Abstract
Oncogenic NRAS and KRAS mutations are prevalent in human juvenile and chronic myelomonocytic leukemia (JMML/CMML). However, additional genetic mutations cooperating with oncogenic RAS in JMML/CMML progression and/or their transformation to acute myeloid leukemia (AML) remain largely unknown. Here, we tested the potential genetic interaction of DNMT3A mutations and oncogenic RAS mutations in leukemogenesis. We found that Dnmt3a−/− induces multiple hematopoietic phenotypes after a prolonged latency, including T cell expansion in peripheral blood, stress erythropoiesis in spleen, and myeloid malignancies in liver. Dnmt3a−/− significantly promoted JMML/CMML progression and shortened the survival of KrasG12D/+ mice in a cell-autonomous manner. Similarly, downregulating Dnmt3a also promoted myeloid malignancies in NrasG12D/+ mice. Further studies show that Dnmt3a deficiency rescues KrasG12D/+-mediated depletion of hematopoietic stem cells and increases self-renewal of KrasG12D/+ myeloid progenitors. Moreover, ~33% of animals developed an AML-like disease, which is driven by KrasG12D/+; Dnmt3a−/− myeloid progenitors. Consistent with our result, COSMIC database mining demonstrates that the combination of oncogenic RAS and DNMT3A mutations exclusively occurred in patients with JMML, CMML, or AML. Our results suggest that DNMT3A mutations and oncogenic RAS cooperate to regulate hematopoietic stem and progenitor cells and promote myeloid malignancies.
Keywords: Dnmt3a, oncogenic Kras, juvenile/chronic myelomonocytic leukemia, acute myeloid leukemia
Introduction
Constitutively active mutations in NRAS and KRAS genes are identified in human hematopoietic malignancies at significant frequencies (1). In particular, oncogenic NRAS and KRAS mutations are predominant in juvenile myelomonocytic leukemia (JMML), myeloproliferative variant of chronic myelomonocytic leukemia (MP-CMML), and the M4 and M5 monocytic subtypes of acute myeloid leukemia (AML) (including both de novo AML and secondary AML with antecedent JMML/CMML). Consistently, mice expressing endogenous oncogenic Kras or Nras develop highly penetrant JMML/MP-CMML-like phenotypes (2–8). Although these animals rarely develop AML spontaneously, acquisition of other mutations does promote their malignant transformation to monocytic AML (7). These findings indicate that JMML, MP-CMML, and M4/M5 AML are related malignancies in which oncogenic Ras signaling plays an essential role.
Although genetic mutations in a few genes are reported to be concurrent with oncogenic RAS mutations (9) in myeloid malignancies, their functional significances remain largely unknown. Acquisition of two copies of oncogenic RAS alleles, including NRASG12D/G12D and KRASG13D/G13D, is associated with JMML/CMML progression in human and mouse (COSMIC database and (6, 10–12)). Consistently, NrasG12D/G12D mice develop JMML/MP-CMML phenotypes much more rapidly than NrasG12D/+ mice (8), indicating that incremental activation of Ras signaling is a pathological mechanism contributing to JMML/CMML development. In contrast, knocking down Tet2 expression in NrasG12D/+ bone marrow cells does not seem to promote JMML/CMML progression or its malignant transformation (13). These data suggest that the potential genetic interaction between oncogenic RAS and other concurrent mutations have to be validated in vivo on a case-by-case basis.
Recent work focusing on AML with a normal karyotype identified mutations in DNA methyltransferase 3A (DNMT3A) with biological, clinical, and potential therapeutic relevance in AML and other types of myeloid malignancies (9, 14). Approximately 90% of DNMT3A mutations occur as a single copy mutation over wild-type DNMT3A (15). Although the predominant mutation at the codon R882 has been shown to be a dominant-negative mutation (16, 17), loss of Dnmt3a in the mouse hematopoietic system does not induce leukemogenesis up to 6 months of age (18). In contrast, recipients transplanted with Dnmt3a deficient hematopoietic stem cells (HSCs) developed both myeloid and lymphoid malignancies (19). Furthermore, loss of Dnmt3a promotes lung tumor progression in oncogenic Kras mice (20). Consistent with this finding, a group of AML patients were identified who carried both oncogenic RAS and DNMT3A mutations (9, 21). However, it remains unclear whether these two mutations cooperate in myeloid leukemia development.
Here, we show that loss of Dnmt3a promotes multiple hematopoietic defects after a prolonged latency, which are distinct from recipients transplanted with Dnmt3a−/− HSCs. Downregulation of Dnmt3a (deleting a single copy or both copies) in oncogenic Ras models not only significantly promote JMML/MP-CMML progression but also leads to transformation to acute myeloid diseases in a cell autonomous manner. Our finding is consistent with COSMIC database mining results showing that oncogenic RAS and DNMT3A mutations were only concurrent in myeloid malignancies, including JMML, CMML, and AML. Further mechanistic studies demonstrate that Dnmt3a deficiency promotes myeloid diseases in oncogenic Kras model through rescuing KrasG12D/+-mediated depletion of HSCs and increasing self-renewal of KrasG12D/+ myeloid progenitor cells. These mutant myeloid progenitors could initiate myeloid malignancies in recipients and thus serve as leukemia initiating cells. Our results suggest that changes in epigenetic landscapes and signaling networks co-regulate hematopoietic stem and progenitor cells to promote myeloid leukemias.
Materials and Methods
Mice
All mouse lines were maintained in a pure C57BL/6 genetic background (>N10). Dnmt3a conditional knockout mice (Dnmt3afl/fl (22); provided by Dr. Qiang Chang) were crossed to mice bearing a conditional oncogenic Kras (KrasLox-stop-Lox (LSL) G12D/+) or Mx1-Cre mice to generate mice carrying both alleles (KrasLSL G12D/+; Dnmt3afl/+ and Dnmt3afl/+; Mx1-Cre, respectively). KrasLSL G12D/+; Dnmt3afl/+ mice were further crossed to Dnmt3afl/+; Mx1-Cre mice to generate our experimental mice, including KrasLSL G12D/+; Dnmt3afl/fl; Mx1-Cre, KrasLSL G12D/+; Mx1-Cre, Dnmt3afl/fl; Mx1-Cre, and Mx1-Cre mice. CD45.1-positive congenic C57BL/6 recipient mice were purchased from NCI. Cre expression was induced through intraperitoneal injection of 2.5 μg/g body weight (GE Healthcare) of polyinosinic-polycytidylic acid (pI-pC) every other day for two times. All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by an Animal Care and Use Committee at UW-Madison. The program is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Flow cytometric analysis of hematopoietic tissues
For lineage analysis of peripheral blood, bone marrow, and spleen, flow cytometric analyses were performed as previously described (4). Myeloid progenitors in bone marrow and spleen were analyzed as previously described (8). HSCs in bone marrow and spleen were analyzed as described in (23). Because hind limb bone marrow represents ~25% of total bone marrow, the number of HSCs in total bone marrow is calculated as 4 X the number of HSCs in hind limb bone marrow. The stained cells were analyzed on a FACS Calibur (BD Biosciences) or a MACSQuant Analyzer (Miltenyi Biotec Inc.). Antibodies specific for the following surface antigens were purchased from eBioscience: CD45.2 (104), B220 (RA3-6B2), CD19 (eBio1D3), Thy1.2 (53-2.1), Mac-1 (M1/70), Gr-1 (RB6-8C5), CD4 (GK1.5), CD8 (53-6.7), CD3 (145-2C11), IgM (II/41), IL7Rα (A7R34), Sca-1 (D7), TER119(TER-119), CD34 (RAM34), cKit (2B8). FcγRII/III (2.4G2) was purchased from BD Biosciences. CD150 (TC15-12F12.2) was purchased from Biolegend.
Additional methods are described in Supplemental Methods.
Results
Somatic deletion of Dnmt3a promotes acute myeloid diseases in KrasG12D/+ mice
To determine whether Dnmt3a plays an important role in KrasG12D/+-mediated hematopoietic malignancies, we generated Dnmt3afl/fl; Mx1-Cre, KrasLSL G12D/+; Mx1-Cre and KrasLSL G12D/+; Dnmt3afl/fl; Mx1-Cre mice. Administration of polyinosinic-polycytidylic acid (pI-pC) in these compound mice stimulates endogenous interferon (IFN) production and thus induces Cre expression from the IFN-α/β-inducible Mx1 promoter, which in turn leads to the expression of oncogenic Kras from its endogenous locus and/or somatic deletion of Dnmt3a (24). We refer to these pI-pC-treated compound mice as Dnmt3a−/−, KrasG12D/+, and KrasG12D/+; Dnmt3a−/− mice respectively, and pI-pC-treated Mx1-Cre or wild-type mice as control mice throughout this manuscript.
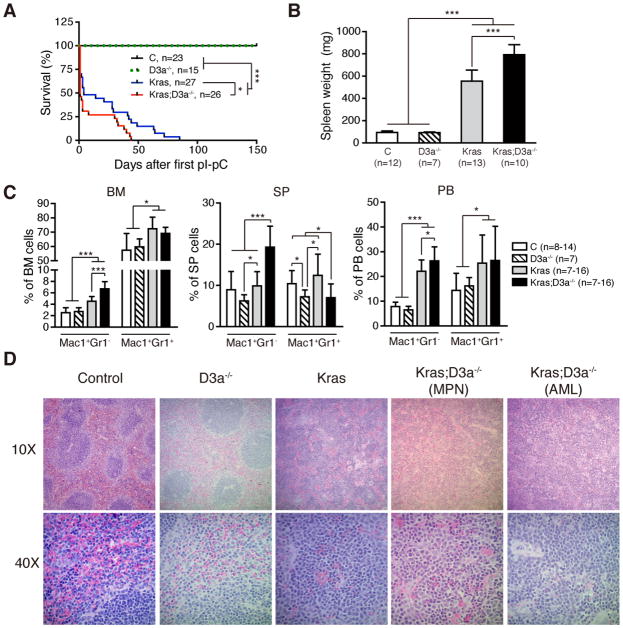
Somatic deletion of Dnmt3a significantly enhanced leukemia phenotypes and shortened the life-span of KrasG12D/+ mice (Fig. 1). Two days after the 2nd pI-pC injection, control, Dnmt3a−/−, KrasG12D/+, and KrasG12D/+; Dnmt3a−/− mice were sacrificed and assessed for hematopoietic phenotypes (Fig. 1B–1D). Consistent with previous reports (2–5), KrasG12D/+ mice developed an acute myeloproliferative neoplasm (MPN), closely resembling human JMML/MP-CMML. This disease was manifested by splenomegaly (Fig. 1B) with significant extramedullary hematopoiesis (Fig. 1D), expanded monocyte and neutrophil compartments in various hematopoietic tissues (Fig. 1C), elevated white blood cell counts in peripheral blood, and defective erythroid and megakaryocyte development (Table S1). Loss of Dnmt3a not only significantly enhanced the MPN phenotypes in KrasG12D/+ mice (Fig. 1B–1C and Table S1), but also promoted the development of acute myeloid leukemia (AML)-like phenotypes in about one third of the animals, as demonstrated by accumulation of immature myeloblast cells in the spleen (Fig. 1D).
Figure 1. Loss of Dnmt3a promotes myeloid diseases in KrasG12D/+ mice.
Control (C), Dnmt3a−/− (D3a−/−), KrasG12D/+ (Kras), and KrasG12D/+; Dnmt3a−/− (Kras; D3a−/−) mice were treated with pI-pC as described in Materials and Methods. We define the day of 1st pI-pC injection as Day 1. Treated mice were sacrificed on Day 5 for analysis of different hematopoietic tissues. (A) Kaplan-Meier survival curves of different groups of mice were plotted against days after first pI-pC injection. P values were determined by the Log-rank test. (B,C) Quantification of spleen weight (B) and myeloid cell compartment in bone marrow (BM), spleen (SP) and peripheral blood (PB) (C). (D) Representative spleen histologic H&E sections from different groups of animals are shown. The results are presented as means ± SD. * P<0.05 and *** P<0.001. MPN, myeloproliferative neoplasm; AML, acute myeloid leukemia.
Dnmt3a deficiency induces multiple hematopoietic defects
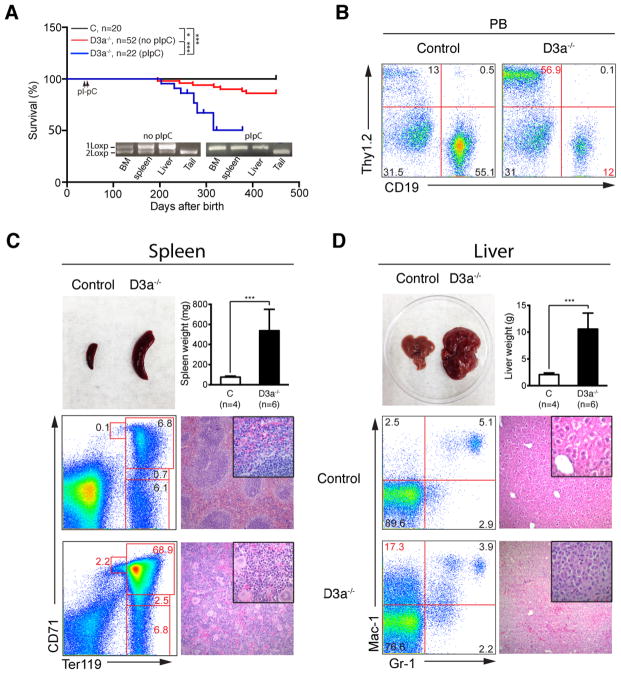
Although all Dnmt3a−/− mice survived in an overtly healthy condition for 6 months, ~14% of Dnmt3afl/fl; Mx1-Cre mice without pI-pC treatment died with multiple hematopoietic defects within 13 months due to the leaky expression of Cre over time (Fig. 2A). In these moribund mice, the fraction of T cells in peripheral blood was significantly elevated but the thymus size and structure were essentially normal, indicating a chronic T cell expansion (Fig. 2B). Our result is consistent with a recent report that Dnmt3a prevents malignant mouse lymphopoiesis (25). In the enlarged spleen (~4–5 fold over control spleens), the normal architecture was completely effaced by extramedullary hematopoietic tissue that was dominated by erythroid and megakaryocytic elements with fewer numbers of maturing myeloid cells. Sheets of blasts were not evident. Consistent with the H&E sections, flow cytometric analysis of splenocytes also demonstrated the dominance of erythroid lineage cells, suggesting a stressed erythropoiesis (Fig. 2C). The markedly enlarged livers (~10–15 fold over control livers) were dramatically altered with only small islands of hepatocytes present separated by sheets of discohesive cells. Based on the histological evaluation, this is most consistent with a histiocytic or myeloid sarcoma (Fig. 2D).
Figure 2. Loss of Dnmt3a induces multiple hematopoietic defects in vivo.
The untreated and pI-pC treated Dnmt3afl/fl; Mx1-Cre mice were monitored for an extended period of time until a moribund stage. (A) Kaplan-Meier survival curves of different groups of mice were plotted against days after birth. Bone marrow (BM), spleen (SP), and liver cells from moribund mice were genotyped to evaluate recombination efficiency of Dnmt3a. P value was determined by the Log-rank test: *P<0.05. (B) Representative analyses of peripheral blood T cells (Thy1.2) and B cells (CD19) from moribund Dnmt3a−/− mice and age-matched control mice are shown. (C) Splenomegaly and representative histologic H&E sections and erythroblast analysis of spleen from moribund Dnmt3a−/− mice and age-matched control mice are shown. (D) Hepatomegaly and representative histologic H&E sections and myeloid cell analysis of liver from moribund Dnmt3a−/− mice and age-matched control mice are shown. The results are presented as means ± SD. *** P<0.001.
To validate our result, we set up another cohort of Dnmt3afl/fl; Mx1-Cre mice injected with pI-pC and monitored them closely. They consistently displayed a transient anemia two months after pI-pC injections with significantly lower red blood cell count, hemoglobin, and hematocrit (Fig. S1A). However, the other aspects of hematopoiesis were indistinguishable from those of control mice (Fig. S1B). Further analysis of erythropoiesis in bone marrow and spleen showed that the red pulp in Dnmt3a−/− spleen was moderately enlarged (Fig. S2A). Consistent with this result, flow cytometric analysis using CD71 and TER119 demonstrated that the erythroid compartment in Dnmt3a−/− spleen was moderately but significantly expanded with increased Region II cells and decreased Region IV cells (Fig. S2B–S2D). In vitro colony assay revealed that the number of CFU-E (colony forming unit-erythroid) progenitors in Dnmt3a−/− spleen was significantly increased (Fig. S2E) without detectable change of their sensitivity to Erythropoietin (EPO) stimulation (Fig. S2F). These results suggest that loss of Dnmt3a affects erythroid differentiation, which can be compensated by stress erythropoiesis in spleen. Compared with Dnmt3afl/fl; Mx1-Cre mice without pI-pC treatment, Dnmt3a−/− mice succumbed to similar hematopoietic defects (including T cell expansion in peripheral blood, stress erythropoiesis in spleen, and myeloid malignancies in liver) with a much higher penetrance (Fig. 2A).
To investigate cell autonomous role of Dnmt3a deficiency in leukemogenesis, we transplanted 2.5 × 105 Dnmt3a−/− bone marrow cells along with same number of competitor cells into individual lethally irradiated mice (Fig. S3). To our surprise, only 2 out of 23 recipients died of hematopoietic maliganacies 300 days after transplantation (Fig. S3A). The first one was found dead with enlarged thymus and spleen, while the second one was sacrificed at the moribund stage and also showed enlarged spleen and thymus (Fig. S3B). Detailed flow cytometric analysis of the second mouse revealed a lethal acute T-cell lymphoblastic leukemia/lymphoma (T-ALL) throughout the entire hematopoietic system (Fig. S3C). Together, our results suggest that somatic downregulation of Dnmt3a does promote leukemogenesis as a first genetic hit.
Downregulation of Dnmt3a promotes myeloid diseases in NrasG12D/+ mice
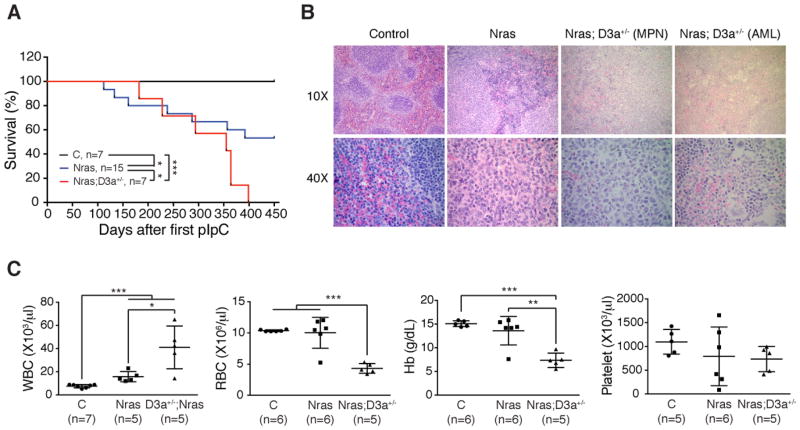
Approximately 90% of DNMT3A mutations in human leukemia patients occur as a single copy mutation over wild-type DNMT3A (15). Although the predominant mutation at the codon R882 has been shown to be a dominant-negative mutation (16, 17), other mutations have not been characterized in detail. We investigated whether loss of a single copy of Dnmt3a would promote oncogenic Ras-induced leukemogenesis. As was seen in oncogenic Kras-induced non-small cell lung carcinoma (20), we did not observe significant acceleration or enhancement of MPN development in KrasG12D/+; Dnmt3a+/− mice compared with KrasG12D/+ mice (our unpublished data). This could be due to the strong MPN phenotypes in KrasG12D/+ mice. Therefore, we generated NrasG12D/+; Dnmt3afl/+; Mx1-Cre mice to address this question (referred as NrasG12D/+; Dnmt3a+/− mice after pI-pC treatment). Compared with NrasG12D/+ mice, NrasG12D/+; Dnmt3a+/− mice indeed showed significantly shortened life-span and developed more severe MPN phenotypes (Fig. 3A and 3C). As with KrasG12D/+; Dnmt3a−/− mice, about one third of NrasG12D/+; Dnmt3a+/− mice developed AML-like phenotypes (Fig. 3B). Thus, our data indicate that downregulating Dnmt3a and oncogenic Nras cooperate to promote myeloid diseases in vivo.
Figure 3. Downregulation of Dnmt3a expression promotes NrasG12D/+-induced myeloid diseases.
Control (C), NrasG12D/+ (Nras), and NrasG12D/+; Dnmt3a+/− (Nras; D3a+/−) mice were injected with pI-pC as described in Methods. (A) Kaplan-Meier survival curves of different groups of mice were plotted against days after first pI-pC injection. P values were determined by the Log-rank test. (B) Representative spleen histologic H&E sections from moribund NrasG12D/+ and NrasG12D/+; D3a+/− mice and from age-matched control mice. (C) Complete blood count (CBC) from moribund NrasG12D/+; D3a+/− mice and from 60-week old control and NrasG12D/+ mice. The results are presented as means ± SD. * P<0.05, ** P<0.01 and *** P<0.001.
Loss of Dnmt3a rescues KrasG12D/+-mediated HSC depletion and increases self-renewal of KrasG12D/+ myeloid progenitors
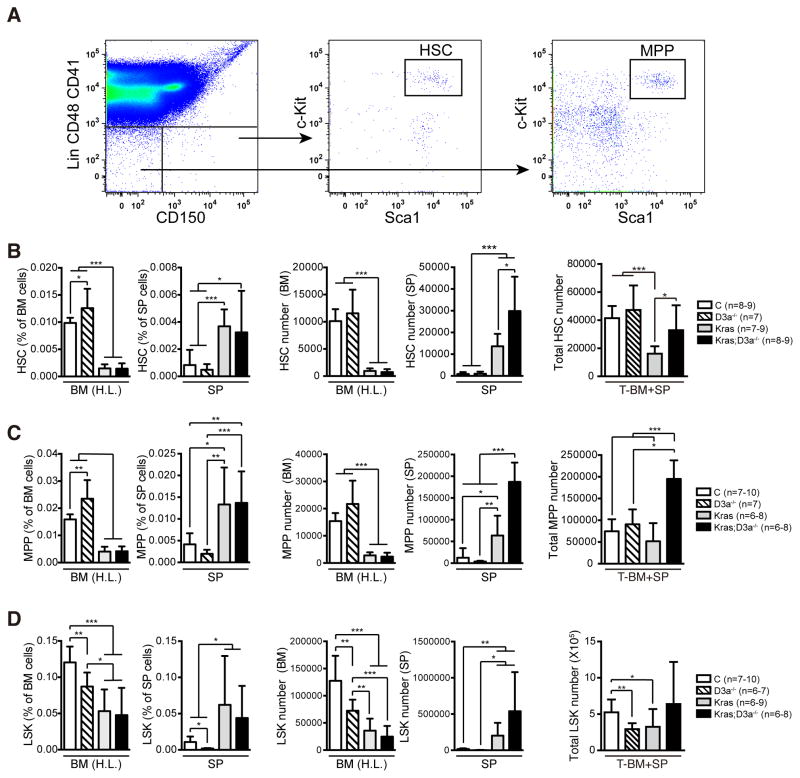
To study the mechanisms of how Dnmt3a deficiency promotes myeloid diseases in oncogenic Kras mice, we first examined the HSC compartment because mutant HSCs serve as JMML/MP-CMML initiating cells in these animals (4, 26). HSCs are defined as Lin− CD41− CD48− c-Kit+ Sca-1+ CD150+ cells throughout the manuscript (Fig. 4A). Although the frequency of Dnmt3a−/− HSCs was moderately increased in bone marrow compared to that of control HSCs, the absolute number was comparable in the whole body (total bone marrow + spleen) (Fig. 4B). Consistent with previous reports (5, 26), the HSC number in KrasG12D/+ mice was significantly less than that in control mice, indicating an oncogenic Kras-mediated HSC depletion. In contrast, the HSC compartment in KrasG12D/+; Dnmt3a−/− mice was comparable to that in control mice, suggesting that depletion of KrasG12D/+ HSCs was rescued by loss of Dnmt3a. The total number of multipotent progenitors (MPPs, defined as Lin− CD41− CD48− c-Kit+ Sca-1+ CD150− cells; Fig. 4A) in KrasG12D/+; Dnmt3a−/− mice was concomitantly increased compared to that in control and KrasG12D/+ mice (Fig. 4C), whereas the LSK (Lin− Sca1+ cKit+) compartment was comparable to controls (Fig. 4D).
Figure 4. Loss of Dnmt3a rescues KrasG12D/+-mediated HSC depletion.
Control (C), Dnmt3a−/− (D3a−/−), KrasG12D/+ (Kras), and KrasG12D/+; Dnmt3a−/− (Kras; D3a−/−) mice were injected with pI-pC as described in Methods. Treated mice were sacrificed on Day 5 for analysis of hematopoietic stem cells (HSCs) (B), multipotential progenitor cells (MPPs) (C) and Lin− Sca-1+ c-Kit+ cells (LSKs) (D) in hind limb bone marrow (BM (H.L.)) and spleen (SP). (A) Representative example of staining and gating for HSCs and MPPs. (B–D) The absolute HSC, MPP and LSK numbers in BM (H.L.) and SP were calculated based on bone marrow or spleen cell numbers and frequencies. Because BM (H.L.) represents 25% of whole body bone marrow, the total number of stem/progenitor cells per animal was calculated as the sum of stem/progenitor cell number in spleen and four fold of stem/progenitor cell number in BM (H.L.). The results are presented as means ± SD. * P<0.05, ** P<0.01 and *** P<0.001.
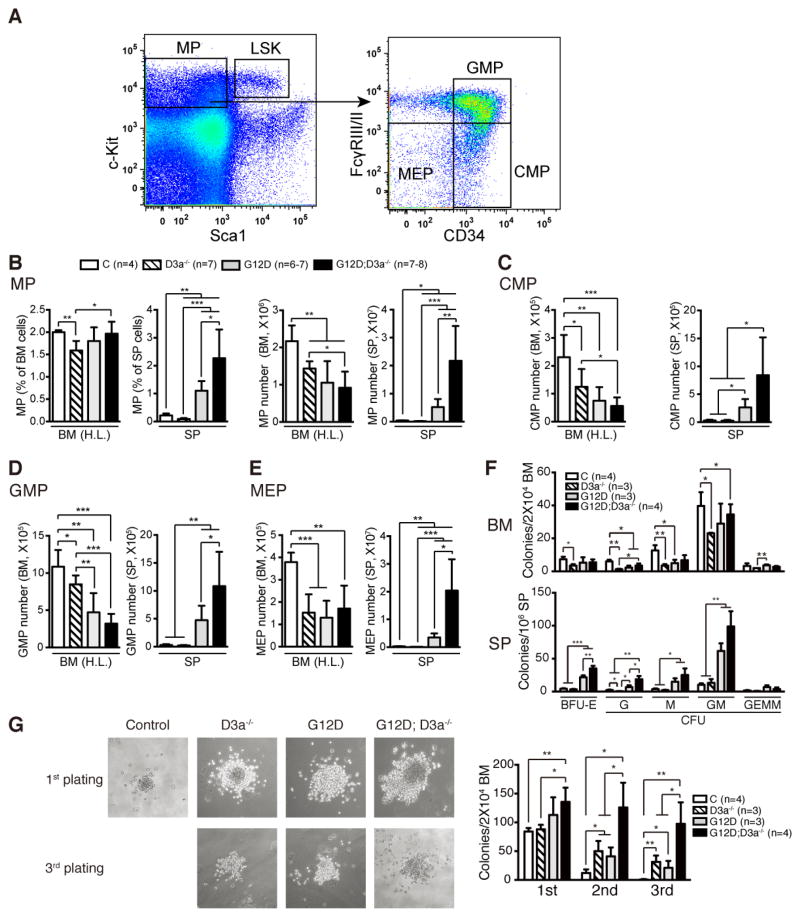
We then analyzed the myeloid progenitors (MPs) in all groups of animals (Fig. 5). In Dnmt3a−/− mice, the number of MPs, including common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs), was significantly decreased in bone marrow compared to that in control mice (Fig. 5A–5E). Consistently, Dnmt3a−/− bone marrow cells formed significantly fewer colonies in semi-solid cultures (Fig. 5F). In contrast, Dnmt3a deficiency promoted further expansion of MP compartment in KrasG12D/+-expressing spleens (Fig. 5A–5F). Moreover, although loss of Dnmt3a or expression of endogenous oncogenic Kras led to a moderate increase of transient self-renewal of MPs, KrasG12D/+; Dnmt3a−/− MPs demonstrated significantly higher self-renewal capability than KrasG12D/+ or Dnmt3a−/− MPs in the replating assay (Fig. 5G). Together, our data suggest that loss of Dnmt3a promotes myeloid diseases in KrasG12D/+ mice through rescuing KrasG12D/+-mediated HSC depletion and enhancing self-renewal of KrasG12D/+ MPs.
Figure 5. Loss of Dnmt3a increases transient self-renewal capability of KrasG12D/+ myeloid progenitor cells.
Control (C), Dnmt3a−/− (D3a−/−), KrasG12D/+ (Kras), and KrasG12D/+; Dnmt3a−/− (Kras; D3a−/−) mice were injected with pI-pC as described in Methods. Treated mice were sacrificed on Day 5 for analysis of myeloid progenitor cells. (A) Representative example of staining and gating for myeloid progenitors (MPs), common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), and megakaryocyte-erythrocyte progenitors (MEPs). (B–E) Quantitative analysis of MPs (B), CMPs (C), GMPs (D), and MEPs (E) in hind limb bone marrow (BM (H.L.)) and spleen (SP). (F) Quantification of various types of hematopoietic colonies cultured for 7 days in methylcellulose-based medium M3434. (G) Assessment of transient self-renewal capability of myeloid progenitors. The results are presented as means ± SD. * P<0.05, ** P<0.01 and *** P<0.001.
Deletion of Dnmt3a promotes myeloid malignancies in KrasG12D/+ mice in a cell autonomous manner
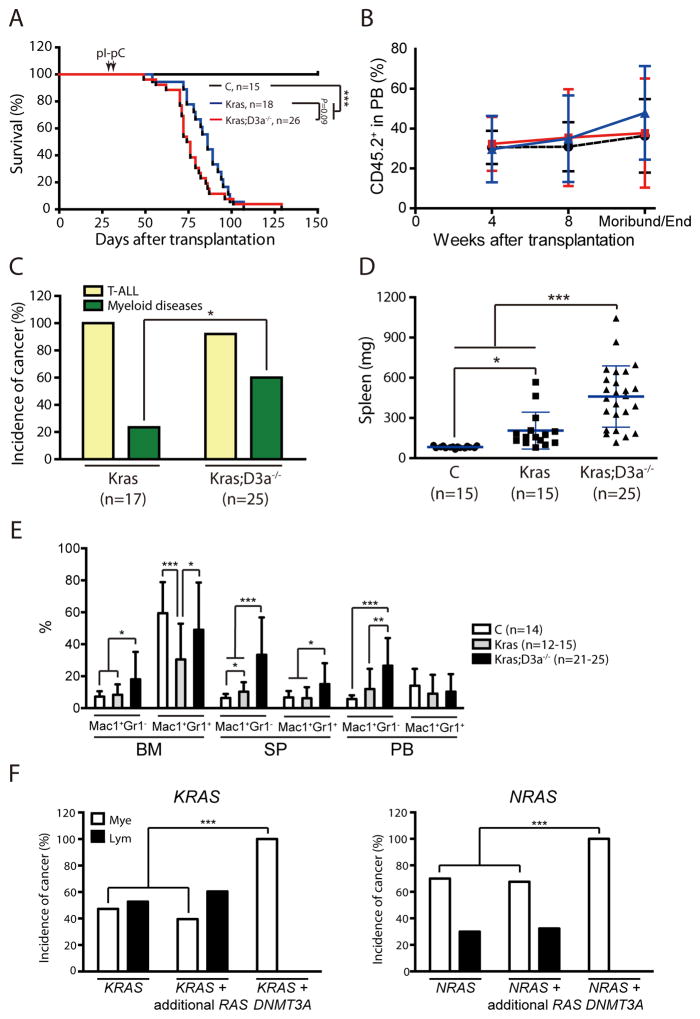
To determine whether Dnmt3a deficiency plays a cell autonomous role in promoting leukemogenesis in KrasG12D/+ model, we transplanted control, KrasLSL G12D/+; Mx1-Cre, or KrasLSL G12D/+; Dnmt3afl/fl; Mx1-Cre bone marrow cells into lethally irradiated mice (Fig. 6). Four weeks after transplantation, recipient mice were injected with pI-pC as before. We did not observe significant change of life span between recipients with KrasG12D/+ cells and those with KrasG12D/+; Dnmt3a−/− cells (Fig. 6A). This could be due to the leaky expression of oncogenic Kras but not efficient Dnmt3a deletion before the pI-pC induction.
Figure 6. Recipients transplanted with KrasG12D/+; Dnmt3a−/− cells develop myeloid diseases with a high penetrance.
Lethally irradiated mice were transplanted with 2.5×105 bone marrow cells from different groups of mice along with same number of competitor cells. Four weeks after transplantation, Cre expression was induced by pI-pC injections. (A) Kaplan-Meier survival curves of different groups of recipient mice were plotted against days after transplantation. P values were determined by the Log-rank test. (B) The percentages of donor-derived cells in different groups of recipients. (C) Disease distribution patterns in recipient mice transplanted with KrasG12D/+ or KrasG12D/+; Dnmt3a−/− cells. We define the mice with a myeloid disease when donor-derived monocytes consist >20% of white blood cells in peripheral blood. Chi-square analysis was performed. (D–E) Quantitative analysis of spleen weight (D), and donor-derived monocytes and neutrophils in bone marrow (BM), spleen (SP), and peripheral blood (PB) (E) from moribund recipient mice. The results are presented as means ± SD. * P<0.05, ** P<0.01 and *** P<0.001. (F) Human patients with either oncogenic KRAS or NRAS mutations from COSMIC database v67 were divided into 3 groups: (1) samples with only one copy of KRAS or NRAS mutation, (2) samples with additional RAS mutations, including more than one copy of KRAS or NRAS mutations, or containing mutations in both NRAS and KRAS genes, (3) samples with concurrent DNMT3A and one copy of KRAS or NRAS mutation. The patients were further stratified based on their disease types, lymphoid or myeloid malignancies. P values were determined by the Chi-square analysis. *** P<0.001.
Although different groups of donor cells engrafted recipients at a comparable level (Fig. 6B), recipients of KrasG12D/+; Dnmt3a−/− cells developed an acute myeloid disease with a much higher incidence than those with KrasG12D/+ cells (60% vs 20%; Fig. 6C). Consistently, the myeloid disease phenotypes characteristic in KrasG12D/+ mice, including splenomegaly and expanded myeloid compartments in various hematopoietic tissues (Fig. 6D–6E) as well as defective phenotypes in erythoid and megakaryocytic lineages of cells (Table S2), were more prominent in recipients of KrasG12D/+; Dnmt3a−/− cells than those of KrasG12D/+ cells. Similarly to primary non-transplanted mice, about one third of myeloid diseases that developed in recipients of KrasG12D/+; Dnmt3a−/− cells were AML-like, and the rest of them closely resembled a MPN (Fig. S4). In contrast, all the myeloid diseases that developed in recipients with KrasG12D/+ cells were MPN. Genotyping of myeloid disease cells from recipient bone marrow revealed that WT Kras allele was significantly downregulated in 50–75% of samples (Fig. S5), which might be caused by clonal expansion of leukemia cells with uniparental disomy of the oncogenic Kras allele or deletion of WT Kras allele. In conclusion, our results indicate that loss of Dnmt3a and oncogenic Ras cooperate to promote myeloid malignancies, which is consistent with the identification of activating RTK/RAS pathway mutations in Dnmt3a−/− induced AML in mice (19, 27).
To test the human relevance of our finding, we conducted a COSMIC database search to quantify the incidence of myeloid versus lymphoid malignancies in leukemia patients with oncogenic RAS mutations (Fig. 6F). We found that ~50% of patients with an oncogenic KRAS mutation and ~70% of patients with an oncogenic NRAS mutation had a myeloid malignancy. Acquisition of additional copies of oncogenic RAS mutations did not significantly change the disease incidence. However, 100% of patients with both oncogenic RAS and DNMT3A mutations had developed myeloid malignancies, including JMML, CMML, and AML. This result suggests that oncogenic RAS and DNMT3A mutations intrinsically interact or cooperate to promote leukemogenesis in myeloid cells.
In addition to myeloid neoplasms, all the recipients of KrasG12D/+; Dnmt3a−/− cells developed T-ALL (Fig. S6). In comparison to the T-ALL that developed in recipients of KrasG12D/+ cells, approximately 30% of KrasG12D/+; Dnmt3a−/− derived tumors contained a significant percentage of CD4− CD8− T-cells, suggesting a more immature phenotype. T-ALL developed in both groups of recipients carried Type 1 deletions in the Notch1 locus, which render ligand-independent cleavage of Notch1 and elevated expression of intracellular C-terminus Notch1. Our results suggest that active Notch1 signaling promotes T-ALL development in both genetic contexts.
KrasG12D/+; Dnmt3a−/− myeloid progenitors are capable of initiating myeloid malignancies in recipient mice
Because KrasG12D/+; Dnmt3a−/− bone marrow cells promoted highly penetrant myeloid malignancies in a cell-autonomous manner (Fig. 6) and KrasG12D/+; Dnmt3a−/− MPs displayed enhanced self-renewal in vitro (Fig. 5F), we investigated whether these mutant progenitors could initiate myeloid diseases in recipient mice and thus serve as leukemia initiating cells. Mutant LSK cells and LK Sca1− MPs were sorted and transplanted with competitor cells into lethally irradiated recipients (Table 1). All the recipients with mutant LSK cells succumbed to hematopoietic malignancies, one with T-ALL and MPN, one with MPN, and one with AML. Six out of seven recipients with mutant MPs died and careful analysis of four mice revealed that three died with MPN and one with AML. Our results indicate that some of KrasG12D/+; Dnmt3a−/− MPs are transformed to leukemia initiating cells.
Table 1.
Summary of diseased recipient mice transplanted with Kras; D3a−/− LSK or MP cells
| Donor cells (#/recipient) | Competitors (#/recipient) | Recipients (#) | Diseased animals (%) | Disease diagnosis(#) |
|---|---|---|---|---|
| LSK (1,000) | BM (200,000) | 3 | 100 | T-ALL and MPN (1) MPN(1) AML (1) |
| MP (15,000) | BM (200,000) | 7 | 86 | MPN (3) AML (1) |
LSK (Lin− Sca1+ cKit+) and MP (myeloid progenitor) cells were flow sorted from KrasG12D/+; Dnmt3a−/− bone marrow and transplanted with congenic competitor cells (BM: bone marrow) into lethally irradiated recipients. Mice were monitored closely until a moribund stage. Note: some animals died before we could catch and analyze them. T-ALL, acute T-cell lymphoblastic leukemia/lymphoma; MPN, myeloproliferative neoplasm; AML, acute myeloid leukemia.
Discussion
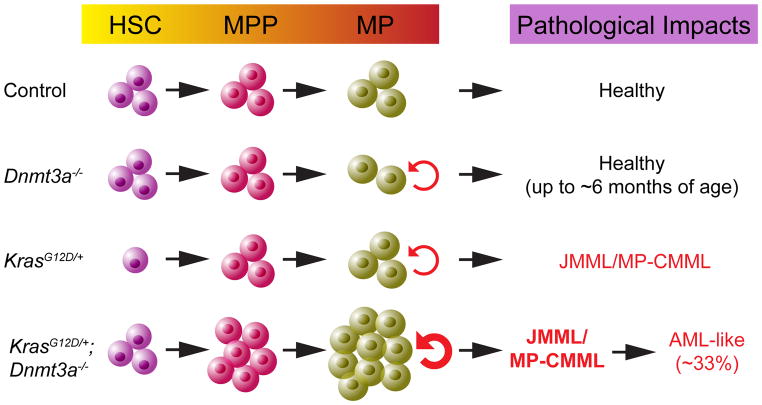
In this manuscript, we demonstrate that somatic downregulation of Dnmt3a induces leukemogenesis as a first genetic hit. Downregulating Dnmt3a cooperates with oncogenic Ras to promote myeloid malignancies in a cell autonomous manner in mice, consistent with the genetic findings in human leukemia patients. Furthermore, our data suggest that Dnmt3a deficiency promotes myeloid diseases through regulating the functions of KrasG12D/+ HSCs and MPs (Fig. 7).
Figure 7.
Schematic illustration summarizes the phenotypes in different genetic groups of animals.
Although the predominant mutation at the codon R882 of DNMT3A is a dominant-negative mutation (16, 17), loss of Dnmt3a in the mouse hematopoietic system does not induce leukemogenesis up to 6 months of age (18). However, we found that Dnmt3a−/− mice developed multiple hematopoietic defects after a prolonged latency (Fig. 2). Our results are consistent with human studies reporting that DNMT3A mutations in HSCs lead to clonal hematopoietic expansion and thus act as the first/early genetic event during AML development (28–31). Interestingly, the hematopoietic phenotypes developed in Dnmt3a−/− mice (T cell expansion in peripheral blood, stress erythropoiesis in spleen, and myeloid malignancies in liver) are distinct from those in recipients transplanted with Dnmt3a−/− HSCs (19, 27) or total bone marrow cells (Fig. S3), which primarily developed MDS/AML and T-ALL. This could be due to both cell-autonomous and cell-nonautonomous effects of Dnmt3a deficiency in non-transplanted Dnmt3a−/− mice. For example, we previously showed that pI-pC injection could sufficiently induce Cre expression in liver (6). Noticeably, the survival of our BMT cohort mice is significantly prolonged than those reported (19, 27). This difference might be due to the lower number of Dnmt3a−/− HSCs we transplanted and the presence of competitor cells.
We found that Dnmt3a deficiency and KrasG12D/+ cooperate to induce myeloid malignancies in a cell autonomous manner in vivo. Dnmt3a−/− significantly promoted JMML/CMML progression and shortened the survival of KrasG12D/+ mice (Fig. 1). Recipients transplanted with KrasG12D/+; Dnmt3a−/− bone marrow cells developed JMML/CMML at a much higher incidence than those transplanted with KrasG12D/+ cells (Fig. 6). Moreover, ~33% of animals developed an AML-like disease. Similarly, loss of one copy of Dnmt3a also promoted myeloid diseases in NrasG12D/+ mice, consistent with the previous study using a retroviral construct to overexpress oncogenic Nras in Dnmt3a−/− bone marrow cells (19). However, the survival data from our genetic model is very different from that of RasG12D overexpression study, which could be mainly explained by the expression levels of oncogenic Nras (endogenous level versus overexpression). Indeed, although recipients transplanted with KrasG12D/+ or NrasG12D/+ cells do not develop AML spontaneously (4, 6, 26, 32), recipients with bone marrow cells overexpressing NrasG12D develop a highly penetrant CMML/AML rapidly (33). The in vivo genetic interaction between oncogenic Ras and Dnmt3a−/− in myeloid diseases is also supported by the identification of activating RTK/RAS pathway mutations (c-KitV750M and KrasG12D) in Dnmt3a−/− induced AML in mice (19, 27). We believe that our results are highly relevant to human physiology because the combination of oncogenic RAS (including both NRAS and KRAS) and DNMT3A mutations exclusively occurred in patients with myeloid leukemias, including JMML, CMML, and AML (Fig. 6F).
Our results demonstrate that genetic interaction between Dnmt3a deficiency and KrasG12D/+ regulates the functions of HSCs and MPs to drive myeloid leukemogenesis. We recently showed that stronger oncogenic Ras signaling (e.g. NrasG12D/G12D) shifts HSC self-renewal to differentiation and leads to HSC exhaustion (23). Similar mechanisms might be also applicable to KrasG12D/+ HSCs. However, KrasG12D/+ -mediated HSC depletion is antagonized by loss of Dnmt3a, which is likely to contribute to the significantly increased incidence of myeloid malignancies in recipients with KrasG12D/+; Dnmt3a−/− cells. Our result is consistent with a previous study reporting that Dnmt3a−/− maintains HSC self-renewal in the presence of oncogenic Ras (19).
We found that expression of endogenous oncogenic Ras (e.g. KrasG12D/+ or NrasG12D/+)promotes progenitor cell growth in vivo and in vitro (2, 3, 5–7), in sharp contrast to the negative effect of RasG12D overexpression on progenitor growth in vitro (19). This is likely due to different expression levels of oncogenic Ras used in the studies. Consistent with a previous report (27), we observed moderately increased self-renewal of Dnmt3a−/− MPs in a re-plating assay. Moreover, KrasG12D/+ and Dnmt3a−/− act synergistically to promote self-renewal in MPs, which are transformed to leukemia initiating cells and drive AML-like diseases in vivo. Our data provide a strong rationale to target both epigenetic regulators and aberrant cytokine signaling in treating myeloid leukemias.
Supplementary Material
Acknowledgments
We are grateful to Dr. Qiang Chang for providing the Dnmt3a conditional knockout mice. We would like to thank the University of Wisconsin Carbone Comprehensive Cancer Center (UWCCC) for use of its Shared Services to complete this research. This work was supported by R01 grants R01CA152108 and R01HL113066, and a Scholar Award from the Leukemia & Lymphoma Society to J.Z. Y.-I.C. was supported by the Ministry of Science and Technology (MOST 103-2320-B-010-047) and a grant from Ministry of Education, Aim for the Top University Plan. This work was also supported in part by NIH/NCI P30 CA014520--UW Comprehensive Cancer Center Support.
Mouse genotype abbreviations
- Control
pI-pC treated Mx1-Cre or wild-type
- KrasG12D/+
recombined KrasG12D/+ heterozygous (pI-pC treated LSL KrasG12D/+; Mx1-Cre)
- Dnmt3a−/−
recombined Dnmt3a knockout (pI-pC treated Dnmt3afl/fl; Mx1-Cre)
- KrasG12D/+; Dnmt3a−/−
recombined KrasG12D/+ heterozygous deficient for Dnmt3a (pI-pC treated LSL KrasG12D/+; Dnmt3afl/fl; Mx1-Cre)
- NrasG12D/+; Dnmt3a+/−
recombined NrasG12D/+ and Dnmt3a+/− double heterozygous (pI-pC treated LSL NrasG12D/+; Dnmt3afl/+; Mx1-Cre).
Footnotes
Conflict of interest:
We declare that no conflict of interest exists.
Supplementary information is available at Leukemia’s website.
References
- 1.Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012 Aug 16; doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, Rozmus J, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004 Jan 13;101(2):597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, Shigematsu H, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004 Feb;113(4):528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF, et al. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009 Feb 5;113(6):1304–1314. doi: 10.1182/blood-2008-01-134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Liu Y, Meline B, Kong G, Tan LX, Lo JC, et al. Loss of CD44 attenuates aberrant GM-CSF signaling in Kras G12D hematopoietic progenitor/precursor cells and prolongs the survival of diseased animals. Leukemia. 2013;27(3):754–757. doi: 10.1038/leu.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JY, Liu YG, Li ZY, Du J, Ryu MJ, Taylor PR, et al. Endogenous oncogenic Nras mutation leads to aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood. 2010;116(26):5991–6002. doi: 10.1182/blood-2010-04-281527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011 Feb 10;117(6):2022–2032. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JY, Liu YG, Li ZY, Wang ZD, Tan LX, Ryu MJ, et al. Endogenous oncogenic Nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood. 2011;118(2):368–379. doi: 10.1182/blood-2010-12-326058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012 Sep;12(9):599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 10.Dunbar AJ, Gondek LP, O’Keefe CL, Makishima H, Rataul MS, Szpurka H, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008 Dec 15;68( 24):10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, et al. Next-Generation Sequencing Technology Reveals a Characteristic Pattern of Molecular Mutations in 72.8% of Chronic Myelomonocytic Leukemia by Detecting Frequent Alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010 Jul 19; doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 12.Kato M, Yasui N, Seki M, Kishimoto H, Sato-Otsubo A, Hasegawa D, et al. Aggressive transformation of juvenile myelomonocytic leukemia associated with duplication of oncogenic KRAS due to acquired uniparental disomy. The Journal of pediatrics. 2013 Jun;162(6):1285–1288. 1288 e1281. doi: 10.1016/j.jpeds.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Chang YI, Damnernsawad A, Allen LK, Yang D, Ranheim EA, Young KH, et al. Evaluation of allelic strength of human TET2 mutations and cooperation between Tet2 knockdown and oncogenic Nras mutation. Br J Haematol. 2014 Apr 3; doi: 10.1111/bjh.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010 Dec 16;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li KK, Luo LF, Shen Y, Xu J, Chen Z, Chen SJ. DNA methyltransferases in hematologic malignancies. Seminars in hematology. 2013 Jan;50(1):48–60. doi: 10.1053/j.seminhematol.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, et al. The R882H DNMT3A Mutation Associated with AML Dominantly Inhibits Wild-Type DNMT3A by Blocking Its Ability to Form Active Tetramers. Cancer Cell. 2014 Apr 14;25(4):442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013 Dec 12;122(25):4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012 Jan;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2014 Nov 21; doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Q, Steine EJ, Barrasa MI, Hockemeyer D, Pawlak M, Fu D, et al. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci U S A. 2011 Nov 1;108(44):18061–18066. doi: 10.1073/pnas.1114946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Network TCGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 May 30;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007 Jun;236(6):1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 23.Kong G, Wunderlich M, Yang D, Ranheim EA, Young KH, Wang J, et al. Combined MEK and JAK inhibition abrogates murine myeloproliferative neoplasm. J Clin Invest. 2014 Jun 2;124(6):2762–2773. doi: 10.1172/JCI74182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995 Sep 8;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 25.Peters SL, Hlady RA, Opavsky J, Klinkebiel D, Pirruccello SJ, Talmon GA, et al. Tumor suppressor functions of Dnmt3a and Dnmt3b in the prevention of malignant mouse lymphopoiesis. Leukemia. 2014 doi: 10.1038/leu.2013.364. Epub on Jan 7, 2014. [DOI] [PubMed] [Google Scholar]
- 26.Sabnis AJ, Cheung LS, Dail M, Kang HC, Santaguida M, Hermiston ML, et al. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol. 2009 Mar 17;7(3):e59. doi: 10.1371/journal.pbio.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celik H, Mallaney C, Kothari A, Ostrander EL, Eultgen E, Martens A, et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood. 2014 Nov 21; doi: 10.1182/blood-2014-08-594564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014 Feb 20;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014 Dec;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014 Dec 25;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014 Dec 25;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Kong G, Liu Y, Du J, Chang Y-I, Zhang X, et al. Nras G12D/+ promotes leukemogenesis by aberrantly regulating haematopoietic stem cell functions. Blood. 2013;121(26):5203–5207. doi: 10.1182/blood-2012-12-475863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parikh C, Subrahmanyam R, Ren R. Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood. 2006 Oct 1;108(7):2349–2357. doi: 10.1182/blood-2004-08-009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.