1% and ~0.8% of the transfected cells, respectively (Fig. 3B).
To analyze the recombination events in the colonies that express just EGFP, just DsRed, or both, we expanded 4 colonies each of the first two types and 10 colonies of the third type and subjected them to the PCR analyses and sequencing of the resultant PCR products. The analyses confirmed the expected recombination events in all expanded colonies. The representative data of the analyses of an expanded green/red colony are shown in Fig. 3C, D.
FV7 is Active in Human Cells
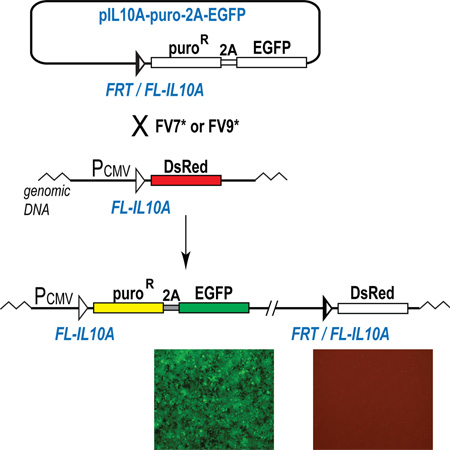
Having established that FV7 is active in the integration and replacement reactions in CHO cells, we next tested the ability of FV7 to utilize the FL-IL10A sequence as a substrate in human cells in model setting. For this, we integrated an FL-IL10A bearing platform reporter into human embryonic kidney 293 cells, HEK293. In the integrated platform reporter, FL-IL10A is located between the CMV promoter and the DsRed gene (Fig. 4A). In the incoming reporter, the hybrid FRT/FL-IL10A sequence is located upstream of the promoterless puro-2A-EGFP gene. Upon integration of the incoming reporter into the platform reporter, the expression of the puro-2A-EGFP gene is activated so the cells become green and resistant to puromycin.
Fig. 4.
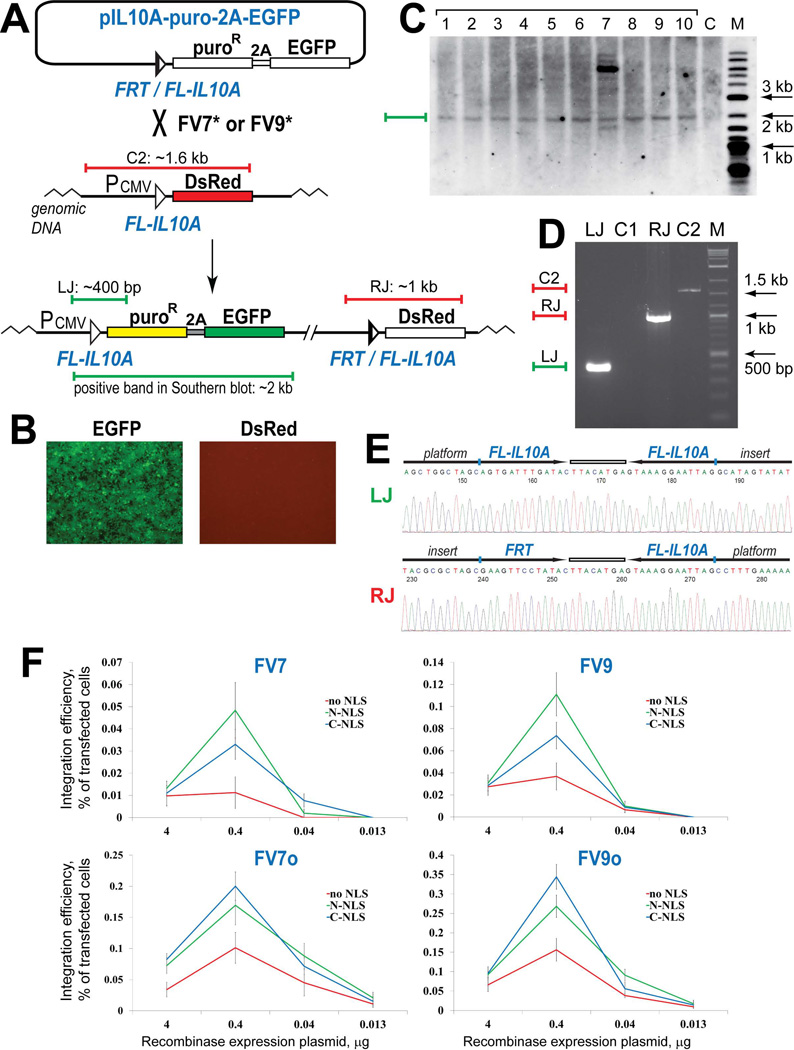
Flp variants FV7, FV9 and their derivatives can integrate a reporter into the FL-IL10A site pre-integrated into the human genome. (A) Schematic outline of the integration assay. The horizontal blue and red bars in panels (A) and (D) schematically represent the PCR products at the left junction [LJ] and the right junction [RJ] of the integration product and the control PCR product of the platform reporter [C2]. (B) Expanded green, puromycin-resistant cells do not express DsRed. (C) Southern blot analysis of the expanded green, puromycin-resistant colonies. The green bar in panels (A) and (C) shows the location of the integration-specific DNA fragment. M, biotinylated DNA marker. (D) PCR analyses of a typical green, puromycin-resistant colony. LJ and C1 lanes show the results of the PCR analysis of the green cells and the original platform cells, respectively, using the same primers that anneal on the CMV promoter and the puroR gene. RJ and C2, the PCR analysis of the green cells and the control platform cells using the primers that anneal on the vector backbone and the DsRed gene. M, DNA marker. (E) Representative sequencing snapshots of the recombination site regions in the diagnostic PCR products LJ and RJ (panel D). The schematics of the recombination sites and the surrounding sequences are shown above the sequencing snapshots. (F) The integration activity of the Flp variants FV7, FV9, FV7o, and FV9o as a function of the location of the NLS sequence and the input of the corresponding recombinase expression vectors at transfection. The results are represented by the data points connected by a line of the respective color. The data points show the mean value of five experiments; the error bars indicate standard deviation.
The pilot integration experiments in model platform HEK293 cells were performed in 6-well plates by co-transfecting the cells with the incoming reporter and the FV7 expression plasmid. When the one-sixth of the expanded transfected cells was expanded, we observed, on average, 1 green colony. This integration efficiency corresponded to ~0.01% of the transfected cells, which was about 10 fold lower than that in CHO cells (Fig. 2C). When the transfected cells were expanded in the media supplemented with puromycin, approximately 90% of the puromycin-resistant colonies that formed were green.
To test what fraction of the green colonies represents the correct integration of the incoming reporter into the platform reporter, we subjected 10 green colonies to the PCR and Southern blot analyses. All 10 colonies were positive for the integration specific PCR bands (Fig. 4D), the identity of which was confirmed by sequencing (Fig. 4E). The Southern blot analysis showed that in all but one colony the incoming reporter was singly integrated into the platform reporter (Fig. 4C).
The Activity of FV7 in Human Cells Can be Enhanced by ‘Humanizing’ the FV7 Gene and by Adding NLS to its Termini
Since the activity of FV7 in human cells was low, we explored two approaches to increase it: (1) codon optimization of the FV7 gene to maximize its expression in human cells and (2) addition of the SV40 nuclear localization sequence, NLS, to its N- or C-termini to facilitate the crossing of FV7 into the nucleus. These modifications were successfully applied to increase the activity of Cre and Flp in mammalian cells [42, 43]. In the optimization experiments, in addition to FV7, we used a de novo evolved Flp variant, FV9, which recombines FL-IL10A in the bacterial deletion assay slightly better than FV7. FV9 differs from FV7 at position 166: FV9 has wild-type Lys at this position while FV7 has Glu.
We tested the integration activity of the original and the modified recombinases at the different inputs of the respective expression plasmids. FV7 was able to generate green colonies only at the higher inputs: 4 µg and 0.4 µg of the expression plasmids per transfection (Fig. 4F, top panel). FV9 was more active than FV7 but it was not able to generate green colonies at the lowest input of the expression plasmids tested: 0.013 µg (Fig. 4F, top panel). The ‘humanized’ versions of FV7 and FV9 – FV7o and FV9o – had higher integration activity than FV7 and FV9 and were able to generate green colonies even at the lowest input of the expression plasmids (Fig. 4F, bottom panel). Unlike FV7 and FV9 that had essentially the same activity at the higher inputs of the expression plasmids (4 µg and 0.4 µg), FV7o and FV9o had a pronounced peak of activity at 0.4 µg.
Although the addition of NLS to the N- or C-termini of both non-optimized and optimized recombinase variants increased their integration activity, this modification was not powerful enough to allow the not-optimized FV7 and FV9 to generate green colonies at the lowest input of the expression plasmids (Fig. 4F, top panel). The variants with added NLS followed the FV7o and FV9o pattern and had a pronounced peak of activity at 0.4 µg of the respective expression plasmids added at transfection (Fig. 4F). For FV7 and FV9, it was the N-terminally placed NLS that helped them to achieve their highest integration activity. In contrast, the ‘humanized’ FV7o and FV9o benefited more from the C-terminal NLS. It was FV9o with this modification, FV9o-C-NLS, which showed the highest integration activity of all recombinase variants tested: on average, this variant was able to generate around 35 green colonies in the one-sixth of the expanded transfected cells; this integration efficiency corresponded to ~0.35% of the transfected cells (Fig. 4F).
Discussion
The power of the tyrosine site-specific recombinases to engineer genomes is widely realized but only in the approaches that employ the pre-introduced cognate wild-type (or essentially wild-type) recombination targets. The approaches that would utilize genomic target-like sequences in their native chromosomal environment could add a valuable tool to the genome engineering toolbox but are yet to be developed. Our present work shows that the field can be advanced if the genome manipulation methods are developed around target-specific recombinase variants that are evolved to be active in bacteria and further modified to optimize their expression level and their delivery into the nucleus in the cells of interest.
We found hamster CHO cells to be convenient and reliable in modeling genome engineering approaches that utilize target-specific Flp variants. Using these cells we were able to establish and optimize the parameters of the integration and dual RMCE reactions mediated by Flp variant FV7 evolved to recognize human genomic FRT-like sequence FL-IL10A (Figs. 2, 3).
The performance of FV7 in human HEK293 cells did not mirror its performance in hamster CHO cells: the integration efficiency of FV7 in CHO was ~0.1% of the transfected cells whereas only ~0.01 % in HEK293. To improve the integration activity of FV7 in human cells, we optimized the codons of the FV7 gene to maximize its expression in human cells and/or added the nuclear localization sequence to the variant to enhance its ability to get into nucleus. The use of the de novo evolved Flp variant, FV9, which recombines FL-IL10A in E. coli slightly better than FV7, was also helpful in increasing the efficiency of FL-IL10A targeting.
The effect of the codon optimization on the integration activity of FV7 and FV9 was quite positive allowing the optimized variants, FV7o and FV9o, to have the integration activity over almost three orders of magnitude versus one order of magnitude for FV7 and two orders of magnitude for FV9 (Fig. 4F). The activity increase was not uniform over the range of the expression plasmid inputs: the peaks of the integration activity were detected at the moderate input of the recombinase expression vectors. The codon optimization effect was more pronounced for FV7: under optimal conditions, FV7o and FV9o were about 10 fold and 5 fold more active than FV7 and FV9, respectively (Fig. 4F).
The addition of NLS to the original and to the codon-optimized recombinase variants increased their integration activity: FV7 and FV9 showed a higher relative increase than their ‘humanized’ versions while FV7o and FV9o showed higher absolute integration activity (Fig. 4F). The integration activity of the NLS modified recombinase variants, as well as the unmodified FV7o and FV9o, exhibited sharp dependence on the input of the respective recombinase expression vectors. The sharp peaks of the integration activity that depend on the recombinase input are not unusual for the tyrosine recombinases and were noted before in the targeting experiments with the purified Cre recombinase [44]. Such sharp peaks may reflect a narrow window of the optimal combination of the efficiency of the reversible integration-excision reactions and the input of a recombinase.
The original and the ‘humanized’ NLS-modified Flp variants had different activity patterns at the moderate input of the recombinase expression vectors (the one that permits the highest integration activity of the recombinase variants, Fig. 4F). At this input, FV7 and FV9 with N-terminal NLS showed higher integration activity than their C-terminal versions. In contrast, FV7o and FV9o variants showed the inverse activity pattern: the variants with C-terminal NLS were more active than their N-terminal counterparts (Fig. 4F). Since the integration activity of a recombinase in a cell was found to depend not only on the actual ability of this enzyme to recombine a particular DNA sequence but also on the concentration of the recombinase [44] and the timing of its expression [45], the observed differences in the activity of the NLS-modified variants may be explained by the differences in the expression of the variants and the timing of their appearance in the nucleus. Indeed, the codon optimization and the addition of NLS can affect these parameters. Another factor to consider is a potential effect of NLS and its location on the actual enzymatic activity of a recombinase variant.
Taken together, the codon optimization of a recombinase gene, the addition of NLS, and the use of a more active Flp variant resulted in about 35-fold increase in the targeting efficiency of FL-IL10A in human cells in the model setting: FV7 was able to integrate a reporter in ~0.01% of the transfected cells while the ‘humanized’ version of FV9 with C-terminally placed NLS performed the task in ~0.35% of the transfected cells.
In the model setting of human cells FV7 is capable of targeting FL-IL10A accurately: out of ten randomly expanded integration-positive clones, only one clone had vector integrated somewhere else in the genome, in addition to the correctly integrated vector (Fig. 4C).
The results of our previous work indicate that in E. coli FV7 is not as efficient as Flp [32, 35]: Flp is able to delete a reporter DNA fragment in almost 100% of the bacterial cells within about an hour while FV7 generates the deletion phenotype in ~80% of the cells in 2.5 hours. In the present work we did not compare the recombination activity of FV7 and Flp directly but indirect evidence suggests that FV7 is not as active as Flp also in mammalian cells. Indeed, when Flp is paired with Cre, the pair is able to mediate very efficient dual RMCE reaction: the replacement can be seen in ~45% of the transfected cells [40]. Flp is also quite efficient when paired with HK022 Int: this recombinase pair is able to exchange the reporter cassettes in about 12% of the transfected cells [41]. In contrast, in a similar reporter setting, the FV7/Cre pair is able to perform replacement in only 0.3% of the transfected cells (Fig. 3B).
We cannot reliably compare the integration activity of FV7 and Flp in mammalian cells. In model CHO cells, FV7 integrates the incoming reporter in ~0.1% of the transfected cells (Fig. 2C). We often use Flp as an integration control in similar model settings and note about the same integration efficiency. On the other hand, we did not extensively analyze the integration activity of Flp so it is possible that Flp can mediate integration more efficiently.
In principle, Flp variants that can target the pre-integrated FL-IL10A site in HEK293 cells could target the endogenous FL-IL10A site in these cells. The later targeting events, however, are expected to be less frequent than the former events since the IL10 gene is not active in HEK293 cells and is likely located in the tightly packed chromatin. Consequently, endogenous FL-IL10A may not be as easily accessible as FL-IL10A located in the integrated platform reporter. The incoming reporter that we used in the experiments described in Fig. 4 is not suited for the detection of the integration events into endogenous FL-IL10A and cannot be used to enrich the cells, in which this integration occurred. Future research will focus on developing incoming reporters that are able to enrich the cells in which the reporter integrates into endogenous FRT-like sequences. Future research will also focus on establishing and optimizing parameters that improve targeting of the endogenous FRT-like sequences.
Materials and Methods
Cell lines and transfection
Hamster CHO TD-In cells [40] and human embryonic kidney HEK 293 cells (ATCC, CRL-1573) were used as model mammalian cells. The cells were propagated in F-12K and DMEM, respectively. Cell transfections were performed using Polyfect (Qiagen).
Vectors
The reporter vectors for the integration and the replacement experiments were based on the pcTD vector described before [40]. The platform reporter pCMV-IL10A-DsRed and the incoming reporter pIL10A-EGFP, which were used in the integration experiments in CHO TD-In cells, were generated by subcloning the DsRed and EGFP genes from pIRES2-DsRed-Express and pIRES2-EGFP (Clontech), respectively, into pcTD downstream from the CMV promoter to obtain the platform reporter pCMV-IL10A-DsRed and pCMV-IL10A-EGFP. FL-IL10A and FRT/FL-IL10A were introduced upstream of the DsRed and EGFP genes, respectively, when the genes were PCR amplified. The CMV promoter in pCMV-IL10A-EGFP was deleted to obtain the incoming reporter pIL10A-EGFP.
The platform reporter pIL10A-loxP/plt and the incoming reporter pIL10A-loxP/inc were constructed by modifying the replacement reporters described earlier [41]. For this, the neoR-STOP cassette of the platform reporter was flanked by the FL-IL10A/loxP sites and the EGFP-CMV cassette of the incoming reporter was flanked by the FRT-FL-IL10A/loxP sites.
The platform reporter pCMV-IL10A-DsRed-neo and the incoming reporter pIL10A-puro-2A-EGFP were generated as follows. The DsRed-neoR cassette from pIRES2-DsRed-Express (Clontech) was PCR amplified and subcloned into pcTD downstream from the CMV promoter to obtain pCMV-IL10A-DsRed-neo. FL-IL10A upstream of the DsRed-neoR cassette was introduced during the amplification of the cassette. The puroR gene from pCAGGS-Flpe-puro (Addgene plasmid 20733) and the EGFP gene from pIRES2-EGFP (Clontech) were PCR-assembled into one gene through the connecting 2A peptide and cloned into pcTD downstream from the CMV promoter to obtain pCMV-IL10A-puro-2A-EGFP. FRT/FL-IL10A upstream of the puro-2A-EGFP gene was introduced during its amplification. The CMV promoter in this plasmid was then deleted to obtain pIL10A-puro-2A-EGFP.
To express Flp variants in mammalian cells, the respective genes were subcloned into pOG44 (Invitrogen) in place of the Flp-F70L variant gene as described before [40]
Construction of platform reporter cell lines
To construct CHO TD-IL10A-DsRed cell line, CHO TD-In cells [40] were co-transfected, in 6-well plates, with 0.2 µg of the pCMV-IL10A-DsRed reporter and 2 µg of the pOG-TD1-40 vector [40], which expresses TD1-40 variant of TD recombinase [46]. 48 hours post-transfection, 1/6 of the cells were transferred into a 100 mm plate containing F-12K medium supplemented with hygromycin (550 mg/l). About 10 days later, several hygromycin resistant, red colonies were transferred into 96-well plate and their sensitivity to zeocin was tested; zeocin sensitive colonies were expanded and used in the integration experiments.
CHO TD-IL10A-loxP/plt cell line was constructed similarly, by co-transfecting CHO TD-In cells with pCMV-IL10A-loxP/plt and pOG-TD1-40 and selecting for the colonies that are sensitive to zeocin and resistant to hygromycin and neomycin.
HEK 293-IL10A-DsRed-neo cell line was constructed by transfecting HEK 293 cells with pCMV-IL10A-DsRed-neo, propagating them in the medium supplemented with neomycin (800 mg/l) and selecting for the red colonies. Several such colonies were expanded and tested for the singly integrated platform reporter vector by Southern blotting; the positive colonies were used in the integration experiments.
Integration experiments
The integration experiments were performed in the same general way for both integration reporter pairs. The reactions were done in 6-well plates. The incoming reporters (0.5 µg) were co-transfected into the respective platform reporter cells with the vectors that express Flp variants analyzed (2 µg in the integration experiments in hamster cells; the amount of the expression vectors added at transfection in human cells is indicated in Fig. 4). 48 hours post transfection (72 hours for human cells), 1/6 of the cells were transferred into 100 mm plates, the cells were allowed to become confluent, and the number of the green colonies was counted. Several integration positive colonies from these plates were expanded and analyzed.
Dual recombinase-mediated cassette exchange experiments
The cassette exchange reaction between the incoming reporter pIL10A-loxP/inc and the platform reporter pIL10A-loxP/plt integrated into CHO TD-In was performed similar to the integration experiments. The incoming reporter (0.5 µg) and the expression vectors pOG-FV7 (2 µg) and pOG-Cre (0.03 µg) were co-transfected into CHO TD-IL10A-loxP/plt. 48 hours post transfection, 1/6 of the cells were transferred into 100 mm plates, the cells were allowed to become confluent, and the number of the green, red, and green/red colonies was counted. Several green/red colonies from these plates were expanded and analyzed.
Southern blotting
Approximately 15 µg of genomic DNA were digested overnight followed by incubation with RNase A for 1 hr. The DNA fragments were separated on 0.7% TAE agarose (molecular biology grade, BioRad). The gel was then treated with the depurination and the alkaline denaturing solutions and the DNA fragments were transferred to the positively charged nylon membrane (Amersham Hybond-N+, GE Healthcare) in the neutral transfer buffer. The DNA was fixed to the membrane, pre-hybridized in the ULTRAhyb hybridization buffer (Ambion) and then hybridized with the biotinylated EGFP probe (~1 µg) overnight. The DNA bands were visualized using the Phototope-Star Detection Kit (New England Biolabs) according to the manufacturer’s protocol.
Other methods
Plasmid DNA was isolated using GeneJET Plasmid Miniprep Kit (Thermo). Amplification of the DNA fragments used for cloning was performed using Pfu-Ultra polymerase (Agilent Technologies). PCR analysis of the mammalian genomic DNA was performed using Taq polymerase (New England Biolabs). Genomic DNA from cultured mammalian cells was isolated using GeneJET Genomic DNA Purification Kit (Thermo).
Acknowledgements
This work was supported in part by grants from Louisiana Tech University, NSF/LEQSF(2008)-PFUND-113, and NIGMS (R01GM085848) to Y.V.
Footnotes
The authors declare no conflict of interest.
Author Contributions
Y.V. planned experiments; R.S., F.L, E.V, and Y.V. performed experiments; Y.V. analyzed data and wrote the paper.
References
- 1.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 3.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 10.Kilby NJ, Snaith MR, Murray JA. Site-specific recombinases: tools for genome engineering. Trends Genet. 1993;9:413–421. doi: 10.1016/0168-9525(93)90104-p. [DOI] [PubMed] [Google Scholar]
- 11.Buchholz F, Refaeli Y, Trumpp A, Bishop JM. Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse. EMBO Rep. 2000;1:133–139. doi: 10.1093/embo-reports/kvd027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills AA, Bradley A. From mouse to man: generating megabase chromosome rearrangements. Trends Genet. 2001;17:331–339. doi: 10.1016/s0168-9525(01)02321-6. [DOI] [PubMed] [Google Scholar]
- 13.Glaser S, Anastassiadis K, Stewart AF. Current issues in mouse genome engineering. Nat Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 14.Calos MP. The phiC31 integrase system for gene therapy. Current gene therapy. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- 15.Wirth D, Gama-Norton L, Riemer P, Sandhu U, Schucht R, Hauser H. Road to precision: recombinase-based targeting technologies for genome engineering. Curr Opin Biotechnol. 2007;18:411–419. doi: 10.1016/j.copbio.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods. 2010;7:893–895. doi: 10.1038/nmeth.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J Mol Biol. 2011;407:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, De Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaj T, Sirk SJ, Barbas CF., 3rd Expanding the scope of site-specific recombinases for genetic and metabolic engineering. Biotechnol Bioeng. 2014;111:1–15. doi: 10.1002/bit.25096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu F, Gamboa M, Farruggio AP, Hippenmeyer S, Tasic B, Schule B, Chen-Tsai Y, Calos MP. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 2014;42:e34. doi: 10.1093/nar/gkt1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagil E, Dorgai L, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of chimeric bacteriophage integrases. J Mol Biol. 1995;252:163–177. doi: 10.1006/jmbi.1995.0485. [DOI] [PubMed] [Google Scholar]
- 22.Dorgai L, Yagil E, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of mutant bacteriophage integrases. J Mol Biol. 1995;252:178–188. doi: 10.1006/jmbi.1995.0486. [DOI] [PubMed] [Google Scholar]
- 23.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 24.Lorimer IA, Pastan I. Random recombination of antibody single chain Fv sequences after fragmentation with DNaseI in the presence of Mn2+ Nucleic Acids Res. 1995;23:3067–3068. doi: 10.1093/nar/23.15.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Arnold FH. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 1997;25:1307–1308. doi: 10.1093/nar/25.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo F, Gopaul DN, Van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 28.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 29.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci U S A. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rufer AW, Sauer B. Non-contact positions impose site selectivity on Cre recombinase. Nucleic Acids Res. 2002;30:2764–2771. doi: 10.1093/nar/gkf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 32.Voziyanov Y, Stewart AF, Jayaram M. A dual reporter screening system identifies the amino acid at position 82 in Flp site-specific recombinase as a determinant for target specificity. Nucleic Acids Res. 2002;30:1656–1663. doi: 10.1093/nar/30.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voziyanov Y, Konieczka JH, Stewart AF, Jayaram M. Stepwise manipulation of DNA specificity in Flp recombinase: progressively adapting Flp to individual and combinatorial mutations in its target site. J Mol Biol. 2003;326:65–76. doi: 10.1016/s0022-2836(02)01364-5. [DOI] [PubMed] [Google Scholar]
- 34.Konieczka JH, Paek A, Jayaram M, Voziyanov Y. Recombination of hybrid target sites by binary combinations of Flp variants: mutations that foster interprotomer collaboration and enlarge substrate tolerance. J Mol Biol. 2004;339:365–378. doi: 10.1016/j.jmb.2004.03.060. [DOI] [PubMed] [Google Scholar]
- 35.Bolusani S, Ma CH, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shultz JL, Voziyanova E, Konieczka JH, Voziyanov Y. A genome-wide analysis of FRT-like sequences in the human genome. PLoS One. 2011;6:e18077. doi: 10.1371/journal.pone.0018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 38.Turan S, Zehe C, Kuehle J, Qiao J, Bode J. Recombinase-mediated cassette exchange (RMCE) - a rapidly-expanding toolbox for targeted genomic modifications. Gene. 2013;515:1–27. doi: 10.1016/j.gene.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 39.Lauth M, Spreafico F, Dethleffsen K, Meyer M. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 2002;30:e115. doi: 10.1093/nar/gnf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson RP, Voziyanova E, Voziyanov Y. Flp and Cre expressed from Flp-2A-Cre and Flp-IRES-Cre transcription units mediate the highest level of dual recombinase-mediated cassette exchange. Nucleic Acids Res. 2012;40:e62. doi: 10.1093/nar/gks027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voziyanova E, Malchin N, Anderson RP, Yagil E, Kolot M, Voziyanov Y. Efficient Flp-Int HK022 dual RMCE in mammalian cells. Nucleic Acids Res. 2013;41:e125. doi: 10.1093/nar/gkt341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- 44.Baubonis W, Sauer B. Genomic targeting with purified Cre recombinase. Nucleic Acids Res. 1993;21:2025–2029. doi: 10.1093/nar/21.9.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Logie C, Stewart AF. Ligand-regulated site-specific recombination. Proc Natl Acad Sci U S A. 1995;92:5940–5944. doi: 10.1073/pnas.92.13.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blaisonneau J, Sor F, Cheret G, Yarrow D, Fukuhara H. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid. 1997;38:202–209. doi: 10.1006/plas.1997.1315. [DOI] [PubMed] [Google Scholar]