Abstract
SH3 domains are evolutionarily conserved protein interaction domains that control nearly all cellular processes in eukaryotes. The current model is that most SH3 domains bind discreet PxxPxR motifs with weak affinity and relatively low selectivity. However, the interactions of full-length SH3 domain-containing proteins with ligands are highly specific and have much stronger affinity. This suggests that regions outside of PxxPxR motifs drive these interactions. In this study, we observed that PxxPxR motifs were required for the binding of the adaptor protein GRB2 to short peptides from its ligand SOS1. Surprisingly, PxxPxR motifs from the proline rich region of SOS1 or CBL were neither necessary nor sufficient for the in vitro or in vivo interaction with full-length GRB2. Together, our findings show that regions outside of the consensus PxxPxR sites drive the high affinity association of GRB2 with SH3 domain ligands, suggesting that the binding mechanism for this and other SH3 domain interactions may be more complex than originally thought.
Keywords: SH3 domains, GRB2, SOS1, CBL, signal transduction
1. Introduction
SH3 domain-containing proteins regulate a large number of cellular functions in humans from signal transduction to metabolism to differentiation [1, 2]. SH3 domains are evolutionally conserved in eukaryotes from budding yeast, which produce 28 proteins with SH3 domains, to humans, which express over 300 proteins with SH3 domains [1, 3]. Although sequence conservation is relatively low, SH3 domains have a conserved structure of 5 or 6 beta strands that form two anti-parallel beta sheets. The current model is that vast majority of SH3 domains bind to discrete sequences of (R/K)xPxxP (class I) or PxxPx(R/K) (class II) in a shallow groove between the beta sheets [2, 3]. Peptide ligands containing these motifs associate in vitro with isolated SH3 domains with relatively weak affinities, ranging from 1 µM to 1 mM [2, 4]. There are other atypical interaction motifs that associate with a small number of SH3 domains. The most well studied of these is these atypical interactions are class III binding reactions driven by RxxK motifs. In contrast to class I or class II SH3 domain binding reactions, the affinity of class III SH3 domain interaction appears to be much stronger [1, 3, 5]. In addition to relatively low affinities, isolated SH3 domains also have variable specificity for peptide ligands. Some isolated domains have relatively high specificity for individual ligands, notably the class III C-terminal SH3 domain of GADS and RxxK peptides from SLP-76, but the majority of class I and class II SH3 domains bind multiple PxxP containing peptides [2, 4]. Based primarily on studies using isolated domains and peptide ligands, the current model is that SH3 domains are evolutionally and structurally conserved protein interaction domains that have weak affinity and low specificity for PxxP containing ligands.
In contrast to isolated SH3 domain/peptide interactions, the binding of full-length SH3 domain containing proteins to larger regions of their ligands have substantially stronger affinity. The binding affinity of the isolated C-terminal SH3 domain of GADS to a peptide derived from SLP-76 was ~250 nM, while the interaction of full-length GADS with the complete proline-rich region of SLP-76 is ~10-fold stronger [5, 6]. Similarly, the SH3 domain of p67PHOX binds with 1000-fold increased affinity to a 32 amino acid peptide derived from p47PHOX compared to shorter peptides due to molecular interactions outside of the PxxP motif [7, 8]. Finally, peptides derived from PxxP motifs found in SOS1interact with individual SH3 domains from GRB2 with affinities ranging from 20 µM to 1 mM [9–13], whereas the association of full-length GRB2 with full-length SOS1 or the complete proline rich regions (PRR) of SOS1 and CBL has affinities of 300–400 nM [14–16]. Together, these studies suggest that the interaction of full-length SH3 domain-containing proteins with in vivo ligands may have substantially stronger affinity than previously thought due to interactions outside of the conserved PxxP motifs.
To more fully address this prediction, we utilized quantitative biophysical and imaging techniques to examine the binding of GRB2 to its physiological ligands SOS1 and CBL. GRB2 is an adaptor protein with a central SH2 domain, which binds to phosphorylated proteins, and two flanking SH3 domains [17, 18]. GRB2 facilitates the interaction of phosphorylated receptors and adaptor proteins with SH3 domain ligands, including the RAS guanine nucleotide exchange factor SOS1 and the E3 ubiquitin ligase, CBL [17, 18]. In this study, we found that similar to previous observations PxxPxR motifs were absolutely required for the interaction of full-length GRB2 with peptides derived from the proline rich C-terminal tail of SOS1. In contrast, these same PxxPxR motifs were neither necessary nor sufficient for the high affinity interaction of SOS1 or CBL with full-length GRB2 or required for the in vivo recruitment of SOS1 to the plasma membrane in activated T cells. These studies show conclusively that regions outside of the PxxPxR motifs are critical for the high affinity interaction of GRB2 with full-length ligands.
2. Material and Methods
2.1. Protein purification
The bacterial expression constructs for full-length human GRB2, the complete proline rich region of murine SOS1 or human CBL were described previously in [6, 15]. Deletions of individual sites in these constructs were performed with the QuickChange II XL Site Directed Mutagenesis kit from Stratagene using standard manufacturer protocol. Rosetta 2 cells expressing 6X-His tagged GRB2 were shaken for 36 hours at 25°C in Superbroth. Rosetta 2 cells expressing 6X-His tagged proline rich regions of SOS1 or CBL were shaken 24–48 hours at 37°C in ZYM-5052 [19]. The cells were pelleted, and pellets were then resuspended in His-tag purification buffer (50 mM NaPO4, 250 mM NaCl, pH 7.4) and lysed using sonication. The supernatant was applied to a Ni2+ HisTrap HP affinity column and bound proteins were eluted using imidazole. Proteins were further purified by size exclusion chromatography using a 16/60 Superdex 75 gel filtration column and then concentrated using 10,000 MWCO centrifugal filters. As determined by dynamic light scattering, little aggregation was observed for any protein (data not shown).
2.2. Peptide Array
The peptide array was produced as previously described in [20]. The array scans the entire proline rich region of SOS1 (amino acids 1117–1319) and contains 12 amino acid peptides that have 9 amino acid overlaps with the peptides on either side (see key for the array in Supplemental Figure 1). The membrane was blocked with 5% milk in TBST and then incubated with His-tagged GRB2 for 1.5 hours. The amount of bound GRB2 was examined by immunoblotting with an anti-GRB2 antibody diluted in 5% milk as previously described in [21] and the array was visualized by chemiluminescence using a Fuji imager.
2.3. Isothermal titration calorimetry (ITC)
The protein samples were degassed and ITC measurements recorded using a MicroCal VP-ITC System with SOS1 or CBL proline rich regions as the injected sample and GRB2 as the cell sample. For the peptide inhibition studies, GRB2 was incubated with varying molar ratios of a SOS1 site 1 peptide (EVPVPPPVPPRRRPE) purchased from EZ Biolabs. The chamber was kept under constant stirring at 350 rpm and all experiments were performed at 25 °C. Injection of SOS1 or CBL into buffer showed constant heats of dilution. The heat of dilution was determined by averaging the last 3–5 injections and subtracted from the raw value. The data are analyzed using the single site binding model using the Origin ITC analysis package. The values for affinity, stoichiometry and ΔH were averaged from at least four separate injections and statistically analyzed via ANOVA using GraphPad Prism. Outliers in the data were determined via the ROUT method using GraphPad Prism, with Q = 0.1 %.
2.4. Dynamic Light Scattering
Dynamic light scattering (DLS) data was collected at 25 °C using a DynaPro NanoStar (Wyatt Technology). The data was analyzed using Dynamic (version 7.1.7, Wyatt Technology). Each acquisition was an average of 10 scans and the values for radius and percent polydispersity were averaged from four individual acquisitions. The data was plotted and statistically analyzed via ANOVA with Tukey’s multiple comparison test using GraphPad Prism.
2.5. Cell lines and transfections
SOS1 wild-type, SOS1 Δ1234Z, and SOS1 scrambled cDNA were PCR amplified from plasmid constructs and inserted into lentiviral expression vectors (Gift from Dr. Stephen Bunnell). The lentiviral expression vectors and packaging vectors were cotransfected into 293T cells using lipofectamine 2000. After 48 hours, the virus-containing medium was removed and virus was concentrated using Lenti-X concentrator (Clontech). Jurkat E6.1 T cells were grown at 37°C with 5% CO2 in complete RPMI (RPMI 1640 supplemented with 10% FBS, 2 mM L-glutamine, 50 U/mL penicillin, 50 ug/mL streptomycin). They were transduced with the described vectors and selected using purimycin. YFP expression on stable cell lines was analyzed by flow cytometry using an Accuri C6 flow cytometer.
2.6. Immunoblotting
Proteins in cellular lysates were separated by polyacrylamide gel electrophoresis using 4–15% Criterion Precast polyacrylamide gels. The separated proteins were then transferred to polyvinylidene difluoride and the membrane was blocked for 1 hour at room temperature in SEA BLOCK Blocking Buffer. The membranes were incubated overnight at 4°C with anti-YFP followed by 30 minutes of incubation at room temperature with the respective secondary antibody. The blots were then developed using the Licor Odyssey Infrared Imager.
2.7. TIRF Microscopy
Imaging was performed using a Leica AM TIRF MC imaging system as previously described [22, 23]. Briefly, Jurkat E6.1 T cells stably expressing SOS1 wild-type PRR-YFP, SOS1 Δ1234Z PRR-YFP, or YFP alone were activated for 5 minutes on glass chamber slides coated with 10 ug/ml of anti-CD3 antibody. Cells were fixed with 3% paraformaldehyde and permeabilized with 0.25% Triton X-100. After blocking, cells were stained with anti-phospho LAT Y226 antibodies followed by incubation with secondary antibodies. The YFP fluorescence and LAT phosphorylation occurring at the membrane was imaged using TIRF microscopy. The total YFP fluorescence in the same cells was imaged by EPI fluorescence. All images were acquired using Leica AF software and processed using Fiji software package. To quantify YFP recruitment, the TIRF YFP fluorescence was normalized to the total YFP fluorescence in the EPI fluorescence channel in the imaged cell. The ratio of the values of TIRF YFP fluorescence over total YFP fluorescence in the EPI fluorescence channel was calculated for 40 randomly selected cells for each of the two independent experiments. The individual values and the mean ± 95% confidence intervals were plotted using GraphPad Prism. The statistical analysis was performed using GraphPad Prism via ANOVA analysis.
3. Results
3.1. PxxPxR motifs are required for the binding of GRB2 to peptide ligands
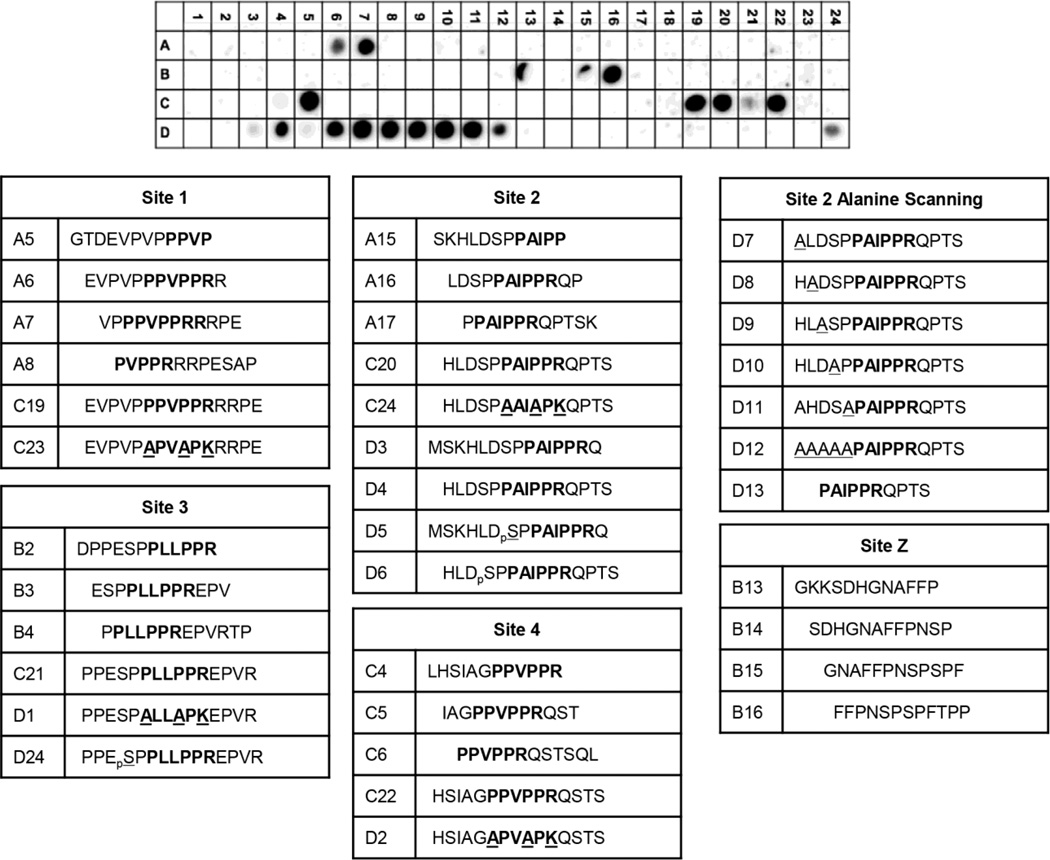
The proline rich C-terminal tail of SOS1 PRR contains four consensus PxxPxR SH3 domain binding motifs, and peptides containing these motifs bind individual GRB2 SH3 domains with affinities ranging from 20 µM to 1 mM [9–13]. Although these studies have shown that GRB2 interacts with the PxxPxR motifs in SOS1, no unbiased screens have been performed to determine if these consensus motifs are necessary for full-length GRB2 binding or to identify novel binding sites. To address these questions, we produced a synthetic peptide array with 12 amino acid long peptides derived from the PRR of SOS1 that have nine amino acid overlaps with adjacent peptides (Supplemental Figure 1). Full-length GRB2 bound to peptides that contained PxxPxR motifs from site 1 (Figure 1A, A7) and site 4 (Figure 1A, C5). We also identified a previously unknown atypical proline rich region with the sequence of FFPNSPSPFTPP christened site Z (Figure 1A, B16). GRB2 did not interact with 12 amino acid peptides containing site 2 (Figure 1A, A15–A17) or site 3 (Figure 1A, B2–B4). However, a 15 amino acid site 2 peptide bound well to GRB2 (Figure 1A, C20 and D4), while a 15 amino acid site 3 peptide had only minimal binding (Figure 1A, C21). Mutations of the PxxPxR motif to AxxAxK abolished binding to sites 1–4 (Figure 1A, C23–D2). Importantly, GRB2 did not associate with the class 1 SH3 domain binding site with the consensus sequence of RxxPxxP (Figure 1A, B22 and B23). Similarly, GRB2 did not associate with other proline rich peptides including PESAPAESSP (Figure 1A, A10), DPPES (Figure 1A, A24 and B1), PLHLQPPPL (Figure 1A, B9, B10), PPPPQTPSP (Figure 1A, B18 and B19), PSPPL (Figure 1A, B22 and B23), PKLPPKT (Figure 1A, C9 and C10), and PSMHRDGPP (Figure 1A, C14 and C15).
Figure 1. PxxPxR motifs are required for binding of full-length GRB2 to SOS1 peptides.
Full-length human GRB2 was incubated with a peptide array derived from the PRR of SOS1. The binding of GRB2 to the array was assessed by immunoblotting with an anti-GRB2 antibody. Peptides containing PxxPxR motifs are described below the blot.
The 15 amino acid site 2 peptide that bound well with GRB2 contains an HLxSPP motif found in the N-terminal and C-terminal halves of the complete PRR of SOS1. The two halves of the PRR of SOS1 have previously been shown to bind with GRB2 in a 1:1 stiochiometry [15]. Since the HLxSPP motifs are the only conserved sequences outside of the PxxPxR motifs on both halves of the PRR of SOS1, the role of the HLxSPP region on the binding to the site 2 peptide was examined. Serine phosphorylation or individual alanine mutations of the HLxSPP motif did not disrupt binding of the site 2 peptide to GRB2 (Figure 1A, D6–D11), whereas alanine mutation of all sites on this region reduced GRB2 binding (Figure 1A, D12). These data show that in the context of short, 12–15 amino acid long peptides, the PxxPxR motifs found in sites 1, 2 and 4 and a novel interaction region, site Z, bind to full-length GRB2. Importantly, no other short 10–12 amino acid GRB2 binding sites outside of PxxPxR motifs were observed in the complete PRR of SOS1.
3.2. PxxPxR-containing peptides do not inhibit the interaction of GRB2 with the complete SOS1 PRR
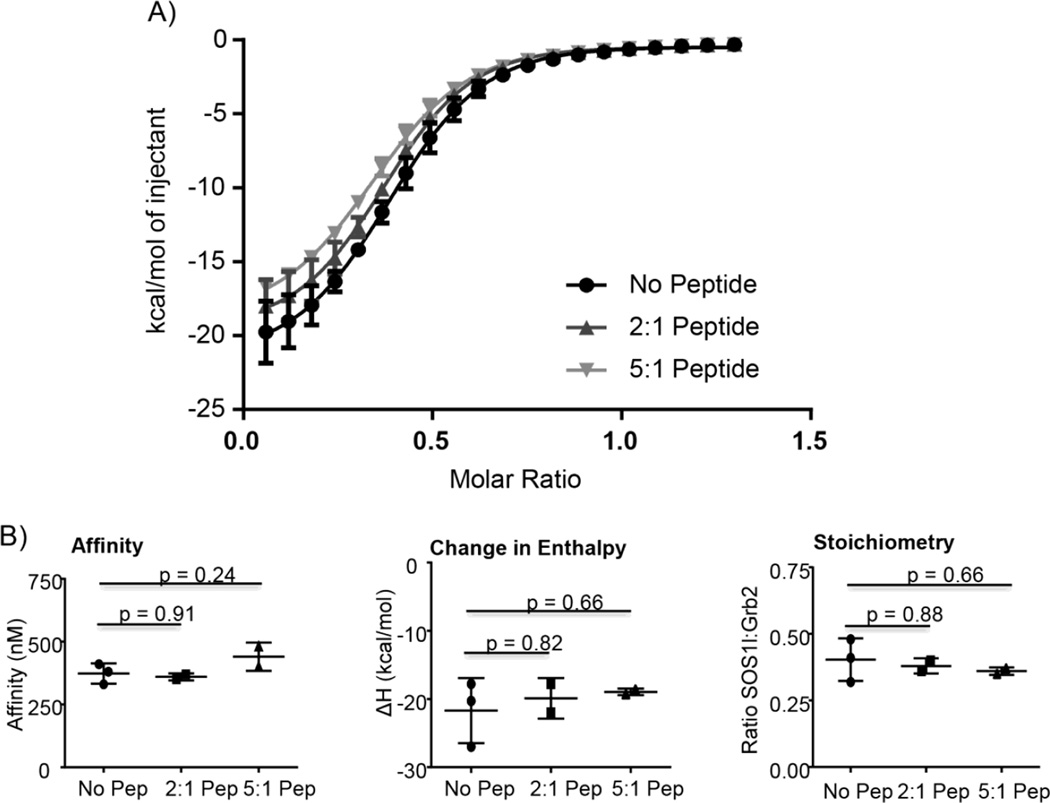
SOS1 site 1 peptides inhibit the interaction of GRB2 with SH3 domain ligands in cellular lysates [9, 24, 25]. This provided direct evidence that the GRB2:SOS1 is driven by the PxxPxR motifs, but the molar ratio of the peptides to endogenous proteins was unknown calling into question these conclusions. To address this issue, full-length GRB2 was mixed with varying molar ratios of a SOS1 site 1 peptide (EVPVPPPVPPRRRPE) and the subsequent binding of wild-type complete PRR of SOS1 was analyzed by isothermal titration calorimetry. This technique is widely utilized for measuring the affinity, stoichiometry and thermodynamics of a proteinprotein interaction [26, 27]. Addition of two or five molar equivalents of the SOS1 peptide had no significant effect on the affinity, change in enthalpy (ΔH) or stoichiometry of the interaction of GRB2 with the complete PRR of SOS1 (Figure 2). The lack of suppression is especially striking in the first injections, where the SOS1 peptide is at a 33x and 83x molar excess compared to the complete PRR of SOS1 in the experiment with two or five molar peptide equivalents, respectively. Thus when the molar ratios are similar, a SOS1 peptide containing PxxPxR motif is incapable of significantly inhibiting the GRB2:SOS1 interaction. Unlike previous studies, this suggests that the PxxPxR motifs are not a major component of the high affinity interaction of the full-length proteins.
Figure 2. SOS1 peptide with a PxxPxR motif does not significantly inhibit the interaction of GRB2 with the complete proline rich region of SOS1.
A) Full-length GRB2 was pre-incubated with a SOS1 site 1 peptide. The ability of the peptide to inhibit the binding of GRB2 to the complete proline rich region of SOS1 was then assessed via ITC. Each point on the curve is an average ± 95% confidence interval of 2 or 3 replicates. The curves were fit using the Boltzmann sigmoidal equation in GraphPad Prism. B) The values for affinity, change in enthalpy (ΔH) and stoichiometry from three independent experiments are shown. The statistical analysis was performed using GraphPad Prism via ANOVA analysis and p values for the comparison of the no peptide control to 2:1 or 5:1 molar ratio of peptide are shown.
3.3. Regions outside of the PxxPxR motifs drive high affinity in vitro interactions of GRB2 with SH3 domain ligands
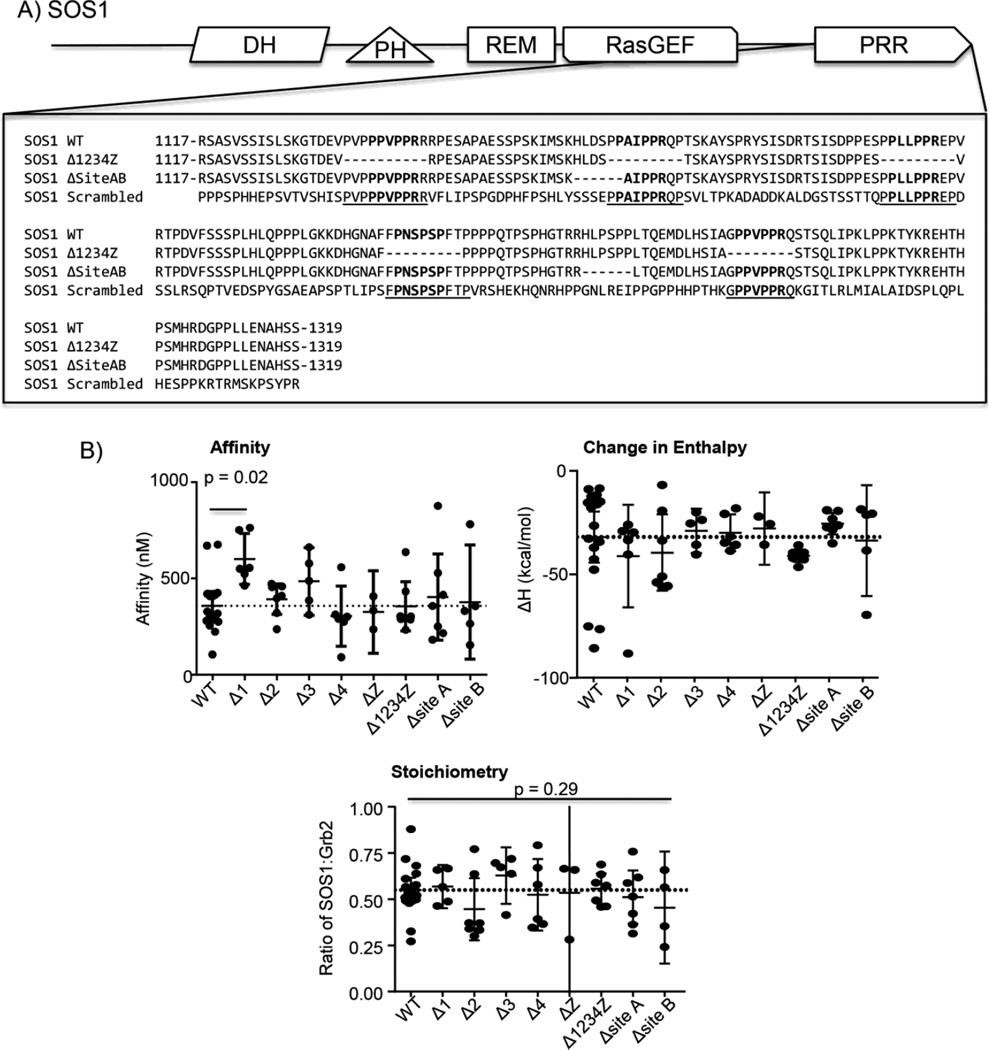
To further address this paradigm shifting possibility, the ability of PxxPxR motifs to drive the high-affinity interaction of GRB2 with the PRR of SOS1 was assessed. The complete PRR of SOS1 and mutants with individual or combined deletions of sites 1–4 and site Z (Figure 3A) were isolated and the affinity, ΔH or stiochiometry of these proteins for GRB2 was determined by ITC. The association of GRB2 with the complete PRR of SOS1 is enthalpically driven with an affinity of ~360 nM and a 2 GRB2:1 SOS1 stoichiometry [15]. Surprisingly, there were no statistically significant differences in the affinity for the interaction of GRB2 with SOS1 proteins with sites 2, 3, Z or 4 deleted alone or together compared to WT SOS1 or ΔH and stoichiometry for the binding of GRB2 to SOS1 proteins with sites 1, 2, 3, Z or 4 deleted alone or together compared to WT SOS1 (Table 1 and Figure 3B). There was a statistically significant difference in the affinity for the interaction of GRB2 with SOS1 Δsite1 (Table 1 and Figure 3B). However, the difference in affinity was only 1.7 fold, which unlikely to physiologically relevant. Additionally, deletion of the two HLxSPP motifs conserved in the N-terminal and C-terminal sides of the complete proline rich region of SOS1 (N-terminal: ΔSite A and C terminal: ΔSite B; Figure 2A) had no significant effect on the GRB2 binding reaction (Table 1 and Figure 3B). To test if the PxxPxR sites are sufficient for the binding of GRB2 to SOS1, a mutant SOS1 PRR was produced where sites 1, 2, 3, Z and 4 were conserved in sequence and position but all other amino acids were randomly repositioned (Scrambled; Figure 3A). The scrambled version of the SOS1 PRR had no detectable binding with GRB2 (Table 1 and Figure 4A). Based on the concentrations used for these studies, it is estimated that the affinity of SOS1 scrambled for GRB2 is greater than 50 µM. These experiments suggest that the PxxPxR motifs or sequences needed for interactions with these motifs in the context of short peptides are not required for the high-affinity binding of GRB2 to SOS1.
Figure 3. The consensus PxxPxR motifs of SOS1 are not required for the high affinity interaction with GRB2.
A) The sequence for SOS1 wild-type, SOS1 Δ1234Z, SOS1 ΔSite AB and SOS1 scrambled are shown. B) The binding of GRB2 to the complete proline rich regions of SOS1 were assessed by ITC. The values for affinity, ΔH and stoichiometry were determined using the Origin ITC analysis package. The individual values for affinity, ΔH and stoichiometry were plotted with the average ± 95% confidence interval using GraphPad Prism. The statistical analysis was performed using GraphPad Prism via ANOVA analysis and the p values < 0.50 are shown.
Table 1.
Affinity, stoichiometry and change in enthalpy (ΔH) values for the interaction of GRB2 with WT and mutant complete proline rich regions from SOS1 and CBL. Each point is shown as a mean ± SEM for at least four separate injections.
| Affinity (nM) |
Stoichiometry (SOS1/GRB2) |
Enthalpy (Kcal/mol) |
|
|---|---|---|---|
| SOS1 Deletions | |||
| SOS1 WT | 360 ± 40 | 0.55 ± 0.03 | −32 ± 6 |
| ΔSite 1 | 600 ± 50 | 0.53 ± 0.05 | −32 ± 3 |
| ΔSite 2 | 390 ± 30 | 0.45 ± 0.07 | −40 ± 8 |
| ΔSite 3 | 490 ± 60 | 0.63 ± 0.06 | −29 ± 4 |
| ΔSite 4 | 310 ± 60 | 0.53 ± 0.08 | −30 ± 4 |
| ΔSite Z | 330 ± 50 | 0.54 ± 0.13 | −28 ± 4 |
| ΔSite 1,2,3,4,Z | 360 ± 50 | 0.54 ± 0.04 | −41 ± 1 |
| ΔSite A | 400 ± 90 | 0.51 ± 0.06 | −26 ± 2 |
| ΔSite B | 380 ± 110 | 0.44 ± 0.08 | −34 ± 10 |
| Scrambled | >50,000 | N.D. | N.D. |
| CBL Deletions | |||
| CBL WT | 520 ± 120 | 0.45 ± 0.04 | −16 ± 1 |
| ΔSite 1,2,3 | 800 ± 70 | 0.34 ± 0.03 | −10 ± 2 |
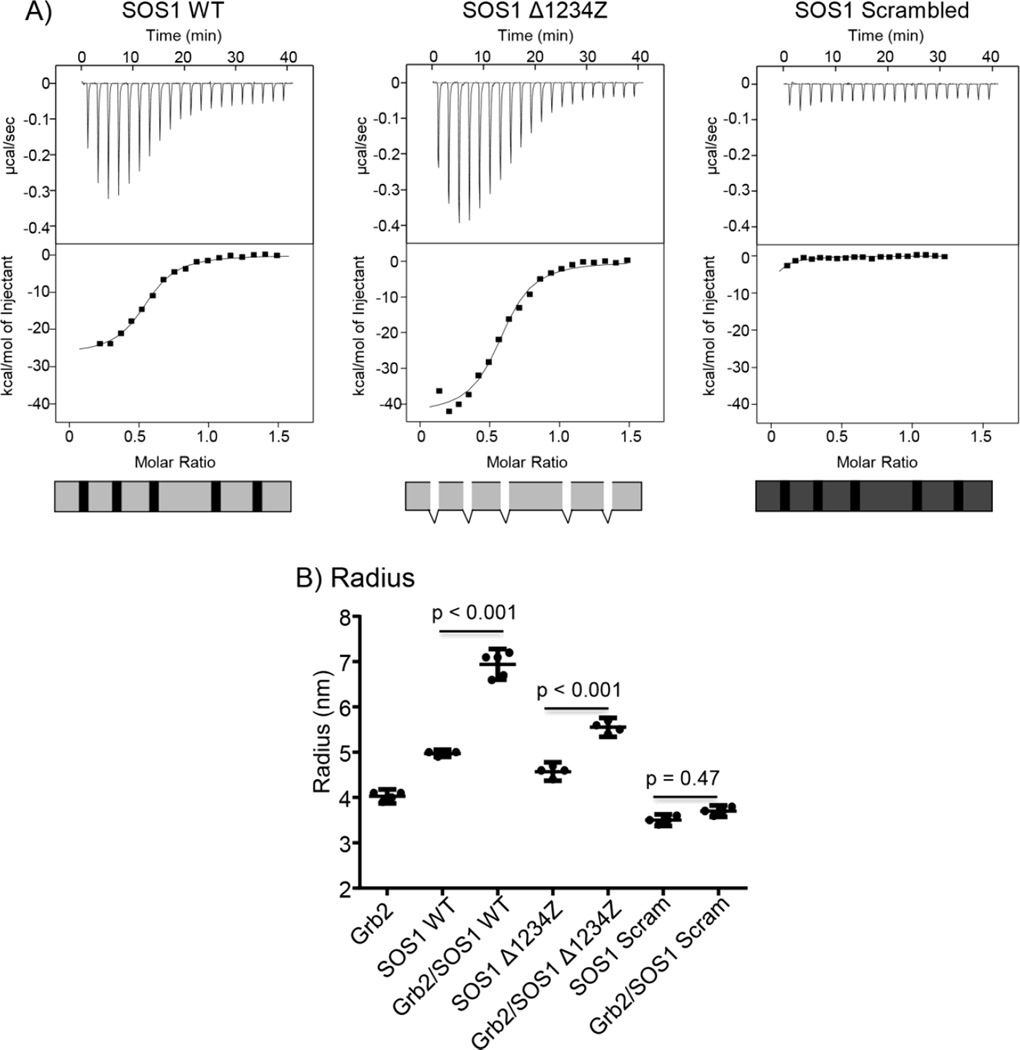
Figure 4. Regions outside of the consensus PxxPxR motifs are required for the high affinity interaction of GRB2 with SOS1.
A) Raw ITC tracings for the interaction of GRB2 with SOS1 WT, SOS1 Δ1234Z and SOS1 scrambled. Each trace is representative of at least 4 separate injections. B) The hydrodynamic radius of GRB2, SOS1 WT, SOS1 Δ1234Z or SOS1 scrambled, both alone and in combination, was determined using DLS. The individual values, with the mean ± 95% confidence intervals were plotted using GraphPad Prism. Statistical analysis was performed using GraphPad Prism via ANOVA analysis and the p values are shown.
Since these observations directly challenges the current model of SH3 domain binding reactions, we further examined the requirement of PxxPxR motifs for high affinity GRB2:SOS1 binding using dynamic light scattering, which determines the hydrodynamic radius of a protein complex. GRB2 had a radius of 4.0 nM, whereas SOS1 WT, Δ1234Z and scrambled had radii of 5.0, 4.6, 3.5 nM, respectively (Figure 4B). Interestingly, the 2:1 stoichimetric mixture of GRB2 with either SOS1 wild-type or SOS1 Δ1234Z had a significantly larger radii of 6.9 and 5.6 nM, respectively, than the unbound GRB2 or SOS1 proteins (Figure 4B), indicating that SOS1 proteins with and without PxxPxR motifs form larger 2:1 complexes with GRB2. The smaller radius of the SOS1 Δ1234Z compared to SOS1 wild-type is likely due to the molecular weight difference, but could also be due to changes in the overall structure of the complex. In contrast, the 2:1 mixture of GRB2 with SOS1 scrambled had a radius of 3.7 nM (Figure 4B), which is the average of the radii for unbound proteins. Unlike SOS1 wild-type and SOS1 Δ1234Z, SOS1 scrambled is incapable of interacting with GRB2. These results confirmed our finding that high affinity binding between SOS1 and GRB2 is independent of PxxPxR motifs.
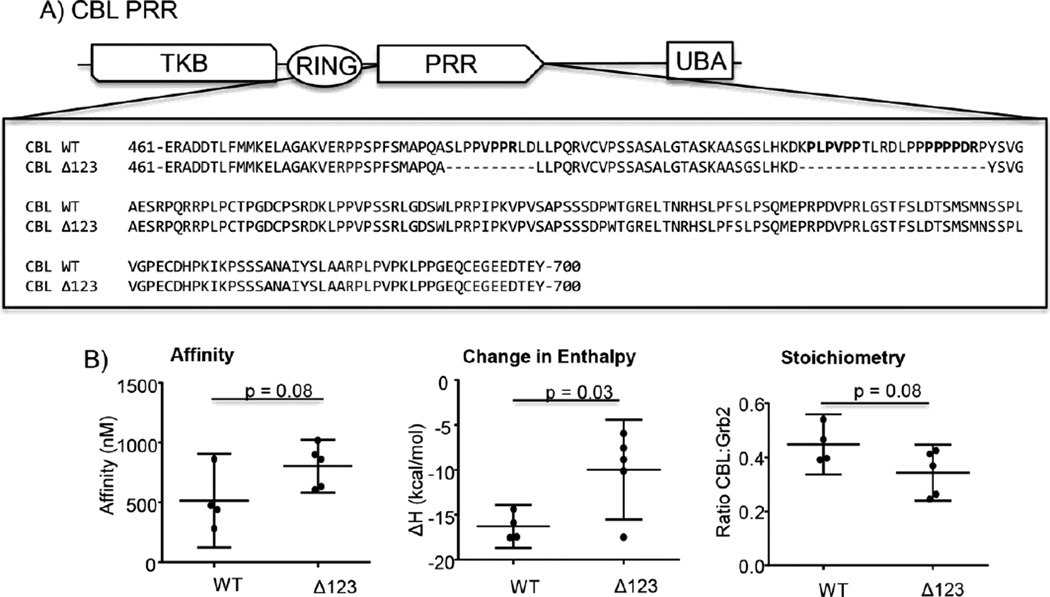
It is possible that the inability of the PxxPxR motifs to drive the high-affinity binding reaction is limited to the GRB2:SOS1 interaction. To examine this prospect, the binding of GRB2 to wild-type and mutated versions of the complete PRR of CBL, which contains three identified PxxPxR GRB2 binding motifs (Figure 5A) [28], was characterized by ITC. Again, deletion of the PxxPxR motifs did not significantly alter the affinity or stoichiometry for the binding of GRB2 compared to the wild-type CBL PRR (Table 1 and Figure 5B). There was a significant difference in the ΔH value for these reactions, but a ~40% change is unlikely to have any meaningful physiological effect.
Figure 5. The consensus PxxPxR motifs of CBL are not required for the high affinity interaction with GRB2.
A) The sequence for CBL wild-type and CBLΔ123 are shown. B) The binding of GRB2 to the complete proline rich regions of CBL were assessed by ITC. The values for affinity, ΔH and stoichiometry were determined using the Origin ITC analysis package. The individual values for affinity, ΔH and stoichiometry were plotted with the average ± 95% confidence interval using GraphPad Prism. The statistical analysis was performed using GraphPad Prism via ANOVA analysis and the p values < 0.50 are shown.
3.4. PxxPxR motifs are not required for the in vivo interactions of GRB2 with SOS1
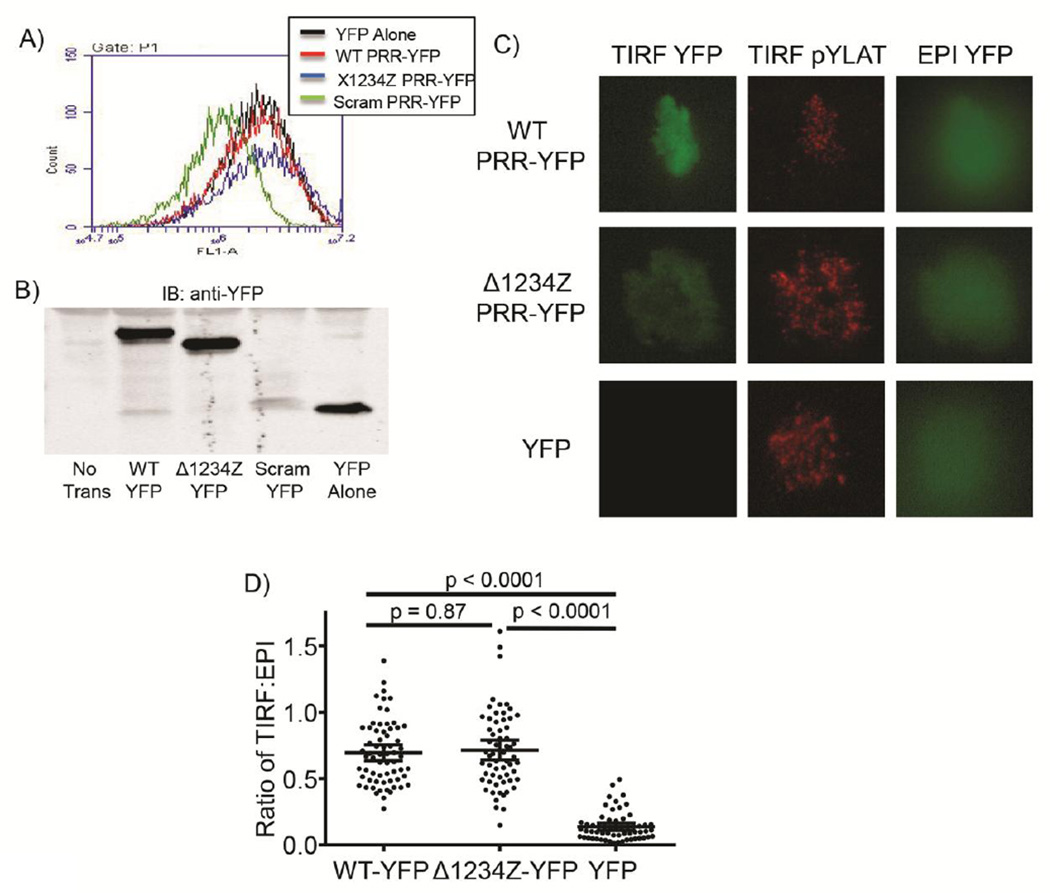
Upon T cell receptor (TCR) activation, GRB2 recruits SOS1 to the membrane adaptor protein LAT. Both GRB2 and SOS1 are then critical for the clustering of LAT into large megadalton sized signaling complexes [15, 29, 30]. To translate the biophysical studies to a cellular context, stable Jurkat E6.1 T cell lines were produced that had similar levels of YFP alone (YFP), wild-type complete proline rich region SOS1-YFP (WT-YFP) and Δ1234Z proline rich region SOS1-YFP (Δ1234Z-YFP) expression as assessed by flow cytometry and immunoblotting (Figure6A and 6B). Unfortunately, SOS1 scrambled-YFP could not be transiently or stably expressed (Figure6A and 6B). Recombinant SOS1 scrambled aggregated more quickly than other SOS1 proteins and had higher polydispersity in DLS experiments compared to WT SOS1 (data not shown). Together this suggests that SOS1 scrambled is unstable in human cells. T cell activation by anti-TCR-coated coverslips results in LAT phosphorylation and the recruitment of GRB2 and SOS1 to the plane of the coverslip [15, 31]. The recruitment of YFP fusion proteins to LAT was assessed by total internal reflection (TIRF) microscopy, which lowers background fluorescence by exciting molecules within 150 nm of the coverslip [32]. TCR stimulation induced substantial phosphorylation of LAT and the recruitment of WT SOS1-YFP and Δ1234Z SOS1-YFP to the plasma membrane (Figure 6C) in Jurkat T cells expressing SOS1-YFP fusion proteins. In contrast, activated Jurkat T cells expressing YFP alone had increased LAT phosphorylation but no YFP recruitment to the membrane (Figure 6C). To quantify YFP recruitment, the TIRF YFP fluorescence was normalized to the total YFP fluorescence in the EPI fluorescence channel in the imaged cell. There was no significant difference in the recruitment of WT SOS1-YFP and Δ1234Z SOS1-YFP to the membrane, while YFP had significantly lower recruitment compared to the SOS1 proteins (Figure 6D). These data conclusively show that the complete PRR of SOS1 missing all PxxPxR motifs is recruited to membrane signaling complexes in activated T cells to a similar level as wild-type SOS1 PRR.
Figure 6. SOS1 wild-type and SOS1 Δ1234Z are recruited to the plasma membrane upon TCR activation in human T cells.
The expression of YFP fusion proteins in Jurkat E6.1 T cells stably expressing SOS1 wild-type PRR-YFP (WT-YFP), SOS1 Δ1234Z PRR-YFP (Δ1234Z-YFP), SOS1 scrambled PRR-YFP (Scram-YFP) or YFP alone (YFP) was assessed by A) flow cytometry or B) immunoblotting. C) Jurkat E6.1 T cells stably expressing SOS1 wild-type PRRYFP (WT-YFP), SOS1 Δ1234Z PRR-YFP (Δ1234Z-YFP) or YFP alone (YFP) were activated using coverslips coated with a stimulatory anti-TCR antibody. The YFP fluorescence and LAT phosphorylation occurring at the membrane was imaged using TIRF microscopy. LAT phosphorylation was assessed by immunofluorescence using a phospho-specific LAT Y226 antibody. The total YFP fluorescence in the cell was imaged by EPI fluorescence. D) The fluorescence in the TIRF and EPI fluorescence channel was quantified for individual cells using NIH Image. The individual values and the mean ± 95% confidence intervals were plotted using GraphPad Prism. The statistical analysis was performed using GraphPad Prism via ANOVA analysis and the p value for each comparison is shown.
4. Discussion
Similar to previous studies [9–13], our data show that PxxPxR motifs can facilitate the SH3 domain-mediated binding of GRB2 to SOS1 peptides. Importantly, these sites were the only discreet 10–12 amino acid sequences that can mediate the interaction of GRB2 with SOS1. In contrast to the peptide studies, the same PxxPxR motifs are neither necessary nor sufficient for the high affinity in vitro binding of GRB2 to the complete PRR of SOS1 or CBL. Similarly, these sites are not required for the recruitment of SOS1 PRR to activated LAT, an excellent model for the receptor stimulation-dependent recruitment of the GRB2/SOS1 complex [17]. Instead, as shown by our studies with the SOS1 scrambled mutant, regions outside of the consensus PxxPxR motifs drive the interaction of the complete proline rich region of SOS1 and CBL with full-length GRB2. These findings are in contrast to work by McDonald and coworkers, who showed that PxxPxR motifs were critical for the binding of GRB2 to the PRR or SOS1. However, these investigators used a truncated PRR for SOS1 that had an affinity of only 7 µM for GRB2 [33]. The SOS1 PRR used in this study bound to GRB2 with an affinity of 350 nM, which is nearly identical to the affinity of GRB2 for full length SOS1 [14]. Ultimately, our studies show that in the context the full length proline rich regions of SOS1 and CBL, the PxxPxR motifs are not required for the high affinity in vivo and in vitro interaction with GRB2.
These observations fit well with previous studies showing that SOS1 peptides bind to individual SH3 domains from GRB2 with affinities ranging from 20 µM to 1 mM [9–13], while the affinity for the binding of full-length GRB2 with full-length SOS1 or the complete PRR of SOS1 and CBL is between 300 and 400 nM (Table 1) and [14–16]. Several studies have suggested that the differences in the affinity GRB2 for peptides versus full length ligands were due to avidity effects, where both SH3 domains of an individual GRB2 bind to two PxxPxR motifs on the same ligand. In silico modeling suggests that multiple interactions can strengthen the affinity of the GRB2/SOS1 interaction [34] and peptides with two linked PxxPxR motifs had enhanced binding of GRB2 compared to peptides with a single motif [35, 36]. However, our data conclusively show that avidity effects derived from the interaction of a single GRB2 with multiple PxxPxR motifs on an SH3 domain ligand cannot explain the 2–3 orders of magnitude differences in affinity for peptides versus full-length ligands.
If PxxPxR motifs on SOS1 and CBL are not high affinity binding sites for GRB2, what is the molecular mechanism for this binding reaction? As shown in Figure 1, there are no discrete binding sites outside of PxxPxR motifs that bind GRB2. Since the interaction of these peptides with GRB2 SH3 domains have mid to high µM affinities, this suggests that no other short sequences on SOS1 have affinities for GRB2 SH3 domains stronger than 100 µM. Thus, the current paradigm that GRB2:SH3 domain interactions are controlled by short discreet binding sites is incorrect (Figure 7). Instead, these binding reactions are likely driven by large number of low affinity contacts across the entire PRR of individual ligands. What the actual contacts are that drive the interaction of GRB2 with either SOS1 or CBL are outside of the scope of this study. The crystal structure of full length GRB2 has been solved, but cysteine mutations on GRB2 were required for crystallization [37, 38]. Additionally, small angle X-ray scattering studies have suggested that this structure does not match conformation of the solution conformation of GRB2 [36]. We have not been able to successfully produce diffractable crystals using non-mutated full-length GRB2, due largely to the flexible conformation of this protein (data not shown). Similarly, the only assigned HSQC spectra for full-length GRB2 was performed using a cysteine mutated GRB2 [36]. It is unlikely that the interaction of non-mutated, full length GRB2 with the PRR of SOS1 or CBL will be easily investigated by NMR due to the large size of the complex and difficulty with the structurally flexible GRB2. Extensive studies using multiple genetic, biochemical and biophysical techniques will be required to conclusively identify the exact residues regulating the high affinity interaction of GRB2 with SH3 domain ligands.
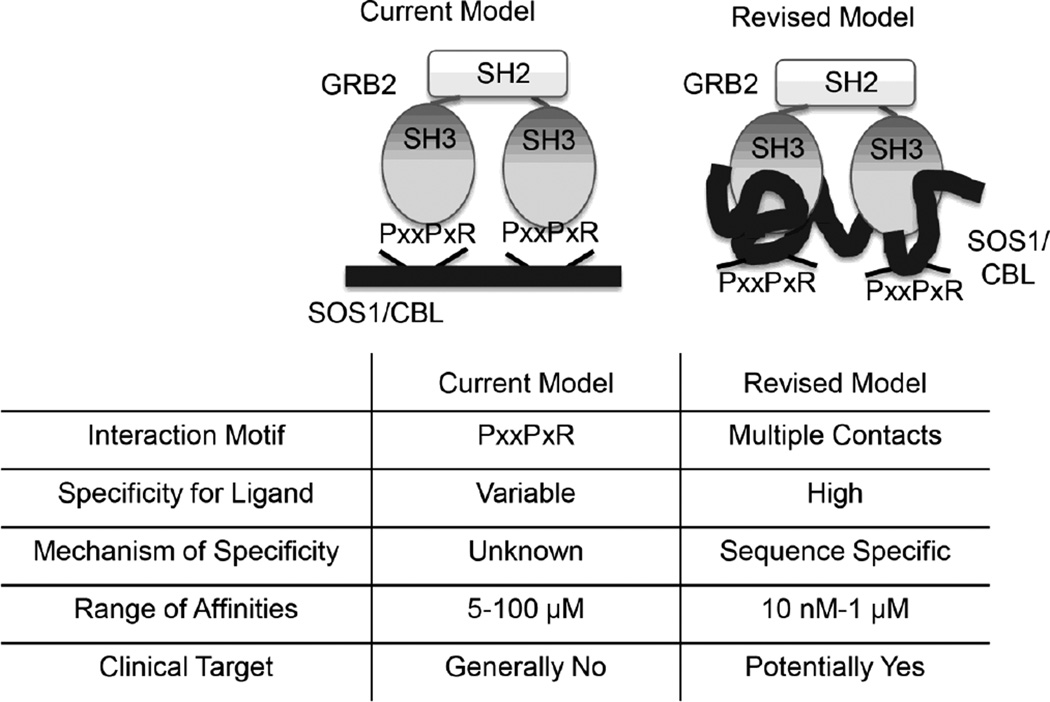
Figure 7. Comparison of the current and revised model for the interaction of GRB2 with SH3 domain ligands.
The current model suggests that PxxPxR motifs are critical for the interaction of GRB2 with SH3 domain ligands. These interactions are low affinity and have variable ligand specificity driven by an unknown mechanism, making the poor clinical targets. Our revised model indicates that the interaction of GRB2 with SH3 domain ligands is driven by multiple contacts outside of the PxxPxR motifs. These interactions are high affinity and have ligand specificity based on sequence differences; thus, these interactions could potentially be targeted for clinical intervention.
If the PxxPxR motifs do not drive the high affinity interaction of GRB2 with SH3 domain ligands, what is their cellular function? The PxxPxR motifs are conserved in SH3 domain ligands and are capable of low affinity interactions with SH3 domains. This strongly suggests that these sites have some capacity to regulate the binding and/or function of SH3 domain-mediated interactions. One possibility is that PxxPxR motifs could facilitate structural stability in the SH3 domain-containing proteins. In support of this idea, recent studies have shown that the linker regions surrounding SH3 and other interaction domains in Abl and PLC-γ1 regulate the dynamics of the interaction cassettes [39–41]. The PxxPxR motifs could be critical for the stable binding with SH3 domains. Deletion of the PxxPxR motifs may shift the on and off rates of the interaction in a linked manner such that equilibrium affinity does not change compared to the wild-type protein. An additional possibility is that the PxxPxR motifs are low affinity interaction regions that serve as promiscuous binding sites for a large number of SH3 domains. Upon binding, other binding contacts that are present only ligands with high specificity then drive the high affinity interaction. Essentially, this model would suggest that the PxxPxR motifs serve as “landing lights” that guide SH3 domain-ligand interactions, but then do not play a substantial role in the final binding mechanism. ITC and DLS are equilibrium systems that cannot identify changes in the kinetics or multistep mechanism of protein-protein interactions; thus, these subtle changes would not be identified by these methods. An additional possibility is that the PxxPxR motifs could be critical for the formation of the correct structure needed for connecting to downstream signaling. The PxxPxR motifs may be vital for correctly orientating different regions of SH3 domain ligands to facilitate further protein-protein interactions needed for enzymatic function. The substantial difference in the hydrodynamic radius of the GRB2-SOS1WT compared to the GRB2-SOS1Δ1,2,3,4,Z mutant would support such a model. Together, our studies show that the main function of the PxxPxR motifs in the proline rich regions of SOS1 and CBL are not to drive the high affinity interaction with GRB2, but instead are likely to have more subtle effects that regulate cellular function of these proteins. The separate question from the focus of this manuscript, specifically how PxxPxR motifs alter the cellular function of SH3 domain ligands, will be the goal of future studies.
5. Conclusions
The canonical model is that SH3 domains associate with ligands via low specificity and affinity interactions driven by PxxPxR motifs and that selectivity for ligands is driven by an unknown mechanism (Figure 7). Our studies question the validity of this model. GRB2 may be unique in requirement for PxxPxR motifs, but this is highly unlikely. Thirty-five signaling proteins in humans contain adjacent SH2 and SH3 domains [42]. At a minimum, the SH3 domain binding mechanism in these proteins is likely conserved, but it is possible that the mechanism for all SH3 domains is similar to GRB2. SH3 domains are generally thought to be poor drug targets since the PxxPxR motifs are relatively low affinity and specificity [43, 44]. If SH3 domain binding is driven by interactions that are unique for each ligand, then this model needs to be modified. However, the drug targeting screens must be performed using large regions of the SH3 domaincontaining protein and its ligand. Additionally, SH3 domain-driven reactions will need to be investigated in the context of full-length proteins and ligands, since the use of peptide ligands and isolated SH3 domains may not fully model the binding of the full length proteins. Our data suggest that PxxPxR motifs are not required for the high affinity binding of GRB2 with SH3 domain ligands, suggesting that the binding mechanism of SH3 domain-containing proteins may be more complex than originally thought.
Supplementary Material
Highlights.
PxxPxR motifs are required for the binding of GRB2 to peptide ligands
PxxPxR-containing peptides do not inhibit high affinity GRB2:SOS1 interactions
PxxPxR motifs are not required for GRB2 to bind complete proline rich regions
Regions outside of PxxPxR motifs drive high affinity GRB2 interactions with SOS1
PxxPxR motifs are not required for in vivo binding of GRB2 with SOS1
Acknowledgements
The authors thank Mikaela Tremblay, Mahmood Bilal and Michael Zhang for their careful reading of the manuscript. These studies were partially funded by a Scientist Development Grant #0830244 from the American Heart Association (JCDH) and R01 CA136729 from the National Institutes of Health (JCDH).
Abbreviations
- DLS
dynamic light scattering
- ITC
isothermal titration calorimetry
- PRR
proline rich region
- TIRF
total internal reflection fluorescence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaneko T, Li L, Li SS. The SH3 domain--a family of versatile peptide- and protein-recognition module. Front Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 2.Saksela K, Permi P. SH3 domain ligand binding: What's the consensus and where's the specificity? FEBS Lett. 2012;586:2609–2614. doi: 10.1016/j.febslet.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 3.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ladbury JE, Arold S. Searching for specificity in SH domains. Chem Biol. 2000;7:R3–R8. doi: 10.1016/s1074-5521(00)00067-3. [DOI] [PubMed] [Google Scholar]
- 5.Berry D, Nash P, Liu S, Pawson T, McGlade CJ. A high-affinity Arg-X-X-Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T Cell signaling. Curr Biol. 2002;12:1336. doi: 10.1016/s0960-9822(02)01038-2. [DOI] [PubMed] [Google Scholar]
- 6.Houtman JCD, Higashimoto Y, Dimasi N, Cho S, Yamaguchi H, Bowden B, Regan C, Malchiodi EL, Mariuzza R, Schuck P, Appella E, Samelson LE. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry. 2004;43:4170–4178. doi: 10.1021/bi0357311. [DOI] [PubMed] [Google Scholar]
- 7.Dutta K, Shi H, Cruz-Chu ER, Kami K, Ghose R. Dynamic influences on a high-affinity, high-specificity interaction involving the C-terminal SH3 domain of p67phox. Biochemistry. 2004;43:8094–8106. doi: 10.1021/bi030268d. [DOI] [PubMed] [Google Scholar]
- 8.Kami K, Takeya R, Sumimoto H, Kohda D. Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. EMBO J. 2002;21:4268–4276. doi: 10.1093/emboj/cdf428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 10.Lemmon MA, Ladbury JE, Mandiyan V, Zhou M, Schlessinger J. Independent binding of peptide ligands to the SH2 and SH3 domains of Grb2. J Biol Chem. 1994;269:31653–31658. [PubMed] [Google Scholar]
- 11.Cussac D, Frech M, Chardin P. Binding of the Grb2 SH2 domain to phosphotyrosine motifs does not change the affinity of its SH3 domains for Sos proline-rich motifs. Embo J. 1994;13:4011–4021. doi: 10.1002/j.1460-2075.1994.tb06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald CB, Seldeen KL, Deegan BJ, Farooq A. SH3 domains of Grb2 adaptor bind to PXpsiPXR motifs within the Sos1 nucleotide exchange factor in a discriminate manner. Biochemistry. 2009;48:4074–4085. doi: 10.1021/bi802291y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald CB, Seldeen KL, Deegan BJ, Farooq A. Structural basis of the differential binding of the SH3 domains of Grb2 adaptor to the guanine nucleotide exchange factor Sos1. Archives of biochemistry and biophysics. 2008;479:52–62. doi: 10.1016/j.abb.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Chook YM, Gish GD, Kay CM, Pai EF, Pawson T. The Grb2-mSos1 complex binds phosphopeptides with higher affinity than Grb2. J Biol Chem. 1996;271:30472–30478. doi: 10.1074/jbc.271.48.30472. [DOI] [PubMed] [Google Scholar]
- 15.Houtman JCD, Yamaguchi H, Barda-Saad M, Braiman A, Bowden B, Appella E, Schuck P, Samelson LE. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat Struct Mol Biol. 2006;13:798–805. doi: 10.1038/nsmb1133. [DOI] [PubMed] [Google Scholar]
- 16.Sastry L, Lin W, Wong WT, Di Fiore PP, Scoppa CA, King CR. Quantitative analysis of Grb2-Sos1 interaction: the N-terminal SH3 domain of Grb2 mediates affinity. Oncogene. 1995;11:1107–1112. [PubMed] [Google Scholar]
- 17.Bartelt RR, Houtman JC. The adaptor protein LAT serves as an integration node for signaling pathways that drive T cell activation. Wiley interdisciplinary reviews. Systems biology and medicine. 2013;5:101–110. doi: 10.1002/wsbm.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giubellino A, Burke TR, Jr, Bottaro DP. Grb2 signaling in cell motility and cancer. Expert Opin Ther Targets. 2008;12:1021–1033. doi: 10.1517/14728222.12.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Liu BA, Jablonowski K, Shah EE, Engelmann BW, Jones RB, Nash PD. SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol Cell Proteomics. 2010;9:2391–2404. doi: 10.1074/mcp.M110.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartelt RR, Cruz-Orcutt N, Collins M, Houtman JC. Comparison of T cell receptor-induced proximal signaling and downstream functions in immortalized and primary T cells. PLoS ONE. 2009;4:e5430. doi: 10.1371/journal.pone.0005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman NM, Connolly SF, Reinl EL, Houtman JC. Focal adhesion kinase negatively regulates Lck function downstream of the T cell antigen receptor. J Immunol. 2013;191:6208–6221. doi: 10.4049/jimmunol.1301587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruz-Orcutt N, Vacaflores A, Connolly SF, Bunnell SC, Houtman JC. Activated PLC-gamma1 is catalytically induced at LAT but activated PLC-gamma1 is localized at both LAT- and TCR-containing complexes. Cell Signal. 2014 doi: 10.1016/j.cellsig.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal M, Montiel JL, Cussac D, Cornille F, Duchesne M, Parker F, Tocque B, Roques BP, Garbay C. Differential interactions of the growth factor receptor-bound protein 2 N-SH3 domain with son of sevenless and dynamin. Potential role in the Ras-dependent signaling pathway. J Biol Chem. 1998;273:5343–5348. doi: 10.1074/jbc.273.9.5343. [DOI] [PubMed] [Google Scholar]
- 25.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 26.Ladbury JE. Calorimetry as a tool for understanding biomolecular interactions and an aid to drug design. Biochem Soc Trans. 2010;38:888–893. doi: 10.1042/BST0380888. [DOI] [PubMed] [Google Scholar]
- 27.Velazquez-Campoy A, Ohtaka H, Nezami A, Muzammil S, Freire E. Isothermal titration calorimetry. Curr Protoc Cell Biol. 2004;Chapter 17(Unit 17):18. doi: 10.1002/0471143030.cb1708s23. [DOI] [PubMed] [Google Scholar]
- 28.Donovan JA, Ota Y, Langdon WY, Samelson LE. Regulation of the association of p120cbl with Grb2 in Jurkat T cells. J Biol Chem. 1996;271:26369–26374. doi: 10.1074/jbc.271.42.26369. [DOI] [PubMed] [Google Scholar]
- 29.Kortum RL, Balagopalan L, Alexander CP, Garcia J, Pinski JM, Merrill RK, Nguyen PH, Li W, Agarwal I, Akpan IO, Sommers CL, Samelson LE. The ability of Sos1 to oligomerize the adaptor protein LAT is separable from its guanine nucleotide exchange activity in vivo. Science signaling. 2013;6:ra99. doi: 10.1126/scisignal.2004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilal MY, Houtman JC. GRB2 Nucleates T Cell Receptor-Mediated LAT Clusters That Control PLC-gamma1 Activation and Cytokine Production. Frontiers in immunology. 2015;6:141. doi: 10.3389/fimmu.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell. Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Fernandez ML, Tynan CJ, Webb SE. A 'pocket guide' to total internal reflection fluorescence. J Microsc. 2013;252:16–22. doi: 10.1111/jmi.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald CB, Balke JE, Bhat V, Mikles DC, Deegan BJ, Seldeen KL, Farooq A. Multivalent binding and facilitated diffusion account for the formation of the Grb2-Sos1 signaling complex in a cooperative manner. Biochemistry. 2012;51:2122–2135. doi: 10.1021/bi3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethi A, Goldstein B, Gnanakaran S. Quantifying intramolecular binding in multivalent interactions: a structure-based synergistic study on Grb2-Sos1 complex. PLoS computational biology. 2011;7:e1002192. doi: 10.1371/journal.pcbi.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vidal M, Liu WQ, Lenoir C, Salzmann J, Gresh N, Garbay C. Design of peptoid analogue dimers and measure of their affinity for Grb2 SH3 domains. Biochemistry. 2004;43:7336–7344. doi: 10.1021/bi030252n. [DOI] [PubMed] [Google Scholar]
- 36.Yuzawa S, Yokochi M, Hatanaka H, Ogura K, Kataoka M, Miura K, Mandiyan V, Schlessinger J, Inagaki F. Solution structure of Grb2 reveals extensive flexibility necessary for target recognition. Journal of molecular biology. 2001;306:527–537. doi: 10.1006/jmbi.2000.4396. [DOI] [PubMed] [Google Scholar]
- 37.Guilloteau JP, Fromage N, Ries-Kautt M, Reboul S, Bocquet D, Dubois H, Faucher D, Colonna C, Ducruix A, Becquart J. Purification, stabilization, and crystallization of a modular protein: Grb2. Proteins. 1996;25:112–119. doi: 10.1002/(SICI)1097-0134(199605)25:1<112::AID-PROT9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 38.Maignan S, Guilloteau JP, Fromage N, Arnoux B, Becquart J, Ducruix A. Crystal structure of the mammalian Grb2 adaptor. Science. 1995;268:291–293. doi: 10.1126/science.7716522. [DOI] [PubMed] [Google Scholar]
- 39.Bunney TD, Esposito D, Mas-Droux C, Lamber E, Baxendale RW, Martins M, Cole A, Svergun D, Driscoll PC, Katan M. Structural and functional integration of the PLCgamma interaction domains critical for regulatory mechanisms and signaling deregulation. Structure. 2012;20:2062–2075. doi: 10.1016/j.str.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corbi-Verge C, Marinelli F, Zafra-Ruano A, Ruiz-Sanz J, Luque I, Faraldo-Gomez JD. Two-state dynamics of the SH3-SH2 tandem of Abl kinase and the allosteric role of the N-cap. Proc Natl Acad Sci U S A. 2013;110:E3372–E3380. doi: 10.1073/pnas.1303966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Oliveira GA, Pereira EG, Ferretti GD, Valente AP, Cordeiro Y, Silva JL. Intramolecular dynamics within the N-Cap-SH3-SH2 regulatory unit of the c-Abl tyrosine kinase reveal targeting to the cellular membrane. J Biol Chem. 2013;288:28331–28345. doi: 10.1074/jbc.M113.500926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Luccarelli J, Thompson S, Hamilton AD. SH3 Domains as Drug Targets. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2013. [Google Scholar]
- 44.Lu XL, Cao X, Liu XY, Jiao BH. Recent progress of Src SH2 and SH3 inhibitors as anticancer agents. Curr Med Chem. 2010;17:1117–1124. doi: 10.2174/092986710790827861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.