Abstract
Macroautophagy (hereafter termed autophagy) is a highly evolutionarily conserved pathway that degrades intracellular components such as damaged organelles in lysosome. Autophagy occurs at low basal levels in virtually all types of cells, which is required for the maintenance of cellular homeostasis. Beclin 1 protein, encoded by the beclin 1 gene, plays a central role in the regulation of autophagy. Beclin 1 primarily functions as a scaffolding protein assembling Beclin 1 interactome to regulate Class III PI3K/VPS34 activity, which in turn, tightly controls autophagy at multiple stages. In addition to autophagy, Beclin 1 participates in the regulation of other biological processes such as endocytosis, apoptosis and phagocytosis. Fine-tuning of Beclin 1 protein levels, intracellular localization and the assembly of its interactome is pivotal for the proper execution of these biological functions. Deregulation of Beclin 1 contributes to the pathogenesis of a variety of human diseases. In this review, we summarize biology of Beclin 1 and its role in human pathology, with an emphasis on heart disease.
Keywords: Autophagy, Beclin 1, gene expression, heart, microRNA, post-translational modification, protein interaction.
INTRODUCTION
Macroautophagy (hereafter termed autophagy) is an evolutionally conserved lysosomal pathway responsible for the turnover of cytoplasmic components such as damaged organelles. In yeast, autophagy is identified to function as a cell self-protective mechanism under conditions of nitrogen starvation by recycling of proteins or organelles [1, 2]. In mammalian cells, this pro-survival function is well preserved. In addition, autophagy gets involved in multiple physiological processes such as development, differentiation and immunity. Deregulation of autophagy contributes to the pathogenesis of many human diseases such as cancer, neurodegeneration, myopathy, viral infection and cardiovascular diseases [3, 4]. The dissection of molecular mechanisms of autophagy and its physiological and pathological roles is largely dependent upon the discovery and characterization of genes involved in autophagy. To date, more than 30 autophagy-related genes (ATG genes) have been identified by yeast genetic screenings [5]. Most of the ATG genes have orthologs in higher eukaryotes. The beclin 1 gene, a mammalian ortholog of yeast ATG6, is the first identified and most extensively studied mammalian gene involved in autophagy. Human beclin 1 gene (BECN1), localized to chromosome 17q21, contains 12 exons and 11 introns. The transcript of BECN1 is 2098 base pairs (bp) in length and comprising a 5' UTR of 120 bp, a coding region of 1353 bp and a 3' UTR of 625 bp [6]. The 60-kDa Beclin 1 protein encoded by BECN1 shares 24% amino acid (AA) identity with yeast ATG6 protein and 31% identity with Arabidopsis ATG6 [7].
Structurally, Beclin 1 is a BH3-only protein consisting of an N-terminal Bcl-2 homology domain 3 (BH3, AA 108-127), a coiled-coil domain (CCD, AA 174-266) and an evolutionarily conserved domain (ECD, AA 244-337), which allows Beclin 1 to bind a number of partners [8-10]. Beclin 1 forms homodimer through CCD domain. Atg14 and UVRAG promote the transition of Beclin 1 homodimer to Beclin 1-Atg14L/UVRAG heterodimer assembly through imperfect interface of Beclin1 coiled-coil domain [11]. The carboxyl-terminal domain of Beclin 1 comprises three β-sheet-α-helix repeats, which is specifically required for autophagy through the targeting of ATG14-Beclin 1-VPS34 complex to the pre-autophagosomal structure. On the other hand, the N-terminal region of Beclin 1 has been shown to be specifically required for vacuolar protein sorting [12]. The tip of a surface loop in Beclin 1 ECD, comprised of three aromatic amino acids, acts as a hydrophobic finger to associate with lipid membrane, consequently results in the deformation of membrane and liposomes [13]. Although implicated in various biological processes, the role of Beclin 1 in autophagy is the best established. Beclin 1 interacts with PI3KC3/VPS34 lipid kinase and modulates its activity. PI3KC3/VPS34 produces phosphatidylinositol-3-phosphate (PI3P), which is a critical regulator for autophagy induction and intracellular membrane trafficking [14]. Beclin 1 tightly controls PI3KC3/VPS34 activity through interaction with its binding partners [15, 16]. Fine-tuning of Beclin 1 protein abundance, intracellular localization and its interaction with other proteins is important to strictly regulate autophagy and some other biological processes. Deregulation of Beclin 1 contributes to the pathophysiology of many types of disorders. In this review, we focus on the transcriptional, post-transcriptional and post-translational regulation of Beclin 1 and its role in heart diseases.
THE BECLIN 1 INTERACTOME
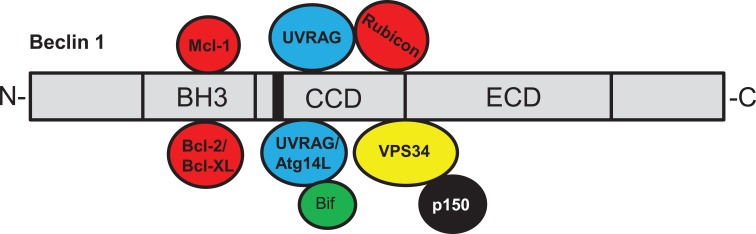
In mammalian cells, Beclin 1 binds PI3KC3/VPS34 to form a Beclin 1- PI3KC3/VPS34 core complex through the CCD and ECD domains and regulates its activity. A number of proteins interact with Beclin 1 and modulate PI3KC3/ VPS34 activity (Fig. 1). Atg14L (also called Barkor) binds Beclin 1-PI3KC3/VPS34, forming Beclin 1-PI3KC3/VPS34-Atg14L complex to enhance PI3KC3/VPS34 activity and in turn, autophagy activation [17]. Ultraviolet irradiation resistance-associated gene (UVRAG), a putative homologue of yeast Vps38, interacts with Beclin 1 through the CCD domain, forming Beclin 1-PI3KC3/VPS34-UVRAG complex, which promotes PI3KC3/VPS34 activity [18, 19]. Phosphorylated Bif is required for UVRAG to enhance PI3KC3/VPS34 activity [20]. Atg14L and UVRAG compete for the interaction with Beclin 1 and exert functions in distinct cellular membrane trafficking steps [18, 21]. Beclin 1-PI3KC3/VPS34-Atg14L stimulates the synthesis of phosphatidylinositol 3-phosphate (PI3P) and induces autophagic initiation, while Beclin 1-PI3KC3/VPS34-UVRAG complex promotes autophagosome maturation and endocytic trafficking [18, 22]. Rubicon interacts with UVRAG and forms the Beclin 1- PI3KC3/VPS34-UVRAG-Rubicon complex, which suppresses autophagosome maturation and endocytic trafficking [23, 24]. Beclin 1-PI3KC3/VPS34-UVRAG has also been suggested to regulate autophagosome formation [19]. The stability of the components in these Beclin 1 complexes is co-dependent upon each other [25, 26]. The activating molecule in Beclin 1-regulated autophagy (Ambra1), a positive regulator of Beclin 1-dependent autophagy, enhances PI3KC3/VPS34 activity by ubiquitinating Beclin 1 [27, 28]. Moreover, it has been reported that the BH3 domain of Beclin 1 interacts with three anti-apoptotic proteins Bcl-2, Bcl-XL and MCL-1, leading to suppression of autophagy [29-31]. The findings that anti-apoptotic proteins inhibit autophagy provide a molecular link between apoptosis and autophagy, two distinct biological processes [15]. Ambra1 and Rhes have been shown to compete with Bcl-2 for the interaction with Beclin 1, leading to activation of autophagy [32, 33].
Fig. (1).
The human Beclin 1 interactome. The Beclin 1 protein consists of a BH3 domain, a CCD domain and an ECD domain. A nuclear export signal is harbored in the CCD domain (aa180-190). Beclin 1 interacts with PI3KC3/VPS34 through the CCD and the ECD domains, resulting in PI3KC3/VPS34 activation, which promotes autophagy, endocytosis and phagocytosis. Bcl-2, Bcl-XL and MCL-1 interact with Beclin 1 through the BH3 domain, which inhibits autophagy and promotes apoptosis. Atg14 and UVRAG compete for the binding to Beclin 1 through the CCD domain and form distinct complexes functioning at different stages of autophagy, with Beclin 1-PI3KC3/VPS34-Atg14L complex promoting autophagosome formation and Beclin 1-PI3KC3/VPS34-UVRAG complex promoting both autophagosome formation and maturation. Bif interacts with Beclin 1 through UVRAG, promoting PI3KC3/VPS34 activity and autophagy. Rubicon binds UVRAG and suppresses autophagosome maturation and endocytic trafficking.
BECLIN 1 PROTEIN ABUNDANCE
Beclin 1 is widely expressed in mammalian cells and numerous studies have demonstrated that Beclin 1 protein abundance is crucial to its biological functions. The beclin 1 gene has initially been shown frequently monoallelically deleted in human breast, ovarian and prostate cancer [34]. Subsequently, beclin 1 knockout mice are generated to study its physiological functions [35, 36]. Beclin 1 heterozygous knockout mice display reduced autophagy in multiple organs and are more susceptible to spontaneous cancers [35]. In addition, Beclin 1 expression is abnormal in multiple types of cancers including breast, prostate, gastric, brain, hepatic, ovarian, colorectal and nasopharyngeal carcinomas, which is correlated with the malignancy and prognosis [37-39]. These studies suggest a tumor suppression function for Beclin 1. Beclin 1 homozygous knockout mice exhibit defective autophagy and are embryonic lethal at E7.5-8.5 [35, 36]. However, mice deficient for other autophagy-related genes such as ATG5 and ATG7 survive until birth [40, 41], suggestive of additional functions beyond autophagy for Beclin 1 during development. Abnormal Beclin 1 protein levels are also observed in human pathology other than cancer such as neurodegeneration and heart disease (see below), which contribute to the pathogenesis of these diseases possibly through deregulation of autophagy, apoptosis and phagocytosis [42-44]. For instance, Beclin 1 is reduced in several neurodegenerative disorders such as Alzheimer’s disease [42]. Over-expression of Beclin 1 ameliorates the pathogenesis in animal models of certain forms of neurodegeneration [43].
INTRACELLULAR LOCALIZATION OF BECLIN 1
Beclin 1 contains a leucine-rich nuclear export signal located within the CCD domain [45]. The nuclear export signal is responsible for the predominant cytoplasmic localization of Beclin 1. In the cytoplasm, Beclin 1 is primarily localized to endoplasmic reticulum, mitochondria, trans-Golgi network and perinuclear membrane [14, 29]. Autophagy is deregulated by intracellular mislocalization of Beclin 1, which alters its interactome. For example, Beclin 1 is sequestered into mutant Huntingtin protein in cultured neuronal cells, transgenic mice models of Huntington’s disease and patients with Huntington disease, leading to impaired autophagic activity and compromised degradation of mutant Huntingtin [46, 47]. Bim, a pro-apoptotic protein, also regulates autophagy by altering Beclin 1 localization. Under physiological conditions, the majority of Bim interact with dynein light chain 1, a subunit of dynein motor complex. Upon cell death stimuli, Bim recruits Beclin 1 to dynein motor complex and suppresses autophagy [48]. The intracellular localization of Beclin 1 is regulated by kinase Akt. Akt-phosphorylated Beclin 1 is more capable of interacting with 14-3-3 adaptor protein and cytoskeletal components vimentin intermediate filament proteins, resulting in suppression of autophagy [49]. Prion protein stimulates autophagy in neurons through regulation of Beclin 1 subcellular localization. In the presence of amyloid β1–42 (Aβ42), prion interacts with Beclin 1 through the BH3 domain and recruits it into lipid rafts in the plasma membrane, where PI3KC3/VPS34 activity is promoted, leading to enhanced autophagy [50].
TRANSCRIPTIONAL REGULATION
Beclin 1 promoter harbors p65/RelA concensus sites and NF-κB positively regulates Beclin 1 expression [51]. E2F is another transcription factor promoting Beclin 1 expression [52]. The signal transducer and activator of transcription 3 (Stat3) directly binds beclin 1 promoter and represses its transcription through recruiting histone deacetylase 3 (HDAC3) in lung cancer cells [53]. Sustained activation of X-box-binding protein 1 (XBP1), a member of basic region/leucine zipper transcription factor family, has a spliced 56 kDa variant, which induces beclin 1 transcription and enhances autophagic response in endothelial cell [54]. HDAC6 has been shown to activate JNK, which in turn, stimulates Beclin 1 expression [55]. In a study conducted by Wang et al. AMPK-MEK/ERK-TSC-mTOR signaling pathway regulates Beclin 1 and autophagy [56]. Li et al. have observed that beclin 1 promoter and/or intron 2 is abnormally methylated in breast cancer, which is associated with down-regulation of Beclin 1 expression, suggesting that Beclin 1 expression is regulated through an epigenetic mechanism [57].
POST-TRANSCRIPTIONAL REGULATION
In recent years, there has been increasing evidence suggesting that microRNAs (miRNAs) regulate Beclin 1 expression. miR-376a, a microRNA from the DLK1/GTL2 gene cluster, blocks Beclin 1 expression under starvation conditions [58]. miR-376b targets Beclin 1 to control starvation and mTOR inhibition-related autophagy [59]. miR-21 reduces Beclin 1 and autophagy induction in Leukemia cells [60]. miR-216b regulates both autophagy and apoptosis by modulating Beclin 1 in hydroxycamptothecin-treated human Tenon’s fibroblasts [61]. miR-17-5p has been indicated to inhibit Beclin 1 expression in lung cancer cells, which is related to paclitaxel resistance [62]. miR-130a regulates Beclin 1 and autophagy in endothelial progenitor cells, which is mediated by Runx3 [63]. microRNA-30a has recently been shown to down-regulate Beclin 1, leading to reduced autophagic activity in cancer cells, cardiomyocytes and neurons [64]. The microRNAs that regulate Beclin 1 expression are summarized in Table 1.
Table 1.
The microRNAs that regulate Beclin 1 expression.
| microRNA | Expression | Function | Reference |
|---|---|---|---|
| miR-376a | MCF-7 and Huh7 cells | reduce autophagy | [58] |
| miR-376b | MCF-7 and Huh7 cells | reduce autophagy | [59] |
| miR-21 | Leukemia cells | reduce autophagy | [60] |
| miR-216b | human Tenon’s fibroblasts | reduce autophagy | [61] |
| miR-17-5p | lung cancer cells | reduce autophagy | [62] |
| miR-130a | endothelial progenitor cells | reduce autophagy | [63] |
| microRNA-30a | cancer cells, cardiomyocytes and neurons | reduce autophagy | [64] |
POST-TRANSLATIONAL REGULATION
Post-translational regulation including phosphorylation, ubiquitination and proteolytic cleavage of Beclin 1 has been shown to modulate PI3KC3/VPS34 activity and in turn, autophagy by altering Beclin 1 protein abundance, intracellular localization and/or its interaction with binding partners.
PHOSPHORYLATION
A number of kinases are able to phosphorylate Beclin 1, which either promotes or suppresses autophagy. Death-associated protein kinase (DAPK) phosphorylates human Beclin 1 Thr-119 residue in the BH3 domain, which dissociates Beclin 1 from Bcl-XL and induces autophagy [65]. ULK1, one of the homologues of yeast ATG1 kinase, is activated in response to starvation and mTOR inhibition. Activated ULK1 phosphorylates mouse Beclin 1 at Ser-14, which enhances PI3KC3/VPS34 activity and promotes autophagy. The ULK1-mediated phosphorylation of Beclin 1 is essential for full autophagic induction in mammalian cells and C. elegans [66]. Fogel et al. have shown that human Beclin 1 is phosphorylated at Ser-90 and Ser-93. Phosphorylation of Beclin 1 on these two sites is Atg14L-dependent and required for complete activation of autophagy [67]. Epidermal growth factor receptor (EGFR), a receptor tyrosine kinase, has been demonstrated to regulate autophagy by phosphorylating Beclin 1. Active EGFR interacts with Beclin 1 through the BH3 and ECD domains, resulting in the phosphorylation of Tyr-229, Tyr-233 and Tyr-352. Multisite tyrosine phosphorylation of Beclin 1 enhances its binding with Bcl-2 and reduces autophagic activation [68]. Beclin 1 is also a target for Akt. Akt phosphorylates human Beclin 1 at Ser-234 and Ser-295, creating binding sites for 14-3-3. Vimentin is in complexes with 14-3-3 and Beclin 1, which inhibits autophagy [48]. AMPK phosphorylates mouse Beclin 1 at Ser-91 and Ser-94, leading to autophagy induction under nutrient stress conditions [69].
UBIQUITINATION
The stability and pro-autophagic activity of Beclin 1 is regulated by ubiquitination. The ubiquitin ligase Nedd4 (neural-precursor-cell-expressed developmentally down-regulated 4) binds Beclin 1 and polyubiquitinates it with Lys11- and Lys63-linked chains. Nedd4-mediated post-translational modification of Beclin 1 with Lys11-linked ubiquitin chains controls its stability [70]. Tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) ubiquitinates Beclin 1 at Lys-117 located in the BH3 domain through K63 linkage and induces autophagy in response to Toll-like receptor 4 (TLR4) signaling in macrophages. A20, a deubiquitinating enzyme, on the contrary, decreases K63-linked Beclin 1 ubiquitination and suppresses autophagy [71]. Beclin 1 is also ubiquitinated at Lys-437 through K63 linkage by Ambra1, an E3 ubiquitin ligase binding to Beclin 1. Ambra1-mediated Beclin 1 ubiquitination is required to promote PI3KC3/VPS34 activity and enhance autophagy. WASH (Wiskott–Aldrich syndrome protein (WASP) and SCAR homologue) suppresses Ambra1-mediated Beclin 1 ubiquitination and inhibits autophagy [28]. USP10 and USP13, two ubiquitin-specific peptidases, stabilize Beclin 1 by targeting Beclin 1 complex for de-ubquitination. Interestingly, Beclin 1 stabilizes USP10 and USP13 by regulating their deubiquitination activity, leading to deubiquitination of tumor suppressor p53, which results in enhanced p53 protein levels [25].
BECLIN 1 CLEAVAGE
Several lines of evidence have shown that caspases and calpain cleave Beclin 1 and link apoptosis to autophagy. Cleavage of Beclin 1 by caspases was first observed by Cho et al. [72]. Subsequently, Zhu et al. show that Beclin 1 is cleaved by caspase 3 at Asp-124 and Asp-149, which abrogates its interaction with Bcl-2. The proteolytic cleavage of Beclin 1 reduces autophagy and enhances apoptosis in Hela cells [73]. Wirawan et al. have identified TDVD133 and DQLD149 as caspase-mediated Beclin 1 cleavage sites in pro-B-cell line Ba/F3. The fragments resulting from cleavage are unable to induce autophagy. However, the C-terminal fragment is translocated to mitochondria membrane, stimulates cytochrome C release and sensitizes the cells to apoptosis [74]. Rohn et al. have demonstrated caspase 3-mediated cleavage of Beclin 1 at Asp-149 and generation of a C-terminal fragment of 35-kDa in the Alzheimer's disease (AD) brain. The proteolytic cleavage is at least partially responsible for the diminished levels of Beclin 1 and autophagy in the AD brain [75]. Li et al. have observed that activated caspase 8 resulting from chemotherapy cleaves Beclin 1 at Asp-133 and Asp-146 in colon cancer cells, leading to autophagy suppression [76, 77]. Recently, the study conducted by You et al. has shown that TRAIL induces Beclin 1 cleavage by caspases in melanoma cell lines deprived of arginine [78]. Active caspase 8 is more capable of cleaving Beclin 1 than other caspases and the mechanisms still remain unclear. It has been shown that calpain cleaves Beclin 1 in retina under ischemia conditions, resulting in the generation of a 50-kDa fragment and down-regulation of autophagy [79]. These studies suggest that autophagy-related protein Beclin 1 promotes the crosstalk between autophagy and apoptosis by proteolytic cleavage. However, the selection of cleavage sites and enzymes for use may be cell type and cell context dependent.
BECLIN 1 IN HEART DISEASE
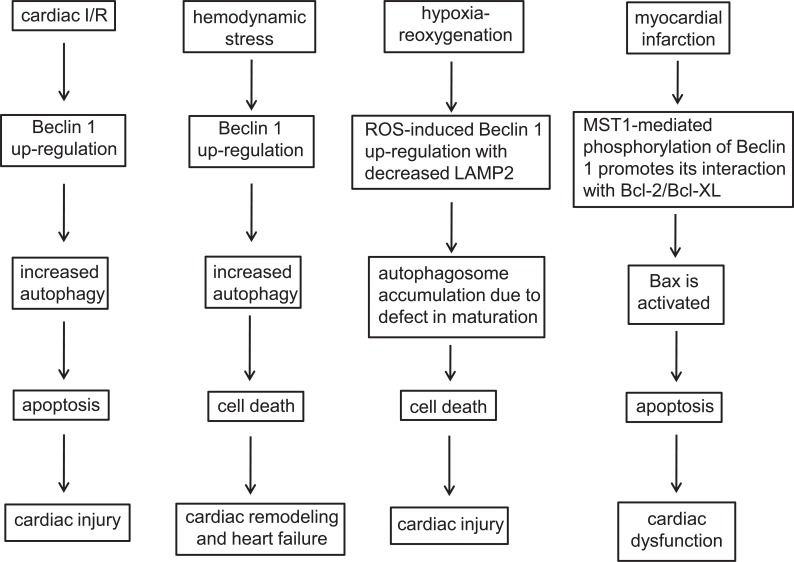
Beclin 1 is aberrantly expressed or post-translationally modified in a number of heart diseases including ischemia-reperfusion, myocardial infarction, cardiac hypertrophy and heart failure. Deregulation of Beclin 1 contributes to the pathogenesis of heart diseases, likely through altered myocardial autophagy and apoptosis. The role of Beclin 1 in heart disease is summarized in (Fig. 3).
Fig. (3).
The role of Beclin 1 in heart disease including cardiac ischemia-reperfusion, myocardial infarction, cardiac hypertrophy and heart failure.
BECLIN 1 IN MYOCARDIAL ISCHEMIA-REPERFUSION
It has been shown that during perfusion phase after cardiac ischemia, Beclin 1 protein levels are elevated and autophagy is enhanced in vivo. Under the identical stress conditions, autophagy is attenuated in the heart from Beclin 1 heterozygous knockout mice, which is associated with decreased apoptosis and amelioration of cardiac injury [80]. This study suggests a detrimental role of Beclin 1 in cardiac reperfusion following ischemia. Valentim et al. have shown that Beclin 1 knockdown reduces autophagy, which is associated with increased cell survival in cultured cardiomyocytes. In addition, urocortin inhibits cell death partially mediated by Beclin 1 reduction during ischemia-reperfusion [81]. Recently, Xu et al. find that in cardiac hypoxia-reoxygenation injury, reactive oxygen species (ROS)-induces Beclin 1 up-regulation accompanied by decreased LAMP2, leading to defect in autophagosome maturation and enhanced cell death. Remarkably, autophagosome clearance is recovered and cell death is attenuated by partial reduction in Beclin 1 coupled with restoration of LAMP2 after hypoxia-reoxygenation [82]. These data suggest that autophagosome accumulation due to impaired autophagosome clearance contributes to cardiac injury during hypoxia-reoxygenation. Zeng et al. have shown that in a rabbit model of ischemia reperfusion, NF-κB activity is enhanced, which promotes Beclin 1 expression and autophagy, leading to myocardial injury [83]. Taken together, these studies demonstrate that Beclin 1 plays a pivotal role in cardiac injury during ischemia-reperfusion, which involves several distinct mechanisms depending upon the experimental models and cellular contexts.
BECLIN 1 IN CARDIAC HYPERTROPHY AND HEART FAILURE
Beclin 1 has also been shown to play an important role in cardiac hypertrophy and heart failure. In severe pressure overload-induced heart failure, Beclin 1 protein abundance and autophagy are increased in the heart. Beclin 1 heterozygous knockout mice show improved pressure overload-induced cardiac dysfunction, which is associated with attenuated autophagy. Conversely, Beclin 1 transgenic mice with cardiac specific over-expression of Beclin 1 exhibit more pronounced cardiac remodeling and systolic dysfunction in response to moderate pressure overload stress, which is associated with enhanced autophagy [84]. These results suggest that Beclin 1 contributes to the transition from hypertrophy to heart failure. Consistently, in a study conducted by Lin et al. activating transcription factor 3 (ATF3), a member of cAMP response element-binding protein/ATF family transcription factors, negatively regulates Beclin 1 expression in cardiomyocytes. ATF3 knockout mice show exacerbated heart failure in response to pressure overload. Mechanistically, ATF3 binds to the ATF/cAMP element of beclin 1 promoter and suppresses Beclin 1 expression and autophagy [85]. These results suggest a detrimental role of enhanced Beclin 1 levels in pressure-overloaded heart failure possibly through regulation of autophagy. Pan et al. [86] showed that beclin 1 gene is a target of miR-30a in cardiomyocytes. miR-30a mimic attenuates angiotensin II-induced cardiomyocyte hypertrophy, while miR-30a inhibitor promotes angiotensin II-induced cardiomyocyte hypertrophy. Intriguingly, circulating miR-30a is elevated in patients with left ventricular hypertrophy and positively correlated with left ventricular wall thickness. Therefore, miR-30a may contribute to the development of cardiac hypertrophy through regulating Beclin 1 expression and circulating miR-30a is implicated as a biomarker for left ventricular hypertrophy. Collectively, Beclin 1 contributes to the pathogenesis of cardiac hypertrophy and heart failure at least partially through regulation of autophagy. Tert-butylhydroquinone (tBHQ), an ATF3 inducer can be potentially used in the treatment of pressure overload-induced heart failure [86]. In addition, cardiac gene transfer of miR-30a expressing vector or miR-30a mimics may serve as therapeutic strategies for pressure-overloaded cardiac hypertrophy and heart failure.
BECLIN 1 IN MYOCARDIAL INFARCTION
Maejima et al. have recently shown that activated pro-apoptotic kinase mammalian Sterile 20-like kinase 1 (MST1) phosphorylates Beclin 1 on Thr108 located within the BH3 domain. MST1-mediated Beclin 1 phosphorylation promotes its interaction with Bcl-2/Bcl-XL and suppresses PI3KC3/VPS34 activity and autophagy in cardiomyocytes. What’s more, Bax is activated as a result of enhanced interaction between phosphorylated Beclin 1 and Bcl-2/Bcl-XL, which induces apoptosis. In a mouse model of myocardial infarction, cardiac dysfunction is exacerbated by MST1-mediated suppression of autophagy, which is associated with increased phosphorylation of Beclin 1 at Thr108. Interestingly, in the failing hearts from patients with end-stage dilated cadiomyopathy, Beclin 1 with Thr108-phosphorylation is increased, which is associated with autophagy suppression. These observations demonstrate that Mst1-mediated Beclin 1 phosphorylation contributes to cardiac dysfunction in myocardial infarction [87]. Beclin 1-Bcl-2 interaction governs autophagy levels and links autophagy to apoptosis. Thus, fine-tuning of cardiomyocyte autophagy by regulating Beclin 1 and Bcl-2 interaction is a strategy in the treatment of heart failure. As a regulator of Beclin 1and Bcl-2 interaction, Mst1 is a potential therapeutic target and Mst1 inhibitors such as 9E1 [88], have the potential application in the treatment of certain forms of heart failure.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
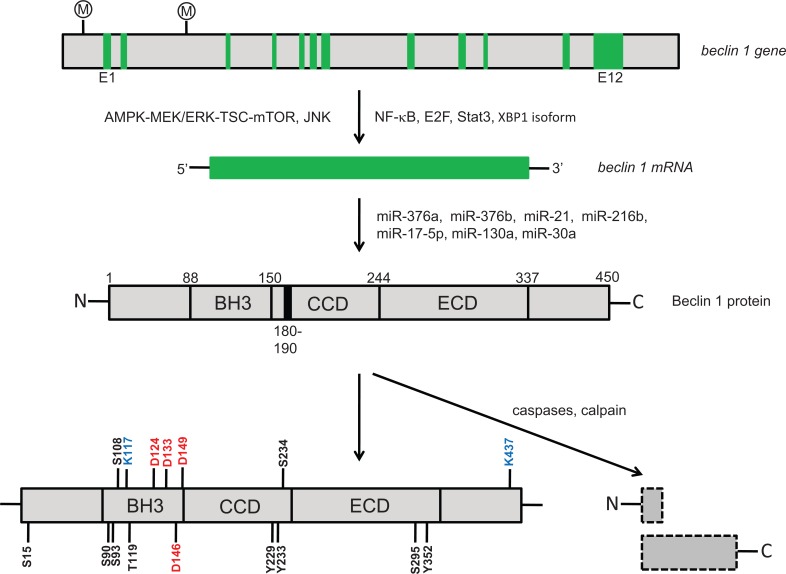
Fig. (2).
Transcriptional, post-transcriptional and post-translational regulation of Beclin 1. Transcription factor including NF-κB, E2F, Stat3 as well as XBP1 isoform and signaling pathways such as AMPK-MEK/ERK-TSC-mTOR and JNK regulate beclin 1 gene transcription. miR- 376a, miR-376b, miR-21, miR-216b, miR-17-5p, miR-130a and miR-30a regulate beclin 1 gene expression at the post-transcriptional level. Human Beclin 1 protein is phosphorylated at S15 (ULK1), S90 (AMPK), S93 (AMPK), S108 (MST1), T119 (DAPK), Y229 (EGFR), Y233 (EGFR), S234 (Akt), S295 (Akt) and Y352 (EGFR) by the kinases indicated in the bracket. TRAF6 and Ambra1 ubiquitinate Beclin 1 protein at K117 and K437, respectively. Beclin 1 protein is cleavable by caspases at D124, D133, D146 and D149, generating an N-terminal and a C-terminal fragments. The C-terminal fragment resulting from cleavage at Asp-133 and Asp-149 has been demonstrated to promote apoptosis. Beclin 1 is also cleaved by calpain. S: Ser; T: Thr; Y: Tyr; D: Asp; K: Lys. Ⓜ: methylation.
ACKNOWLEDGEMENTS
Declared none.
ABBREVIATIONS
- UTR =
untranslated region
- bp =
base pair
- kDa =
kilodaltons
- VPS34 =
vacuolar protein sorting 34
- PI3KC3 =
Class III phosphatidylinositol 3-kinase
- ATG gene =
autophagy-related gene
- UVRAG =
ultraviolet irradiation resistance-associated gene
- ULK1 =
Unc-51-like kinase 1
- Thr =
threonine
- Ser =
serine
- Tyr =
tyrosine
- Lys =
lysine
- Asp =
aspartic acid
- USP =
ubiquitin-specific protease
- AD =
Alzheimer’s disease
- NF-κB =
nuclear factor-κB
- JNK =
c-Jun N-terminal kinase
- RUNX3 =
runt-related transcription factor 3
- TRAIL =
TNF-related apoptosis-inducing ligand
- AMPK =
adenosine monophosphate-activated protein kinase
- DAPK =
death-associated protein kinase
- EGFR =
the epidermal growth factor receptor
- LAMP2 =
lysosome-associated membrane protein 2
- Bif =
Bax interacting factor 1
- PI3P =
phosphatidylinositol 3-phosphate
- MCL-1 =
myeloid cell leukemia 1
- Bcl-2 =
B cell lymphoma 2
- Bcl-XL =
B cell lymphoma-extra large
- Bim =
Bcl-2 interacting mediator of apoptosis
- miR =
microRNA
- MST1 =
mammalian Sterile 20-like kinase 1
- TSC =
tuberous sclerosis complex
- mTOR =
mammalian target of rapamycin
References
- 1.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yorimitsu T., Klionsky D.J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl. 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arroyo D.S., Gaviglio E.A., Peralta Ramos J.M., Bussi C., Rodriguez-Galan M.C., Iribarren P. Autophagy in inflammation, infection, neurodegeneration and cancer. Int. Immunopharmacol. 2014;18(1):55–65. doi: 10.1016/j.intimp.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi A.M., Ryter S.W., Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368(19):1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 5.Klionsky D.J., Cregg J.M., Dunn W.A., Jr, Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5(4):539–545. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 6.Aita V.M., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59(1):59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 7.Fujiki Y., Yoshimoto K., Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143(3):1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oberstein A., Jeffrey P.D., Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007;282(17):13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 9.Li X., He L., Che K.H., Funderburk S.F., Pan L., Pan N., Zhang M., Yue Z., Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuya N., Yu J., Byfield M., Pattingre S., Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1(1):46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 11.Li X., He L., Che K.H., Funderburk S.F., Pan L., Pan N., Zhang M., Yue Z., Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang W., Choi W., Hu W., Mi N., Guo Q., Ma M., Liu M., Tian Y., Lu P., Wang F.L., Deng H., Liu L., Gao N., Yu L., Shi Y. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22(3):473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noda N.N., Kobayashi T., Adachi W., Fujioka Y., Ohsumi Y., Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J. Biol. Chem. 2012;287(20):16256–16266. doi: 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2(4):330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He C., Levine B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010;22(2):140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B.H., Jung J.U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8(7):688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y., Coppola D., Matsushita N., Cualing H.D., Sun M., Sato Y., Liang C., Jung J.U., Cheng J.Q., Mulé J.J., Pledger W.J., Wang H.G. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itakura E., Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5(4):534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 22.Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10(7):776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong Y., Wang Q.J., Li X., Yan Y., Backer J.M., Chait B.T., Heintz N., Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Xia H., Kim M., Xu L., Li Y., Zhang L., Cai Y., Norberg H.V., Zhang T., Furuya T., Jin M., Zhu Z., Wang H., Yu J., Li Y., Hao Y., Choi A., Ke H., Ma D., Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Z., An L., Ye Y., Wu J., Zou Y., He L., Zhu H. Essential role for UVRAG in autophagy and maintenance of cardiac function. Cardiovasc. Res. 2014;101(1):48–56. doi: 10.1093/cvr/cvt223. [DOI] [PubMed] [Google Scholar]
- 27.Fimia G.M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., Gruss P., Piacentini M., Chowdhury K., Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 28.Xia P., Wang S., Du Y., Zhao Z., Shi L., Sun L., Huang G., Ye B., Li C., Dai Z., Hou N., Cheng X., Sun Q., Li L., Yang X., Fan Z. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32(20):2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Maiuri M.C., Le Toumelin G., Criollo A., Rain J.C., Gautier F., Juin P., Tasdemir E., Pierron G., Troulinaki K., Tavernarakis N., Hickman J.A., Geneste O., Kroemer G. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26(10):2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Germain M., Nguyen A.P., Le Grand J.N., Arbour N., Vanderluit J.L., Park D.S., Opferman J.T., Slack R.S. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J. 2011;30(2):395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mealer R.G., Murray A.J., Shahani N., Subramaniam S., Snyder S.H. Rhes, a striatal-selective protein implicated in Huntington disease, binds beclin-1 and activates autophagy. J. Biol. Chem. 2014;289(6):3547–3554. doi: 10.1074/jbc.M113.536912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strappazzon F., Vietri-Rudan M., Campello S., Nazio F., Florenzano F., Fimia G.M., Piacentini M., Levine B., Cecconi F. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30(7):1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 35.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou W.H., Tang F., Xu J., Wu X., Yang S.B., Feng Z.Y., Ding Y.G., Wan X.B., Guan Z., Li H.G., Lin D.J., Shao C.K., Liu Q. Low expression of Beclin 1, associated with high Bcl-xL, predicts a malignant phenotype and poor prognosis of gastric cancer. Autophagy. 2012;8(3):389–400. doi: 10.4161/auto.18641. [DOI] [PubMed] [Google Scholar]
- 38.Nicotra G., Mercalli F., Peracchio C., Castino R., Follo C., Valente G., Isidoro C. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Mod. Pathol. 2010;23(7):937–950. doi: 10.1038/modpathol.2010.80. [DOI] [PubMed] [Google Scholar]
- 39.Fu L.L., Cheng Y., Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int. J. Biochem. Cell Biol. 2013;45(5):921–924. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 41.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickford F., Masliah E., Britschgi M., Lucin K., Narasimhan R., Jaeger P.A., Small S., Spencer B., Rockenstein E., Levine B., Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J. Clin. Invest. 2008;118(6):2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nascimento-Ferreira I., Nóbrega C., Vasconcelos-Ferreira A., Onofre I., Albuquerque D., Aveleira C., Hirai H., Déglon N., Pereira de Almeida L. Beclin 1 mitigates motor and neuropathological deficits in genetic mouse models of Machado-Joseph disease. Brain. 2013;136(Pt 7):2173–2188. doi: 10.1093/brain/awt144. [DOI] [PubMed] [Google Scholar]
- 44.Lucin K.M., O’Brien C.E., Bieri G., Czirr E., Mosher K.I., Abbey R.J., Mastroeni D.F., Rogers J., Spencer B., Masliah E., Wyss-Coray T. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer’s disease. Neuron. 2013;79(5):873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang X.H., Yu J., Brown K., Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61(8):3443–3449. [PubMed] [Google Scholar]
- 46.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281(20):14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 47.Salminen A., Kaarniranta K., Kauppinen A., Ojala J., Haapasalo A., Soininen H., Hiltunen M. Impaired autophagy and APP processing in Alzheimer’s disease: The potential role of Beclin 1 interactome. Prog. Neurobiol. 2013;106-107:33–54. doi: 10.1016/j.pneurobio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Luo S., Garcia-Arencibia M., Zhao R., Puri C., Toh P.P., Sadiq O., Rubinsztein D.C. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol. Cell. 2012;47(3):359–370. doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang R.C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., White M., Reichelt J., Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338(6109):956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nah J., Pyo J.O., Jung S., Yoo S.M., Kam T.I., Chang J., Han J., Soo A An S., Onodera T., Jung Y.K. BECN1/Beclin 1 is recruited into lipid rafts by prion to activate autophagy in response to amyloid β 42. Autophagy. 2013;9(12):2009–2021. doi: 10.4161/auto.26118. [DOI] [PubMed] [Google Scholar]
- 51.Copetti T., Demarchi F., Schneider C. p65/RelA binds and activates the beclin 1 promoter. Autophagy. 2009;5(6):858–859. doi: 10.4161/auto.8822. [DOI] [PubMed] [Google Scholar]
- 52.Weinmann A.S., Bartley S.M., Zhang T., Zhang M.Q., Farnham P.J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 2001;21(20):6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao L.J., Huang F.X., Sun Z.T., Zhang R.X., Huang S.F., Wang J. Stat3 inhibits Beclin 1 expression through recruitment of HDAC3 in nonsmall cell lung cancer cells. Tumour Biol. 2014;35(7):7097–7103. doi: 10.1007/s13277-014-1961-6. [DOI] [PubMed] [Google Scholar]
- 54.Margariti A., Li H., Chen T., Martin D., Vizcay-Barrena G., Alam S., Karamariti E., Xiao Q., Zampetaki A., Zhang Z., Wang W., Jiang Z., Gao C., Ma B., Chen Y.G., Cockerill G., Hu Y., Xu Q., Zeng L. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J. Biol. Chem. 2013;288(2):859–872. doi: 10.1074/jbc.M112.412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung K.H., Noh J.H., Kim J.K., Eun J.W., Bae H.J., Chang Y.G., Kim M.G., Park W.S., Lee J.Y., Lee S.Y., Chu I.S., Nam S.W. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56(2):644–657. doi: 10.1002/hep.25699. [DOI] [PubMed] [Google Scholar]
- 56.Wang J., Whiteman M.W., Lian H., et al. Regulating Beclin 1 Pathway Regulates Autophagy via a non-canonical MEK/ERK signaling. J. Biol. Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z., Chen B., Wu Y., Jin F., Xia Y., Liu X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. doi: 10.1186/1471-2407-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korkmaz G., Tekirdag K.A., Ozturk D.G., Kosar A., Sezerman O.U., Gozuacik D. MIR376A is a regulator of starvation-induced autophagy. PLoS One. 2013;8(12):e82556. doi: 10.1371/journal.pone.0082556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korkmaz G., le Sage C., Tekirdag K.A., Agami R., Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8(2):165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 60.Seca H., Lima R.T., Lopes-Rodrigues V., Guimaraes J.E., Almeida G.M., Vasconcelos M.H. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr. Drug Targets. 2013;14(10):1135–1143. doi: 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]
- 61.Xu X., Fu Y., Tong J., Fan S., Xu K., Sun H., Liang Y., Yan C., Yuan Z., Ge Y. MicroRNA-216b/Beclin 1 axis regulates autophagy and apoptosis in human Tenon’s capsule fibroblasts upon hydroxycamptothecin exposure. Exp. Eye Res. 2014;123:43–55. doi: 10.1016/j.exer.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 62.Chatterjee A., Chattopadhyay D., Chakrabarti G. miR-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. PLoS One. 2014;9(4):e95716. doi: 10.1371/journal.pone.0095716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu Q., Meng S., Liu B., Li M.Q., Li Y., Fang L., Li Y.G. MicroRNA-130a regulates autophagy of endothelial progenitor cells through Runx3. Clin. Exp. Pharmacol. Physiol. 2014;41(5):351–357. doi: 10.1111/1440-1681.12227. [DOI] [PubMed] [Google Scholar]
- 64.Wang P., Liang J., Li Y., Li J., Yang X., Zhang X., Han S., Li S., Li J. Down-regulation of miRNA-30a alleviates cerebral ischemic injury through enhancing beclin 1-mediated autophagy. Neurochem. Res. 2014;39(7):1279–1291. doi: 10.1007/s11064-014-1310-6. [DOI] [PubMed] [Google Scholar]
- 65.Zalckvar E., Berissi H., Mizrachy L., Idelchuk Y., Koren I., Eisenstein M., Sabanay H., Pinkas-Kramarski R., Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10(3):285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.Y., Kim J., Kim H., Neufeld T.P., Dillin A., Guan K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fogel A.I., Dlouhy B.J., Wang C., Ryu S.W., Neutzner A., Hasson S.A., Sideris D.P., Abeliovich H., Youle R.J. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell. Biol. 2013;33(18):3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei Y., Zou Z., Becker N., Anderson M., Sumpter R., Xiao G., Kinch L., Koduru P., Christudass C.S., Veltri R.W., Grishin N.V., Peyton M., Minna J., Bhagat G., Levine B. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154(6):1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J., Kim Y.C., Fang C., Russell R.C., Kim J.H., Fan W., Liu R., Zhong Q., Guan K.L. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1-2):290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Platta H.W., Abrahamsen H., Thoresen S.B., Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012;441(1):399–406. doi: 10.1042/BJ20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi C.S., Kehrl J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010;3(123):ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho D.H., Jo Y.K., Hwang J.J., Lee Y.M., Roh S.A., Kim J.C. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009;274(1):95–100. doi: 10.1016/j.canlet.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 73.Zhu Y., Zhao L., Liu L., Gao P., Tian W., Wang X., Jin H., Xu H., Chen Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1(5):468–477. doi: 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wirawan E., Vande Walle L., Kersse K., Cornelis S., Claerhout S., Vanoverberghe I., Roelandt R., De Rycke R., Verspurten J., Declercq W., Agostinis P., Vanden Berghe T., Lippens S., Vandenabeele P. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rohn T.T., Wirawan E., Brown R.J., Harris J.R., Masliah E., Vandenabeele P. Depletion of Beclin-1 due to proteolytic cleavage by caspases in the Alzheimer’s disease brain. Neurobiol. Dis. 2011;43(1):68–78. doi: 10.1016/j.nbd.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Wang P., Sun Q., Ding W.X., Yin X.M., Sobol R.W., Stolz D.B., Yu J., Zhang L. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res. 2011;71(10):3625–3634. doi: 10.1158/0008-5472.CAN-10-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H., Wang P., Yu J., Zhang L. Cleaving Beclin 1 to suppress autophagy in chemotherapy-induced apoptosis. Autophagy. 2011;7(10):1239–1241. doi: 10.4161/auto.7.10.16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You M., Savaraj N., Kuo M.T., Wangpaichitr M., Varona-Santos J., Wu C., Nguyen D.M., Feun L. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Mol. Cell. Biochem. 2013;374(1-2):181–190. doi: 10.1007/s11010-012-1518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Russo R., Berliocchi L., Adornetto A., Varano G.P., Cavaliere F., Nucci C., Rotiroti D., Morrone L.A., Bagetta G., Corasaniti M.T. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2:e144. doi: 10.1038/cddis.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matsui Y., Takagi H., Qu X., Abdellatif M., Sakoda H., Asano T., Levine B., Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 2007;100(6):914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 81.Valentim L., Laurence K.M., Townsend P.A., Carroll C.J., Soond S., Scarabelli T.M., Knight R.A., Latchman D.S., Stephanou A. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J. Mol. Cell. Cardiol. 2006;40(6):846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 82.Ma X., Liu H., Foyil S.R., Godar R.J., Weinheimer C.J., Hill J.A., Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125(25):3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeng M., Wei X., Wu Z., Li W., Li B., Zhen Y., Chen J., Wang P., Fei Y. NF-κB-mediated induction of autophagy in cardiac ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2013;436(2):180–185. doi: 10.1016/j.bbrc.2013.05.070. [DOI] [PubMed] [Google Scholar]
- 84.Zhu H., Tannous P., Johnstone J.L., Kong Y., Shelton J.M., Richardson J.A., Le V., Levine B., Rothermel B.A., Hill J.A. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Invest. 2007;117(7):1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin H., Li H.F., Chen H.H., Lai P.F., Juan S.H., Chen J.J., Cheng C.F. Activating transcription factor 3 protects against pressure-overload heart failure via the autophagy molecule Beclin-1 pathway. Mol. Pharmacol. 2014;85(5):682–691. doi: 10.1124/mol.113.090092. [DOI] [PubMed] [Google Scholar]
- 86.Pan W., Zhong Y., Cheng C., Liu B., Wang L., Li A., Xiong L., Liu S. MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy. PLoS One. 2013;8(1):e53950. doi: 10.1371/journal.pone.0053950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maejima Y., Kyoi S., Zhai P., Liu T., Li H., Ivessa A., Sciarretta S., Del Re D.P., Zablocki D.K., Hsu C.P., Lim D.S., Isobe M., Sadoshima J. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat. Med. 2013;19(11):1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anand R., Maksimoska J., Pagano N., Wong E.Y., Gimotty P.A., Diamond S.L., Meggers E., Marmorstein R. Toward the development of a potent and selective organoruthenium mammalian sterile 20 kinase inhibitor. J. Med. Chem. 2009;52(6):1602–1611. doi: 10.1021/jm8005806. [DOI] [PMC free article] [PubMed] [Google Scholar]