Abstract
Peroxidases are enzymes catalyzing redox reactions that cleave peroxides. Their active redox centers have heme, cysteine thiols, selenium, manganese, and other chemical moieties. Peroxidases and their mimetic systems have several technological and biomedical applications such as environment protection, energy production, bioremediation, sensors and immunoassays design, and drug delivery devices. The combination of peroxidases or systems with peroxidase-like activity with nanostructures such as nanoparticles, nanotubes, thin films, liposomes, micelles, nanoflowers, nanorods and others is often an efficient strategy to improve catalytic activity, targeting, and reusability.
Keywords: antioxidants, antioxidant enzymes, improved and reusable peroxidase activity, self-assembly, particles, nanotubes, micelles
The oxygen paradox in metabolism and the role of peroxidases
The majority of complex organisms on Earth require oxygen for their existence. The use of oxygen in metabolism allows a high-energy output accompanied of the deleterious oxygen effects due to oxygen partial reduction (Halliwell and Gutteridge, 2007). The deleterious effects of oxygen include the oxidative damage to essential biomolecules in the cell such as DNA, proteins and lipids (Sies, 1997; Vertuani et al., 2004). Oxidants are normal products of the aerobic metabolism that reduces molecular oxygen to water. The escape of electrons from the respiratory chain produces the superoxide anion radical, which can form hydrogen peroxide. Hydrogen peroxide can yield the hydroxyl radical, the most reactive pro-oxidant species (Augusto and Miyamoto, 2011). Organic peroxides also derive from the oxidation of lipids and proteins (Sies, 1997). The reactive oxygen species (ROS) are peroxides and free radicals derived from oxygen that are highly reactive toward biomolecules. Free radicals are any atom or molecule that contains unpaired electrons. Hence, as part of the defense against the ROS, organisms developed an intricate network of antioxidants that inhibits the oxidation of other molecules by terminating the reactions leading to the production of free radicals. Antioxidants can remove the free radicals intermediates by oxidizing themselves so that they are often reducing agents such as poly-phenols, tocopherols, carotenes, vitamin A, ubiquinols, thiols, ascorbic acid, and others (Sies, 1997). The antioxidants act in concert with the enzymatic antioxidant defense represented by superoxide dismutases, catalases, glutathione peroxidases, and others (Sies, 1997). An imbalance between oxidants and antioxidants in favor of the oxidants is termed “oxidative stress.” Oxidative stress has been related to the development of several diseases such as Alzheimer's disease (Christen, 2000; Nunomura et al., 2006), diabetes (Giugliano et al., 1996; Davi et al., 2005), rheumatoid arthritis (Hitchon and El-Gabalawy, 2004), Parkinson's disease (Wood-Kaczmar et al., 2006), and neuronal degeneration (Cookson and Shaw, 1999). For example, low density-lipoprotein (LDL) oxidation triggers atherosclerosis leading to cardiovascular disease (Aviram, 2000; Van Gaal et al., 2006). Oxidative damage in DNA can cause cancer (Maynard et al., 2009; Khan et al., 2010). The dietary intake of vitamins E and C, and β-carotene can lower the risk of Alzheimer's disease (Li et al., 2012). However, ROS have also useful cellular functions such as redox signaling (Collins et al., 2012; Reczek and Chandel, 2014; Sies, 2014) or defense against invasive pathogens (Delattin et al., 2014). Hydrogen peroxide (produced from reactions catalyzed by the NADPH oxidases or complex III of the mitochondrial respiratory chain) is a messenger under fine control of peroxiredoxins, glutathione peroxidases and catalase signaling major processes such as cell proliferation, tissue repair, differentiation, inflammation, and aging (Sies, 2014). The main ROS involved in redox signaling is hydrogen peroxide; however, other forms of ROS also contribute as the nitric oxide (NO) generated by NO synthases, which diffuses into mitochondria and modulates mitochondrial function by competing with O2 at respiratory complex IV thereby slowing respiration (Collins et al., 2012). ROS are not only a toxic by-product of mitochondrial respiration but also play a role in cellular signaling as for example the role of H2O2 in thiol oxidation modulating the function of proteins (Collins et al., 2012; Reczek and Chandel, 2014). However, the mechanism by which the ROS signal reaches its target protein in the face of highly reactive and abundant antioxidants in the cell is not fully understood (Reczek and Chandel, 2014). The function of antioxidant systems is not the complete removal of oxidants, but instead the optimization of their intracellular concentrations (Rhee, 2006). Against fungus, azoles, echinocandins and liposomal amphotericin B, besides their specific mode of action, also induce ROS in planktonic and biofilm cells (Delattin et al., 2014). The three main classes of bactericidal antibiotics, namely, the quinolones, the beta-lactams, and the aminoglycosides also induce ROS (Delattin et al., 2014).
The protection available for cells under oxidative stress comes from natural antioxidants and several antioxidant enzymes (Davies, 1995; Sies, 1997). Superoxide released by oxidative processes yields firstly the hydrogen peroxide in a step catalyzed by superoxide dismutase. Then, the second step is the reduction of hydrogen peroxide by catalases and other peroxidases. Since these enzymes play in concert, it is often difficult to ascertain their individual role in the antioxidant defense. The generation of transgenic mice deficient in just one antioxidant enzyme has been a major approach to get further insight on the individual role of each peroxidase (Ho et al., 1998).
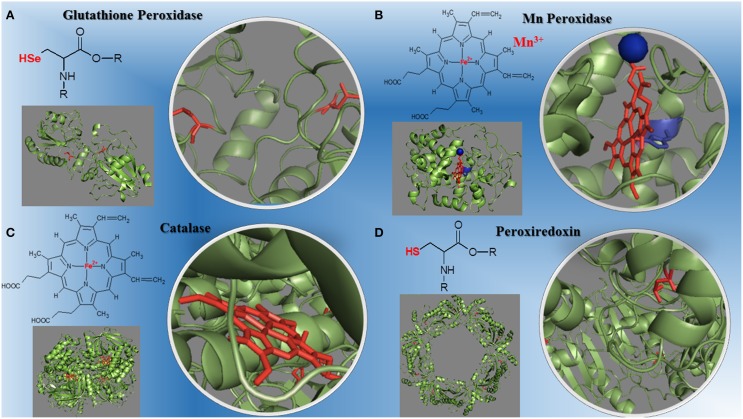
The peroxidases catalyze redox reactions involving peroxides cleavage (Mhamdi et al., 2010; Al Ghouleh et al., 2011; Hall et al., 2011; Nantes et al., 2011; Mishra and Imlay, 2012; Rhee et al., 2012; Shao, 2012; Whittaker, 2012; Zámockı et al., 2012; Rodríguez-Rodríguez et al., 2014). The molecular evolution of peroxidases required for efficient removal of peroxides led to three metallo-enzyme families that differ in oligomers organization, monomer architecture, active site geometry, and catalytic residues. They are: (1) the highly conserved structures of mono-functional heme catalases found in all domains of life; (2) the bi-functional catalase-peroxidases, members of a protein family predominantly present among eubacteria and archaea with only two evolutionary branches found in eukaryotic organisms; (3) the non-heme manganese catalases, a small protein family with old roots, only present in bacteria and archaea (Zámockı et al., 2012). These non-heme manganese catalases represent an environmentally important alternative to heme-containing catalases in antioxidant defense. Manganese catalases contain binuclear manganese complexes in their catalytic site rather than a heme, and cycle between Mn(2)(II,II) and Mn(2)(III,III) states during turnover (Whittaker, 2012). Plants contain several types of H2O2 –metabolizing proteins; catalases are highly active and do not require cellular reductants as they primarily catalyze a dismutase reaction (Mhamdi et al., 2010). In mammalian cells, six peroxiredoxins isoforms expressed and localized to various cellular compartments function as a peroxidase and contain an active site cysteine that can be oxidized by H2O2 (Rhee et al., 2012). The peroxiredoxins use a conserved Cys residue to reduce peroxides and are highly expressed in organisms from all kingdoms with 72 structures already determined covering much of the diversity of the family (Hall et al., 2011). Other peroxidases play major roles in cardiovascular, lung and brain diseases (Al Ghouleh et al., 2011; Shao, 2012; Rodríguez-Rodríguez et al., 2014) such as myeloperoxidase, a heme enzyme secreted by human artery wall macrophages which oxidizes apolipoprotein A-I (apoA-I), the major HDL protein, diminishing apoA-I ability to promote cellular cholesterol efflux. The oxidation of specific tyrosine and methionine residues in apoA-I contributes to its loss of activity (Shao, 2012). Peroxiredoxin 6 (Prdx6) is the only mammalian 1-Cys member of the Prdx family. This peroxidase uses the antioxidant tripeptide glutathione (GSH) instead of thioredoxin as the physiological reductant to reduce the oxidized sn-2 fatty acyl group of phospholipids (peroxidase activity) or to hydrolyze the sn-2 ester (alkyl) bond of phospholipids (phospholipase A(2) [PLA(2)] activity). This bi-functional protein has separate active sites for peroxidase and phospholipase PLA activities. These activities are dependent on binding of the protein to phospholipids at acidic pH and to oxidized phospholipids at cytosolic pH (Fisher, 2011). The structures of different peroxidases and their redox centers is on Figure 1.
Figure 1.
Structural diversity of peroxidases and their redox centers. (A) The glutathione peroxidase (1GP1-SeH) monomer and its selenium cysteine SeCys-35 (Epp et al., 1983). (B) The manganese peroxidase (1YYD) monomer from Phanerochaete chrysosporium and its heme iron plus Mn3+ (Sundaramoorthy et al., 2005). (C) The dimeric catalase (1DGB) structure from human erythrocytes and its heme iron (Putnam et al., 2000). (D) The decameric peroxiredoxin (1QMV) from human erythrocytes and its cysteine thiol residues (Schröder et al., 2000). The PDB ID for each protein is inbetween parentheses.
Applications for peroxidases and their assemblies
Horseradish peroxidase (HRP) is a much studied and versatile peroxidase with about 600 articles presently available on its assemblies. HRP certainly is the analyst's friend (Ryan et al., 1994; Krainer and Glieder, 2015). This oxidoredutase accepts a wide variety of hydrogen donors to reduce H2O2, a property applicable to a range of colorimetric, fluorimetric, chemiluminescent, and electrochemical and immuno assays based on HRP activity. However, HRP isolation and purification gives low yields and recombinant HRP obtained in the yeast Pichia pastoris is often hyper-glycosylated though an unglycosylated, active and stable HRP variant is now available from site-directed mutagenesis (Capone et al., 2014). The many difficulties inherent to HRP production and purification (Spadiut and Herwig, 2013) essentially emphasize the importance of its functional immobilization in nanostructures for improving activity and allowing reusability.
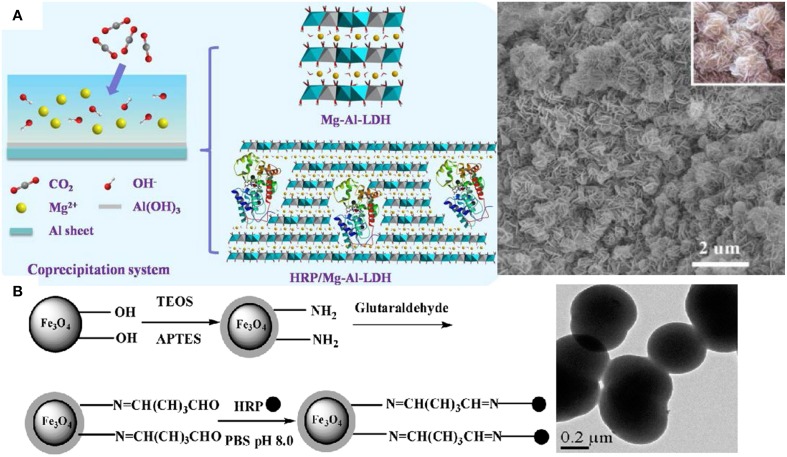
The water-soluble heme-containing peptides obtained by proteolytic digestion of cytochrome c are the microperoxidases (MP) often used to explore aspects of the chemistry of iron porphyrins, and as mimics for some reactions catalyzed by peroxidases (Prieto et al., 2006; Araujo et al., 2007; Marques, 2007). MP are not only model compounds but also useful molecules for applications in biosensors as electron carriers, photoreceptors, microzymes, and drugs. In a systematic study to define the minimal requirements for covalent attachment of hemes to c-type cytochromes, some artificial MPs were produced in vivo by exploiting the secretion and cyt c apparatuses of E. coli (Braun and Thöny-Meyer, 2004). MP-11, a MP with 11 aminoacids residues, assembles to boron nitride nanotubes which enhance catalysis due to a strong electron coupling between the active center of MP-11 and the nanotube showing that nanostructures often modulate peroxidases activity (Li et al., 2014b). The assembly of peroxidases into micelles (Prieto et al., 2006), reversed micelles (Das and Das, 2009; Maiti et al., 2012; Das et al., 2013), liposomes (Dotsikas and Loukas, 2012), supported bilayers (Leão-Silva et al., 2011), lipid monolayers at the air-water interface (Schmidt et al., 2008), organic, inorganic, metallic, magnetic or composite nanoparticles (Xu and Han, 2004; Kim et al., 2005; Araujo et al., 2007; Khaja et al., 2007; Silva et al., 2007; Chen et al., 2011; Klyachko et al., 2012; Li et al., 2012; Pan et al., 2013; Duan et al., 2014), hydrogels and microgels (Nakashima et al., 2009; González-Sánchez et al., 2011; Wu et al., 2012; Bruns et al., 2013), nanotubes (Li et al., 2014b), multilayers and nanostructured films (Kim et al., 2010; Pallarola et al., 2012; Cortez et al., 2013) has been explored for important applications such as immunoassays design (Marquette and Blum, 2009; Dotsikas and Loukas, 2012), drug delivery (Allen et al., 2011; Kotchey et al., 2013), cancer therapy (Ibañez et al., 2015), environment protection and bioremediation (Bansal and Kanwar, 2013; Duan et al., 2014), construction of sensors (Xu and Han, 2004; Kim et al., 2005; Chen et al., 2011; Pallarola et al., 2012; Chen and Chatterjee, 2013; Cortez et al., 2013), and energy production (Ramanavicius et al., 2005; Ramanavicius and Ramanaviciene, 2009). HRP adsorbs onto silicon wafers providing reusable films for emulsion polymerization (Naves et al., 2007) and is useful for tissue engineering applications by forming hydrogels in situ via crosslinking (Bae et al., 2014) and for atom transfer radical polymerization (Bruns et al., 2013). Although the enzyme-catalyzed polymerizations are environmentally advantageous, the high cost, large quantity of enzymes required for polymerization and formation of relatively low molecular weight polymers obstruct their employment in the industry (Albertsson and Srivastava, 2008). Catalase and superoxide dismutase modified by poly (ethylene glycol) (PEG) or encapsulated in PEG-coated liposomes have increased bioavailability and enhanced protection of the enzymatic activity in animal models. Pluronic-based micelles formed with antioxidant enzymes or PEG copolymers also protect catalase and superoxide dismutase from proteolysis improving delivery of these enzymes to vascular endothelial cells. Liposomes, protein conjugates and magnetic nanoparticles also successfully deliver these antioxidant enzymes to sites of vascular oxidative stress (Hood et al., 2011). Nanoparticles of polyethylene glycol and poly-lactic/poly-glycolic acid encapsulate catalase and HRP into ~300 nm diameter nanospheres combating the oxidative stress both in cell culture and in animals (Dziubla et al., 2008). Different cross-linking strategies and certain reaction conditions of pH and polycation/proteincharge ratio allow functional immobilization of superoxide dismutase (SOD1) and catalase in cross-linked nanoparticles made of cationic block copolymers such as polyethyleneimine-PEG (PEI-PEG) or poly(L-lysine)-PEG (Klyachko et al., 2012). In mice, 125I-labeled SOD1-containing nanoparticles display increased stability in blood and in the brain, and improved accumulation in brain tissues, in comparison with non-cross-linked complexes and native SOD1 (Klyachko et al., 2012). Catalase self-assembles in a cationic block copolymer of PEI-PEG yielding stable complexes with ca. 60–100 nm in size that retain antioxidant activity with negligible cytotoxicity (Batrakova et al., 2007). These particles are rapidly, in 40–60 min, taken up by bone-marrow-derived macrophages and retain catalytic activity for more than 24 h being released in active form whereas “naked” catalase is quickly degraded; about 0.6% of the injected dose locates in the brain (Batrakova et al., 2007). Catalytically active enzyme aggregates of HRP cross-linked by glutaraldehyde yield 83% of activity recovery when compared with the native enzyme; the advantage of this procedure is the possibility of including other stabilizing proteins in the aggregate such as albumin (Šulek et al., 2011). Eventually HRP immobilization by physical adsorption can be more effective than immobilization by crosslinking (Tatsumi et al., 1996). Figure 2 shows examples of immobilized HRP by entrapment in an inorganic, flower-like inorganic matrix (Figure 2A), or by covalent attachment to hybrid particles (Figure 2B).
Figure 2.
Nanostructures for immobilizing and reusing peroxidases. (A) Entrapment of HRP in inorganic matrix of flowers like nanomaterial (Wang et al., 2014). (B) Covalent attachment of HRP on NH2 –modified Fe3O4/SiO2 magnetic nanoparticles (Chang and Tang, 2014).
A strategy receiving a wide spread interest is the use of peroxidase mimetic systems with intrinsic peroxidase-like activities associated to reliable reactivity and facile production and storage. Hemin, hematin and porphyrin (Johnstone et al., 1997; Wang et al., 2007; Griffith et al., 2012), magnetic nanoparticles and nanocomposites (Dai et al., 2009; Park et al., 2011; Gao et al., 2013), metals and alloys (Bernsmann et al., 2011; Sun et al., 2013; Zhou et al., 2013), metal oxides and sulfides (Asati et al., 2009; He et al., 2012; Hong et al., 2013), carbon materials (Shi et al., 2011; Li et al., 2013), and others (Wang et al., 2013) have been described and praised for their intrinsic peroxidase-like activities in a variety of assays. For non-aqueous applications, the peroxidase-mimetic materials are more convenient and versatile than the natural enzymes. For example, tungsten carbide (WC) as catalyst for electron-transfer reactions has been extensively investigated for applications in fuel cells and oxygen reduction (Palanker et al., 1976; Rosenbaum et al., 2006), because it exhibits catalytic properties similar to those of noble metals (Levy and Boudart, 1973). Recently, the intrinsic catalytic activity of tungsten carbide nanorods toward typical peroxidase substrates was described in comparison to the one of natural HRP leading to the demonstration that the WC NRs have excellent catalytic behavior in various organic media, are stable and reusable representing a promising peroxidase mimic (Li et al., 2014a).
Layer-by-layer deposition of anionic and cationic polyelectrolytes plus catalase—gold nanoparticles composites originates multilayers onto electrodes that effectively permit electron transfer between catalase and the electrode at high loading of the composites (Kim et al., 2010). Following a similar approach the polyelectrolyte DNA organizes an enzyme cascade of discrete glucose-oxidase and HRP pairs with controlled inter-enzyme distance resulting in enhanced enzymatic activity at short distances (Fu et al., 2012). DNA also directs HRP immobilization onto porous SiO2 thin films resulting in high enzymatic activity (Shtenberg et al., 2012). Peroxidases degrade carbon nanotubes in vitro and in vivo so that carbon-based nanocarriers specifically designed to target organs and cells can deliver their cargo, and biodegrade via peroxidase-driven mechanism, a really attractive therapeutic delivery option in nanomedicine (Shvedova et al., 2012; Kotchey et al., 2013). Similarly, peroxidases can also release the hydrophobic cargo of poly (propylene sulfide) nanoparticles in the presence of H2O2 (Allen et al., 2011).
Remediation of phenol waste water by immobilized HRP in magnetic nanoparticles involves the oxidation of phenols in the presence of H2O2 and the reaction of the phenoxy radicals with each other in a non-enzymatic process forming polymers; the polymers are then removed by precipitation with salts or condensation and the reusable magnetic nanoparticles with improved HRP activity are rescued (Duan et al., 2014). The importance of peroxidases for treating and removing pollutants such as phenols and halogenated phenols, polycyclic aromatic hydrocarbons (PAH), endocrine disruptive chemicals (EDC), pesticides, dioxins, polychlorinated biphenyls (PCB), industrial dyes, and other xenobiotics has been recognized and recently reviewed (Bansal and Kanwar, 2013). The recent efforts on the rational design of supramolecular hemoprotein assemblies have also been recently reviewed (Oohora and Hayashi, 2014).
For biomedical and technological applications, peroxidases in nanostructures represent a rapidly growing area for innovative research improving enzyme activity, protection, targeting, and reusability. Free peroxidases have some inherent disadvantages including loss of catalytic activity in solution during the reaction, high cost and lack of reusability. To overcome these issues, several supports including various nanomaterials for instance metal and magnetic nanomaterials, polymer based nanofibers, silicon and carbon based nanomaterials, have been used for peroxidases immobilization (Lei et al., 2002; Dulay et al., 2005; Sheldon, 2007; Wang et al., 2007; Lee et al., 2009; Cao et al., 2010; Zhu et al., 2013). In general, enhancement in enzyme activity and stability is desirable for given reactions or applications. A decrease in activity compared to the one of the free enzyme is due to the following reasons: (1) use of organic solvent for the immobilization procedure, which deactivates some parts of enzyme; (2) partially blocking of enzyme active sites during the immobilization process, which makes enzyme less accessible to the substrate; (3) unfavorable enzyme conformations on external supports; (4) mass-transfer limitations between enzymes and substrates (Luckarift et al., 2004; Kim et al., 2008; Ge et al., 2009). Nanoparticles and other nanomaterials have been the driving force behind the development of sophisticated biosensors in recent years (Bhakta et al., 2015). The use of metal nanoparticles, such as gold and platinum, results in improved rate of electron transfer at the interface. Nanofilms also detect several analytes and several are useful in the rapidly growing biosensors area. The tunable feature of nanotubes also represents a way for improving the analytical performance of biosensors. The importance of nanomaterials is growing as the demand for quick, selective, inexpensive, stable, and reproducible analytical devices continues to surge. Bioactivity and compatibility of the enzyme/material nanocomposites is a critical consideration to further the applicability of biosensors (Bhakta et al., 2015). A immobilization approach to obtain protein-inorganic hybrid nanoflowers greatly increases enzyme activity and stability (Ge et al., 2012). HRP and Fe2+ ions together yield flowerlike hybrid nanostructures, the hybrid nanoflowers (HNF) that enhance HRP activity by 5–7 times when compared to free HRP; this is due to high local HRP concentration, appropriate HRP conformation, less mass transfer limitations, and Fe2+ activation (Ocsoy et al., 2015). Therefore, peroxidases and their mimetic systems in nanostructures (like the nanoflowers) represent a flourishing field for biomedical and technological research.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Financial support from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) to AMC-R (grant 302352/2014-7) is gratefully achnowledged. TP is the recipient of a postdoctoral fellowship grant PNPD 2867/2011 from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes).
References
- Albertsson A. C., Srivastava R. K. (2008). Recent developments in enzyme-catalyzed ring-opening polymerization. Adv. Drug Deliv. Rev. 60, 1077–1093. 10.1016/j.addr.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Al Ghouleh I., Khoo N. K., Knaus U. G., Griendling K. K., Touyz R. M., Thannickal V. J., et al. (2011). Oxidases and peroxidases in cardiovascular and lung disease: new concepts in reactive oxygen species signaling. Free Radic. Biol. Med. 51, 1271–1288. 10.1016/j.freeradbiomed.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen B. L., Johnson J. D., Walker J. P. (2011). Encapsulation and enzyme-mediated release of molecular cargo in polysulfide nanoparticles. ACS Nano 5, 5263–5272. 10.1021/nn201477y [DOI] [PubMed] [Google Scholar]
- Araujo J. C., Prieto T., Prado F. M., Trindade F. J., Nunes G. L., dos Santos J. G., et al. (2007). Peroxidase catalytic cycle of MCM-41-entrapped microperoxidase-11 as a mechanism for phenol oxidation. J. Nanosci. Nanotechnol. 7, 3643–3652. 10.1166/jnn.2007.853 [DOI] [PubMed] [Google Scholar]
- Asati A., Santra S., Kaittanis C., Nath S., Perez J. M. (2009). Oxidase-like activity of polymer-coated cerium oxide nanoparticles. Angew. Chem. Int. Ed. Engl. 48, 2308–2312. 10.1002/anie.200805279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto O., Miyamoto S. (2011). Oxygen radicals and related species in Principles of Free Radical Biomedicine, Vol. 1, eds Pantopolos K., Schipper H. M. (New York, NY: Nova Science Publishers, Inc.), 19–42. [Google Scholar]
- Aviram M. (2000). Review of human studies on oxidative damage and antioxidant protection related to cardiovascular diseases. Free Radic. Res. 33, S85–S97. [PubMed] [Google Scholar]
- Bae J. W., Choi J. H., Lee Y., Park K. D. (2014). Horseradish peroxidase-catalysed in situ-forming hydrogels for tissue-engineering applications. J. Tissue Eng. Regen. Med. [Epub ahead of print]. 10.1002/term.1917 [DOI] [PubMed] [Google Scholar]
- Bansal N., Kanwar S. S. (2013). Peroxidase(s) in environment protection. Sci. World J. 2013:714639. 10.1155/2013/714639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batrakova E. V., Li S., Reynolds A. D., Mosley R. L., Bronich T. K., Kabanov A. V., et al. (2007). A macrophage-nanozyme delivery system for Parkinson's disease. Bioconjug. Chem. 18, 1498–1506. 10.1021/bc700184b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsmann F., Ball V., Addiego F., Ponche A., Michel M., Gracio J. J. D., et al. (2011). Dopamine-melanin film deposition depends on the used oxidant and buffer solution. Langmuir 27, 2819–2825. 10.1021/la104981s [DOI] [PubMed] [Google Scholar]
- Bhakta S. A., Evans E., Benavidez T. E., Garcia C. D. (2015). Protein adsorption on to nanomaterials for the development of biosensors and analytical devices: a review. Anal. Chim. Acta. 872, 7–25. 10.1016/j.aca.2014.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M., Thöny-Meyer L. (2004). Biosynthesis of artificial microperoxidases by exploiting the secretion and cytochrome c maturation apparatuses of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 12830–12835. 10.1073/pnas.0402435101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns N., Lörcher S., Makyła K., Pollarda J., Renggli K., Spulber M. (2013). Combining polymers with the functionality of proteins: new concepts for atom transfer radical polymerization, nanoreactors and damage self-reporting materials. Chimia 67, 777–781. 10.2533/chimia.2013.777 [DOI] [PubMed] [Google Scholar]
- Cao A., Ye Z., Cai Z., Dong E., Yang X., Liu G., et al. (2010). A facile method to encapsulate proteins in silica nanoparticles: encapsulated green fluorescent protein as arobust fluorescence probe. Angew. Chem. Int. Ed. Engl. 49, 3022–3025. 10.1002/anie.200906883 [DOI] [PubMed] [Google Scholar]
- Capone S., Pletzenauer R., Maresch D., Metzger K., Altmann F., Herwig C., et al. (2014). Glyco-variant library of the versatile enzyme horseradish peroxidase. Glycobiology 24, 852–863. 10.1093/glycob/cwu047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q., Tang H. (2014). Immobilization of horseradish peroxidase on NH2-modified magnetic Fe3O4/SiO2 particles and its application in removal of 2, 4-dichlorophenol. Molecules 19, 15768–15782. 10.3390/molecules191015768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Chatterjee S. (2013). Nanomaterials based electrochemical sensors for biomedical applications. Chem. Soc. Rev. 42, 5425–5438. 10.1039/c3cs35518g [DOI] [PubMed] [Google Scholar]
- Chen Z., Sun D., Zhou Y., Zhao J., Lu T., Huang X., et al. (2011). Nano polyurethane-assisted ultrasensitive biodetection of H2O2 over immobilized microperoxidase-11. Biosens. Bioelectron. 29, 53–59. 10.1016/j.bios.2011.07.065 [DOI] [PubMed] [Google Scholar]
- Christen Y. (2000). Oxidative stress and Alzheimer disease. Am. J. Clin. Nutr. 71, 621–629. [DOI] [PubMed] [Google Scholar]
- Collins Y., Chouchani E. T., James A. M., Menger K. E., Cochemé H. M., Murphy M. P. (2012). Mitochondrial redox signalling at a glance. J. Cell Sci. 125, 801–806. 10.1242/jcs.098475 [DOI] [PubMed] [Google Scholar]
- Cookson M. R., Shaw P. J. (1999). Oxidative stress and motor neurone disease. Brain Pathol. 9, 165–186. 10.1111/j.1750-3639.1999.tb00217.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez M. L., Pallarola D., Ceolín M., Azzaroni O., Battaglini F. (2013). Electron transfer properties of dual self-assembled architectures based on specific recognition and electrostatic driving forces: its application to control substrate inhibition in horseradish peroxidase-based sensors. Anal. Chem. 85, 2414–2422. 10.1021/ac303424t [DOI] [PubMed] [Google Scholar]
- Dai Z., Liu S., Bao J., Ju H. (2009). Nanostructured FeS as a mimic peroxidase for biocatalysis and biosensing. Chem. Eur. J. 15, 4321–4326. 10.1002/chem.200802158 [DOI] [PubMed] [Google Scholar]
- Das D., Das P. K. (2009). Superior activity of structurally deprived enzyme-carbon nanotube hybrids in cationic reverse micelles. Langmuir 25, 4421–4428. 10.1021/la803753g [DOI] [PubMed] [Google Scholar]
- Das K., Maiti S., Ghosh M., Mandal D., Das P. K. (2013). Graphene oxide in cetyl trimethylammonium bromide (CTAB) reverse micelle: a befitting soft nanocomposite for improving efficiency of surface-active enzymes. J. Colloid Interface Sci. 395, 111–118. 10.1016/j.jcis.2012.12.064 [DOI] [PubMed] [Google Scholar]
- Davi G., Falco A., Patrono C. (2005). Lipid peroxidation in diabetes mellitus. Antioxid. Redox Signal. 7, 256–268. 10.1089/ars.2005.7.256 [DOI] [PubMed] [Google Scholar]
- Davies K. J. (1995). Oxidative stress: the paradox of aerobic life. Biochem. Soc. Symp. 61, 1–31. 10.1042/bss0610001 [DOI] [PubMed] [Google Scholar]
- Delattin N., Cammue B. F., Thevissen K. (2014). Reactive oxygen species-inducing antifungal agents and their activity against fungal biofilms. Future Med. Chem. 6, 77–90. 10.4155/fmc.13.189 [DOI] [PubMed] [Google Scholar]
- Dotsikas Y., Loukas Y. L. (2012). Improved performance of antigen-HRP conjugate-based immunoassays after the addition of anti-HRP antibody and application of a liposomal chemiluminescence marker. Anal. Sci. 28, 753–757. 10.2116/analsci.28.753 [DOI] [PubMed] [Google Scholar]
- Duan X., Corgié S. C., Aneshansley D. J., Wang P., Walker L. P., Giannelis E. P. (2014). Hierarchical hybrid peroxidase catalysts for remediation of phenol wastewater. Chem. Phys. Chem. 15, 974–980. 10.1002/cphc.201300808 [DOI] [PubMed] [Google Scholar]
- Dulay M. T., Baca Q. J., Zare R. N. (2005). Enhanced proteolytic activity of covalently bound enzymes in photopolymerized sol gel. Anal. Chem. 77, 4604–4610. 10.1021/ac0504767 [DOI] [PubMed] [Google Scholar]
- Dziubla T. D., Shuvaev V. V., Hong N. K., Hawkins B., Muniswamy M., Takano H., et al. (2008). Endothelial targeting of semi-permeable polymer nanocarriers for enzyme therapies. Biomaterial 29, 215–227. 10.1016/j.biomaterials.2007.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp O., Ladenstein R., Wendel A. (1983). The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur. J. Biochem. 133, 51–69. 10.1111/j.1432-1033.1983.tb07429.x [DOI] [PubMed] [Google Scholar]
- Fisher A. B. (2011). Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxid. Redox Signal. 15, 831–844. 10.1089/ars.2010.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Liu M., Liu Y., Woodbury N. W., Yan H. (2012). Inter-enzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 134, 5516–5519. 10.1021/ja300897h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Xu M., Hou L., Chen G., Tang D. (2013). Magnetic bead-based reverse colorimetric immunoassay strategy for sensing biomolecules. Anal. Chem. 85, 6945–6952. 10.1021/ac401433p [DOI] [PubMed] [Google Scholar]
- Ge J., Lei J., Zare R. N. (2012). Protein–inorganic hybrid nanoflowers. Nat. Nanotechnol. 7, 428–432. 10.1038/nnano.2012.80 [DOI] [PubMed] [Google Scholar]
- Ge J., Lu D., Liu Z. X., Liu Z. (2009). Recent advances in nanostructured biocatalysts. Biochem. Eng. J. 44, 53–59. 10.1016/j.bej.2009.01.002 [DOI] [Google Scholar]
- Giugliano D., Ceriello A., Paolisso G. (1996). Oxidative stress and diabetic vascular complications. Diabetes Care 19, 257–267. 10.2337/diacare.19.3.257 [DOI] [PubMed] [Google Scholar]
- González-Sánchez M. I., Rubio-Retama J., López-Cabarcos E., Valero E. (2011). Development of an acetaminophen amperometric biosensor based on peroxidase entrapped in polyacrylamide microgels. Biosens. Bioelectron. 26, 1883–1889. 10.1016/j.bios.2010.03.024 [DOI] [PubMed] [Google Scholar]
- Griffith M. J., Sunahara K., Wagner P., Wagner K., Wallace G. G., Officer D. L., et al. (2012). Porphyrins for dye-sensitised solar cells: new insights into efficiency-determining electron transfer steps. Chem. Commun. 48, 4145–4162. 10.1039/c2cc30677h [DOI] [PubMed] [Google Scholar]
- Hall A., Nelson K., Poole L. B., Karplus P. A. (2011). Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 15, 795–815. 10.1089/ars.2010.3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J. M. C. (2007). Free Radicals in Biology and Medicine. New York, NY: Oxford University Press. [Google Scholar]
- He W., Jia H., Li X., Lei Y., Li J., Zhao H., et al. (2012). Understanding the formation of CuS concave superstructures with peroxidase-like activity. Nanoscale 4, 3501–3506. 10.1039/c2nr30310h [DOI] [PubMed] [Google Scholar]
- Hitchon C., El-Gabalawy H. (2004). Oxidation in rheumatoid arthritis. Arthritis Res. Ther. 6, 265–278. 10.1186/ar1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. S., Magnenat J. L., Gargano M., Cao J. (1998). The nature of antioxidant defense mechanisms: a lesson from transgenic studies. Environ. Health Perspect. 106, S1219–S1228. 10.1289/ehp.98106s51219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Liu A., Li G., Chen W., Lin X. (2013). Chemiluminescent cholesterol sensor based on peroxidase-like activity of cupric oxide nanoparticles. Biosens. Bioelectron. 43, 1–5. 10.1016/j.bios.2012.11.031 [DOI] [PubMed] [Google Scholar]
- Hood E., Simone E., Wattamwar P., Dziubla T., Muzykantov V. (2011). Nanocarriers for vascular delivery of antioxidants. Nanomedicine 6, 1257–1272. 10.2217/nnm.11.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez I. L., Notcovich C., Catalano P. N., Bellino M. G., Durán H. (2015). The redox-active nanomaterial toolbox for cancer therapy. Cancer Lett. 359, 9–19. 10.1016/j.canlet.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Johnstone R. A. W., Stocks P. A., Simpson A. J. (1997). Porphyrins in aqueous amphiphilic polymers as peroxidase mimics. Chem. Commun. 23, 2277–2278. 10.1039/a705934e [DOI] [Google Scholar]
- Khaja S. D., Lee S., Murthy N. (2007). Acid-degradable protein delivery vehicles based on metathesis chemistry. Biomacromolecules 8, 1391–1395. 10.1021/bm061234z [DOI] [PubMed] [Google Scholar]
- Khan M. A., Tania M., Zhang D., Chen H. (2010). Antioxidant enzymes and cancer. Chin. J. Cancer Res. 22, 87–92. 10.1007/s11670-010-0087-716475022 [DOI] [Google Scholar]
- Kim S. H., Kim B., Yadavalli V. K., Pishko M. V. (2005). Encapsulation of enzymes within polymer spheres to create optical nanosensors for oxidative stress. Anal. Chem. 77, 6828–6833. 10.1021/ac0507340 [DOI] [PubMed] [Google Scholar]
- Kim S., Park J., Cho J. (2010). Layer-by-layer assembled multilayers using catalase-encapsulated gold nanoparticles. Nanotechnology 21:375702. 10.1088/0957-4484/21/37/375702 [DOI] [PubMed] [Google Scholar]
- Kim J., Grate J. W. Wang, P. (2008). Nanobiocatalysis and its potential applications. Trends Biotechnol. 26, 639–646. 10.1016/j.tibtech.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Klyachko N. L., Manickam D. S., Brynskikh A. M., Uglanova S. V., Li S., Higginbotham S. M., et al. (2012). Cross-linked antioxidant nanozymes for improved delivery to CNS. Nanomedicine 8, 119–129. 10.1016/j.nano.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchey G. P., Zhao Y., Kagan V. E., Star A. (2013). Peroxidase-mediated biodegradation of carbon nanotubes in vitro and in vivo. Adv. Drug Deliv. Rev. 65, 1921–1932. 10.1016/j.addr.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer F. W., Glieder A. (2015). An updated view on horseradish peroxidases: recombinant production and biotechnological applications. Appl. Microbiol. Biotechnol. 99, 1611–1625. 10.1007/s00253-014-6346-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leão-Silva A. C., Naves A. F., Pereira E. M., Petri D. F., Carmona-Ribeiro A. M. (2011). Assembly of horseradish peroxidase within supported cationic bilayers. Biotechnol. Prog. 27, 1433–1441. 10.1002/btpr.640 [DOI] [PubMed] [Google Scholar]
- Lee C. H., Lin T. S., Mou C. Y. (2009). Mesoporous materials for encapsulating enzymes. Nano Today 4, 165–179. 10.1016/j.nantod.2009.02.001 [DOI] [Google Scholar]
- Lei C., Shin Y., Liu J., Ackerman E. J. (2002). Entrapping enzyme in a functionalized nanoporous support. J. Am. Chem. Soc. 124, 11242–11243. 10.1021/ja026855o [DOI] [PubMed] [Google Scholar]
- Levy R. B., Boudart M. (1973). Platinum-like behavior of tungsten carbide in surface catalysis. Science 181, 547–549. 10.1126/science.181.4099.547 [DOI] [PubMed] [Google Scholar]
- Li N., Yan Y., Xia B., Wang J., Wang X. (2014a). Novel tungsten carbide nanorods: an intrinsic peroxidase mimetic with high activity and stability in aqueous and organic solvents. Biosens. Bioelectron. 54, 521–527. 10.1016/j.bios.2013.11.040 [DOI] [PubMed] [Google Scholar]
- Li R., Liu J., Li L., Wang H., Weng Z., Lam S. K., et al. (2014b). Non-covalent surface modification of boron nitride nanotubes for enhanced catalysis. Chem. Commun. 50, 225–227. 10.1039/C3CC45667F [DOI] [PubMed] [Google Scholar]
- Li R., Zhen M., Guan M., Chen D., Zhang G., Ge J., et al. (2013). A novel glucose colorimetric sensor based on intrinsic peroxidase-like activity of C60-carboxyfullerenes. Biosens. Bioelectron. 47, 502–507. 10.1016/j.bios.2013.03.057 [DOI] [PubMed] [Google Scholar]
- Li X., Wen F., Creran B., Jeong Y., Zhang X., Rotello V. M. (2012). Colorimetric protein sensing using catalytically amplified sensor arrays. Small 8, 3589–3592. 10.1002/smll.201201549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckarift H. R., Spain J. C., Naik R. R., Stone M. O. (2004). Enzyme immobilization in abiomimetic silica support. Nat. Biotechnol. 22, 211–213. 10.1038/nbt931 [DOI] [PubMed] [Google Scholar]
- Maiti S., Das K., Dutta S., Das P. K. (2012). Striking improvement in peroxidase activity of cytochrome c by modulating hydrophobicity of surface-functionalized gold nanoparticles within cationic reverse micelles. Chemistry 18, 15021–15030. 10.1002/chem.201202398 [DOI] [PubMed] [Google Scholar]
- Marques H. M. (2007). Insights into porphyrin chemistry provided by the microperoxidases, the haempeptides derived from cytochrome c. Dalton Trans. 21, 4371–4385. 10.1039/b710940g [DOI] [PubMed] [Google Scholar]
- Marquette C. A., Blum L. J. (2009). Chemiluminescent enzyme immunoassays: a review of bioanalytical applications. Bioanalysis 1, 1259–1269. 10.4155/bio.09.69 [DOI] [PubMed] [Google Scholar]
- Maynard S., Schurman S. H., Harboe C., de Souza-Pinto N. C., Bohr V. A. (2009). Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 30, 2–10. 10.1093/carcin/bgn250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220. 10.1093/jxb/erq282 [DOI] [PubMed] [Google Scholar]
- Mishra S., Imlay J. (2012). Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 525, 145–160. 10.1016/j.abb.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Kamiya N., Koda D., Maruyama T., Goto M. (2009). Enzyme encapsulation in microparticles composed of polymerized ionic liquids for highly active and reusable biocatalysts. Org. Biomol. Chem. 7, 2353–2358. 10.1039/b823064a [DOI] [PubMed] [Google Scholar]
- Nantes I. L., Durán N., Pinto S. M. S., Silva F. B., Souza J. S., Isoda N., et al. (2011). Modulation of the catalytic activity of porphyrins by lipid- and surfactant-containing nanostructures. J. Braz. Chem. Soc. 22, 1621–1633. 10.1590/s0103-50532011000900002 [DOI] [Google Scholar]
- Naves A. F., Carmona-Ribeiro A. M., Petri D. F. (2007). Immobilized horseradish peroxidase as a reusable catalyst for emulsion polymerization. Langmuir 23, 1981–1987. 10.1021/la061884o [DOI] [PubMed] [Google Scholar]
- Nunomura A., Castellani R., Zhu X., Moreira P., Perry G., Smith M. (2006). Involvement of oxidative stress in Alzheimer disease. J. Neuropathol. Exp. Neurol. 65, 631–641. 10.1097/01.jnen.0000228136.58062.bf [DOI] [PubMed] [Google Scholar]
- Ocsoy I., Dogru E., Usta S. (2015). A new generation of flowerlike horseradish peroxidasesas a nanobiocatalyst for superior enzymatic activity. Enzyme Microb. Technol. 75–76, 25–29. 10.1016/j.enzmictec.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Oohora K., Hayashi T. (2014). Hemoprotein-based supramolecular assembling systems. Curr. Opin. Chem. Biol. 19, 154–161. 10.1016/j.cbpa.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Palanker V. S., Sokolsky D. V., Mazulevsky E. A., Baybatyrov E. N. (1976). Highly dispersed tungsten carbide for fuel cells with an acidic electrolyte. J. Power Sources 1, 169–176. 10.1016/0378-7753(76)80019-5 [DOI] [Google Scholar]
- Pallarola D., Von Bildering C., Pietrasanta L. I., Queralto N., Knoll W., Battaglini F., et al. (2012). Recognition-driven layer-by-layer construction of multiprotein assemblies on surfaces: a biomolecular toolkit for building up chemoresponsive bioelectrochemical interfaces. Phys. Chem. Chem. Phys. 14, 11027–11039. 10.1039/c2cp41225j [DOI] [PubMed] [Google Scholar]
- Pan M., Sun Y., Zheng J., Yang W. (2013). Boronic acid-functionalized core-shell-shell magnetic composite microspheres for the selective enrichment of glycoprotein. ACS Appl. Mater. Interfaces 5, 8351–8358. 10.1021/am401285x [DOI] [PubMed] [Google Scholar]
- Park K. S., Kim M. I., Cho D.-Y., Park H. G. (2011). Label-free colorimetric detection of nucleic acids based on target-induced shielding against the peroxidase-mimicking activity of magnetic nanoparticles. Small 7, 1521–1525. 10.1002/smll.201001886 [DOI] [PubMed] [Google Scholar]
- Prieto T., Marcon R. O., Prado F. M., Caires A. C., Di Mascio P., Brochsztain S., et al. (2006). Reaction route control by microperoxidase-9/CTAB micelle ratios. Phys. Chem. Chem. Phys. 8, 1963–1973. 10.1039/b601671e [DOI] [PubMed] [Google Scholar]
- Putnam C. D., Arvai A. S., Bourne Y., Tainer J. A. (2000). Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism. J. Mol. Biol. 296, 295–309. 10.1006/jmbi.1999.3458 [DOI] [PubMed] [Google Scholar]
- Ramanavicius A., Kausaite A., Ramanaviciene A. (2005). Biofuel cell based on direct bioelectro catalysis. Biosens. Bioelectron. 20, 1962–1967. 10.1016/j.bios.2004.08.032 [DOI] [PubMed] [Google Scholar]
- Ramanavicius A., Ramanaviciene A. (2009). Hemoproteins in design of biofuel cells. Fuel Cells 1, 25–36. 10.1002/fuce.200800052 [DOI] [Google Scholar]
- Reczek C. R., Chandel N. S. (2014). ROS-dependent signal transduction. Curr. Opin. Cell Biol. 33C, 8–13. 10.1016/j.ceb.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. G. (2006). Cell Signaling: H2O2, a Necessary Evil for Cell Signaling. Science 312, 1882–1883. 10.1126/science.1130481 [DOI] [PubMed] [Google Scholar]
- Rhee S. G., Woo H. A., Kil I. S., Bae S. H. (2012). Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 287, 4403–4410. 10.1074/jbc.R111.283432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rodríguez A., Egea-Guerrero J. J., Murillo-Cabezas F., Carrillo-Vico A. (2014). Oxidative stress in traumatic brain injury. Curr. Med. Chem. 21, 1201–1211. 10.2174/0929867321666131217153310 [DOI] [PubMed] [Google Scholar]
- Rosenbaum M., Zhao F., Schröder U., Scholz F. (2006). Interfacing electrocatalysis and biocatalysis with tungsten carbide: a high-performance, noble-metal-free microbial fuel cell. Angew. Chem. Int. Ed. Engl. 45, 6658–6661. 10.1002/anie.200602021 [DOI] [PubMed] [Google Scholar]
- Ryan O., Smyth M. R., Fágáin C. O. (1994). Horseradish peroxidase: the analyst's friend. Essays Biochem. 28, 129–146. [PubMed] [Google Scholar]
- Schmidt T. F., Caseli L., Viitala T., Oliveira O. N., Jr. (2008). Enhanced activity of horseradish peroxidase in Langmuir-Blodgett films of phospholipids. Biochim. Biophys. Acta 1778, 2291–2297. 10.1016/j.bbamem.2008.05.012 [DOI] [PubMed] [Google Scholar]
- Schröder E., Littlechild J. A., Lebedev A. A., Errington N., Vagin A. A., Isupov M. N. (2000). Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution. Structure 8, 605–615. 10.1016/S0969-2126(00)00147-7 [DOI] [PubMed] [Google Scholar]
- Shao B. (2012). Site-specific oxidation of apolipoprotein A-I impairs cholesterol export by ABCA1, a key cardioprotective function of HDL. Biochim. Biophys. Acta 1821, 490–501. 10.1016/j.bbalip.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon R. A. (2007). Enzyme immobilization: the quest for optimum performance. Adv. Synth. Catal. 349, 1289–1307. 10.1002/adsc.200700082 [DOI] [Google Scholar]
- Shi W. B., Wang Q., Long Y., Cheng Z., Chen S., Zheng H., et al. (2011). Carbon nanodots as peroxidase mimetics and their applications to glucose detection. Chem. Commun. 47, 6695–6697. 10.1039/c1cc11943e [DOI] [PubMed] [Google Scholar]
- Shtenberg G., Massad-Ivanir N., Engin S., Sharon M., Fruk L., Segal E. (2012). DNA-directed immobilization of horseradish peroxidase onto porous SiO2 optical transducers. Nanoscale Res. Lett. 7:443. 10.1186/1556-276X-7-443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedova A. A., Kapralov A. A., Feng W. H., Kisin E. R., Murray A. R., Mercer R., et al. (2012). Impaired clearance and enhanced pulmonary inflammatory/fibrotic response to carbon nanotubes in myeloperoxidase -deficient mice. PLoS ONE 7:e30923. 10.1371/journal.pone.0030923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H. (1997). Oxidative stress: oxidants and antioxidants. Exp. Physiol. 82, 291–295. 10.1113/expphysiol.1997.sp004024 [DOI] [PubMed] [Google Scholar]
- Sies H. (2014). Role of metabolic H2O2 generation: redox signaling and oxidative stress. J. Biol. Chem. 289, 8735–8741. 10.1074/jbc.R113.544635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva R. A., Carmona-Ribeiro A. M., Petri D. F. (2007). Adsorption behavior and activity of horseradish peroxidase onto polysaccharide-decorated particles. Int. J. Biol. Macromol. 41, 404–409. 10.1016/j.ijbiomac.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Spadiut O., Herwig C. (2013). Production and purification of the multifunctional enzyme horseradish peroxidase. Pharm. Bioprocess 1, 283–295. 10.4155/pbp.13.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šulek F., Fernández D. P., Knez Z., Habulin M., Sheldon R.A. (2011). Immobilization of horseradish peroxidase as crosslinked enzyme aggregates (CLEAs). Process Biochem. 46, 765–769. 10.1016/j.procbio.2010.12.001 [DOI] [Google Scholar]
- Sun X., Guo S., Chung C., Zhu W., Sun S. (2013). A sensitive H2O2 assay based on Dumbell-like PtPd-Fe3O4 nanoparticles. Adv. Mater. 25, 132–136. 10.1002/adma.201203218 [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy M., Youngs H. L., Gold M. H., Poulos T. L. (2005). High-resolution crystal structure of manganese peroxidase: substrate and inhibitor complexes. Biochemistry 44, 6463–6470. 10.1021/bi047318e [DOI] [PubMed] [Google Scholar]
- Tatsumi K., Wada S., Ichikawa H. (1996). Removal of chlorophenols from wastewater by immobilized horseradish peroxidase. Biotechnol. Bioeng. 51, 126–130. [DOI] [PubMed] [Google Scholar]
- Van Gaal L., Mertens I., De Block C. (2006). Mechanisms linking obesity with cardiovascular disease. Nature 444, 875–880. 10.1038/nature05487 [DOI] [PubMed] [Google Scholar]
- Vertuani S., Angusti A., Manfredini S. (2004). The antioxidants and pro-oxidants network: an overview. Curr. Pharm. Des. 10, 1677–1694. 10.2174/1381612043384655 [DOI] [PubMed] [Google Scholar]
- Wang H., Jiang W., Wang Y., Liu X., Yao J., Yuan L., et al. (2013). Catalase-like and peroxidase-like catalytic activities of silicon nanowire arrays. Langmuir 29, 3–7. 10.1021/la304378w [DOI] [PubMed] [Google Scholar]
- Wang M., Bao W. J., Wang J., Wang K., Xu J. J., Chen H. Y., et al. (2014). A green approach to the synthesis of novel “Desert rose stone”-like nanobiocatalytic system with excellent enzyme activity and stability. Sci. Rep. 4:6606. 10.1038/srep06606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Yang Z., Zhang X., Xiao X., Chang C. K., Xu B., et al. (2007). A supramolecular-hydrogel-encapsulated hemin as an artificial enzyme to mimic peroxidase. Angew. Chem. Int. Ed. Engl. 46, 4285–4289. 10.1002/anie.200700404 [DOI] [PubMed] [Google Scholar]
- Whittaker J. W. (2012). Non-heme manganese catalase-the ‘other’ catalase. Arch. Biochem. Biophys. 525, 111–120. 10.1016/j.abb.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood-Kaczmar A., Gandhi S., Wood N. (2006). Understanding the molecular causes of Parkinson's disease. Trends Mol. Med. 12, 521–528. 10.1016/j.molmed.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Wu Y., Hu H., Hu J., Liu S. (2012). Glucose-regulated insulin release from acid-disintegrable microgels covalently immobilized with glucose oxidase and catalase. Macromol. Rapid Commun. 33, 1852–1860. 10.1002/marc.201200411 [DOI] [PubMed] [Google Scholar]
- Xu S., Han X. (2004). A novel method to construct a third-generation biosensor: self-assembling gold nanoparticles on thiol-functionalized poly (styrene-co-acrylic acid) nanospheres. Biosens. Bioelectron. 19, 1117–1120. 10.1016/j.bios.2003.09.007 [DOI] [PubMed] [Google Scholar]
- Zámockı M., Gasselhuber B., Furtmüller P. G., Obinger C. (2012). Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys. 525, 131–144. 10.1016/j.abb.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., He W., Wamer W. G., Hu X., Wu X., Lo Y. M., et al. (2013). Enzyme-mimetic effects of gold@platinum nanorods on the antioxidant activity of ascorbic acid. Nanoscale 5, 1583–1591. 10.1039/c2nr33072e [DOI] [PubMed] [Google Scholar]
- Zhu J. Y., Zhang Y. F., Lu D. N., Zare R. N., Ge J., Liu Z. (2013). Temperature-responsive enzyme–polymer nanoconjugates with enhanced catalytic activities in organic media. Chem. Commun. 49, 6090–6092. 10.1039/c3cc42493f [DOI] [PubMed] [Google Scholar]