Abstract
The stunning diversity of cichlid fishes has greatly enhanced our understanding of speciation and radiation. Little is known about the evolution of cichlid parasites. Parasites are abundant components of biodiversity, whose diversity typically exceeds that of their hosts. In the first comprehensive phylogenetic parasitological analysis of a vertebrate radiation, we study monogenean parasites infecting tropheine cichlids from Lake Tanganyika. Monogeneans are flatworms usually infecting the body surface and gills of fishes. In contrast to many other parasites, they depend only on a single host species to complete their lifecycle. Our spatially comprehensive combined nuclear-mitochondrial DNA dataset of the parasites covering almost all tropheine host species (N = 18), reveals species-rich parasite assemblages and shows consistent host-specificity. Statistical comparisons of host and parasite phylogenies based on distance and topology-based tests demonstrate significant congruence and suggest that host-switching is rare. Molecular rate evaluation indicates that species of Cichlidogyrus probably diverged synchronically with the initial radiation of the tropheines. They further diversified through within-host speciation into an overlooked species radiation. The unique life history and specialisation of certain parasite groups has profound evolutionary consequences. Hence, evolutionary parasitology adds a new dimension to the study of biodiversity hotspots like Lake Tanganyika.
Elucidating speciation mechanisms is considered crucial for understanding the dynamics and function of biodiversity. Alternative speciation modes, such as allopatric, sympatric and parapatric speciation are increasingly understood with the help of phylogenetics1. A special process in this field is adaptive radiation, the phenomenon in which rapid speciation is combined with niche differentiation of the evolving species. Studying radiations has proven to be particularly promising to shed light on the causes and mechanisms driving speciation especially when dealing with species confined to a relatively closed system such as lakes2. One of the most prolific vertebrate radiations are the cichlid fishes (Teleostei, Cichlidae) of the East African Great Lakes3.
Lake Tanganyika, the oldest and deepest of these lakes, harbours the genetically and phenotypically most diverse cichlid community of these African lakes4. Its cichlid assemblage is subdivided into 12 to 17 mostly endemic tribes5. One of these tribes, the monophyletic Tropheini, is phylogenetically nested within the tribe Haplochromini and represents the sister group of the species flocks of Lake Malawi and the Lake Victoria region, and of several East African riverine lineages6. Tropheini consists of 23 endemic nominal species. Although considerable knowledge gaps exist regarding their taxonomy and distribution7, their phylogeny is well-resolved and updated8,9. Most species are adapted to rocky shores, and representatives of most genera occur sympatrically7,8. Tropheini contains generalist as well as specialist species that exhibit variable levels of genetic and phenotypic structuring, related to differences in habitat preference, dispersal ability and territoriality8. All these factors sparked substantial scientific interest and rendered the Tropheini radiation a “natural experiment” for species formation.
However, regardless of this showcase of biodiversity, the most spectacular radiations are found among parasites10. Mutual evolutionary pressures maintain genetic diversity in host and parasite, and fuel the rate of genetic diversification11. Moreover, the availability of numerous niches across a host’s body is an additional factor fostering parasite within-host diversification12 and hence speciation. Organisms with a parasitic lifestyle account for most of Earth’s biodiversity13. However, biodiversity studies tend to focus on conspicuous faunas, ignoring the vast biomass and species-richness of helminths and other less sizeable animals14,15. As such, the potential to understand speciation through the study of parasite evolution remains almost unexplored12,16 and the contribution of parasites to the species richness of the African Great Lakes has remained largely overlooked17,18.
We combine speciation research on cichlid hosts and their monogenean flatworm parasites. Monogeneans are mostly ectoparasites of cold-blooded aquatic or amphibious vertebrates although some infect aquatic invertebrates or exhibit an endoparasitic lifestyle19. Cichlid monogeneans provide a good model for elucidating parasite speciation20,21. Their direct (single-host) life cycle makes them particularly interesting, as it may be difficult to discern host factors that influence parasite evolution for parasites with an intermediate host22. Previous studies on Lake Tanganyika monogeneans uncovered a diverse and largely endemic fauna belonging to Gyrodactylus von Nordmann, 1832 and Cichlidogyrus Paperna, 196018,23,24,25. The latter gill parasites represent the most abundant and prevalent monogenean genus on Tanganyika cichlids23. In most tropheine cichlid populations screened to this end, over two-thirds of fish individuals were infected by representatives of this genus26,27,28. Eggs of Cichlidogyrus develop and hatch on the bottom, after which a free-living ciliated larvae infects a host fish29.
We want to understand speciation by reconstructing the phylogenetic history of parasites belonging to Cichlidogyrus, retrieved from almost all tropheine host species with nuclear ITS rDNA and mitochondrial COI sequences. Taxonomic coverage is important in phylogenetic studies in general1 and in cophylogenetic work in particular30,31. The taxonomical and phylogenetic background that exists for the tropheine hosts (see above) is essential for a successful analysis of parasite diversification31. (i) We hypothesize that species of Cichlidogyrus infecting tropheine cichlids are host-specific, with a higher species richness on more stenotopic hosts. The generality of these patterns will be assessed to complement the few morphology-based reports23,28. (ii) The morphology of Cichlidogyrus suggests an influence of tropheine phylogeny on host choice24 and therefore we hypothesize that host and parasite phylogenies are to a certain extent congruent. However, following the results of Mendlová et al.32 for West African species of Cichlidogyrus, we expect that other speciation modes have also contributed to parasite diversification. Possible mechanisms of species formation in parasites include host-switching (ecological transfer between host species), cospeciation (concomitant speciation of host and parasite), duplication (within-host parasite speciation) and sorting (parasite extinction).
Results
Sequence diversity and host-specificity
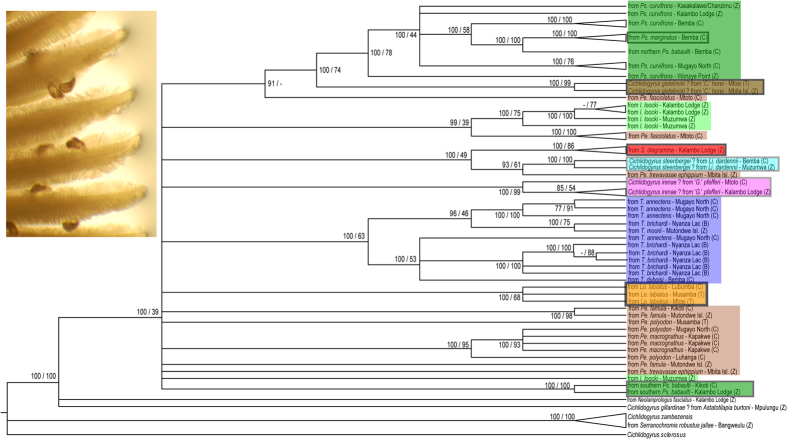
The dataset based on a rDNA fragment of 1399 bp length contained 82 haplotypes and 575 variable sites, of which 323 were parsimony-informative. As two rDNA sequences each from parasite of Petrochromis trewavasae trewavasae and from Petrochromis trewavasae ephippium and three from parasites of Tropheus brichardi differed to the extent that meaningful alignment was hampered, they were excluded from further analyses. For COI, the dataset amounted to 583 bp, 73 haplotypes, 278 variable sites and 230 parsimony-informative sites. The concatenated dataset included 62 haplotypes and totaled 1935 nucleotide positions. This alignment contained 621 bp of ITS-1, 157 bp of 5.8S rDNA, 574 bp of ITS-2 and 583 bp of COI. There were 861 variable sites, of which 659 parsimony-informative. Table 1 provides an overview of the number of sequences and unique haplotypes retrieved for parasites of each host species, of corrected pairwise genetic distances between parasites of the respective host species, and of the estimated number of species sampled based on the species-level cut-offs proposed for the respective nuclear and mitochondrial sequences (see Materials and Methods). Our largest ITS rDNA dataset covers parasites of Lobochilotes labiatus, Simochromis diagramma and ‘Ctenochromis’ horei. Using ITS based species richness estimates, these cichlids harbor seven, three and one species of Cichlidogyrus, respectively. Identical parasite haplotypes were consistently retrieved from conspecific hosts. This host-specificity pattern is stronger than obvious from the phylogenetic tree (Fig. 1) as most haplotypes represent multiple individuals (Table 2).
Table 1. Summary of the sequence dataset of Cichlidogyrus flatworms from Lake Tanganyika.
| Host species | ITS rDNA |
COI |
estimated #species |
|||||
|---|---|---|---|---|---|---|---|---|
| #sequences | #haplotypes | distances | #sequences | #haplotypes | distances | Ziętara & Lumme | Hansen et al. | |
| C. horei | 30 | 4 | 0.1–0.7 | 2 | 2 | 11.6 | 1 | 2 |
| G. pfefferi | 5 | 3 | 0.1–0.3 | 3 | 3 | 0.5–2.2 | 1 | 2 |
| I. loocki | 8 | 3 | 1.3–2.0 | 5 | 5 | 0.3–17.8 | 3 | 3 |
| Li. dardennii | 5 | 3 | 0.1–0.3 | 3 | 3 | 0.5–17.0 | 1 | 2 |
| Lo. labiatus | 46 | 26 | 0.1–3.9 | 3 | 3 | 12.6–14.7 | 7 | 3 |
| Pe. famula | 6 | 3 | 0.2–1.5 | 3 | 3 | 6.5–16.8 | 2 | 3 |
| Pe. fasciolatus | 3 | 2 | 0.9 | 3 | 3 | 0.2–15.8 | 1 | 2 |
| Pe. macrognathus | 3 | 1 | / | 6 | 6 | 1.1–18.0 | 1 | 5 |
| Pe. polyodon | 8 | 6 | 0.6–4.3 | 5 | 5 | 1.8–19.3 | 5 | 4 |
| Pe. trewavasae trewavasae | 2* | / | / | 5 | 5 | 0.4–20.1 | ||
| Pe. trewavasae ephippium | 10 (+2*) | 3 | 2.4–3.2 | 2 | 2 | 19.3 | 3 | 2 |
| northern Ps. babaulti | 2 | 1 | / | 3 | 2 | 0.7 | 1 | 1 |
| southern Ps. babaulti | 3 | 2 | 0.3 | 5 | 4 | 0.7–22.0 | 1 | 3 |
| Ps. curvifrons | 12 | 6 | 0.1–6.3 | 7 | 7 | 0.4–14.7 | 4 | 2 |
| Ps. marginatus | 13 | 5 | 0.1–2.5 | 2 | 2 | 0.2 | 2 | 1 |
| S. diagramma | 23 | 4 | 0.1–1.5 | 2 | 2 | 14.4 | 3 | 2 |
| T. annectens | 5 | 3 | 0.2–5.7 | 4 | 4 | 0.2–27.8 | 2 | 2 |
| T. brichardi | 6 (+3*) | 2 | 3.4 | 9 | 6 | 0.7–18.4 | 2 | 2 |
| T. duboisi | 3 | 1 | / | 1 | 1 | / | 1 | 1 |
| T. moorii | 5 | 2 | 4.8 | 1 | 1 | / | 2 | 1 |
| A. burtoni | / | / | / | 2 | 1 | / | 1 | |
| Se. robustus | 1 | 1 | / | 3 | 3 | 1.1–1.9 | 1 | 1 |
| N. fasciatus | 1 | 1 | / | / | / | / | 1 | / |
Gamma-corrected pairwise genetic distances (in %) between Cichlidogyrus haplotypes retrieved within respective host species, with indication of the number of sequences and unique haplotypes obtained for each sequenced region. The estimated number of Cichlidogyrus species per host species in this dataset is given according to the rules-of-thumb mentioned (for ITS) in Ziętara & Lumme57 and (for COI) in Hansen et al.69.
(*not included in the analysis–non-alignable).
Figure 1. Consensus cladogram of flatworms belonging to the genus Cichlidogyrus infecting Lake Tanganyika cichlids.
Cladogram based on the combined nuclear ITS-1, 5.8S rDNA, ITS-2 and mitochondrial COI sequences of Cichlidogyrus parasitizing Lake Tanganyika tropheine cichlids and the outgroups mentioned in Table 2. Statistical support is shown as posterior probability under BI/ML bootstrap. Clades that neither yield a support value of 85 nor of 70 under BI or ML, respectively, were collapsed; “–” indicates that a clade was not recovered in a particular analysis. Tip labels indicate host species with sampling locality and country (C: Democratic Republic of Congo; B: Burundi; T: Tanzania and Z: Zambia) and are coloured according to host genus consistent with Fig. 4. Monophyletic assemblages infecting one host species are boxed. Inlet: Cichlidogyrus parasites (300–400 μm in length) on the gills of Sarotherodon melanotheron Rüppell, 1852 (photograph taken by author A.P.).
Table 2. Overview of cichlids, parasites and locations sampled.
| LT |
BW | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DRC |
B | T |
Z |
||||||||||||||||
| Luhanga, 27/3/2010 3°31′04′′S, 29°08′57′′E | Bemba, 26/3/2010 3°37′22′′S, 29°08′56′′E | Lubumba, 24/3/2010 3°58′54′′S, 29°06′32′′E | Mugayo North, 11/4/2010 6°46′42′′S, 29°33′30′′E | Mtoto, 15/4/2010 6°58′03′′S, 29°43′50′′E | Kapakwe, 15/4/2010 6°58′27′′S, 29°44′05′′E | Mufazi, 13/4/2010 7°05′12′′S, 29°54′45′′E | Kikoti, 20/4/2010 7°11′28′′S, 30°04′01′′E | Nyanza Lac, 2/5/2010 4°14'38" S, 29°33'17" E | Mtosi, 24/4/2008 7°35'27''S, 30°38´29′′E | Musamba, 25/4/2008 7°49'54′′S, 30°56´49′′E | Kalambo Lodge, 15–19/4/2008 8°37'22''S, 31°12'02''E | Muzumwa, 3/9/2011 8°42'06"S, 31°11'60''E | Wonzye Point, 12/4/2008 8°43'31''S, 31°08'00''E | Mutondwe Island, 11/4/2008 08°42'09S'', 31°07'12''E | Kalambo River Delta, Chipwa, 9/2011 8° 36' 6'' S, 31°11' 12'' E | Mbita Island, 9–10/4/2008 8°44'55''S, 31°05'28''E | Kasakalawe/Chanzimu, 13/4/2008 8°46'52''S,31°05'25''E | Fiwili, 18/7/2010 11°57'S 30°15'E | |
| Tropheini | |||||||||||||||||||
| ‘C.’ horei (Günther, 1894) | (1)3/1/1 | (9)24/0/- | (1)3/1/1 | ||||||||||||||||
| ‘G.’ pfefferi (Boulenger, 1898) | (1)1/1/1 | (3)4/2/2 | |||||||||||||||||
| I. loocki (Poll, 1949) | (1)2/2/2 | (2)6/3/3 | |||||||||||||||||
| Li. dardennii (Boulenger, 1899) | (1)2/1/1 | (1)2/0/- | (1)1/2/1 | ||||||||||||||||
| Lo. labiatus (Boulenger, 1898) | (1)4/1/1 | (1)2/1/1 | (1)2/1/1 | (5)36/0/- | (1)2/0/- | ||||||||||||||
| Pe. famula Matthes and Trewavas, 1960 | (1)1/1/1 | (2)5/2/2 | |||||||||||||||||
| Pe. fasciolatus Boulenger, 1914 | (1)3/3/3 | ||||||||||||||||||
| Pe. macrognathus Yamaoka, 1983 | (1)0/3/- | (1)3/3/3 | |||||||||||||||||
| Pe. polyodon Boulenger, 1898 | (1)1/1/1 | (1)0/1/- | (1)1/2/1 | (2)5/1/1 | (1)1/0/- | ||||||||||||||
| Pe. trewavasae trewavasae Poll, 1948 | (3)2/5/2 | ||||||||||||||||||
| Pe. trewavasae ephippium Brichard, 1989 | (1)6/0/- | (2)6/2/2 | |||||||||||||||||
| northern Ps. babaulti (Pellegrin, 1927) | (1)2/3/2 | ||||||||||||||||||
| southern Ps. babaulti | (2)0/2/- | (1)1/1/1 | (2)2/2/2 | ||||||||||||||||
| Ps. curvifrons (Poll, 1942) | (1)1/0/- | (1)3/2/2 | (1)2/2/2 | (2)4/1/1 | (1)1/1/1 | (1)1/1/1 | |||||||||||||
| Ps. marginatus (Poll, 1956) | (1)2/2/2 | (2)11/0/- | |||||||||||||||||
| S. diagramma (Günther, 1894) | (6)21/2/2 | (1)2/0/- | |||||||||||||||||
| T. annectens Boulenger, 1900 | (2)4/4/4 | (1)1/0/- | |||||||||||||||||
| T. brichardi Nelissen and Thys van den Audenaerde, 1975 | (4)9/9/6 | ||||||||||||||||||
| T. duboisi Marlier, 1959 | (1)3/1/1 | ||||||||||||||||||
| T. moorii Boulenger, 1898 | (2)5/1/1 | ||||||||||||||||||
| Haplochromini | |||||||||||||||||||
| A. burtoni (Günther, 1894) | (1)0/2/- | ||||||||||||||||||
| Se. robustus (Günther, 1864) | (1)1/3/- | ||||||||||||||||||
| Lamprologini | |||||||||||||||||||
| N. fasciatus (Boulenger, 1898) | (1)1/0/- | ||||||||||||||||||
Numbers between brackets indicate the number of host fish specimens used; other numbers represent the number of Cichlidogyrus specimens sequenced as follows: number of nuclear ITS rDNA sequences obtained/number of mitochondrial COI sequences obtained/number of specimens of which both sequences were obtained. LT: Lake Tanganyika; BW: Bangweulu Wetlands; DRC: Democratic Republic of Congo; B: Burundi; T: Tanzania; Z: Zambia.
Phylogenetic analyses
Tree reconstruction on the basis of the concatenated dataset (Fig. 1) showed that all Tanganyika monogeneans are grouped in a well-supported clade. In the tree, the parasites of the following taxa are grouped together: the non-tropheine Tanganyika endemic Neolamprologus fasciatus, the haplochromine Astatotilapia burtoni, which is endemic to the Tanganyika Basin but not to the lake proper, and the Tropheini. Within this group, Cichlidogyrus infecting tropheines is supported as a monophyletic group. Cichlidogyrus sclerosus and its congener C. zambezensis, the latter hosted on the riverine haplochromine Serranochromis robustus jallae, are clearly separated from the Tanganyikan parasites. Well-supported clusters of Cichlidogyrus are organised according to host species. Irrespective of sampling locality, ‘Ctenochromis’ horei, ‘Gnathochromis’ pfefferi, Limnotilapia dardennii, Lobochilotes labiatus, Simochromis diagramma and southern Pseudosimochromis babaulti all harbour monophyletic parasite clades. The parasites of Ps. curvifrons are paraphyletic with respect to the lineages infecting northern Ps. babaulti and Ps. marginatus. This is in agreement with the hosts’ affinities within one genus and with the recent removal of Ps. babaulti and Ps. marginatus from Simochromis25. The Tropheus parasites cluster at the host genus level. The clades of T. annectens and T. brichardi parasites do not strictly follow host species boundaries. However, haplotypes were never shared between Tropheus species. Petrochromis does not host monophyletic parasite assemblages, which is in agreement with its paraphyly and need for taxonomic revision8,24.
A decrease in speciation rate towards the present can be observed on the LTT plot (Fig. 2). A positive value (17.41) for the difference in AIC score between the best-fit rate-constant and rate-variable model indicates that the speciation rate of Cichlidogyrus changed with time. This difference was significant as it outnumbered all differences in AIC scores comparing the same models for 5000 randomly generated trees of the same size.
Figure 2. Lineages-through-time plot for Cichlidogyrus parasites of Tropheini.
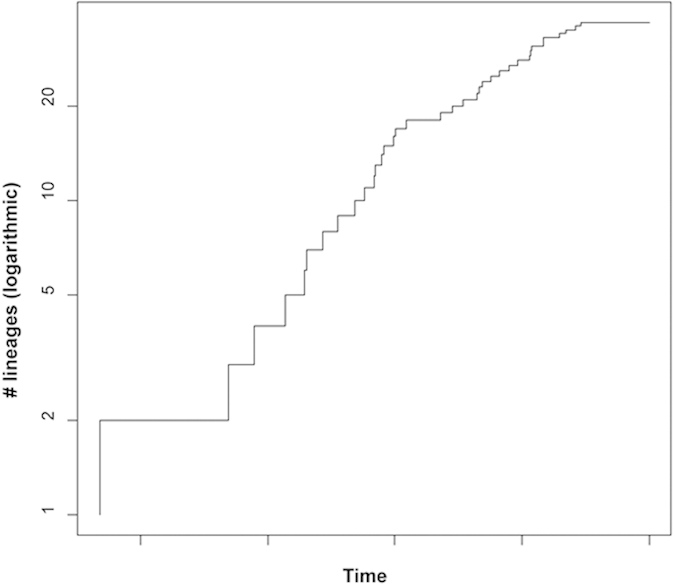
Lineages-through-time plot based on an ultrametric Bayesian ITS rDNA tree, constructed under a relaxed clock model, of Cichlidogyrus living on tropheine hosts; x-axis: time; y-axis: number of lineages (logarithmic scale).
Cophylogenetic analyses
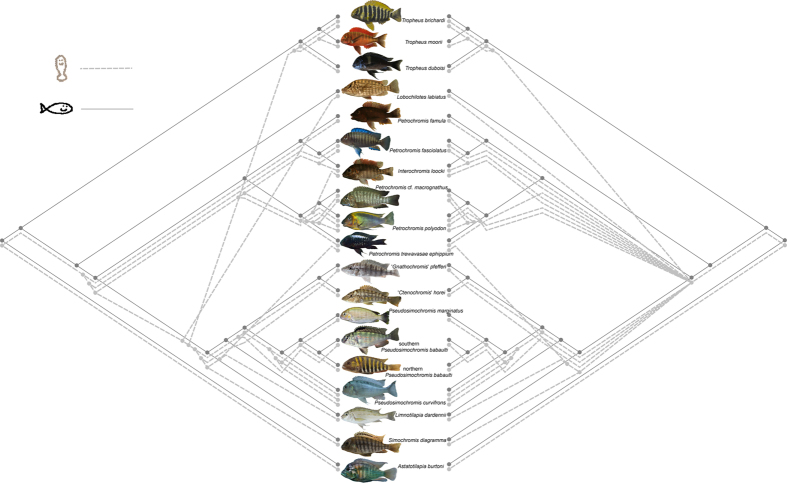
The two best solutions proposed by the CoRe-Pa software, either based on a resolved parasite tree or a parasite tree with a basal polytomy, are presented in Table 3. The best solution for the resolved tree and the second best solution in the polytomy case invoked no host-switching and an unrealistically high cost for it, in comparison to the costs inferred for other events. These scenarios are not biologically plausible. Cost schemes should not only be judged on a purely statistical basis but also include the biological context33. Hence these reconstructions were not further considered and, in case of the resolved tree scenario, replaced by the second best proposal (Fig. 3). The two retained solutions, with and without a polytomy in the parasite tree, proposed comparable numbers for all events, with a high number (33–40) of sortings, similar frequencies of cospeciation (11–13) and within-host speciation events (12-11) and a low number of host-switches (4-3) (Table 3; Fig. 3). In the software package TreeMap the inferred number of 12 cospeciation events was found statistically significant at a level of 0.05, because only 421 out of 104 random reconciliations included 12 or more cospeciation events. This indicates topological congruence between host and parasite trees. Overall congruence between the host and parasite mitochondrial genotypes was significant in a distance-based cophylogenetic analysis (P < 0.01) although only 11 out of 47 links were reported as significant at a level of 0.05.
Table 3. Cophylogenetic reconciliations proposed by CoRe-Pa.
| Quality | Total cost | Cospeciation | Sorting | Duplication | Host-switch | |
|---|---|---|---|---|---|---|
| Fully resolved | 0.0054 | 0.61 | 12 (0.017) | 51 (0.0040) | 15 (0.013) | 0 (0.97) |
| 0.04 | 9.53 | 11 (0.25) | 33 (0.074) | 12 (0.20) | 4 (0.47) | |
| Basal polytomy | 0.012 | 7.98 | 13 (0.16) | 40 (0.051) | 11 (0.19) | 3 (0.60) |
| 0.018 | 0.032 | 10 (0.000998) | 65 (0.00017) | 17 (0.00067) | 0 (0.998) |
Quality value, total value, and number of cospeciation, sorting, duplication and host-switch events invoked (with estimated cost in brackets) for the two best CoRe-Pa reconciliations of Tropheini (based on the AFLP markers as published by Koblmüller et al.8) and Cichlidogyrus (based on the concatenated nuclear-mitochondrial dataset) trees. Topology-based cophylogenetic analyses used either a fully resolved parasite ML tree or a parasite ML tree with nodes supported by a bootstrap value under 70 collapsed. Solutions depicted in bold are visualized in Fig. 3.
Figure 3. Co-phylogenetic reconciliations of Cichlidogyrus and Tropheini trees.
CoRe-Pa reconciliations of Tropheini (based on AFLP as published by Koblmüller et al.8) and Cichlidogyrus (based on the concatenated nuclear-mitochondrial dataset) trees. Left: second best reconciliation based on a fully resolved parasite ML tree; right: best reconciliation based on a parasite ML tree where nodes with bootstrap support under 70 were collapsed. Branches and tips represent hosts (dark gray) or parasites (dashed/light gray). Drawings made by author T.H.; photographs taken by authors P.I.H. (I. loocki, L. labiatus, Pe. famula, T. moorii), M.P.M.V. (Ps. curvifrons) and M.V.S. (L. dardennii) and reproduced with kind permission from Radim Blažek (‘C.’ horei, ‘G.’ pfefferi, northern Ps. babaulti, S. diagramma, T. duboisi) and Ad Konings (A. burtoni, Pe. fasciolatus, Pe. macrognathus, Pe. polyodon, Pe. trewavasae ephippium, Ps. marginatus, southern Ps. babaulti, T. brichardi).
Discussion
The well-studied cichlid tribe Tropheini of Lake Tanganyika, a lineage of endemic and mainly rock-dwelling species, was used as a framework to study parasite diversity and speciation. Phylogenetic reconstruction of its monogenean parasites belonging to Cichlidogyrus covered nearly all nominal tropheine host species and resulted in a clear pattern of host-specificity and congruence between host and parasite trees. We explore how parasite diversity relates to the biology of the respective host species, and how parasite speciation mechanisms relate to the radiation within Tropheini.
Representatives of Cichlidogyrus are abundant on tropheine hosts: our sampling shows a picture throughout the tribe’s populations, similar to previous case studies26,27,28, of two-thirds to all of the hosts infected (unpublished data). Many species within the genus infect just one (or a set of closely related) host species21. However, host-specificity varies among species and lineages of Cichlidogyrus, and the degree of host-specificity may correlate with the biology of the host34. In Lake Tanganyika, a rather generalist species of Cichlidogyrus infects pelagic bathybatines18. Conversely, based on the few morphology-based case-studies, representatives of Cichlidogyrus seemed to be host-specific on littoral Tanganyika cichlids (overview in Pariselle et al.18). We confirm this host-specificity genetically for parasites belonging to Cichlidogyrus of the entire tribe Tropheini. Conspecific cichlid populations host the same parasite species even when geographically separated by hundreds of kilometers. This is clearly exemplified by the monophyletic and monospecific parasite clades of ‘C.’ horei, ‘G.’ pfefferi, L. dardennii and southern Ps. babaulti (Table 1; Fig. 1). Conversely, sympatric host species had their unique set of parasite species. On several localities in the D.R. Congo and Zambia, sampling comprised representatives of the four main clades within Tropheini, namely Lobochilotes, Petrochromis/Interochromis, Tropheus and the ‘substrate dwellers’ including ‘Ctenochromis’, ‘Gnathochromis’, Limnotilapia, Pseudosimochromis and Simochromis8. Their parasite fauna never overlapped. As eggs of Cichlidogyrus develop away from their parental host and infective larvae have to actively colonize a new fish, each parasite individual in the survey may be considered an independent observation. Hence there is little chance of overestimating host-specificity, unlike species with clonal reproduction on the host (e.g. within Gyrodactylus35).
This consistent host-specificity is remarkable in view of tropheine ecology; with many species sympatrically inhabiting shallow rocky habitat, opportunities for parasite transfer are plenty. Monogeneans recognize their host using the species-specific chemical and physical properties of the fish’ integument36. Interindividual and interspecific variation in chemical cues characterize cichlids, as evidenced by tests for olfaction-based mate recognition37. Chemical cues emitted by the tropheine hosts might hence explain how flatworms belonging to Cichlidogyrus discern between host species. The life history of Cichlidogyrus seems to select for successful colonization through host specialisation, as larvae are shortlived and therefore have to find a suitable host soon. Moreover they do not have a second chance because they cannot switch hosts after attachment29. It should be noted that factors other than colonization of the host may also explain host-specificity. For example, differential survival after reaching an ant host colony is considered important in the specificity of “cuckoo species” of myrmecophilous lycaenid butterflies38. Competition can also mediate host-specificity39. Anyhow, the narrow host-range of Cichlidogyrus from the tropheine system is striking, given that some congeners display a much wider host range, both within18 and outside Lake Tanganyika34,40. More specific monogeneans tend to be found on larger-bodied or longer-lived fishes and their specialisation is considered to be a consequence of higher predictability of host resources41,42. While Mendlová and Šimková34 did not find evidence of this predictability hypothesis in Cichlidogyrus with regard to host body size or longevity, we assume that Lake Tanganyika tropheine littoral cichlids are predictable resources as regards their ecology because their abundance and species-richness are higher than for cichlids in the pelagic realm5,17. It has been observed in many systems that abundant hosts harbor more specialised parasite species43.
Our estimates of species richness (Table 1) have to be regarded with caution because sample size plays a role when estimating species numbers. The risk of underestimating parasite diversity is especially valid in rarely sampled host species. However, for the tropheine hosts of which the species of Cichlidogyrus are well characterized morphologically, ITS based estimates correspond to the number of formally described parasite species: one in ‘Ctenochromis’ horei, ‘Gnathochromis’ pfefferi and Limnotilapia dardennii44 and three in Interochromis loocki24 and Simochromis diagramma25. By focusing on the best-sampled species, a pattern emerges. A stenotopic host such as Lobochilotes labiatus harbors more species of Cichlidogyrus than eurytopic cichlids such as Simochromis species and ‘C.’ horei8. This matches with the morphology-based observation of Grégoir et al.28, which was based on just two tropheine species. Hence, as suggested by Pariselle et al.18, host isolation or migration influences the species richness of the Cichlidogyrus community. Many factors were mentioned to determine the number of congeneric parasites a host supports, but the evolutionary mechanisms remain poorly understood45. Isolation among host populations is suggested as a strong driver of parasite genetic structuring16,46 and is here proposed to also promote parasite speciation.
Parasites provide useful complementary data on their fish host, e.g. in elucidating their hosts’ biogeography, identification or phylogeny47. The affinities between representatives of Cichlidogyrus in closely related hosts corroborate recent findings on the phylogenetic relationships among the Tropheini in several aspects. Firstly, the monophyletic clustering of Tropheus parasites includes parasites found on T. duboisi. The position of this host species within Tropheus was confirmed only recently8. Secondly, Ps. marginatus parasites are phylogenetically nested within those of the closely related Ps. curvifrons, corroborating their shared parasite species25. Thirdly, although they also share a species of Cichlidogyrus25, northern and southern Ps. babaulti are also infected by monogeneans which are not closely related. Hence parasite data agree with the morphological and genetic differentiation of geographically separated Ps. babaulti populations. This corresponds with the historical division of the Lake in subbasins, the influence of which on cichlid diversity is well-documented48.
The significant congruence between host and parasite phylogenies in distance-based and topology-based cophylogenetic analysis suggests that cospeciation played an important role in the diversification of the parasite fauna of the Tropheini. Although host-specificity may promote cospeciation49, it does not preclude speciation through host-switching50. Since the seminal paper of Hafner et al.51 which showed cospeciation between gophers and their ectoparasitic lice, cospeciation has rarely been demonstrated. Many empirical studies have shown that the combination of host and parasite traits and biogeography led to little or no congruence between host and parasite phylogenies, even when there is host-specificity (e.g. for Rhabdomys four-striped mice and Polyplax sucking lice52). Even in the case of phylogenetic congruence, other factors have been shown to be stronger drivers of speciation than coevolutionary interactions, such as geographic isolation (yuccas and associated prodoxid moths53) or phylogenetically constrained host-switching (gobies and Gyrodactylus54). For several systems, including Cichlidogyrus of West African cichlids, host-switching and duplication were suggested to be the underlying mechanisms of parasite diversity32. The difference with our results could be explained by two factors. West African cichlids are infected by a combination of specialist and more generalist monogeneans, and the ecological differences between the lacustrine tropheines and the more generalist cichlids included in Mendlová et al.32 are considerable. Topological reconciliations propose that cospeciation and duplication events are three to four times as frequent as host-switches in the Cichlidogyrus-Tropheini system, and that the number of parasite extinctions is high (Table 3). Frequent extinction is likely in the case of overdispersion, which is common in Monogenea20. In addition, the often small and fluctuating population sizes of monogeneans55 may promote extinction. The fact that the outcome is very similar for fully resolved and unresolved parasite trees (Table 3; Fig. 3) suggests that the proportions are robust and not an artefact of poor phylogenetic resolution. In order to discriminate cospeciation from preferential host-switching, an absolute timeframe is required to establish temporal congruence54,56. Most parasite genetic distances between host species range between 2 and 7% (ITS). Using the 2.4 Mya estimate of Koblmüller et al.8 for the most recent common ancestor of Tropheini, this would translate into an ITS mutation rate of Cichlidogyrus of 0.4–1.5% my−1. This is lower than the rate of 5.5% my−1 calculated for Gyrodactylus57 but this monogenean has a much shorter generation time35, which likely results in a higher mutation rate58. Therefore, four conclusions come to mind. (1) Divergence within Tropheini and the associated Cichlidogyrus fauna was probably concomitant. (2) The basal polytomy in the parasite tree (Fig. 1) represents a true (“hard”) polytomy, congruent with the rapid radiation of the host which led to a similar polytomy8. (3) It is likely that the diversification of these flatworms happened within the confines of Lake Tanganyika in view of the monophyly of the ingroup. (4) Simultaneous diversification of tropheine cichlids and their parasites belonging to Cichlidogyrus might explain the decrease in parasite speciation rate towards the present (Fig. 2), which is indicative of a radiation event1.
Cospeciation has rarely been observed in fish parasites in general22,33,59 or in monogeneans in particular60. In terrestrial systems it is in many instances a by-product of restricted contact between host species (phthirapteran chewing lice of geomyid pocket gophers51 or of seabirds61) or predominantly vertical transmission (Buchnera symbiotic bacteria of Uroleucon aphids30). Neither of them apply to representatives of Cichlidogyrus infecting the many sympatrically occurring8 tropheine cichlids. Our survey covers aquatic hosts occurring in the same lentic microhabitats, with parasites that have a free-living and actively recolonizing larval stage, hence offering plenty of opportunities for host-switching. However, the colonisation mode of the parasite, with a single attachment event to the host, a short survival time away from the host, and sufficient access to the typical host species in the littoral habitat, seems to select for a specific host choice and against ecological transfer. Both the topology-based phylogenetic analysis and the monophyletic host-associated clusters point to the equally important role of within-host speciation. Its importance has been reported in other dactylogyridean monogeneans, e.g. on West African cichlids32, European cyprinids62 and Asian pangasiids63. Hence, we propose that the monogenean fauna diversified as a result of reproductive isolation following host speciation, in combination with within-host duplication resulting in higher parasite than host diversity. When parasites on a shared host species are sister taxa like in the present case, Poulin64 suggests that they may be named parasite species flocks. Hence, we propose that the Cichlidogyrus fauna on tropheine cichlids is an overlooked case of the numerous invertebrate radiations of Lake Tanganyika17.
Methods
Sampling and data collection
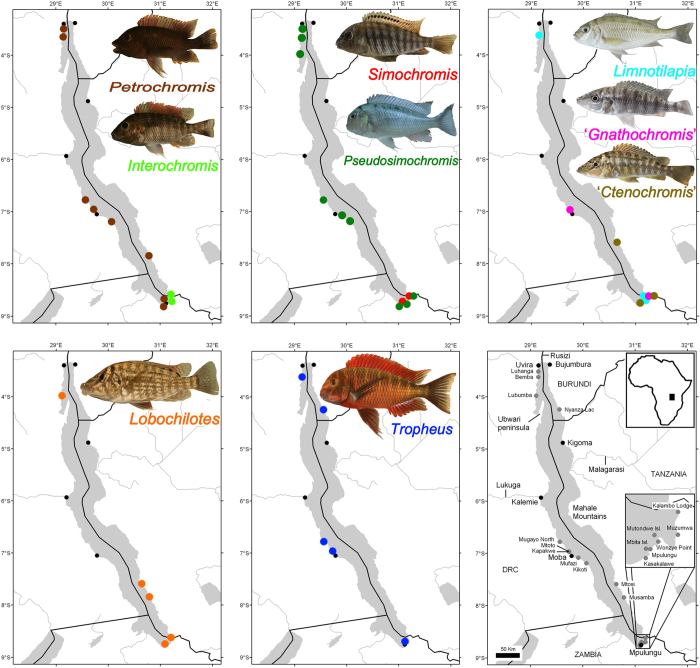
Cichlidogyrus specimens were collected on 18 of the 23 nominal species of Tropheini. Nominal species excluded are Tropheus kasabae Nelissen, 1977, a junior synonym of T. moorii65; T. polli Axelrod, 1977, a junior synonym of T. annectens65; Petrochromis horii Takahashi & Koblmüller, 2014, which was unknown at the time of sampling; Petrochromis orthognathus Matthes, 1959 and Simochromis margaretae Axelrod and Harrison, 1978. The latter species is known only from four museum specimens, none of which suited for molecular analyses; inspection of two of these individuals did not yield gill monogeneans. We distinguish between northern and southern Ps. babaulti. Indeed, the southern populations form a separate clade8 and were classified at the time of sampling as a separate species: Ps. pleurospilus (Nelissen, 1978). The latter species was only recently synonymized with Ps. babaulti25. Hence, host taxon sampling is almost as exhaustive as possible. Given the position of the Tropheini within the haplochromines (see above), two haplochromines were included to use their parasites belonging to Cichlidogyrus as outgroup. These are Astatotilapia burtoni, a derived haplochromine which occurs in Lake Tanganyika tributaries, and a more basal representative of the Haplochromini, Serranochromis robustus jallae from southern Africa. In addition, the lamprologine Neolamprologus fasciatus was sampled as it shares the rocky littoral habitat with many tropheines. Figure 4 and Table 2 provide a detailed overview of species and locations from which samples were retrieved.
Figure 4. Lake Tanganyika localities sampled for monogenean cichlid parasites belonging to Cichlidogyrus.
Colour codes refer to the respective host genera; the bottom right map details the sampling localities and major cities. For details, see Table 2. Photographs were taken by authors P.I.H. (I. loocki, L. labiatus, Pe. famula, T. moorii), M.P.M.V. (Ps. curvifrons) and M.V.S. (L. dardennii) and reproduced with kind permission from Radim Blažek (‘C.’ horei, ‘G.’ pfefferi, S. diagramma). Map created using ArcMap v.10 and reproduced with kind permission by Tobias Musschoot.
Cichlids were collected in the rocky littoral of Lake Tanganyika using gill nets. Sampling protocols were approved by the following competent national authorities, and carried out in accordance with research permit no. 2007-258-CC-2006-151 from the Tanzania Commission for Science and Technology (COSTECH); the memorandum of understanding between the Karl-Franzens University of Graz, the University of Zambia and the Department of Fisheries, Zambian Ministry of Agriculture and Co-operatives; and mission statement no. 013/MNRST/CRHU/2010 from the Ministère de la Recherche Scientifique et Technologique–CRH-Uvira. Newly collected fish were kept alive in aerated tanks until they were sacrificed by severing the spinal cord or with an overdose of MS-222. They were identified to species level in situ and in the laboratories of the RMCA, where host vouchers are kept (Table 4). Host fish or their branchial arches were fixed and stored in pure ethanol. Gills were inspected for monogeneans under an Olympus SZX12 stereomicroscope. Parasites were isolated with a dissection needle and stored in 5 μl of milli-Q H20 at −20 °C awaiting further processing. A small number of flatworms were stored in the field on FTA Classic Cards (Whatman). In total, sequences were obtained from 220 parasite specimens, retrieved from 84 cichlid individuals. DNA extraction, PCR amplification and sequencing followed Vanhove23 (Supplementary Methods S1). Table 4 shows the GenBank accession numbers of the parasite sequences obtained.
Table 4. Accession numbers of parasite sequences and host vouchers used to reconstruct a combined nuclear-mitochondrial phylogeny of Cichlidogyrus infecting Lake Tanganyika tropheine cichlids.
| Host species | Country | Locality | RMCA (MRAC) accession numbers (host vouchers) | GenBank accession numbers (parasite sequences) |
|
|---|---|---|---|---|---|
| rDNA | COI | ||||
| ‘Ctenochromis’ horei* | Tanzania | Mtosi | B2-04-P-117 | KT037139-41 | KT037337 |
| Zambia | Kalambo Lodge | B2-04-P-119-131 (1,4,5,6,7,8,9,10,11) | KT037142-65 | / | |
| Mbita Island | B2-04-P-118 | KT037166-8 | KT037338 | ||
| ‘Gnathochromis’ pfefferi* | D.R. Congo | Mtoto | T10-2024 | KT037169 | KT037339 |
| Zambia | Kalambo Lodge | B2-04-P-149-165 (1,2,3) | KT037170-3 | KT037340-1 | |
| Interochromis loocki* | Zambia | Kalambo Lodge | B3-36-P-1 | KT037174-5 | KT037342-3 |
| Muzumwa | B1-23-P-339-341 (1,6) | KT037176-81 | KT037344-6 | ||
| Limnotilapia dardennii* | D.R. Congo | Bemba | B0-12-P-1205 | KT037182-3 | sequence too short for GenBank |
| Zambia | Kalambo Lodge | B2-04-P-132-148 (4) | KT037184-5 | / | |
| Muzumwa | T11_Lida5 | KT037186 | KT037347-8 | ||
| Lobochilotes labiatus | D.R. Congo | Lubumba | B0-12-P-312-315 (2) | KT037187-90 | KT037349 |
| Tanzania | Mtosi | B2-04-P-184-189 (1) | KT037191-2 | KT037350 | |
| Musamba | B2-04-P-190-194 (2) | KT037193-4 | KT037351 | ||
| Zambia | Kalambo Lodge | B2-04-P-166-181 (1,2,3,4,5) | KT037195-230 | / | |
| Mbita Island | B2-04-P-113 | KT037231-2 | / | ||
| Petrochromis famula | D.R. Congo | Kikoti | B0-12-P-866 | KT037233 | KT037352 |
| Zambia | Mutondwe Island | B2-04-P-195-199 (2,4) | KT037234-8 | KT037353-4 | |
| Pe. fasciolatus | D.R. Congo | Mtoto | B0-12-P-861 | KT037239-41 | KT037355-7 |
| Pe. macrognathus | D.R. Congo | Kapakwe | T10-Pema1 | KT037242-4 | KT037358-60 |
| Luhanga | B0-12-P-1206 | / | KT037361-3 | ||
| Pe. polyodon | D.R. Congo | Bemba | B0-12-P-1208-1210 (788) | / | KT037364 |
| Luhanga | B0-12-P-1207 | KT037245 | KT037365 | ||
| Mugayo North | B0-12-P-1211 | KT037246 | KT037366-7 | ||
| Tanzania | Musamba | B2-04-P-200-201 (1,2) | KT037247-51 | KT037368 | |
| Zambia | Mbita Island | B2-04-P-211 | KT037252 | / | |
| Pe. trewavasae trewavasae | D.R. Congo | Kikoti | B0-12-P-867, B0-12-P-456-457 (1,2) | / | KT037369-72, one sequence too short for GenBank |
| Pe. trewavasae ephippium | Zambia | Mbita Island | B2-04-P-203-204 (1,2) | KT037253-6 | KT037373-4 |
| Mutondwe Island | B2-04-P-202 | KT037257-62 | / | ||
| northern Pseudosimochromis babaulti* | D.R. Congo | Bemba | B0-12-P-846 | KT037263-4 | KT037375-7 |
| southern Ps. babaulti* | D.R. Congo | Kikoti | B0-12-P-426 | KT037265 | KT037378 |
| Mufazi | B0-12-P-816-829 (3,8) | / | KT037379-80 | ||
| Zambia | Kalambo Lodge | B2-04-P-70-91 (1,10) | KT037266-7 | KT037381-2 | |
| Ps. curvifrons* | D.R. Congo | Bemba | B0-12-P-430 | KT037268-70 | KT037383-4 |
| Luhanga | B0-12-P-748 | KT037271 | / | ||
| Mugayo North | B0-12-P-750 | KT037272-3 | KT037385-6 | ||
| Zambia | Kalambo Lodge | B2-04-P-98-110 (1,4) | KT037274-7 | KT037387 | |
| Kasakalawe/Chanzimu | B2-04-P-97 | KT037278 | KT037388 | ||
| Wonzye Point | B2-04-P-95 | KT037279 | KT037389 | ||
| Ps. marginatus* | D.R. Congo | Bemba | B0-12-P-429 | KT037280-1 | KT037390-1 |
| Lubumba | B0-12-P-379-380 | KT037282-92 | / | ||
| Simochromis diagramma* | Zambia | Kalambo Lodge | B2-04-P-52, B2-04-P-58-64 (1,4,5,6,8) | KT037293-313 | KT037392-3 |
| Mbita Island | B2-04-P-114-115 (2) | KT037314-5 | / | ||
| Tropheus annectens | D.R. Congo | Kapakwe | B0-12-P-1235 | KT037316 | / |
| Mugayo North | B0-12-P-1212-1234 (660,673) | KT037317-20 | KT037394-7 | ||
| T. brichardi | Burundi | Nyanza Lac | B0-12-P-1236-1256 (1336,1339,1341,1345,1346) | KT037321-6 | KT037398-406 |
| T. duboisi | D.R. Congo | Bemba | B0-12-P-7 | KT037327-9 | KT037407 |
| T. moorii | Zambia | Mutondwe Island | B2-04-P-205-210 (1,3) | KT037330-4 | KT037408 |
| Astatotilapia burtoni* | Zambia | Mpulungu | T11-CHC2** | / | KT037409-10 |
| Serranochromis robustus jallae*,*** | Zambia | Fiwili | B5-15-P-1 | KT037335 | KT037411-3 |
| Neolamprologus fasciatus | Zambia | Kalambo Lodge | B2-04-P-212 | KT037336 | / |
Phylogenetic analyses of parasites belonging to Cichlidogyrus
The closest BLAST hit from GenBank, the complete ITS-1 sequence of Cichlidogyrus sclerosus Paperna and Thurston, 1969 (DQ537359) was included as additional outgroup for rooting. Sequence alignment was performed by MUSCLE v.3.866 under default distance measures and sequence weighting schemes. The resulting alignments were visually inspected and improved in MEGA v.567. In the case of COI, alignment was straightforward as there were no gaps and translation into amino acids (using the echinoderm and flatworm mitochondrial code) did not result in nonsense or stop codons. These nuclear and mitochrondrial datasets were used separately for an assessment of genetic diversity. jModelTest v.0.1.168 was used to select the optimal molecular evolution model starting from a maximum likelihood (ML) optimized tree. Based on the corrected Akaike information criterion (AICc), the TVM + Γ model was selected for the nuclear alignment and the TIM2 + I + Γ model for the mitochondrial dataset (with gamma shape parameter of 0.40 for ITS rDNA and 0.11 for COI). Gamma-corrected pairwise genetic distances were calculated in PAUP* v.4.01b (Swofford, 2001, Sinauer Associates). As a rough estimate of the species diversity contained in the sample, the number of haplotypes displaying at least 1% (for ITS rDNA) and 2% divergence (for COI) was determined. This follows the ITS divergence cut-off proposed to match morphospecies boundaries in the best-studied monogenean, Gyrodactylus57, and the threshold of sequence divergence between species commonly used in barcoding69.
For tree reconstruction, a concatenated dataset was built on the basis of the specimens that yielded both nuclear and mitochondrial sequences. For an assessment of the phylogenetic content of the dataset, we performed a likelihood mapping analysis based on quartet puzzling70 implemented in TREE-PUZZLE v.5.271. In this combined alignment, the proportion of fully resolved quartets was 85.2%, with 9.4% partly resolved and 5.5% unresolved. In view of its relatively high phylogenetic content and the use of independently evolving (unlinked) markers, such concatenated nuclear-mitochondrial dataset allows for more robust (co-)phylogenetic hypotheses to be put forward (see also the recommendations by de Vienne et al.56).
Bayesian inference of phylogeny (BI) was carried out in MrBayes v.372. Posterior probabilities were calculated over 107 generations. Stationarity of the Markov chain was reached, as evidenced by a standard deviation of split frequencies of 0.008, by a potential scale reduction factor converging to 1 and by the absence of a trend in the plot of log-probabilities as a function of generations. The Markov chain was sampled with a frequency of 102 generations; one-fourth of the samples were discarded as “burn-in”. A ML search was carried out in RAxML v.7.3.073, assessing nodal support through 1000 bootstrap samples. The evolutionary model was optimized for each fragment separately, suggesting HKY + Γ for ITS-1, JC for 5.8S rDNA, TPM1uf + Γ for ITS-2 and GTR + Γ for COI. These models were substituted by GTR + Γ in RAxML and, in the case of ITS-2, also in MrBayes, as this was the implemented model with the best AICc score. In the latter software, all parameter estimates for the various sequence portions were unlinked.
To assess the rate of diversification as a function of time, we started from the largest dataset (ITS rDNA) in order to include a maximal sample size. Identical sequences were removed, as well as haplotypes differing less than 0.01, with the help of the CD-HIT Suite web server74, and additional manual removal in case of length differences. This sequence selection (see above for the rationale behind this cut-off) ensures a focus on speciation rather than on intraspecific variation. For this nuclear alignment, MEGA selected the HKY + Γ model as the optimal model of molecular evolution based on the Bayesian information criterion. Under this model, an ultrametric tree was built in BEAST v.1.8.175 applying four rate categories with the initial and average value of the gamma shape parameter set to 0.29, under the Yule tree prior and an uncorrelated relaxed log-normal clock model. Indeed, a likelihood-ratio test conducted in TREE-PUZZLE had rejected the molecular clock hypothesis. The Markov Chain Monte Carlo run was run for 107 generations with a sample frequency of 103 generations; a burn-in of one-tenth was applied. Based on this tree, a lineages-through-time (LTT) plot was constructed in APE76. In view of the possibly ambiguous interpretation of the course of a LTT, LASER77 was used to quantify possible changes in diversification rate over time. This package compares the likelihood of data under models with a constant versus variable rate of diversification by contrasting the AIC score of the best-fit rate-constant model with that of the best-fit rate-variable model. To assess statistical significance, the difference in AIC scores between the best-fit rate-constant and rate-variables models was calculated for 5000 randomly generated trees, using the same set of models and the same tree size (37 terminal nodes).
Cophylogenetic analyses
A range of methods exists to compare the phylogeny and divergence in host-parasite or other symbiotic systems, inferring the speciation patterns that contributed to the consistencies or inconsistencies in their evolutionary trajectories. Of these, methods reconciling host and parasite tree topologies are often considered to maximize cospeciation events, and to take topological congruence as an evidence for cospeciation, which is not always justified56. To minimize the risks of such assumptions, host and parasite tree were reconciled in the software package CoRe-Pa v.0.578, checking 104 cost sets using a simplex method on the quality function. This software carries out an event-based analysis. A major asset is its parameter-adaptive approach that allows for the automated estimation of proportional event costs, thus avoiding a priori cost assignment to the different categories of parasite speciation mechanisms79,80. Root-to-root mapping was enforced and the chronological consistency of events checked. To work with a fully resolved tree that best approaches the “species tree”, the ML parasite phylogram reconstructed by RAxML for the combined nuclear-mitochondrial dataset, was included in cophylogenetic analysis. The AFLP tree of Koblmüller et al.8 was coded with the help of TreeSnatcher81 to provide a host topology. The selection of optimal trees obviously entails some uncertainty for this topology-based analysis. Therefore, the analysis was repeated with a parasite phylogeny in which all nodes receiving less than 70% of bootstrap support were collapsed. As within-host speciation in terminal taxa can artificially infer cospeciations at the cost of additional duplications56,63, terminal monophyletic clades associated with a single host species were collapsed. For the same reason, the software was set not to bill an additional duplication for a host-switching event. Host and parasite tree were also reconciled in a heuristic search in TreeMap v.1.0a82. Because this topology-based software is known to maximize the number of cospeciation events, the number of the various speciation mechanisms that TreeMap proposes will not be taken into consideration. The test for the significance of the number of cospeciation events inferred is only considered as a measure of topological congruence rather than of cospeciation56. This was assessed by randomizing host and parasite topologies (104 random trees used) under the proportion-to-distinguishable model.
Distance-based cophylogenetic analyses test for correlation between phylogenies without assuming congruence to be produced by cospeciation, and are hence considered less biased than topology-based methods56. Distance-based cophylogenetic analysis was carried out in Copycat v.1.1483 making use of AxParafit and AxPcoords84 using 9999 permutations under default settings. In this analysis, the independence between host and parasite patristic distances is tested. To this end, distance matrices were constructed with the help of T-rex85. For the hosts, the mitochondrial ND2 and control region data from Koblmüller et al.8 were used, as these mitochondrial fragments are better suited for distance calculations than AFLP data (long terminal and short internal branches in the AFLP tree). Because of the large number of insertions/deletions in the nuclear fragment which inevitably bias genetic distance estimates over the whole dataset, only all COI mitochondrial fragments, omitting the third codon position, were used to infer parasite patristic distances.
Additional Information
How to cite this article: Vanhove, M. P. M. et al. Hidden biodiversity in an ancient lake: phylogenetic congruence between Lake Tanganyika tropheine cichlids and their monogenean flatworm parasites. Sci. Rep. 5, 13669; doi: 10.1038/srep13669 (2015).
Supplementary Material
Acknowledgments
J. Bamps, A. F. Grégoir, L. Makasa, J. K. Zimba, C. Katongo, T. Veall, O. R. Mangwangwa, V. Nshombo Muderhwa, T. Mulimbwa N’sibula, D. Muzumani Risasi, F. Muterezi Bukinga, J. Mbirize Ndalozibwa, V. Lumami Kapepula, the Schreyen-Brichard family, F. Willems, S. Dessein, A. Indermaur, A. Theis, B. Egger and W. Salzburger are thanked for their help in collecting samples, R. Bills for permission to inspect Simochromis margaretae specimens for gill parasites, T. Musschoot for Fig. 4, M. Parrent for curatorial services and A. Konings and R. Blažek for fish pictures. D. T. J. Littlewood, E. Verheyen, S. B. Janssens, M. H. D. Larmuseau, F. Van den Broeck, N. Van Steenkiste, L. Rüber, M. Gelnar, D. R. Brooks and W. A. Boeger are gratefully acknowledged for valuable suggestions. This is publication ISE-M 2015-047. M.P.M.V. and M.V.S. were recipients of a PhD fellowship, and T.H. of a post-doctoral fellowship, of the Research Foundation–Flanders (FWO– Vlaanderen). Fieldwork was funded by two travel grants from the Research Foundation–Flanders (to M.P.M.V.) and from the King Leopold III Fund for Nature Conservation and Exploration (to M.P.M.V. and M.V.S.), while molecular analysis was funded by the Joint Experimental Molecular Unit (JEMU) project PARATROPH. C.S. was supported by the Austrian Science Foundation, Grant numbers I48-B06, P20994-B03 and P22737-B09. We gratefully acknowledge the support of KU Leuven–VES/12/005, of FWO-Vlaanderen Research Programme G.0553.10, and of the Czech Science Foundation, Project no. P505/12/G112 (European Centre of Ichthyoparasitology (ECIP)–Centre of excellence).
Footnotes
Author Contributions M.P.M.V., C.S., J.S., F.A.M.V. and T.H. conceived and designed the study; M.P.M.V., C.G., B.H. and T.H. carried out the labwork; M.P.M.V., C.G., S.K. and T.H. analysed the data; M.P.M.V., A.P., M.V.S., J.A.M.R., P.I.H., F.C.B. and C.S. provided samples and/or consumables; M.P.M.V., A.P., M.V.S., J.A.M.R., P.I.H., S.K., C.S., J.S., F.A.M.V. and T.H. wrote the manuscript. All authors checked and agreed with the final manuscript.
References
- Barraclough T. G. & Nee S. Phylogenetics and speciation. Trends Ecol. Evol. 16, 391–399 (2001). [DOI] [PubMed] [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation (Oxford University Press, Oxford, 2000). [Google Scholar]
- Kocher T. D. Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet. 5, 288–298 (2004). [DOI] [PubMed] [Google Scholar]
- Snoeks J. How well known is the ichthyodiversity of the large East African lakes? Adv. Ecol. Res. 31, 17–38 (2000). [Google Scholar]
- Koblmüller S., Sefc K. M. & Sturmbauer C. The Lake Tanganyika cichlid species assemblage: recent advances in molecular phylogenetics. Hydrobiologia 615, 5–20 (2008). [Google Scholar]
- Salzburger W., Mack T., Verheyen E. & Meyer A. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of haplochromine cichlid fishes. BMC Evol. Biol. 5, 17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steenberge M. et al. A recent inventory of the fishes of the north-western and central western coast of Lake Tanganyika (Democratic Republic Congo). Acta Ichthyol. Piscat. 41, 201–214 (2011). [Google Scholar]
- Koblmüller S., Egger B., Sturmbauer C. & Sefc K. M. Rapid radiation, ancient incomplete lineage sorting and ancient hybridization in the endemic Lake Tanganyika cichlid tribe Tropheini. Mol. Phylogenet. Evol. 55, 318–334 (2010). [DOI] [PubMed] [Google Scholar]
- Takahashi T. & Koblmüller S. A new species of Petrochromis (Perciformes: Cichlidae) from Lake Tanganyika. Ichthyol. Res. 61, 252–264 (2014). [Google Scholar]
- Price P. W. Evolutionary Biology of Parasites (Princeton University Press, Princeton, 1980). [Google Scholar]
- Thompson J. N. The evolution of species interactions. Science 284, 2116–2118 (1999). [DOI] [PubMed] [Google Scholar]
- de Meeûs T., Michalakis Y. & Renaud F. Santa Rosalia revisited: or why are there so many kinds of parasites in ‘the garden of early delights’? Parasitol. Today 14, 10–13 (1998). [DOI] [PubMed] [Google Scholar]
- Windsor D. A. Most of the species on Earth are parasites. Int. J. Parasitol. 28, 1939–1941 (1998). [DOI] [PubMed] [Google Scholar]
- Lafferty K. D. et al. Parasites in food webs: the ultimate missing links. Ecol. Lett. 11, 533–546 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca V. G. et al. Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nature Comm. 1, 98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyse T., Poulin R. & Théron A. Speciation in parasites: a population genetics approach. Trends Parasitol. 21, 469–475 (2005). [DOI] [PubMed] [Google Scholar]
- Coulter G. W. Lake Tanganyika and Its Life (Natural History Museum Publications and Oxford University Press, London and New York, 1991). [Google Scholar]
- Pariselle A., Muterezi Bukinga F., Van Steenberge M. & Vanhove M. P. M. Ancyrocephalidae (Monogenea) of Lake Tanganyika: IV: Cichlidogyrus parasitizing species of Bathybatini (Teleostei, Cichlidae): reduced host-specificity in the deepwater realm? Advances in Cichlid Research: Behavior, Ecology and Evolutionary Biology. Koblmüller S., Albertson R. C., Genner M. J., Sefc K. M. & Takahashi T. (eds.) Hydrobiologia 748, 99–119 (2015). [Google Scholar]
- Pugachev O. N., Gerasev P. I., Gussev A. V., Ergens R. & Khotenowsky I. Guide to Monogenoidea of Freshwater Fish of Palaearctic and Amur Regions (Ledizione-LediPublishing, Milan, 2009). [Google Scholar]
- Pariselle A., Morand S., Deveney M. & Pouyaud L. Parasite species richness of closely related hosts: historical scenario and “genetic” hypothesis. Hommage à Louis Euzet—taxonomie, écologie et évolution des métazoaires parasites. Taxonomy, ecology and evolution of metazoan parasites. Combes C. & Jourdan J. (eds.) 147–166 (Presses Universitaires de Perpignan, Perpignan, 2003). [Google Scholar]
- Pouyaud L., Desmarais E., Deveney M. & Pariselle A. Phylogenetic relationships among monogenean gill parasites (Dactylogyridea, Ancyrocephalidae) infesting tilapiine hosts (Cichlidae): systematic and evolutionary implications. Mol. Phylogenet. Evol. 38, 241–249 (2006). [DOI] [PubMed] [Google Scholar]
- Jousson O., Bartoli P. & Pawlowski J. Cryptic speciation among intestinal parasites (Trematoda: Digenea) infecting sympatric host fishes (Sparidae). J. Evol. Biol. 13, 778–785 (2000). [Google Scholar]
- Vanhove M. P. M. Species Flocks and Parasite Evolution. Towards a Co-phylogenetic Analysis of Monogenean Flatworms of Cichlids and Gobies (KU Leuven, Leuven, 2012). [Google Scholar]
- Pariselle A. et al. Ancyrocephalidae (Monogenea) of Lake Tanganyika: does the Cichlidogyrus parasite fauna of Interochromis loocki (Teleostei, Cichlidae) reflect its host’s phylogenetic affinities? Contrib. Zool. 84, 25–38 (2015). [Google Scholar]
- Van Steenberge, M. et al. Morphology, molecules, and monogenean parasites: an example of an integrative approach to cichlid biodiversity. PLoS ONE 10, e0124474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers J. A. M. et al. Contrasting parasite communities among allopatric colour morphs of the Lake Tanganyika cichlid Tropheus. BMC Evol. Biol. 13, 41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hablützel P. I. et al. Intermediate number of major histocompatibility complex class IIB length variants relates to enlarged perivisceral fat deposits in the blunt‐head cichlid Tropheus moorii. J. Evol. Biol. 27, 2177–2190 (2014). [DOI] [PubMed] [Google Scholar]
- Grégoir A. F. et al. A link between host dispersal and parasite diversity in two sympatric cichlid fishes of Lake Tanganyika. Freshwater Biol. 60, 323–335 (2015). [Google Scholar]
- Paperna I. Parasites, infections and diseases of fishes in Africa. An update. CIFA Technical Paper 31 (Food and Agriculture Organization of the United Nations, Rome, 1996). [Google Scholar]
- Clark M. A., Moran N. A., Baumann P. & Wernegreen J. J. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution 54, 517–525 (2000). [DOI] [PubMed] [Google Scholar]
- Page R. D. M., Clayton D. H. & Paterson A. M. Lice and cospeciation: a response to Barker. Int. J. Parasitol. 26, 213–218 (1996). [DOI] [PubMed] [Google Scholar]
- Mendlová M., Desdevises Y., Civáňová K., Pariselle A. & Šimková A. Monogeneans of West African cichlid fish: evolution and cophylogenetic interactions. PLoS ONE 7, e37268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdevises Y., Morand S., Jousson O. & Legendre P. Coevolution between Lamellodiscus (Monogenea; Diplectanidae) and Sparidae (Teleostei): the study of a complex host-parasite system. Evolution 56, 2459–2471 (2002). [DOI] [PubMed] [Google Scholar]
- Mendlová M. & Šimková A. Evolution of host specificity in monogeneans parasitizing African cichlid fish. Parasite. Vector 7, 69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke T. A., Cable J. & Harris P. D. The biology of gyrodactylid monogeneans: The “Russian-doll Killers.” Adv. Parasit. 64, 161–376, 459-460 (2007). [DOI] [PubMed] [Google Scholar]
- Whittington I. D., Cribb B. W., Hamwood T. E. & Halliday J. A. Host-specificity of monogenean (platyhelminth) parasites: a role for anterior adhesive areas? Int. J. Parasitol. 30, 305–320 (2000). [DOI] [PubMed] [Google Scholar]
- Plenderleith M., van Oosterhout C., Robinson R. L. & Turner G. F. Female preference for conspecific males based on olfactory cues in a Lake Malawi cichlid fish. Biol. Lett. 1, 411–414 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli D. et al. Evidence of high larval host ant (Hymenoptera: Formicidae) specificity in the first post-adoption phase for the myrmecophilous butterfly Phengaris (Maculinea) nausithous (Lepidoptera: Lycaenidae). Sociobiology 55, 861–870 (2010). [Google Scholar]
- Adamson M. L., & Caira J. N. Evolutionary factors influencing the nature of parasite specificity. Parasitology 109, S85–S95 (1994). [DOI] [PubMed] [Google Scholar]
- Vanhove M. P. M. et al. Biogeographical implications of Zambezian Cichlidogyrus species (Platyhelminthes: Monogenea: Ancyrocephalidae) parasitizing Congolian cichlids. Zootaxa 3608, 398–400 (2013). [DOI] [PubMed] [Google Scholar]
- Sasal P. & Morand S. Comparative analysis: a tool for studying monogenean ecology and evolution. Int. J. Parasitol. 28, 1637–1644 (1998). [DOI] [PubMed] [Google Scholar]
- Desdevises Y., Morand S. & Legendre P. Evolution and determinants of host specificity in the genus Lamellodiscus (Monogenea). Biol. J. Linn. Soc. 77, 431–443 (2002). [Google Scholar]
- Norton D. A. & Carpenter M. A. Mistletoes as parasites: host specificity and speciation. Trends Ecol. Evol. 13, 101–105 (1998). [DOI] [PubMed] [Google Scholar]
- Gillardin C., Vanhove M. P. M., Pariselle A., Huyse T. & Volckaert F. A. M. Ancyrocephalidae (Monogenea) of Lake Tanganyika: II: description of the first Cichlidogyrus spp. parasites from Tropheini fish hosts (Teleostei, Cichlidae). Parasitol. Res. 110, 305–313 (2012). [DOI] [PubMed] [Google Scholar]
- Poulin R. Speciation and diversification of parasite lineages: an analysis of congeneric parasite species in vertebrates. Evol. Ecol. 13, 455–467 (1999). [Google Scholar]
- McCoy K. D., Boulinier T., Tirard C. & Michalakis Y. Host-dependent genetic structure of parasite populations: differential dispersal of seabird tick host races. Evolution 57, 288–296 (2003). [DOI] [PubMed] [Google Scholar]
- Pariselle A. et al. The monogenean parasite fauna of cichlids: a potential tool for host biogeography. Int. J. Evol. Biol. 2011, 471480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturmbauer C., Bariç S., Salzburger W., Rüber L. & Verheyen E. Lake level fluctuations synchronize genetic divergences of cichlid fishes in African Lakes. Mol. Biol. Evol. 182, 144–154 (2001). [DOI] [PubMed] [Google Scholar]
- Kearn G. C. Evolutionary expansion of the Monogenea. Int. J. Parasitol. 24, 1227–1271 (1994). [DOI] [PubMed] [Google Scholar]
- Brooks D. R. & McLennan D. A. The nature of diversity: an evolutionary voyage of discovery (University of Chicago Press, Chicago and London, 2002). [Google Scholar]
- Hafner M. S. et al. Disparate rates of molecular evolution in cospeciating hosts and parasites. Science 265, 1087–1090 (1994). [DOI] [PubMed] [Google Scholar]
- Du Toit N., van Vuuren B. J., Matthee S. & Matthee C. A. Biogeography and host‐related factors trump parasite life history: limited congruence among the genetic structures of specific ectoparasitic lice and their rodent hosts. Mol. Ecol. 22, 5185–5204 (2013). [DOI] [PubMed] [Google Scholar]
- Althoff D. M., Segraves K. A., Smith C. I., Leebens-Mack J. & Pellmyr O. Geographic isolation trumps coevolution as a driver of yucca and yucca moth diversification. Mol. Phylogenet. Evol. 62, 898–906 (2012). [DOI] [PubMed] [Google Scholar]
- Huyse T. & Volckaert F. A. M. Comparing host and parasite phylogenies: Gyrodactylus flatworms jumping from goby to goby. Syst. Biol. 54, 710–718 (2005). [DOI] [PubMed] [Google Scholar]
- Appleby C. Population dynamics of Gyrodactylus sp. (Monogenea) infecting the sand goby in the Oslo Fjord, Norway. J. Fish Biol. 49, 402–410 (1996). [Google Scholar]
- de Vienne D. M. et al. Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 198, 347–385 (2013). [DOI] [PubMed] [Google Scholar]
- Ziętara M. S. & Lumme J. Speciation by hostswitching and adaptive radiation in a fish parasite genus Gyrodactylus (Monogenea, Gyrodactylidae). Evolution 56, 2445–2458 (2002). [DOI] [PubMed] [Google Scholar]
- Li M., Shi S.-F., Brown C. L. & Yang T.-B. Phylogeographical pattern of Mazocraeoides gonialosae (Monogenea, Mazocraeidae) on the dotted gizzard shad, Konosirus punctatus, along the coast of China. Int. J. Parasitol. 41, 1263–1272 (2011). [DOI] [PubMed] [Google Scholar]
- Gunter N. L. & Adlard R. D. Bivalvulidan (Myxozoa: Myxosporea) parasites of damselfishes with description of twelve novel species from Australia’s Great Barrier Reef. Parasitology 135, 1165–1178 (2008). [DOI] [PubMed] [Google Scholar]
- Littlewood D. T. J., Rohde K. & Clough K. A. Parasite speciation within or between host species? Phylogenetic evidence from site-specific polystome monogeneans. Int. J. Parasitol. 27, 1289–1297 (1997). [DOI] [PubMed] [Google Scholar]
- Paterson A. M., Wallis G. P., Wallis L. J. & Gray R. D. Seabird and louse coevolution: complex histories revealed by 12S rRNA sequences and reconciliation analyses. Syst. Biol. 49, 383–399 (2000). [DOI] [PubMed] [Google Scholar]
- Šimková A., Morand S., Jobet E., Gelnar M. & Verneau O. Molecular phylogeny of congeneric monogenean parasites (Dactylogyrus): a case of intrahost speciation. Evolution 58, 1001–1018 (2004). [DOI] [PubMed] [Google Scholar]
- Šimková A., Serbielle C., Pariselle A., Vanhove M. P. M. & Morand S. Speciation in Thaparocleidus (Monogenea: Dactylogyridae) parasitizing Asian pangasiid catfishes. BioMed Res. Int. 2013, 353956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R. The evolution of monogenean diversity. Int. J. Parasitol. 32, 245–254 (2002). [DOI] [PubMed] [Google Scholar]
- Konings A. Tropheus in their natural habitat (Cichlid Press, El Paso, 2013). [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Hansen H., Bakke T. A. & Bachmann L. DNA taxonomy and barcoding of monogenean parasites: lessons from Gyrodactylus. Trends Parasitol. 23, 363–367 (2007). [DOI] [PubMed] [Google Scholar]
- Strimmer K. & von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. P. Natl. Acad. Sci. USA 94, 6815–6819 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. A., Strimmer K., Vingron M. & von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- Huang Y., Niu B., Gao Y., Fu L. & Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. & Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. Analysis of Phylogenetics and Evolution with R (Second Edition) (Springer, New York, 2012). [Google Scholar]
- Rabosky D. L. LASER: A maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol. Bioinform. Online 2, 247–250 (2006). [PMC free article] [PubMed] [Google Scholar]
- Merkle D., Middendorf M. & Wieseke N. A parameter-adaptive dynamic programming approach for inferring cophylogenies. BMC Bioinformatics 11(Suppl. 1), S60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblueth M., Sayavedra L., Sámano-Sánchez H., Roth A. & Martínez-Romero E. Evolutionary relationships of flavobacterial and enterobacterial endosymbionts with their scale insect hosts (Hemiptera: Coccoidea). J. Evol. Biol. 25, 2357–2368 (2012). [DOI] [PubMed] [Google Scholar]
- Lauron E. J. et al. Coevolutionary patterns and diversification of avian malaria parasites in African sunbirds (Family Nectariniidae). Parasitology 142, 635–647 (2015). [DOI] [PubMed] [Google Scholar]
- Laubach T., von Haeseler A. & Lercher M. J. TreeSnatcher plus: capturing phylogenetic trees from images. BMC Bioinformatics 13, 110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. D. M. Parallel phylogenies: reconstructing the history of host-parasite assemblages. Cladistics 10, 155–173 (1994). [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Huson D. H. & Göker M. Copycat: Cophylogenetic Analysis Tool. Bioinformatics 23, 898–900 (2007). [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Auch A. F., Meier-Kolthoff J. & Göker M. AxPcoords and parallel AxParafit: statistical cophylogenetic analyses on thousands of taxa. BMC Bioinformatics 8, 405 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boc A., Diallo Alpha B. & Makarenkov V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40(W1), W573–W579 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muterezi Bukinga F., Vanhove M. P. M., Van Steenberge M. & Pariselle A. Ancyrocephalidae (Monogenea) of Lake Tanganyika: III: Cichlidogyrus infecting the world’s biggest cichlid and the non-endemic tribes Haplochromini, Oreochromini and Tylochromini (Teleostei, Cichlidae). Parasitol. Res. 111, 2049–2061 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.