SUMMARY
cAMP-dependent protein kinase (PKA) is regulated in part by N-terminal myristylation of its catalytic (C) subunit. Structural information about the role of myristylation in membrane targeting of PKA has been limited. In mammalian cells there are four functionally non-redundant PKA Regulatory subunits (RIα, RIβ, RIIα, and RIIβ). PKA is assembled as an inactive R2C2 holoenzyme in cells. To explore the role of N-myristylation in membrane targeting of PKA holoenzymes, we solved crystal structures of RIα:myrC and RIIβ2:myrC2 and showed that the N-terminal myristylation site in the myrC serves as a flexible “switch” that can potentially be mobilized for membrane anchoring of RII, but not RI, holoenzymes. Furthermore, we synthesized nanodiscs and showed by electron microscopy that membrane targeting through the myristic acid is specific for the RII holoenzyme. This membrane anchoring myristylation switch is independent of A Kinase Anchoring Proteins (AKAPs) that target PKA to membranes by other mechanisms.
INTRODUCTION
Over 500 eukaryotic protein kinases are encoded by the human genome (Manning et al., 2002), and the activity and localization of most are highly regulated by post- and co-translational modifications as well as by adjacent domains and motifs. While the role of phosphorylation in kinase activation has been extensively studied, the mechanistic details of acylation and membrane anchoring is less clear. Myristylation, the irreversible enzymatic attachment of myristic acid onto the N-terminal glycine of target proteins (Farazi et al., 2001; Li et al., 2007; Song et al., 1997), enhances the target's association with membranes (Resh, 1994). Myristylation typically occurs co-translationally although it can also occur in conjunction with caspase cleavage (Harada et al., 1999). The PKA catalytic (C) subunit was the first myristylated protein to be identified (Carr et al., 1982), and most non-receptor tyrosine kinases are also N-myristylated. In the free myristylated subunit (myrC) the acyl group is docked into a hydrophobic allosteric pocket that penetrates into the large lobe of the conserved kinase core (Bastidas et al., 2012; Bossemeyer et al., 1993), and myrC does not strongly associate with membranes. In cells, however, PKA exists not as free C-subunit but as an inactive holoenzyme with regulatory (R) subunit dimers that are bound to two C-subunits. Activation of the R2C2 holoenzyme is mediated by cAMP binding to the R-dimer causing a conformational change that unleashes the catalytic activity, but does not necessarily cause the C-subunits to dissociate (Smith et al., 2013; Vigil et al., 2004). Early fluorescence studies showed that the N-terminus of myrC can become accessible to membranes when it is part of a holoenzyme complex but did not address if this phenomenon was isoform specific (Gangal et al., 1999).
In mammalian cells there are four functionally non-redundant R-subunits (RIα, RIβ, RIIα, and RIIβ), which contribute significantly to PKA signaling specificity (Amieux et al., 2002). In general, RII subunits are targeted at membranes in close proximity to receptors, transporters and channels while RI subunits tend to be more cytosolic and dynamically recruited to membranes (Taylor et al., 2012). RII subunits typically bind with high affinity (low nM) to A Kinase Anchoring Proteins (AKAPs) that also recruit other signaling proteins such as phosphatases and phosphodiesterases to membranes (Taylor et al., 2012). For PKA it is essential that we now think in terms of these “signaolsomes” rather than free R and C-subunits. Here we ask whether myristylation also contributes to membrane anchoring and if so whether this anchoring is isoform-specific. All PKA R-subunits share the same domain organization: an N-terminal Dimerization/Docking (D/D) domain followed by a flexible linker containing an inhibitor site and two conserved cyclic nucleotide binding (CNB) domains (Canaves and Taylor, 2002). The recently solved RIα, RIβ, and RIIβ tetrameric holoenzyme structures show that the quaternary structures of each holoenzyme are distinct (Boettcher et al., 2011; Ilouz et al., 2012; Zhang et al., 2012), and the quaternary constraints imposed by the holoenzyme create somewhat different allosteric mechanisms for activation (Taylor et al., 2012). Given our new appreciation of the importance of the full-length PKA holoenzymes and their ability to nucleate targeted “signalosomes”, we revisited the role of myristylation.
The C-subunit has a conserved core consisting of an N- and C-lobe that is shared by all eukaryotic protein kinases. The conserved core is flanked by N- and C-terminal tails (N-tail and C-tail). While the C-tail is a highly conserved feature of all PKA C-subunits and of every AGC kinase, the N-tail (residues 1–39) is variable even within different mammalian PKA isoforms (Soberg et al., 2013). Residues 1-13 correspond to the first exon and constitute a myristylation motif (Myr motif), which includes the N-terminal Gly as well as basic amino acids that are important for strengthening the membrane anchoring potential. Most C-subunit isoforms are myristylated but some are not and a recent C-subunit in liver cancer was shown to be a fusion protein where the first 14 residues (Exon 1) are replaced with the J domain of DNAJB1 (Honeyman et al., 2014). A dominant feature of the N-Tail (Exons 1 and 2) is a long amphipathic A-helix, which in the active kinase is anchored through its hydrophobic surface to both lobes of the kinase core (Herberg et al., 1997). This N-Tail is rich in signaling information (Figure 1). The A-Helix binds through its hydrophilic surface to another protein, the A-kinase interacting protein (AKIP) that is important for PKA shuttling between the cytoplasm and the nucleus (Sastri et al., 2005). The N-tail also harbors numerous co- and post-translational modifications that, in addition to myristylation, include deamidation of Asn2 and phosphorylation of Ser10, and these modifications can contribute in a variety of ways to PKA function (Bastidas et al., 2012; Bastidas et al., 2013; Gangal et al., 1999; Pepperkok et al., 2000; Tholey et al., 2001b). Here we focus on myristylation.
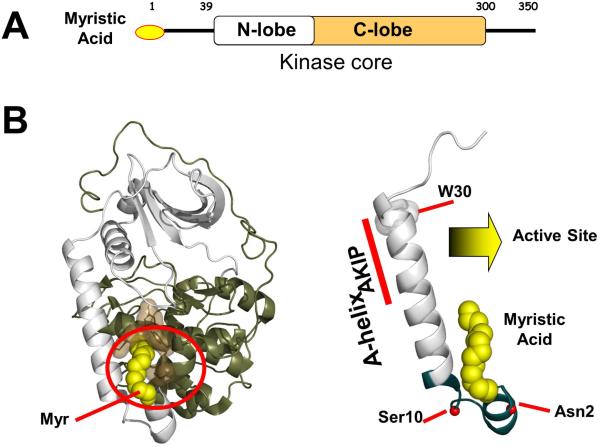
Figure 1. Domain organization and structure of PKA myrC-subunit.
(A)Domain organization of myrC-subunit. (B) The myristic acid (myr) is docked into a hydrophobic pocket of the myrC-subunit (left) (PDB ID: 4DG2). The A-Helix at the N-terminus of C-subunit (right) can serve as an AKIP binding site and harbors numerous co- and post-translational modifications. The Myr motif is shown in teal.
Our structural knowledge about the role of myristylation in membrane targeting of PKA has so far been limited to the free myrC and complexes with nucleotide and/or small peptides (IP20 or SP20) (Bastidas et al., 2012; Bossemeyer et al., 1993). Based on fluorescence anisotropy in solution, Johnson predicted earlier that association with membranes was enhanced in the holoenzyme and that membrane association would be isoform-specific (Gangal et al., 1999). In this study, we further elucidate the role of N-myristylation in membrane targeting of PKA holoenzymes by crystallizing myrC with RIα and RIIβ. We specifically solved crystal structures of an RIα heterodimer (RIα:myrC) and a full length RIIβ holoenzyme (RIIβ2:myrC2). The RIα:myrC heterodimer shows that the Myr motif is sequestered in the acyl pocket similar to myrC. By contrast, the entire Myr motif is expelled from the acyl pocket in the RIIβ complex and is not visible, presumably due to its flexibility. To confirm that this isoform-specific conformational switch also facilitates binding to membranes, we synthesized nanodiscs, and used them to show by gel filtration and electron microscopy that membrane targeting through the myristic acid is specifically associated with the RIIβ holoenzyme. In addition, the C-terminus of the RIIβ holoenzyme, but not the RIα holoenzyme, is flexible. The C-terminus of each R-subunit provides a helical capping motif for the bound cAMP in the activation loop. In the holoenzymes this capping motif is in close proximity to the N-terminal Myr motif in myrC. In RIα the capping motif is stabilized in an inactive conformation whereas in the RIIβ holoenzyme the C-terminal capping motif is also disordered. Together the results define a bivalent conformational membrane-targeting myristylation switch that is mobilized specifically in PKA type II holoenzymes and is independent of AKAPs. This is similar to the calcium mobilized myristylation switch in recoverin and the IP3 induced myristylation switch in HIV-gag. The nearby disordered capping motif at the C-terminus of the RIIβ-subunit provides additional opportunities for protein:membrane interactions at this interface and opens the potential for interactions with other acyl groups that target myristylated-, farnisylated-, and palmitylated-proteins to membranes.
RESULTS
RIα:myrC heterodimer structure
To better define the myristylation site in an RIα complex we crystallized an R:C heterodimer of myrC with a deletion mutant of RIα. As explained in the methods two mutations were introduced. In the deletion mutant of RIα, RIα(91-379), Arg333 in the CNB-B domain was replaced with Lys as previously described (Kim et al., 2007). For myrC we used the K7C mutant because it expressed at higher levels. The crystal structure of the RIα:myrC was solved at 2.4 Å (Figure 2A and Table 1). In the RIα:myrC structure the myristylated N-terminus is well ordered (Figure 2B), while in the same heterodimer structure with the non-myristylated C-subunit the first 12 residues are disordered (Kim et al., 2007) (PDB ID: 2QCS). This is similar to the previous observation that myristylation stabilizes the N-terminal residues of the myrC-subunit no matter whether the myrC-subunit is alone or bound to a small PKA substrate peptide SP20 (Bastidas et al., 2012) or to a high affinity inhibitor peptide (IP20). Specifically the RIα:myrC structure reveals a conformation that includes the entire N-terminus as well as the myristic acid and the hydrophobic side chains that line the acyl pocket; all elements are well resolved in the electron density (Figure 2B). As in the RIα(91-379) complex with the non myristylated C-subunit, both CNB domains are locked into stable inactive conformations where the helical motif that provides a capping residue for the bond cAMP is locked into an inactive conformation.
Figure 2. Crystal structure of RIα:myrC (PDB ID: 4X6R) showing docked myristic acid.
(A) Side view of the RIα:myrC structure. The myristyl acid is shown in yellow. The N-lobe of the C-subunit is white; the C-lobe and C-tail are tan. R-subunit is cyan. (B) Electron density 2Fo-Fc map of the myristic acid and N-terminus of C-subunit at 1.0 sigma level. (C) The myristic acid binds within its previously defined hydrophobic pocket. (D) Overlay of the RIα:myrC (teal, myristic acid is shown in yellow) and myrC:SP20 (tan, myristic acid is shown in sand). These two structures display different conformations at the N-terminus of the C-subunit.
Table 1.
Data Collection and Refinement Statistics
| Data Set | RIα:myrC (PDB ID:4X6R) |
RIIβ2:myrC2 (PDB ID: 4X6Q) |
|---|---|---|
| Data collection | ALS beamline 8.2.1 | ALS beamline 8.2.1 |
| Space group | P 32 2 1 | C222 |
| No. of molecules in one asymmetric unit |
2 | 1/2 |
| Cell constants | ||
| a (Å) | 125.76 | 152.71 |
| b (Å) | 125.76 | 214.04 |
| c (Å) | 140.88 | 61.87 |
| α (°) | 90.0 | 90.0 |
| β (°) | 90.0 | 90.0 |
| γ (°) | 120.0 | 90.0 |
| Average Redundancy | 3.6 | 4.0 |
| No. of unique reflections | 48209 | 31571 |
| Resolution (Å) | 2.4 | 2.5 |
| R sym † | 0.068 (0.47) ‡ | 0.095 (0.49) ‡ |
| Completeness (%) | 99.2 (94.6) ‡ | 95.2 (93.1) ‡ |
| I/sigma | 15.9 (3.4) ‡ | 19.8 (3.3) ‡ |
|
| ||
| Refinement | ||
| Resolution (Å) | 63.0-2.4 | 40.0-2.5 |
| Rwork ∥/Rfree ¶ (%) | 18.4/23.4 | 19.8/26.1 |
| No. of protein atoms | 5269 | 4918 |
| ligand and ion | MgATP | None |
| No. of water | 529 | 76 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.019 | 0.011 |
| Bond angles (°) | 1.15 | 1.35 |
| Average B-factor | 44.6 | 57.0 |
|
| ||
| Ramachandran angles (%) | ||
| most favored (%) | 98.2 | 95.9 |
| disallowed | none | none |
Rsym = ΣhΣi|I(h) - I(h)i|IΣhΣi I(h)i, where I(h) is the mean intensity after rejections.
Numbers in parentheses correspond to the highest resolution shell of data.
Rwork = Σh||Fobs(h)| -|Fcalc(h)||IΣh|Fobs(h)|; no I/σ cutoff was used during refinement.
5.0% of the truncated data set was excluded from refinement to calculate Rfree.
Although the overall RIα:myrC(K7C) heterodimer closely resembles the previously solved myrC(K7C) in the myrC(K7C):SP20 structure, the conformations of the N-termini, nevertheless, differ somewhat in these two structures (Figure 2C and 2D). The N-terminus of the myrC(K7C):SP20 structure displays a helix-turn-helix motif following the myristate with the A-helix starting at Glu1. The RIα:myrC structure is missing the first helix; instead the A-helix starts at Lys8 and is preceded by a loop. The position of the C-terminal region of the myristic acid, including the ester bond to Gly1, is dislodged from the A-helix while the acyl chain binds within its previously defined hydrophobic pocket. We also compared the flexibility of residues at the N-terminus and near the myristyl binding pocket for effects of N-myristylation when binding to the small peptide SP20 vs the pseudo substrate RIα. Even though ordered, the B-factors at the N-terminus are greater for the RIα:myrC structure (105 Å2) than for the myrC(K7C ):SP20 structure (30.2 Å2) despite similar overall B-values (~50 Å2).
RIIβ2:myrC2 holoenzyme structure
Previous fluorescence anisotropy studies suggested that binding of the RIIβ -subunit causes the N-terminal myristylated tail to become more flexible and dynamic in solution, and, in contrast to free C and the RIα tetramer, association with membranes was further enhanced in RII holoenzyme complexes (Gangal et al., 1999). To confirm structurally whether the myristylated N-terminus is ordered or disordered in the RIIβ holoenzyme structure, we thus crystallized the RIIβ holoenzyme with the myristylated C-subunit. The crystal structure of the tetrameric RIIβ(R230K)2:myrC(K7C)2 holoenzyme (labeled as RIIβ2:myrC2) was solved at 2.5 Å (Figure 3A and Table 1). Even though the entire myristylated N-terminus was present in the protein, residues 1-13 of the myrC-subunit (GNAAAAKKGSEQE) as well as the myristic acid are not visible in the RIIβ2:myrC2 structure (Figure 3B). Thus binding to RIIβ selectively displaces the entire Myr motif leaving it more flexible and accessible to solvent. This is unique to the RIIβ2:myrC2 holoenzyme. It is different from the previously reported structures of the myrC-subunit alone, the myrC bound to SP20 or IP20 (Bastidas et al., 2012; Bastidas et al., 2013; Bossemeyer et al., 1993) and myrC bound to RIα which all show a more stable Myr Motif that is anchored into the acyl pocket in the C-Lobe of C-subunit (Bastidas et al., 2013) (Bastidas et al., 2012; Bossemeyer et al., 1993). The overall RIIβ2:myrC2 structure closely resembles the RIIβ2:C2 structure (Zhang et al., 2012)with a Cα RMSD of 0.19Å and the largest matching Cα atoms distance of 1.2 Å. The C-subunit still remains in a closed conformation even though there is no nucleotide bound to the active site and still has low B factors (~50 Å2) which is similar to the overall B-values in both the C and N-lobe. Furthermore, the N-termini of the C-subunits in each of the RIα:myrC and RIIβ2:myrC2 structures have no interactions with neighboring molecules in the crystal packing, thus it is unlikely the N-terminus is ordered or disordered due to crystal contacts.
Figure 3. Crystal structure of the RIIβ2:myrC2 holoenzyme (PDB ID: 4X6Q).
(A) The N-lobe of the C-subunit is shown in white; the C-lobe and C-tail are shown in tan. R-subunit is shown in cyan. (B)2Fo-Fc Electron density maps of the previously defined hydrophobic pocket where the myristic acid is in, at 1.0(top), 0.75(middle) and 0.5(bottom) sigma level, respectively. The density of Myr motif and the myristyl density (yellow) are missing.
Since the RIIβ holoenzyme is typically targeted by AKAPs to membranes, the exposed myristyl chains could thus provide additional opportunities for bivalent membrane anchoring. Based on our structure, we hypothesized that myristylation may in fact provide an alternative and/or complementary mechanism for targeting RII holoenzymes directly to membranes in addition to or independent of AKAPs, which are known to target RII holoenzymes in close proximity to membranes.
C-terminal capping motif in the R-subunit is also disordered in the RIIβ holoenzyme but ordered in the RIα holoenzyme
There is another difference between the RIα holoenzyme complexes and the RIIβ holoenzyme. In the R-subunits major conformational changes are associated with the CNB-domains as the protein toggles between its active (cAMP-bound) state and its inactive (holoenzyme) state (Berman et al., 2005; Kornev and Taylor, 2010)). In the active or cAMP bound conformation, cAMP is bound to the Phosphate Binding Cassette (PBC) and the adenine ring is capped by a hydrophobic residue. In every CNB-B domain, this capping residue (TYR397 in RIIβ) is located in a C-terminal helix (αC”) (Figure 4A). The two CNB domains interact closely in this active conformation whereas the CNB domains undergo a major conformational change in the holoenzyme that causes them to extend away from each other due to an extended αBC helix in CNB-A (Figure 4B and 4C). In the RIα holoenzyme, the reorganized C-terminal helical motif is packed against the PBC and well ordered (Figure 4C). Thus the C-terminal motif of the RIα subunit and N-terminal motif of the myrC, which are in close proximity, are both ordered. In contrast, in the RIIβ holoenzyme, most of the C-terminal helical motif (residue 394-416), including the capping TYR397, is disordered (Figure 4C). These two flexible regions, the N-terminal Myr motif in the myrC subunit and the C-terminal helical motif in RIIβ, are thus in relatively close proximity, and open up the opportunity for interacting with other proteins that may also be anchored to membranes.
Figure 4. The C-terminus of R-subunits.
(A)Up: Sequence alignment of four R-subunits isoforms. Helices observed in crystal structures for RIα (1RGS) and RIIβ (1CX4) are shown as red cylinders. Capping residues in αC" are shaded. Down: Helical propensity for R-subunits predicted by AGADIR(Lacroix et al., 1998) Sequence for RIα is shown along the X-axis. (B) Left: In the active or cAMP bound RIα (left) and RIIβ (right) conformation, cAMP is bound to the PBC and the adenine ring is capped by a hydrophobic residue. In every CNB-B domain, this capping residue (TYP 371 in RIα, TYR 397 in RIIβ) is located in αC”. (C) In the RIα holoenzyme (left), the reorganized C-terminal helical motif is packed against the PBC and well ordered, whereas in the RIIβ holoenzyme (right), most of the C-terminal helical motif (residue 394-416), including the capping TYR397, is disordered. The flexible N-terminal Myr motif in the myrC subunit and C-terminal helical motif in RIIβ are in relatively close proximity.
Effect of C-subunit myristylation and holoenzyme formation on membrane binding
To determine whether the Myr motif in RIIβ selectively serves as a myristylation switch and enables the membrane-anchoring properties of the myristylated C-subunit, we prepared nanodiscs to explore whether the various PKA complexes as well as free myrC-subunit could bind to nanodisc lipid bilayler. Specifically, the nanodiscs (Nath et al., 2007) are discrete ~10nm diameter model membranes that consist of phospholipid bilayers that are encircled by membrane scaffold protein, a modified amphipathic peptide from apolipoprotein A-I (Nath et al., 2007). The PKA myrC, RIα2:myrC2, and RIIβ2:myrC2 were then incubated with the nanodisc MSP and lipds. To confirm that membrane association was due specifically to the myristic acid we also incubated the non-myr C-subunit and the two holoenzymes formed with non-myristylated C-subunit, RIα2:C2 and RIIβ2:C2, with the nanodiscs. Assembly of the PKA with the nanodisc was assessed initially by gel filtration and confirmed by SDS-PAGE (Figure 5A and Figure S1). Among all these tested proteins, only RIIβ2:myrC2 associated with the nanodiscs (Figure 5A and Figure S1). Association of RIIβ2:myrC2 with the nanodisc was further confirmed by imaging negative-stained preparations by electron microscopy (Figure 5B). In negative-stained electron micrographs, we observed a round density with a diameter of ~120Å for the empty nanodiscs, whereas the nanodiscs that assembled with RIIβ2:myrC2 were more heterogeneous with diverse diameters ranging up to 250 Å confirming the binding of the holoenzyme. The distinct difference between the empty nanodiscs and those bound to RIIβ holoenzyme suggests that RIIβ2:myrC2 binding can alter the lipid structure. Since we do not see PKA holoenzyme beside the nanodics, we suggest that the binding is perpendicular to the lipid bilayer surface, though it remains to be determined to how exactly binding of RIIβ2:myrC2 caused the distortion of the nanodiscs.
Figure 5. Membrane anchoring of RIIβ2:myrC2 holoenzyme.

(A) Size exclusion chromatography shows assembly of RIIβ2:myrC2 to nanodiscs. Peak1, peak2 and peaks3 contains protein and lipid aggregation, RIIβ2:myrC2 holoenzyme and nanodiscs, and empty nanodiscs, respectively. The SDS-PAGE gel of these three peaks are shown in Figure S1. (B) Characterization of RIIβ2:myrC2 holoenzyme nanodiscs. On the left, negatively stained electron micrograph of the empty nanodiscs. On the right, negatively stained electron micrograph of the RIIβ2:myrC2 holoenzyme nanodiscs which were separated from the empty one by size exclusion. The scale bar is 50nm.
DISCUSSION
Because myristylation is added co-translationally, the modification is carried throughout the lifetime of a protein in contrast to palmitoylation. Our results described here provide new insights into the role of the myristic acid group and the dynamic properties of the N-terminal tail of the C-subunit. The N-terminal 13 residues including the myristic acid are not visible in the electron density in the RII holoenzyme, indicating that this segment is flexible. By contrast, the N-terminus and myristic acid are ordered and visible in the electron density in the RI holoenzyme and in the myrC-subunit in complex with an inhibitor peptide (IP20/SP20) (Bastidas et al., 2012). Our results therefore suggest that the dynamic properties of the N-terminal Myr motif are isoform specific. While we do not yet know the detailed mechanisms that account for this, we note that the C-terminus of the RIIβ subunit is also much more dynamic in the holoenzyme than in the RI holoenzyme complex, and these two flexible elements are in close proximity. Thus there are opportunities for additional interactions. Little is known about the specific mechanisms for acyl groups interacting with membranes and membrane proteins, but the availability of many new structures of membrane proteins now allow us to explore this phenomenon in more depth.
We hypothesized that the enhanced mobility of the N-terminus in RII subunits could aid in membrane association due to the exposed myristic acid group. Confirming this hypothesis, we showed that the RII subunits can bind to nanodiscs but only in complex with the myristylated C-subunit. Therefore, RII subunits can bind to membranes independent of AKAPs, which was also suggested by previous fluorescence polarization studies (Gangal et al., 1999). The nanodisc binding assays are also consistent with the myrC and RIα:myrC structures where the N-terminus and myristic acid group are ordered and not solvent exposed; neither bind to the nanodics. These results demonstrate a clear functional difference between the RI and RII isoforms. Only the RII holoenzyme has a “myristylation switch” (Figure 6) for membrane anchoring. This expulsion of the myristic acid group poises the RII holoenzyme for interaction with membranes. Furthermore, the dynamic C-terminus of the RIIβ-subunit in the RIIβ holoenzyme suggests that there may be additional factors that contribute to a membrane:protein interface or a protein:protein interface with an as yet unknown binding partner. In contrast the RIα has a type II PDZ motif at its C-terminus and in this holoenzyme only the terminal four residues are accessible for docking to another protein.
Figure 6. PKA has a dynamic and isoform-specific myristylation switch.
(A) Potential role of Ser10 in stabilizing the N-terminus of myrC subunit. Left: The distance between the Ser10 side chain and the backbone carbonyl of residue 7 is displayed, showing that the atoms are in H-bonding distance in the myrC(K7C):SP20 (tan) (PDB ID: 4DFX) and WT myrC:SP20 structures (teal) (PDB ID: 4DG2). These two structures are aligned with the backbone atoms of residue 7 and side chain of Ser10 shown in stick representation. Right: Interactions of Ser10 side chain and Cys7 and Ala6 in the myrC(K7C):RIα(K7C) structure. (B)The three myristylated structures presented here are aligned. The conformation and B-factors analysis of the myristic acid and Myr motif can be in two distinct “IN” and “OUT” modes. The myristic acid and Myr motif in SP20: myrC (PDB ID: 4DFX), RIα: myrC, and RIIβ2:myrC2 are shown. The right bottom is the scale bar for B-factors.
Like Src kinase, association of myristylated proteins with membranes usually requires a patch of basic amino acids (Resh, 1994). Indeed, the C-subunit also has a group of basic residues lining the A-helix and the surface of the C-subunit near the N-terminus. In the RIIβ2:myrC2 holoenzyme, the Lys residues in the N-terminus of the A-helix (GNAAAAKKG) also become very dynamic, which could further aid in the insertion of the myristic acid group into membranes. The N-terminus of the C-subunit contains several other potential post-translational modification sites that could act to regulate this membrane binding property of RII subunits. These include phosphorylation of Ser10 and deamidation of Asn2. Phosphorylation of Ser10, in addition to almost certainly destabilizing the observed ordered conformations of the N-terminus (Figures 6), could prevent membrane binding through the added negative charges. Additionally, deamidation of Asn2 may cause a similar destabilizing effect on the N-terminus due to the close proximity of the added negative charge to the myristic acid group. Furthermore, the deamidated form of PKA purified from tissues can autophosphorylate at Ser10 (Toner-Webb et al., 1992), suggesting Ser10 phosphorylation and Asn2 deamidation may act synergistically to prevent membrane binding. Therefore, there may be interplay between N-terminal myristylation and other N-terminal modifications. While Thr197 and Ser338 are constitutively phophorylated in mammalian cells, phosphorylation of Ser10 tends to be more transient and associated with specific cells (Breitenlechner et al., 2004; Kinzel et al., 2000; Tholey et al., 2001a). Phosphorylation of Ser10 has not been observed from PKA purified from tissues, however, most of the protein is auto-phosphorylated on Ser10 when the non-myristylated C-subunit is expressed in bacteria (Yonemoto et al., 1993). Alignment of the myrC(K7C):SP20 and myrC(WT):SP20 structures (Figure 6A) reveals that they display the same conformation of the N-terminus of the C-subunit. The distance between the Ser10 side chain and backbone carbonyl of residue 7 is around 2.8 Å (Figure 6A). In the RIα:myrC(K7C) structure, between the side chain hydroxyl group of Ser10 and Cys7, Ser10 also forms weak hydrogen-bonding with Cys7 even though the N-terminus conformation is different from the myrC(K7C) and myrC(WT) structures. The H-bonding interaction of Ser10 and residue 7 may help in the ordering of the N-terminus and stabilizing the myristyl acid. Phosphorylation of Ser10 would almost certainly destabilize this conformation by disrupting the interactions between Ser10 and other N-terminal residues. This is consistent with recent NMR studies (Gaffarogullari et al., 2011) on the myristylated C-subunit, which suggested that Ser10 phosphorylation destabilizes the N-terminus of PKA and causes the myristic acid to be dislodged from its hydrophobic pocket. More importantly, Ser10 phosphorylation and myristylation do not occur together in our bacterial co-expression system for recombinant myristylated protein (Bastidas et al., 2012; Yonemoto et al., 1993). This is also consistent with our recent finding that eliminating Ser10 phosphorylation, which is caused by K7C mutation and thus has the PKA recognition sequence abolished, increased the total yield of myristylated C protein (Bastidas et al., 2012). Based on previous studies with Src, phosphorylation at Ser10 would prevent association of the myristylated N-tail with membranes (Resh, 1994). In the RIIβ2:myrC2 holoenzyme structure (Figures 3 and 6), however, Ser10 density is missing together with myristic acid and Myr motif, due to their dynamic properties, possibly making the RIIβ2:myrC2 more accessible to membrane binding. Only the RII holoenzyme has the myristic acid and Myr motif in an “out” conformation.
Localization of the PKA holoenzyme complex to membranes is further mediated by specific AKAPs. The RIα holoenzyme is not capable of contributing to its own localization through myrislytation-membrane interaction because the myristic acid group is docked into the hydrophobic pocket. However, RI holoenzymes can be targeted to the plasma membrane via binding to dual specific or RI specific AKAPs, such as smAKAP. smAKAP is small, has N-terminal myristylation and palmitoylation sites, and selectively bind to RI with high affinity (1-2nM) (Figure 7) (Burgers et al., 2012). When the RIIβ holoenzyme is targeted by AKAPs to membrane proteins, the exposed myristic acid groups will likely be on the same surface as the AKAP. This configuration would provide a mechanism for bivalent membrane anchoring in addition to AKAP targeting (Figure 7).
Figure 7. Membrane anchoring models of RIa2: myrC2 and RIIβ2:myrC2 holoenzymes.
Left: RIα2:myrC2 holoenzyme is not capable of contributing to its own localization though myristylation-membrane interaction as the myristic acid is docked in C-subunit. It can be targeted to plasma membrane via binding to dual specific or RI specific AKAPs such as smAKAP which can be myristylated and palmitoylated at its N-terminus. Right: The exposed myristic acid groups in the RIIβ2:myrC2 holoenzyme and AKAPs are capable of providing a mechanism for bivalent membrane anchoring. Possible position of the D/D domain and interacting AKAP helix is shown. The myristyl groups (yellow) are on the same side as the AKAP (orange)-binding surface of the D/D domain.
Many signaling proteins are myristylated, and myristylation is essential for membrane binding and proper cellular localization of some proteins (Li et al., 2007; Song et al., 1997). The bivalent membrane targeting of RII holoenzymes opens the door to specific lipid interactions with the farnesylation, palmitic acid moiety and the myristylation site of other membrane proteins (Bohm et al., 1997). These potential interactions at membrane sites may provide an additional platform to assemble PKA. The association of the myristyl groups on the C-subunit in the RII holoenzyme with lipids or other lipid anchored proteins may provide a means for the C-subunits to remain bound to or in close proximity to the RII subunits even in the presence of cAMP.
Experimental Procedures
Purification of the R and C-Subunit Proteins
The non-myristylated C-subunit was expressed and purified as described previously (Herberg et al., 1993). Preparation of the myristylated C-subunit by co-expression with yeast N-myristyl transferase (NMT) (Duronio et al., 1990) and the purification protocol were also described previously (Bastidas et al., 2012). We used a cysteine mutation at the N-terminus of the myrC-subunit (K7C). As previously described (Bastidas et al., 2012), this mutation provides several advantages for studying the effects of N-terminal myristylation. It blocks autophosphorylation of Ser10 by eliminating the PKA recognition sequence at Ser10. Eliminating Ser10 phosphorylation increased the total yield of myristylated protein since myristylation and Ser10 phosphorylation typically do not occur together in our bacterial coexpression system, which was also observed previously with the recombinant myristylated protein. The K7C mutation also appears to have a more stable N-terminus based on time-resolved fluorescence anisotropy, which helped to structurally characterize the effects of N-myristylation in crystals. The structure of the K7C mutant was very similar to the wild type C-subunit (Bastidas et al., 2012). Expression and purification of the RIα and RIα(91-379:R333K) mutant as well as formation and purification of the RC heterodimers were described previously (Kim et al., 2007) with the exception that holoenzyme was formed using ATP and magnesium instead of AMP-PNP/Mn. RIIβ was over-expressed in E.coli and purified as described earlier (Zhang et al., 2012). RIIβ and wild type C-subunit were mixed in a 1:1.2 molar ratio and spin dialyzed at 4°C into holoenzyme buffer containing 20 mM MES (pH 5.8), 50 mM NaCl and 1 mM TCEP-HCl. The complex was then gel filtered through Superdex 200 in the same holo buffer to remove excess C-subunit.
Crystallization of RIα(91-379,R333K):myrC(K7C) heterodimer
The same crystallization conditions were utilized for this crystal structure as the previous RIα(91-379,R333K):C heterodimer (Kim et al., 2007). Specifically, the RC heterodimer was concentrated to 14 mg/mL and screened for crystallization using the hanging drop vapor diffusion method against different ammonium sulfate concentrations, which ranged from 0.8-2.5 M in 0.1 M sodium citrate buffer, whilst the pH was varied from 5.0-6.0. The crystal used for structure determination was obtained from a 4 μL drop containing 1:1 protein to well solution with the well solution containing 1.6 M ammonium sulfate and 0.1 M sodium citrate at pH 5.5.
Crystallization of RIIβ(R230K)2:myrC(K7C)2 holoenzyme
The same crystallization conditions were utilized for this crystal structure as the previous RIIβ holoenzyme (Zhang et al., 2012). Specifically, crystals were obtained from RIIβ holoenzyme that was concentrated to 12 mg/mL. The crystal used for structure determination was obtained from a 3 μL drop containing 1:1 protein to well solution with the well solution containing 10% PEG8000, 8% ethylene glycol, 20mM MES, at pH5.8.
Data collection and refinement
Data were collected on the synchrotron beam line 8.2.1 of the Advanced Light Source, Lawrence Berkeley National Labs (Berkeley, CA). The crystals were flash frozen in cryoprotectant conditions (well solution containing 20% glycerol for the RIα:C crystals and 30% PEG8000 for the RIIβ holoenzyme crystals ). The collected datasets were processed using HKL2000 program package(Otwinowski and Minor, 1997). For the RIα(91-379,R333K):myrC(K7C) datasets, phasing was performed using molecular replacement with 2QCS (Kim et al., 2007) as a search model. For the RIIβ(R230K)2:myrC(K7C)2 datasets, phasing was performed using molecular replacement with 3NTP (Zhang et al., 2012) as a search model. Refinement was initially performed using Refmac(Vagin et al., 2004) in the CCP4 program package(Winn et al., 2011). Subsequently, refinement was performed using PHENIX(Adams et al., 2010) to further improve the protein geometry. Model building was done in Coot(Emsley and Cowtan, 2004). The RIα(91-379,R333K): myrC(K7C)structure and RIIβ(R230K)2:myrC(K7C)2 structure were refined to Rwork/ Rfree values of 18.4%/23.4%, and 19.8%/26.1%, respectively. The data collection and refinement statistics are shown in Table 1.
PKA nanodiscs assembly and purification
PKA nanodiscs were assembled based on a protocol adapted from previous publications (Nath et al., 2007; Ye et al., 2010). In brief, DMPC and DMPG were solubilized in chloroform or a chloroform/methanol mixture, mixed thoroughly, and dried onto a glass tube under steady flow of nitrogen. The homogeneous lipid mixture was then solubilized in 100 mM cholate in 10 mM Tris and 100 mM NaCl, pH 7.4, giving a lipid concentration of 50 mM. 72 μl of the lipid solution was then mixed with 200 μl of 200 μM membrane scaffold protein (MSP) in dH2O and 200 μl of 10 μM purified PKA holoenzyme. The final ratio of lipids/MSP/protein is 90:1:0.05 in a total volume of 472 μl. The PKA nanodiscs were assembled by removing the detergents with SM-2 Biobeads overnight at room temperature. The assembled PKA nanodiscs were then purified with a hi-load 16/60 Superdex 200 size exclusion column with 20 mM Tris, 150 mM NaCl, 0.5 mM MgCl2, and 10mM AMPPNP, pH 7.4 for RI and 20 mM MES and 150 mM NaCl, pH 5.8 for RII, as the column buffer. Different buffers were used for RI and RII because of obtaining stable protein. The PKA RII holoenzyme nanodiscs and empty nanodiscs were readily separated and the successful assembly was verified by SDS-PAGE analysis. When necessary, the nanodiscs were further concentrated using Ultracel-30k (Millipore).
Negatively stained EM images of PKA nanodiscs
Purified nanodiscs were diluted to ~10μ g/ml and 3μl of the diluted sample was applied on to a glow-discharged carbon grid. The protein sample was absorbed for 30 seconds and blotted away. The grid was then washed and stained with 2% uranyl acetate. The grids were left air dry and loaded into a Tecnai G2 Sphera microscope (FEI) equipped with a LaB6 electron gun and a Gatan Ultrascan 1000 UHS CCD camera (Gatan, Inc.) for analysis. Images of nanodiscs were taken at a voltage of 200KV and a magnification of 65,000× in Digital Micrograph software (Gatan, Inc.).
Supplementary Material
High lights.
Solved X-ray crystal structure of PKA RIα:myrC
Solved X-ray crystal structure of PKA RIIβ2:myrC2
Isoform specific PKA-membrane anchoring
N-terminal myristylation site in the myrC serves as a flexible “switch”
Acknowledgements
We thank members of the Taylor laboratory for helpful discussions. We thank the Advanced Light Source (beam line 8.2.1) staff of Lawrence Berkeley National Laboratory for beam access and help with data collection. This work was supported by the National Institutes of Health grant GM34921 (S.S.T.) and HL106489 (M.H.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PDB coordinates
Coordinates and structure factors have been deposited in the Protein data bank with accession numbers 4X6Q (RIIβ2:myrC2) and 4X6R (RIα:myrC).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieux PS, Howe DG, Knickerbocker H, Lee DC, Su T, Laszlo GS, Idzerda RL, McKnight GS. Increased basal cAMP-dependent protein kinase activity inhibits the formation of mesoderm-derived structures in the developing mouse embryo. J Biol Chem. 2002;277:27294–27304. doi: 10.1074/jbc.M200302200. [DOI] [PubMed] [Google Scholar]
- Bastidas AC, Deal MS, Steichen JM, Keshwani MM, Guo Y, Taylor SS. Role of N-Terminal Myristylation in the Structure and Regulation of cAMP-Dependent Protein Kinase. J Mol Biol. 2012;422:215–229. doi: 10.1016/j.jmb.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas AC, Pierce LC, Walker RC, Johnson DA, Taylor SS. Influence of N-myristylation and ligand binding on the flexibility of the catalytic subunit of protein kinase A. Biochemistry. 2013;52:6368–6379. doi: 10.1021/bi400575k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Ten Eyck LF, Goodsell DS, Haste NM, Kornev A, Taylor SS. The cAMP binding domain: An ancient signaling module. P Natl Acad Sci USA. 2005;102:45–50. doi: 10.1073/pnas.0408579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher AJ, Wu J, Kim C, Yang J, Bruystens J, Cheung N, Pennypacker JK, Blumenthal DA, Kornev AP, Taylor SS. Realizing the Allosteric Potential of the Tetrameric Protein Kinase A RIalpha Holoenzyme. Structure. 2011;19:265–276. doi: 10.1016/j.str.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm A, Gaudet R, Sigler PB. Structural aspects of heterotrimeric G-protein signaling. Current opinion in biotechnology. 1997;8:480–487. doi: 10.1016/s0958-1669(97)80072-9. [DOI] [PubMed] [Google Scholar]
- Bossemeyer D, Engh RA, Kinzel V, Ponstingl H, Huber R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24) EMBO J. 1993;12:849–859. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenlechner C, Engh RA, Huber R, Kinzel V, Bossemeyer D, Gassel M. The typically disordered N-terminus of PKA can fold as a helix and project the myristoylation site into solution. Biochemistry. 2004;43:7743–7749. doi: 10.1021/bi0362525. [DOI] [PubMed] [Google Scholar]
- Burgers PP, Ma Y, Margarucci L, Mackey M, van der Heyden MA, Ellisman M, Scholten A, Taylor SS, Heck AJ. A small novel A-kinase anchoring protein (AKAP) that localizes specifically protein kinase A-regulatory subunit I (PKA-RI) to the plasma membrane. J Biol Chem. 2012;287:43789–43797. doi: 10.1074/jbc.M112.395970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaves JM, Taylor SS. Classification and phylogenetic analysis of the cAMP-dependent protein kinase regulatory subunit family. J Mol Evol. 2002;54:17–29. doi: 10.1007/s00239-001-0013-1. [DOI] [PubMed] [Google Scholar]
- Carr SA, Biemann K, Shoji S, Parmelee DC, Titani K. n-Tetradecanoyl is the NH2-terminal blocking group of the catalytic subunit of cyclic AMP-dependent protein kinase from bovine cardiac muscle. Proc Natl Acad Sci U S A. 1982;79:6128–6131. doi: 10.1073/pnas.79.20.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, Jackson-Machelski E, Heuckeroth RO, Olins PO, Devine CS, Yonemoto W, Slice LW, Taylor SS, Gordon JI. Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria. Proc Natl Acad Sci U S A. 1990;87:1506–1510. doi: 10.1073/pnas.87.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- Gaffarogullari EC, Masterson LR, Metcalfe EE, Traaseth NJ, Balatri E, Musa MM, Mullen D, Distefano MD, Veglia G. A myristoyl/phosphoserine switch controls cAMP-dependent protein kinase association to membranes. J Mol Biol. 2011;411:823–836. doi: 10.1016/j.jmb.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangal M, Clifford T, Deich J, Cheng X, Taylor SS, Johnson DA. Mobilization of the A-kinase N-myristate through an isoform-specific intermolecular switch. Proc Natl Acad Sci U S A. 1999;96:12394–12399. doi: 10.1073/pnas.96.22.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Becknell B, Wilm M, Mann M, Huang LJ, Taylor SS, Scott JD, Korsmeyer SJ. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- Herberg FW, Bell SM, Taylor SS. Expression of the catalytic subunit of cAMP-dependent protein kinase in Escherichia coli: multiple isozymes reflect different phosphorylation states. Protein Eng. 1993;6:771–777. doi: 10.1093/protein/6.7.771. [DOI] [PubMed] [Google Scholar]
- Herberg FW, Zimmermann B, McGlone M, Taylor SS. Importance of the A-helix of the catalytic subunit of cAMP-dependent protein kinase for stability and for orienting subdomains at the cleft interface. Protein Sci. 1997;6:569–579. doi: 10.1002/pro.5560060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim, Gleason CE, Murphy JM, Rosenberg BR, Teegan L, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilouz R, Bubis J, Wu J, Yim YY, Deal MS, Kornev AP, Ma Y, Blumenthal DK, Taylor SS. Localization and quaternary structure of the PKA RIbeta holoenzyme. Proc Natl Acad Sci U S A. 2012;109:12443–12448. doi: 10.1073/pnas.1209538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Kinzel V, Konig N, Pipkorn R, Bossemeyer D, Lehmann WD. The amino terminus of PKA catalytic subunit--a site for introduction of posttranslational heterogeneities by deamidation: D-Asp2 and D-isoAsp2 containing isozymes. Protein Sci. 2000;9:2269–2277. doi: 10.1110/ps.9.11.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev AP, Taylor SS. Defining the conserved internal architecture of a protein kinase. Biochim Biophys Acta. 2010;1804:440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J Mol Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- Li H, Dou J, Ding L, Spearman P. Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. J Virol. 2007;81:12899–12910. doi: 10.1128/JVI.01280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CWJ, Sweet RM, editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pepperkok R, Hotz-Wagenblatt A, Konig N, Girod A, Bossemeyer D, Kinzel V. Intracellular distribution of mammalian protein kinase A catalytic subunit altered by conserved Asn2 deamidation. J Cell Biol. 2000;148:715–726. doi: 10.1083/jcb.148.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Myristylation and palmitylation of Src family members: the fats of the matter. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- Sastri M, Barraclough DM, Carmichael PT, Taylor SS. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc Natl Acad Sci U S A. 2005;102:349–354. doi: 10.1073/pnas.0408608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FD, Reichow SL, Esseltine JL, Shi D, Langeberg LK, Scott JD, Gonen T. Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. eLife. 2013;2:e01319. doi: 10.7554/eLife.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberg K, Jahnsen T, Rognes T, Skalhegg BS, Laerdahl JK. Evolutionary paths of the cAMP-dependent protein kinase (PKA) catalytic subunits. PLoS One. 2013;8:e60935. doi: 10.1371/journal.pone.0060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KS, Sargiacomo M, Galbiati F, Parenti M, Lisanti MP. Targeting of a G alpha subunit (Gi1 alpha) and c-Src tyrosine kinase to caveolae membranes: clarifying the role of N-myristoylation. Cellular and molecular biology. 1997;43:293–303. [PubMed] [Google Scholar]
- Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nat Rev Mol Cell Bio. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholey A, Pipkorn R, Bossemeyer D, Kinzel V, Reed J. Influence of myristoylation, phosphorylation, and deamidation on the structural behavior of the N-terminus of the catalytic subunit of cAMP-dependent protein kinase. Biochemistry. 2001a;40:225–231. doi: 10.1021/bi0021277. [DOI] [PubMed] [Google Scholar]
- Tholey A, Pipkorn R, Bossemeyer D, Kinzel V, Reed J. Influence of myristoylation, phosphorylation, and deamidation on the structural behavior of the N-terminus of the catalytic subunit of CAMP-dependent protein kinase. Biochemistry. 2001b;40:225–231. doi: 10.1021/bi0021277. [DOI] [PubMed] [Google Scholar]
- Toner-Webb J, van Patten SM, Walsh DA, Taylor SS. Autophosphorylation of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1992;267:25174–25180. [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Vigil D, Blumenthal DK, Brown S, Taylor SS, Trewhella J. Differential effects of substrate on type I and type II PKA holoenzyme dissociation. Biochemistry. 2004;43:5629–5636. doi: 10.1021/bi0499157. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemoto W, Garrod SM, Bell SM, Taylor SS. Identification of phosphorylation sites in the recombinant catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1993;268:18626–18632. [PubMed] [Google Scholar]
- Zhang P, Smith-Nguyen EV, Keshwani MM, Deal MS, Kornev AP, Taylor SS. Structure and Allostery of the PKA RIIbeta Tetrameric Holoenzyme. Science. 2012;335:712–716. doi: 10.1126/science.1213979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.