Abstract
Studying the importance of genetic factors in a desired cell type or tissue necessitates the use of precise genetic tools. With the introduction of bacteriophage Cre recombinase/loxP mediated DNA editing and promoter-specific Cre expression, it is feasible to generate conditional knockout mice in which particular genes are disrupted in a cell type-specific manner in vivo. In cardiac myocytes, this is often achieved through α-myosin heavy chain promoter (αMyHC)-driven Cre expression in conjunction with a loxP-site flanked gene of interest. Recent studies in other cell types demonstrate toxicity of Cre expression through induction of DNA damage. However, it is unclear to what extent the traditionally used αMyHC-Cre line [1] may exhibit cardiotoxicity. Further, the genotype of αMyHC-Cre+/− is not often included as a control group in cardiac myocyte-specific knockout studies. Here we present evidence that these αMyHC-Cre+/− mice show molecular signs of cardiac toxicity by 3 months of age and exhibit decreased cardiac function by 6 months of age compared to wild-type littermates. Hearts from αMyHC-Cre+/− mice also display evidence of fibrosis, inflammation, and DNA damage. Interestingly, some of the early functional changes observed in αMyHC-Cre+/− mice are sexually dimorphic. Given the high level of Cre recombinase expression resulting from expression from the αMyHC promoter, we asked if degenerate loxP-like sites naturally exist in the mouse genome and if so, whether they are affected by Cre in the absence of canonical loxP-sites. Using a novel bioinformatics search tool, we identified 619 loxP-like sites with 4 or less mismatches to the canonical loxP-site. 227 sites overlapped with annotated genes and 55 of these genes were expressed in cardiac muscle. Expression of ~26% of the 27 genes tested was disrupted in αMyHC-Cre+/− mice indicating potential targeting by Cre. Taken together, these results highlight both the importance of using αMyHC-Cre mice as controls in conditional knockout studies as well as the need for a less cardiotoxic Cre driver for the field.
Keywords: Transgenic mice, Cre recombinase, LoxP site, DNA damage, Cardiotoxicity
1. Introduction
When genes were first discovered as the fundamental heritable units the race was on to determine individual gene function. The gold standard for assessing gene function has since been gene deletion, or knockout, and gene overexpression. Increasingly precise genetic engineering techniques enable investigators to perform both of these tasks. Perhaps the most important contributor to this field over the past 30 years has been the introduction of bacteriophage P1 Cre recombinase-mediated recombination of mammalian genomic DNA (reviewed in [2]). Hundreds of studies have since utilized this technology to modify a gene of interest at the cell or organismal level in an effort to study its requirement or sufficiency.
By expressing Cre in a tissue or cell-type specific manner, genes that are engineered with flanking loxP sites can be deleted or overexpressed in an analogous fashion. In the adult cardiac myocyte, this is most often achieved using Cre expression driven by the cardiac myocyte-specific α-myosin heavy chain (αMyHC) promoter/enhancer [1,3]. Cre expression from this promoter has been shown to be both cardiac myocyte-specific and to drive highly efficient recombination [1]. Use of this transgenic mouse circumvents embryonic or perinatal lethality induced by deletion of genes required for other cell types and has therefore uncovered many cardiac myocyte-specific genetic roles. To date, nearly 130 primary research studies (Supplemental Table 1) have utilized this transgenic mouse to conditionally modify a gene of interest in cardiac myocytes. Thus, this tool has become indispensible to the cardiac biology field.
Cre recombinase mediates recombination between pairs of 34-base-pair palindromic loxP sites [4]. Although its preference is for sites consisting of two 13-base-pair inverted repeats separated by an 8-base-pair spacer [5], Cre has been shown to have promiscuous activity and is capable of recombination at sites containing up to 10 mismatches to the canonical loxP site [6]. Such degenerate loxP sites have been identified in mammalian genomes and can serve as legitimate Cre substrates [7]. Further, it has been shown that Cre binding to loxP sites in the absence of recombination has the ability to block downstream transcription [8]. Thus, Cre has the potential for a variety of off-target effects.
Indeed, several instances of off-target Cre effects have been documented over the past decade. In non-cardiac tissue cell-types, Cre expression has been shown to induce DNA damage and apoptosis in the absence of bona fide loxP sites [9,10]. Toxic effects associated with Cre expression have been observed in gastrointestinal cells, neurons, and spermatids [10–12]. Finally, tamoxifen-inducible Cre expression in myocardium has also been demonstrated to induce fibrosis, DNA damage, and cardiac dilation independently of tamoxifen-associated toxicity [13–16]. However a Cre-only control genotype is not often included in studies requiring Cre expression. In fact, the αMyHC-Cre+/− genotype is only included as a control group in ~20% of studies identified that utilize this Cre line (Supplemental Table 1).
Given the extensive use of the αMyHC-Cre mice and anecdotal reports of associated cardiotoxicity [17–19], we sought to formally assess whether cardiac myocyte-restricted Cre expression in the absence of engineered loxP sites modified cardiac biology. We hypothesized that prolonged myocardial Cre expression would lead to cardiotoxicty. To test our hypothesis, we took a combined functional, molecular, and bioinformatics approach. We examined a panel of genes whose expression consistently changes with cardiac pathology and discovered significant changes in αMyHC-Cre+/− mice at 3 months of age that progressed to more profound changes by 6 months of age. These molecular changes were accompanied by a decrease in cardiac function in these mice as well as pathological signaling pathway activation and evidence of an activated DNA damage response. We suggest that during prolonged Cre expression, endogenous, non-canonical loxP sites are targeted, thus activating a DNA damage response that is associated with pathological signaling pathway and gene expression activation, and adverse remodeling of the myocardium.
2. Methods
Mice
All animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Colorado at Boulder. Mice were fed ad libitum standard rodent chow and housed in a facility with a 12 hour light, 12 hour dark cycle. αMyHC-Cre mice were backcrossed into the C57Bl/6J background for at least 10 generations and bred using heterozygote males and wild-type (WT) females. For all experiments, WT mice were compared to αMyHC-Cre+/− mice. Genotyping was performed from tail snip biopsies and re-confirmed post-mortem. Primers for genotyping are listed in Supplemental Table 2. Myocardial Cre expression was confirmed by quantitative real-time reverse transcriptase PCR (qRT-PCR) and western blot for Cre expression (Supplemental Figure 1). For sample collection, animals were sedated using 1–4% inhaled isoflurane and sacrificed by cervical dislocation. Hearts were excised, perfused in PBS, and flash frozen in liquid nitrogen.
Transthoracic echocardiography
Non-invasive echocardiographic images and measurements were made using the Philips Sonos 5500 system. Mice were placed on a heating pad and maintained on 2% isoflurane via spontaneous inhalation. The mouse’s fur was first removed from the ribcage using a depilatory cream, and an image-potentiating gel was then applied for image acquisition. M-(motion) mode images were captured for each animal at the level of the papillary muscles (A2 view). Left ventricular dimensions and function were calculated from the M-mode images, where wall thickness of the anterior and posterior walls and the chamber diameter were measured using the ASE leading edge convention.
Gene expression
Total RNA was purified using TRI Reagent (Ambion) according to the manufacturer's protocol. cDNA was synthesized with Superscript III reverse transcriptase (Invitrogen) and random hexamer primers. Gene expression was determined by qRT-PCR using SYBR Green dye with gene specific primer sets (Supplemental Table 2) and a Bio-Rad CFX-96 Real-Time PCR system.
Western blot
Left ventricles were homogenized in 50mM Tris pH 8.0, 150mM NaCl, 1% NP40, 0.5% Na-deoxycholate, 0.1% SDS, complete protease inhibitor cocktail (Roche) and the following phosphatase inhibitors: 1mM PMSF, 2mM NaF, 2mM NaPPi, 1mM Beta-Glycerophosphate, 1mM Na-molybdate dihydrate, and 1mM Na-Orthovanadate. 10μg of protein was resolved on an SDS-PAGE gel and probed with the following antibodies: GAPDH (Cell Signaling Technologies 2118s), Cre (Cell Signaling Technologies 7803), phospho-ERK (Cell Signaling Technologies 9101s), total ERK (Cell Signaling Technologies 9102s), phospho-p38 (Cell Signaling Technologies 4511s), total p38 (Cell Signaling Technologies 9212s), MCIP1.4 ([20]), Calcineurin (CnA) (Cell Signaling Technologies 2614), γH2AX (Cell Signaling Technologies 9718), p53 (Santa Cruz 6243), Bax (Santa Cruz 526), PARP (Cell Signaling Technologies 9542), and ME3 (Novus NBP1-30525).
Identification of degenerate loxP sites
To identify putative, degenerate loxP sites we used LADMA (Levenshtein Automata based Degenerate Motif Annotator) to annotate the Mus musculus genome (http://hgdownload.cse.ucsc.edu/goldenPath/mm10/bigZips/), allowing up to 4 mismatches based on a conservative input Cre binding motif sequence: ATNACNNCNTATA NNNTANNN TATANGNNGTNAT [6] (http://dowell.colorado.edu/resources.html). Briefly, the Levenshtein distance is defined by the minimal number of insertions, deletions or substitutions required to transform one string to another [21]. Thusly, LADMA accepts a number of mismatches (in our case 4) and builds a Levenshtein non-determinisitc finite state automata (NFA) to find all possible DNA subsequences within some Levenshtein distance. To increase computational efficiency, this NFA is converted to a deterministic finite state automata and inserted into an Aho-Corasick-like data structure in order to find potential matches across the entire genome [22].
LADMA was run on each chromosome and returned 617 total sites finding 0 perfect match, 0 1-mismatch, 12 2-mismatches, 244 3-mismatches, and 361 4-mismatches (Supplemental Table 4). These sites were then intersected with full-length gene bodies extracted from UCSC mm10 Ensembl annotations with the introns included (http://hgdownload.cse.ucsc.edu/goldenPath/mm10/database/) using BedTools IntersectBed (BedTools version 2.16.2) [23]. This resulted in 227 genes that contained one or more degenerate binding site(s).
Transgene copy number analysis
Transgene copy number was estimated as described [24]. Briefly, genomic DNA (gDNA) was isolated using phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. gDNA was then digested overnight with EcoRI (New England Biolabs). Purified, digested DNA was quantified and 10ng were loaded into a qPCR reaction. Standard curves were constructed using known copy numbers of linearized plasmids containing the coding region of Cre or Myh15. Copy number was normalized to Myh15 (2 copies/genome).
Determination of Cre transgene insertion site
To determine insertion site of the αMyHC-Cre transgene, Thermal Asymmetric Interlaced PCR (TAIL-PCR) was performed as described [25,26]. Briefly, we performed three rounds of nested PCR on purified gDNA in which one primer was anchored in the 3’ end of the αMyHC-Cre transgene and degenerate oligonucleotides were used as reverse primers. PCR products were purified, sequenced, and mapped to the mouse genome.
Histology
Hearts were fixed in 10% buffered formalin overnight and then transferred to 70% ethanol. Fixed hearts were embedded in paraffin, sectioned, and stained with Picrosirius Red (Polysciences). The Picrosirius Red positive area was quantified as a fraction of total tissue area using ImageJ using either brightfield microscopy or polarized light microscopy (yellow, type I collagen fibers were quantified). 3–4 images were quantified per heart in 3–4 hearts/group. Tissue edges and vessels were excluded from analysis. To identify inflammatory cells, F4/80 immunostaining (Serotec MCA497) was performed to label marcrophages. F4/80 positive cells was counted across transverse sections in 3–4 hearts per genotype. Analogous quantification of TUNEL-positive cells (Roche 11684809910) was also performed in transverse sections to identify DNA fragmentation and/or apoptosis.
Data and statistical analysis
Data are presented as mean ± SEM. Differences between groups were evaluated for statistical significance using Student's two-tailed t test (two groups) or one-way ANOVA (more than two groups) followed by Tukey’s post-hoc test for pairways comparisons. P values less than 0.05 were considered significant unless otherwise noted.
3. Results
3.1 αMyHC-Cre expression is cardiotoxic in an age-dependent manner
Although a cardiotoxic effect of myocardial Cre expression in the widely used αMyHC-Cre mice has been reported [17–19], to our knowledge no thorough investigation of this phenotype has been published. To that end, we chose to formally characterize the effect of myocardial Cre expression driven by the 5.5 kb murine αMyHC promoter [1]. We characterized these mice at 3 months of age as this has been a commonly used adult time-point in a literature search of studies employing this transgenic line (Supplemental Table 1). Further, we examined males and females separately as it is known that the heart is a sexually dimorphic organ yet many studies in which this line had been utilized did not specify sex [27]. At 3 months, no gross abnormalities were observed in systolic cardiac function (%Ejection Fraction, %EF) in female αMyHC-Cre+/− mice as compared to wild-type (WT) littermates (Figure 1A,B and Supplemental Table 3). In males, however, we observed an increase in cardiac function and an increase in heart rate (HR) that coincided with a trend towards hypertrophy in αMyHC-Cre+/− males (Figure 1B and Supplemental Table 3). Although the increase in HR may mediate the observed change in %EF [28,29], we suspect these changes may be due to a compensatory state preceding functional decline. Further, we observed a correlative relationship between Cre expression and cardiac function at this time point suggesting a dose-dependent effect of myocardial Cre expression (Supplemental Figure 2).
Figure 1. αMyHC-Cre is cardiotoxic by six months of age.
(A) Ventricular contractile function (% Ejection fraction) at 3 months and 6 months. (B) Cardiac mass, heart weight normalized to body weight (HW/BW) at indicated ages. (C) Cardiac gene expression at 3 months in males and females (C) and 6 months, females (D), and males (E). Error bars: SEM, N=4–6/group *P < 0.05, **P < 0.01 vs. age-matched and sex-matched WT control.
We next examined gene expression in the ventricles of 3 month-old mice since such molecular changes frequently precede functional changes [30]. To that end, we examined reactivation of fetal gene expression of naturietic peptides, Anp and Bnp, as well as selective cardiac myosin expression in hearts of male and female mice. We observed statistically significant, though modest, increases in Anp and Bnp in both males and females but did not observe changes in any other genes (Figure 1C).
In light of the observed changes in gene expression observed at 3 months, we next asked whether there was a resultant decline in cardiac function at 6 months in αMyHC-Cre+/− mice. Indeed, cardiac function was significantly decreased at 6 months in both males and females (Figure 1A) and significant hypertrophy was observed in males as compared to WT littermates (Figure 1B). Further, expression of fetal genes was increased and myosin expression was shifted toward a pathological state (elevated βMyHC) at this time-point (Figure 1D,E) thus supporting our initial hypothesis that the gene expression changes and hypertrophy observed at 3 months were likely precursors to the more notable changes observed at 6 months. We therefore conclude that αMyHC-Cre expression in this model is cardiotoxic in an age-dependent manner.
In order to better characterize the observed cardiotoxicity at 6 months, we examined pathological signaling pathway activation. In agreement with the changes in gene expression and cardiac function, we observed activation of the stress-induced, pathological p38 MAPK signaling pathway (Figure 2A). We did not observe activation of pro-hypertrophic P-ERK signaling but other pathological and stress induced signaling molecules such as MCIP1.4 and Calcineurin (CnA) were also upregulated in αMyHC-Cre+/− mice (Figure 2A).
Figure 2. Induction of pathological intracellular signaling and inflammatory or fibrotic genes in female αMyHC Cre+/− ventricles at 6 months.
(A) MAPK phosphorylation status or MCIP1.4 and Calcineurin protein expression (immunoblot). (B) Inflammatory cytokine and fibrotic gene expression (qRT-PCR). (C) Histochemical assessment of fibrosis (Picrosirius Red stain) 10× magnification using either brightfield or polarized light microscopy. Picrosirius Red-positive area was quantified as a percentage of total tissue area. Error bars: SEM, N=4–6/group *P < 0.05, **P < 0.01, ***P < 0.001 vs. age-matched and sex-matched WT control.
3.2 Prolonged αMyHC-Cre expression is associated with mild fibrosis and inflammation
We then asked whether gene expression related to p38 signaling and cardiac pathology was correspondingly modified. In ventricles from female αMyHC-Cre+/− mice we observed increases in several pro-inflammatory markers including Il-1β and Tnfα, as well as increases in pro-fibrotic makers, Col1α1, Col3α1, and Ctgf (Figure 2B). Further we observed a trending increase in expression of a marker of activated, infiltrating macrophages, Cd68 (Figure 2B). These pro-inflammatory, pro-fibrotic gene expression changes correlated with modest increases (2-fold) in left-ventricular fibrosis (Figure 2C) similar to that reported in models of hypertrophic cardiomyopathy and trans-aortic banding [31,32]. A trending increase in inflammatory cell presence was observed in αMyHC-Cre+/− ventricular myocardium (Supplemental Figure 3). Comparable changes in gene expression were also confirmed in 6 month-old male αMyHC-Cre+/− mice (Supplemental Figure 4A). These pro-fibrotic gene expression changes may be directly attributable to activated p38 MAPK signaling (reviewed in [33]).
3.3 Prolonged αMyHC-Cre expression is associated with DNA damage response
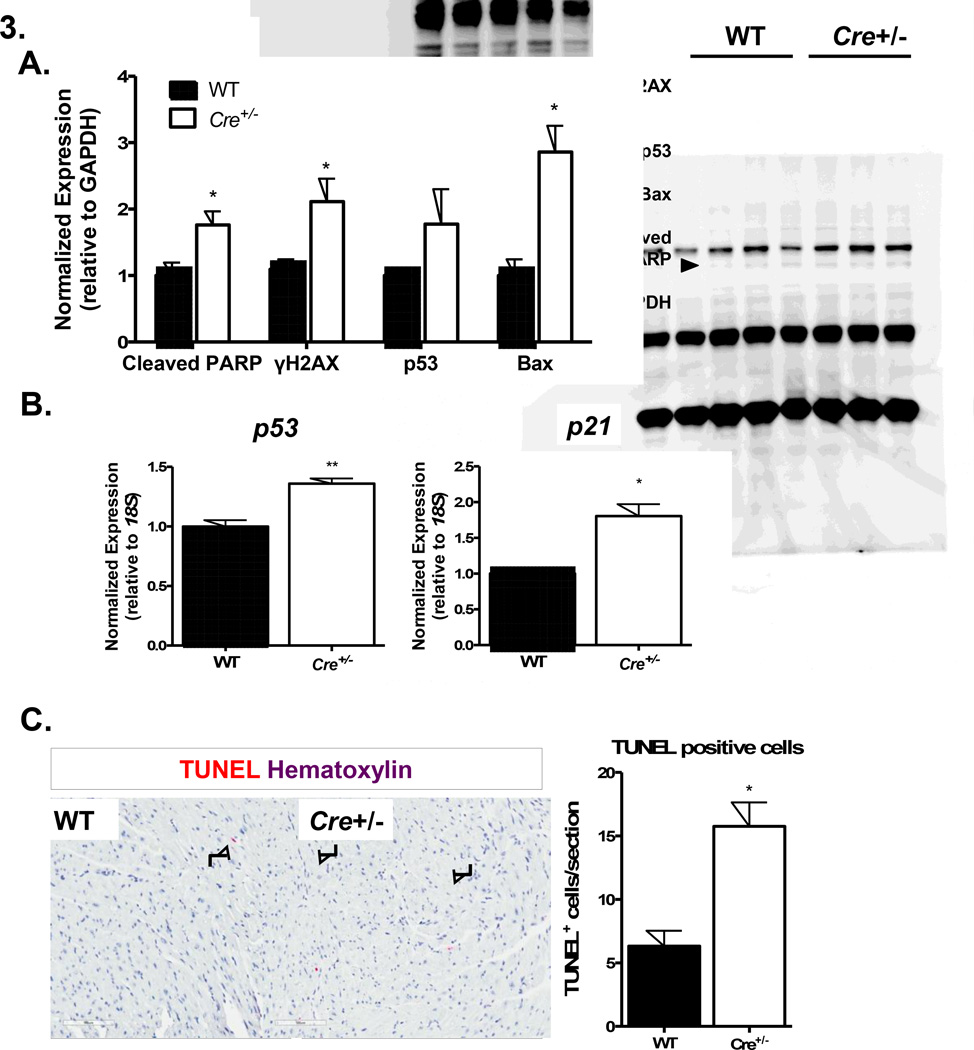
Because Cre is a DNA recombinase, we hypothesized that the observed cardiotoxicity in αMyHC-Cre+/− mice may be attributable to Cre-related DNA damage. To this end, we examined DNA damage response effectors in female αMyHC-Cre+/− mice at 6 months of age. Indeed, we saw modest increases in several factors related to DNA damage and apoptosis including cleaved PARP, γH2A.X, p53, and Bax (Figure 3A). These changes were accompanied by detectable transcriptional upregulation of master DNA damage response effectors, p21 and p53 in both female and male αMyHC-Cre+/− mice (Figure 3B, Supplemental Figure 4B) as well as a ~3-fold increase in cells with DNA fragmentation as identified by TUNEL-positive cells (Figure 3C).
Figure 3. Ventricular DNA damage response in 6-month-old female αMyHC-Cre+/− mice.
(A) mRNA expression of DNA damage response regulators, p21 and p53 (qRT-PCR). (B) DNA damage-related and pro-apoptotic protein levels (immunoblot). (C) Histochemical assessment of TUNEL+ cells in ventricular myocardium. Arrows indicate TUNEL+ cells. Error bars: SEM, N=4–6/group *P < 0.05, **P < 0.01 vs. age-matched and sex-matched WT control.
3.4 Endogenous, degenerate loxP sites may be targeted by Cre following persistent expression under the αMyHC promoter
Given the absence of canonical loxP sites in the αMyHC-Cre+/− mice in our studies and the presence of a general DNA damage response, we next asked whether this response might be a result of off-target Cre action at degenerate, endogenous loxP sites. Using an unbiased bioinformatics approach and a novel motif-finding algorithm, we searched the mouse genome for “loxP-like” sites, tolerating mismatches to the canonical loxP site at positions known to be dispensable for Cre binding and recombination [6,34] (Figure 4). Several hundred of these sites were identified, depending on the number of mismatches to the canonical site tolerated (Supplemental Table 4). Further, a large percentage of these sites in the mouse genome (37%) were located intragenically and many (24%) of these were present in transcriptionally active cardiac genes as determined by transcriptome profiling of ventricular myocardium [35]. We next tested whether expression of 27 of these cardiac genes containing degenerate loxP sites was modified by the presence of αMyHC-Cre by designing qRT-PCR primers downstream of or spanning the putative loxP site. We found expression changes for 27% of the genes tested (Supplemental Figure 5A) and confirmed a change in protein expression consistent with the observed change in transcript expression for one target, ME3 (Supplemental Figure 5B).
Figure 4. Degenerate loxP sites are abundant in the mouse genome.
Flow chart of genome-wide search for degenerate loxP sites in mouse. Degenerate loxP site sequence was based on published Cre binding and recombination data [6,34]. This sequence is a conservative estimate for maximum-tolerated mismatches by Cre. Note that a search for loxP sites harboring just two mismatches identified 12 endogenous sites while a search for loxP sites harboring five mismatches identified more than 7500 endogenous sites. Transcriptionally relevant genes for the heart were identified by intersecting the list of loxP-like containing genes with both the publically available University of Washington (UW) ENCODE dataset and transcriptome data from [35].
3.5 Transgene copy number or insertion site does not likely account for observed cardiotoxicity in αMyHC-Cre mice
Finally, we sought to rule out an effect of cardiotoxicity mediated by transgene insertion site or exceedingly high copy number of the transgene. We used Thermal Asymmetric Interlaced PCR (TAIL-PCR) [25,26] to map the insertion site of the αMyHC-Cre transgene (Supplemental Figure 6A). By this method, 6/10 sequenced PCR products mapped to an intergenic locus on Chromosome 6 and 4/10 products mapped to the 5’ end of the transgene, suggesting tandem insertion (Supplemental Figure 6B). We then used qPCR of gDNA in conjunction with standard curve generation to determine copy number of the αMyHC-Cre transgene [24]. By this method, copy number was estimated to be 6 copies/genome which we believe to be tandem insertions based on our results from TAIL-PCR (Supplemental Figure 7). Taken together, we do not believe insertion site or high copy number of the αMyHC-Cre transgene can account for the progressive cardiotoxicity observed in these mice.
5. Discussion
Here we present evidence that prolonged exposure to Cre recombinase in cardiac myocytes has deleterious effects on cardiac function. Although we are not the first to report cardiotoxicity associated with Cre expression in the heart, we believe this is the first characterization of such in the widely used αMyHC-Cre mouse line [17–19]. Our findings of depressed cardiac function, an activated DNA damage response, modest but statistically significant fibrosis, and mild inflammation in αMyHC-Cre+/− myocardium are consistent with findings in inducible cardiac-specific MerCreMer models and other models of myocardial Cre expression [13–15].
The early molecular changes observed in the αMyHC-Cre+/− mice precede the more robust molecular and functional changes observed at 6 months. Given the dynamic cross-regulation of the DNA damage response pathway, p38 MAPK stress response pathway, and inflammatory response, we cannot conclude that DNA damage is a primary insult [36–38]. However, given prolonged, high Cre expression, Cre’s role as a DNA recombinase with promiscuous activity, and the presence of degenerate loxP sites in the mouse genome (Figure 4), we suggest this as a potential mechanism of cardiotoxicity in conjunction with an inflammatory response activated by long-term exposure to a non-endogenous DNA recombinase. Further, in a skin model of Cre toxicity, DNA damage response was shown to be dependent on Cre recombinase activity as an endonuclease-deficient isoform of Cre did not activate this pathway [9].
Although we did not observe gross changes in gene expression at many of the genes containing putative loxP sites (Supplemental Figure 5A), this does not rule out an age or time-point dependent change in expression or genomic insults at the hundreds of intergenic sites also identified but not examined (Figure 4). Further, genomic changes are likely heterogeneous among cardiac myocytes so detecting changes in individual genes from a population of myocytes may be difficult. It is likely that genomic sites with high accessibility or sites that are more closely matched to the canonical loxP site may be more often targeted and more easily identified by changes in gene expression (as in the case of Me3 or Ptgfr, Supplemental Figure 5A). Although beyond the scope of this report, whole genome sequencing of αMyHC-Cre cardiac myocytes may be a preferred method for addressing Cre’s effect on genomic integrity.
Genetic background, diet, and vivarium conditions may all play a part in the Cre-associated phenotype we observed. Cardiac outcomes can be heavily influenced by genetic background in particular [39–42]. We chose to perform our studies in the C57Bl/6J background given the wide usage of this isogenic strain however it is unclear to what extent other strains are susceptible to Cre-related cardiotoxicity. Interestingly, αMyHC-Cre mice on a mixed genetic background appear more resistant to cardiotoxicity than those observed in this study [43]. Similar results were observed for αMyHC-MerCreMer mice in which animals on a mixed background (129Ola/C57Bl6/J) displayed less fibrosis than pure C57Bl6/J mice [13].
Sexually dimorphic phenotypes were observed at both timepoints examined, both by echocardiography and morphometric analysis (Figure 1 and Supplemental Table 3). Our subsequent analyses focused on female mice, given their demonstrated resistance to cardiac insult compared to males [44,45]. Future studies are needed to more carefully dissect differential hypertrophic and molecular changes in αMyHC-Cre males, although many of the gene expression changes observed in females were also confirmed in males (Supplemental Figure 4). Although we did not further investigate the sexually dimorphic nature of our findings, they support the need for sex-specific genetic studies.
Given the high promoter activity of αMyHC in cardiac myocytes, it is perhaps not surprising that the resulting, persistent expression of Cre eventually leads to pathological remodeling. Interestingly, in another (“alternate”) model of cardiomyocyte-specific Cre expression [46] in which myocardial Cre protein expression is considerably lower than the more commonly used, “conventional” αMyHC mice [1], cardiotoxicity was not observed in age-matched mice (Supplemental Figure 8). This supports our hypothesis that Cre-related toxicity may be time and dose-dependent.
Since the efficiency of Cre to induce recombination of canonical loxP sites is quite high [47] and cardiac myocytes rarely divide, we suggest using a temporally restricted or dose-dependent promoter to drive Cre expression. Others have shown that even one dose of tamoxifen is sufficient to induce recombination in an inducible αMyHC-MerCreMer model [13]. However, some genomic loci are resistant to recombination and require more persistent Cre expression. In light of these issues and the data we present, we propose that a less cardiotoxic model of cardiac myocyte-specific genome editing is needed for the field. Until this is introduced, we strongly urge investigators to be cautious in their choice of Cre driver and to always include a Cre-only control group, even in studies of young animals (< 3 months old). Although we did not observe profound molecular changes at 3 months, it may be that Cre expression in cardiac myocytes sensitizes young mice to other pathological stimuli, especially those characterized by DNA damage (e.g. myocardial infarction [48]). Finally, we strongly encourage the use and separation of both sexes as molecular and phenotypic differences are common and clinically relevant.
Supplementary Material
Highlights.
Widely used αMyHC-Cre transgenic mice display age-dependent cardiotoxicity.
Cardiotoxicity is associated with fibrosis, inflammation, and DNA damage response.
A novel platform was used to find degenerate loxP sites in the mouse genome.
The mouse genome has hundreds of degenerate loxP sites that may be targeted by Cre.
Off-target Cre effects at degenerate sites may contribute to cardiotoxicity.
Acknowledgments
αMyHC-Cre mice were a generous gift from Michael Schneider. This work was supported in part by the American Heart Association 13PRE16380002 and NIH 5-T32 GM007135 (EKP), Marsico Professor of Excellence award and NIH GM29090 (LAL), NSF ABI DBI-12624L0 (RDD, JA), and an NSF IGERT 1144807 (JA). The authors acknowledge the BioFrontiers Computing Core at the University of Colorado Boulder for providing High Performance Computing resources (NIH 1S10OD012300) supported by BioFrontiers IT. The authors thank Dr. Alberto Rossi for reagents, Colin Clark and Amy Perry for technical assistance, and Drs. Kristen Barthel, Stephen Langer, Massimo Buvoli, Jason Magida and Angela Peter for helpful discussion.
Abbreviations
- αMyHC
alpha-myosin heavy chain
- LoxP
locus of crossover P1
- LADMA
Levenshtein Automata based Degenerate Motif Annotator
- NFA
non-determinisitc finite state automata
- PBS
Phosphate Buffered Saline
- %EF
Percent Ejection Fraction
- HR
Heart Rate
- Anp
Atrial Naturietic Peptide
- Bnp
Brain Naturietic Peptide
- p38 MAPK
p38 Mitogen Activated Kinase
- ERK
Extracellular-Signal-Regulated Kinase
- PARP
poly (ADP-ribose) polymerase
- Bax
BCL2-associated X protein
- Ctgf
Connective tissue growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
Contributor Information
Emily K. Pugach, Email: Emily.pugach@coloado.edu.
Phillip A. Richmond, Email: Phillip.richmond@colorado.edu.
Joseph G. Azofeifa, Email: Joseph.azofeifa@colorado.edu.
Robin D. Dowell, Email: Robin.dowell@colorado.edu.
Leslie A. Leinwand, Email: Leslie.leinwand@colorado.edu.
References
- 1.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 3.Subramaniam A, Jones WK, Gulick J, Wert S, Neumann J, Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J Biol Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- 4.Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R. Site-specific Recombination and Its Role in the Life Cycle of Bacteriophage P1. Cold Spring Harb Symp Quant Biol. 1981;45:297–309. doi: 10.1101/sqb.1981.045.01.042. [DOI] [PubMed] [Google Scholar]
- 5.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucl Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thyagarajan B, Guimarães MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 7.Ito M, Yamanouchi K, Naito K, Calos MP, Tojo H. Site-specific integration of transgene targeting an endogenous lox-like site in early mouse embryos. J Appl Genet. 2011;52:89–94. doi: 10.1007/s13353-010-0011-3. [DOI] [PubMed] [Google Scholar]
- 8.Iovino N, Denti MA, Bozzoni I, Cortese R. A loxP-containing pol II promoter for RNA interference is reversibly regulated by Cre recombinase. RNA Biology. 2005;2:86–92. doi: 10.4161/rna.2.3.2045. [DOI] [PubMed] [Google Scholar]
- 9.Janbandhu V, Moik D, Fässler R. Cre recombinase induces DNA damage and tetraploidy in the absence of LoxP sites. Cell Cycle. 2014;13:462–470. doi: 10.4161/cc.27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USa. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harno E, Cottrell EC, White A. Metabolic pitfalls of CNS Cre-based technology. Cell Metab. 2013;18:21–28. doi: 10.1016/j.cmet.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Huh WJ, Mysorekar IU, Mills JC. Inducible activation of Cre recombinase in adult mice causes gastric epithelial atrophy, metaplasia, and regenerative changes in the absence of “floxed” alleles. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2010;299:G368–G380. doi: 10.1152/ajpgi.00021.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lexow J, Poggioli T, Sarathchandra P, Santini MP, Rosenthal N. Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis Model Mech. 2013;6:1470–1476. doi: 10.1242/dmm.010470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bersell K, Choudhury S, Mollova M, Polizzotti BD, Ganapathy B, Walsh S, et al. Moderate and high amounts of tamoxifen in αMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis Model Mech. 2013;6:1459–1469. doi: 10.1242/dmm.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buerger A, Rozhitskaya O, Sherwood MC, Dorfman AL, Bisping E, Abel ED, et al. Dilated Cardiomyopathy Resulting From High-Level Myocardial Expression of Cre-Recombinase. Journal of Cardiac Failure. 2006;12:392–398. doi: 10.1016/j.cardfail.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Koitabashi N, Bedja D, Zaiman AL, Pinto YM, Zhang M, Gabrielson KL, et al. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circulation Research. 2009;105:12–15. doi: 10.1161/CIRCRESAHA.109.198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doetschman T, Azhar M. Cardiac-Specific Inducible and Conditional Gene Targeting in Mice. Circulation Research. 2012;110:1498–1512. doi: 10.1161/CIRCRESAHA.112.265066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis J, Maillet M, Miano JM, Molkentin JD. Lost in transgenesis: a user's guide for genetically manipulating the mouse in cardiac research. Circulation Research. 2012;111:761–777. doi: 10.1161/CIRCRESAHA.111.262717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molkentin JD, Robbins J. With great power comes great responsibility: Using mouse genetics to study cardiac hypertrophy and failure. Journal of Molecular and Cellular Cardiology. 2009;46:130–136. doi: 10.1016/j.yjmcc.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bush E, Fielitz J, Melvin L, Martinez-Arnold M, McKinsey TA, Plichta R, et al. A small molecular activator of cardiac hypertrophy uncovered in a chemical screen for modifiers of the calcineurin signaling pathway. Proc Natl Acad Sci USa. 2004;101:2870–2875. doi: 10.1073/pnas.0308723101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz KU, Mihov S. Fast string correction with Levenshtein automata. Ijdar. 2002;5:67–85. [Google Scholar]
- 22.Aho AV, Corasick MJ. Efficient string matching: an aid to bibliographic search. Communications of the ACM. 1975;18:333–340. [Google Scholar]
- 23.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi MU, Pittman HK, Haisch CE, Verbanac KM. Real-time PCR to determine transgene copy number and to quantitate the biolocalization of adoptively transferred cells from EGFP-transgenic mice. Biotechniques. 2008;45:247–258. doi: 10.2144/000112913. [DOI] [PubMed] [Google Scholar]
- 25.Pillai MM, Venkataraman GM, Kosak S, Torok-Storb B. Integration site analysis in transgenic mice by thermal asymmetric interlaced (TAIL)-PCR: segregating multiple-integrant founder lines and determining zygosity. Transgenic Res. 2008;17:749–754. doi: 10.1007/s11248-007-9161-4. [DOI] [PubMed] [Google Scholar]
- 26.Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 27.Luczak ED, Leinwand LA. Sex-based cardiac physiology. Annu Rev Physiol. 2009;71:1–18. doi: 10.1146/annurev.physiol.010908.163156. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer S, Taylor AL, Lee HR, Niggemann EH, Levine BD, Popma JJ, et al. Effect of increasing heart rate on left ventricular performance in patients with normal cardiac function. Am J Cardiol. 1988;61:617–620. doi: 10.1016/0002-9149(88)90776-x. [DOI] [PubMed] [Google Scholar]
- 29.Higgins CB, Vatner SF, Franklin D, Braunwald E. Extent of regulation of the heart's contractile state in the conscious dog by alteration in the frequency of contraction. 1973;52:1187–1194. doi: 10.1172/JCI107285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villarreal FJ, Dillmann WH. Cardiac hypertrophy-induced changes in mRNA levels for TGF-beta 1, fibronectin, and collagen. Am J Physiol. 1992;262:H1861–H1866. doi: 10.1152/ajpheart.1992.262.6.H1861. [DOI] [PubMed] [Google Scholar]
- 31.Konhilas JP, Watson PA, Maass A, Boucek DM, Horn T, Stauffer BL, et al. Exercise can prevent and reverse the severity of hypertrophic cardiomyopathy. Circulation Research. 2006;98:540–548. doi: 10.1161/01.RES.0000205766.97556.00. [DOI] [PubMed] [Google Scholar]
- 32.Barrick CJ, Dong A, Waikel R, Corn D, Yang F, Threadgill DW, et al. Parent-of-origin effects on cardiac response to pressure overload in mice. Am J Physiol Heart Circ Physiol. 2009;297:H1003–H1009. doi: 10.1152/ajpheart.00896.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clerk A, Sugden PH. Inflame my heart (by p38-MAPK) Circulation Research. 2006;99:455–458. doi: 10.1161/01.RES.0000241053.89089.c3. [DOI] [PubMed] [Google Scholar]
- 34.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 35.Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW. Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Galphaq. Circulation Research. 2010;106:1459–1467. doi: 10.1161/CIRCRESAHA.110.217513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan Z, Baier LD, Morrison AR. p38 Mitogen-activated Protein Kinase Down-regulates Nitric Oxide and Up-regulates Prostaglandin E2 Biosynthesis Stimulated by Interleukin-1β. J Biol Chem. 1997;272:8083–8089. doi: 10.1074/jbc.272.12.8083. [DOI] [PubMed] [Google Scholar]
- 37.Zechner D, Thuerauf DJ, Hanford DS. A role for the p38 mitogen-activated protein kinase pathway in myocardial cell growth, sarcomeric organization, and cardiac-specific gene expression. The Journal of Cell Biology. 1997;139:115–127. doi: 10.1083/jcb.139.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molkentin JD, Lu J-R, Antos CL, Markham B, Richardson J, Robbins J, et al. A Calcineurin-Dependent Transcriptional Pathway for Cardiac Hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu Z, Cang Y, Goff SP. c-Abl tyrosine kinase regulates cardiac growth and development. Pnas. 2010;107:1136–1141. doi: 10.1073/pnas.0913131107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulkey CE, Regmi SD, Magnan RA, Danzo MT, Luther H, Hutchinson AK, et al. The maternal-age-associated risk of congenital heart disease is modifiable. Nature. 2015;520:230–233. doi: 10.1038/nature14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semsarian C, Healey MJ, Fatkin D, Giewat M, Duffy C, Seidman CE, et al. A polymorphic modifier gene alters the hypertrophic response in a murine model of familial hypertrophic cardiomyopathy. Journal of Molecular and Cellular Cardiology. 2001;33:2055–2060. doi: 10.1006/jmcc.2001.1466. [DOI] [PubMed] [Google Scholar]
- 42.Sanford LP, Kallapur S, Ormsby I, Doetschman T. Influence of genetic background on knockout mouse phenotypes. Methods Mol Biol. 2001;158:217–225. doi: 10.1385/1-59259-220-1:217. [DOI] [PubMed] [Google Scholar]
- 43.Bergo MO, Lieu HD, Gavino BJ, Ambroziak P, Otto JC, Casey PJ, et al. On the physiological importance of endoproteolysis of CAAX proteins: heart-specific RCE1 knockout mice develop a lethal cardiomyopathy. J Biol Chem. 2004;279:4729–4736. doi: 10.1074/jbc.M310081200. [DOI] [PubMed] [Google Scholar]
- 44.Cavasin MA, Tao Z, Menon S, Yang X-P. Gender differences in cardiac function during early remodeling after acute myocardial infarction in mice. Life Sci. 2004;75:2181–2192. doi: 10.1016/j.lfs.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 45.Douglas PS, Katz SE, Weinberg EO, Chen MH, Bishop SP, Lorell BH. Hypertrophic remodeling: gender differences in the early response to left ventricular pressure overload. J Am Coll Cardiol. 1998;32:1118–1125. doi: 10.1016/s0735-1097(98)00347-7. [DOI] [PubMed] [Google Scholar]
- 46.Abel ED, Kaulbach HC, Tian R, Hopkins JCA, Duffy J, Doetschman T, et al. Cardiac hypertrophy with preserved contractile function after selective deletion of GLUT4 from the heart. 1999;104:1703–1714. doi: 10.1172/JCI7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rüfer AW, Sauer B. Non-contact positions impose site selectivity on Cre recombinase. Nucl Acids Res. 2002;30:2764–2771. doi: 10.1093/nar/gkf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh G, Tamura J, Suzuki M, Suzuki Y, Ikeda H, Koike M, et al. DNA fragmentation of human infarcted myocardial cells demonstrated by the nick end labeling method and DNA agarose gel electrophoresis. Am J Pathol. 1995;146:1325–1331. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.