Arbuscular mycorrhizal fungi produce a variety of signaling molecules that are shown to promote symbiosis signaling in a range of plant species.
Abstract
Establishment of arbuscular mycorrhizal interactions involves plant recognition of diffusible signals from the fungus, including lipochitooligosaccharides (LCOs) and chitooligosaccharides (COs). Nitrogen-fixing rhizobial bacteria that associate with leguminous plants also signal to their hosts via LCOs, the so-called Nod factors. Here, we have assessed the induction of symbiotic signaling by the arbuscular mycorrhizal (Myc) fungal-produced LCOs and COs in legumes and rice (Oryza sativa). We show that Myc-LCOs and tetra-acetyl chitotetraose (CO4) activate the common symbiosis signaling pathway, with resultant calcium oscillations in root epidermal cells of Medicago truncatula and Lotus japonicus. The nature of the calcium oscillations is similar for LCOs produced by rhizobial bacteria and by mycorrhizal fungi; however, Myc-LCOs activate distinct gene expression. Calcium oscillations were activated in rice atrichoblasts by CO4, but not the Myc-LCOs, whereas a mix of CO4 and Myc-LCOs activated calcium oscillations in rice trichoblasts. In contrast, stimulation of lateral root emergence occurred following treatment with Myc-LCOs, but not CO4, in M. truncatula, whereas both Myc-LCOs and CO4 were active in rice. Our work indicates that legumes and non-legumes differ in their perception of Myc-LCO and CO signals, suggesting that different plant species respond to different components in the mix of signals produced by arbuscular mycorrhizal fungi.
INTRODUCTION
Plants associate with a wide range of microorganisms that facilitate the acquisition of nutrients and protect them against biotic and abiotic stresses. Interactions with arbuscular mycorrhizal (AM) fungi are widespread within land plants, and this association aids in the uptake of nutrients from the soil (Harrison, 2005). In contrast, associations with nitrogen-fixing bacteria leading to nitrogen-fixing nodules are restricted to specific groups of plants, including legumes (Soltis et al., 1995). Both interactions require chemical communication for the establishment of the association that involves the release of diffusible signals from the plant, strigolactones and flavonoids, and the production of diffusible signals from the symbionts (Oldroyd, 2013).
Rhizobial bacteria signal to the plant with Nod factors, which are lipochitooligosaccharides (LCOs) containing a chitin backbone substituted with an N-acyl group and a number of additional groups that vary between Nod factors produced by different species of rhizobia (Dénarié et al., 1996). Perception of Nod factor requires the Nod factor receptors NFR5/NFP and NFR1/LYK3 (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006), in combination with a signaling pathway that is also involved in the establishment of mycorrhizal associations (Oldroyd, 2013). Central to the common symbiosis signaling pathway is the induction of calcium oscillations, which are a feature of rhizobial and mycorrhizal responses (Ehrhardt et al., 1996; Kosuta et al., 2008; Sieberer et al., 2012). Induction of these calcium oscillations requires an additional receptor-like kinase (Endre et al., 2002; Stracke et al., 2002), components of the nuclear pore (Kanamori et al., 2006; Saito et al., 2007; Groth et al., 2010) and nuclear-localized ion channels (Ané et al., 2004; Charpentier et al., 2008), while perception of the calcium oscillations involves a calcium-regulated kinase and its interacting substrate (Lévy et al., 2004; Mitra et al., 2004; Messinese et al., 2007; Yano et al., 2008). Downstream or parallel to this symbiotic signaling pathway are transcription factors with differing roles in rhizobial and mycorrhizal symbioses (Kaló et al., 2005; Smit et al., 2005; Middleton et al., 2007; Gobbato et al., 2012).
AM fungi have also been found to produce diffusible signals that are recognized by the host plant via the common symbiosis signaling pathway, and research suggests that at least two different mycorrhizal signals are active on Medicago truncatula (Kosuta et al., 2003, 2008; Chabaud et al., 2011). This is supported by work in rice (Oryza sativa) that reveals mycorrhizal signaling both dependent and independent of the common symbiosis signaling pathway (Gutjahr et al., 2008). The AM fungus Rhizophagus irregularis produces LCOs (Maillet et al., 2011), some of which are sulfated with a structure very similar to the Nod factor produced by Sinorhizobium meliloti, the symbiont of M. truncatula. These Myc-LCOs activate responses in M. truncatula similar to Nod factor, notably, the induction of a symbiosis reporter, root hair deformation, and the promotion of lateral root outgrowth (Maillet et al., 2011). These responses to the Myc-LCOs were dependent on the common symbiosis signaling pathway and surprisingly also on Nod factor Perception (NFP), a component of the Nod factor receptor of M. truncatula that is not required for the mycorrhizal association (Amor et al., 2003). However, dependencies on downstream transcription factors were more in line with their associated symbionts (Maillet et al., 2011): sulfated S. meliloti Nod factors stimulate lateral root emergence via Nodulation signaling pathway 1 (NSP1; coding a GRAS protein transcription factor predominantly required for nodulation; Smit et al., 2005; Hirsch et al., 2009), whereas nonsulfated Myc-LCOs elicit this response via the Required for Arbuscular Mycorrhization 1 (RAM1) coding for a GRAS protein transcription factor involved in the mycorhizal association (Gobbato et al., 2012).
In addition to the LCOs, AM fungi also produce short-chain chitooligosaccharides (COs), which are capable of activating calcium oscillations in M. truncatula and carrot (Daucus carota) root epidermal cells (Genre et al., 2013). Only the shorter chain COs, such as tetra-acetyl chitotetraose (CO4), but not longer chain COs (e.g., CO8), were found to induce repetitive calcium oscillations (Genre et al., 2013). The mycorrhizal and CO4-induced calcium oscillations have a somewhat different structure to Nod factor-induced calcium oscillations, being less periodic and probably lower amplitude (Kosuta et al., 2008; Chabaud et al., 2011; Genre et al., 2013). It is difficult to explain how the plant discriminates between COs generated by symbiotic versus pathogenic fungi, both of which generate COs as part of the production or breakdown of cell walls. This is further complicated by recent observations that other fungal cell wall fractions from pathogens can activate calcium responses in M. truncatula (Nars et al., 2013). However, these pathogen-induced calcium responses can be clearly discriminated from the much more periodic calcium oscillations observed during symbiotic signaling (Ehrhardt et al., 1996) and even the responses to CO4 (Genre et al., 2013).
While much work has been done on the perception of Myc signals in M. truncatula, these fungi associate with a wide variety of plants and signaling may differ among host species. To better understand the mechanisms by which AM fungi signal to their host, we assessed the induction of symbiotic signaling in legumes and in rice by the AM-produced LCOs and CO4, using two complementary assays: the activation of calcium oscillations and promotion of lateral root emergence. We show that legumes can respond to both Myc-LCOs and CO4 with activation of calcium oscillations, and in M. truncatula, this is a function of the common symbiosis signaling pathway. In contrast, the non-legume rice only appears to respond to CO4 for activation of calcium oscillations, although a mix of CO4 and Myc-LCOs is necessary for induction of calcium oscillations in trichoblasts. Rice responds to Myc-LCOs and CO4 with promotion of lateral root emergence. We conclude that host plants of mycorrhizal fungi are differentially perceptive to the Myc-LCOs and CO4, with differential activation of signaling pathways and downstream developmental responses.
RESULTS
Activation of Calcium Oscillations by the Myc-LCOs and COs in M. truncatula
Two major species of LCOs have been characterized from exudates of R. irregularis: LCO-IV(C16:0, S or C18:1, S), which we will refer to as sulfated (S)-LCO and LCO-IV(C16:0 or C18:1), which we will refer to as nonsulfated (NS)-LCO (Maillet et al., 2011). For this study, we used S-LCO and NS-LCO that were either purified from R. irregularis exudates or were synthesized in genetically modified bacteria as previously described (Maillet et al., 2011). For the synthetic LCOs, a 1:1 mixture of C16:0 and C18:1 N-acyl chains was used for both S-LCO and NS-LCO, as used previously by Maillet et al. (2011) and Czaja et al. (2012). In addition, we used a crude butanol extract from exudates of R. irregularis germinated spores (referred to as M-LCOs) that was concentrated but had no additional purification performed. The LCO content of M-LCOs was previously described (Maillet et al., 2011), and this extract was derived from a butanol extraction that removes COs and therefore should contain the mix of LCOs, but not COs, at the relative concentrations produced by the fungus.
To assess the activation of the symbiosis signaling pathway by the Myc-LCOs and COs, we determined their ability to activate calcium oscillations, the pathway’s earliest measurable event (Oldroyd, 2013). M-LCOs activated calcium oscillations in M. truncatula trichoblasts (root hair cells), as did both purified and synthesized S-LCO and NS-LCO (Figures 1A and 1B). Calcium oscillations activated by R. irregularis (Kosuta et al., 2008) or by the Myc-LCOs were associated with the nuclear region (Figure 1A), which is comparable to what has been described for Nod factors (Sieberer et al., 2009). CO4 was also able to activate nuclear-associated calcium oscillations; however, the structure of these oscillations was very variable (Supplemental Figure 1), and we didn’t observe responses as periodic or as sustained as the responses to Myc-LCOs, consistent with previous reports (Walker et al., 2000; Oldroyd et al., 2001a; Genre et al., 2013).
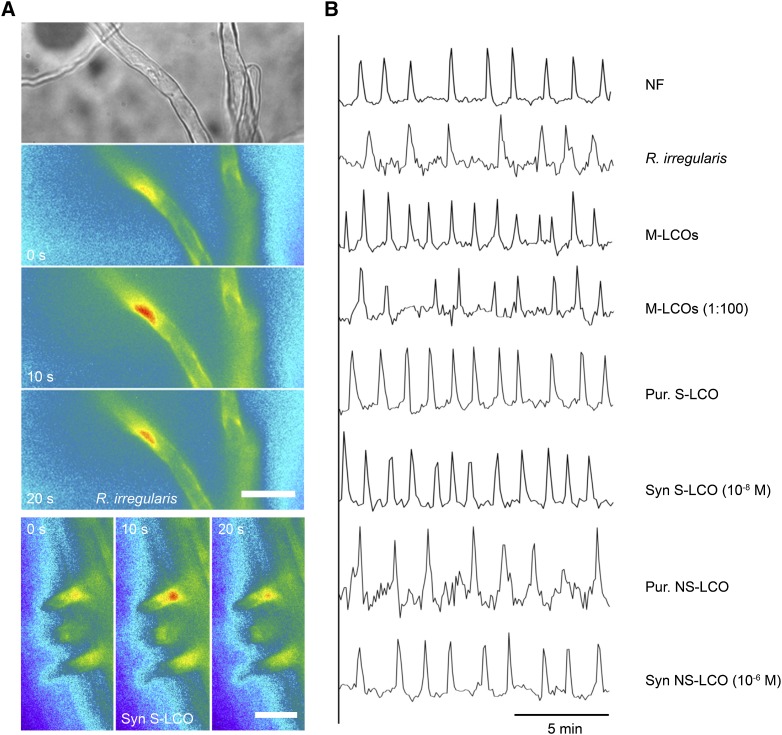
Figure 1.
Myc-LCO-Induced Calcium Responses in M. truncatula.
(A) Images showing a single calcium transient measured using the Yellow Cameleon calcium sensor in a trichoblast of M. truncatula in close proximity to R. irregularis (upper panels) and cells treated with S-LCO at 10−8 M (lower panels). Note that the calcium changes are restricted to the nuclear region. Bars = 50 μm.
(B) Representative traces of M. truncatula trichoblasts responding to Nod factor (NF), R. irregularis, and Myc-LCOs: the LCO mix (M-LCOs), fungal purified (Pur) S-LCOs and NS-LCOs, and synthetic (Syn) S-LCOs and NS-LCOs. The y axis is the ratio of YFP to CFP in arbitrary units.
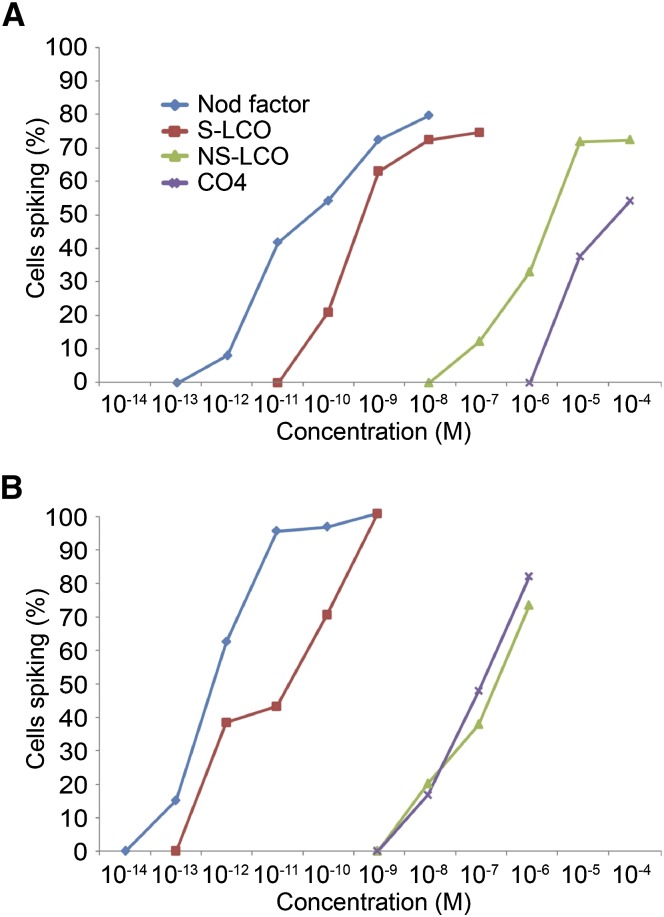
To define physiologically equivalent concentrations for these molecules, we used different concentrations of Nod factor, S-LCO, NS-LCO, and CO4, to assess the induction of calcium oscillations in trichoblasts of M. truncatula. This analysis revealed that S-LCO was ∼10-fold less active than S. meliloti Nod factor, while NS-LCO was ∼10,000-fold less active than S. meliloti Nod factor (Figure 2A). The strong activity of S-LCO might be due to its high structural similarity with S. meliloti Nod factor possibly resulting in the activation of the Nod signaling pathway (Maillet et al., 2011). CO4 activated calcium oscillations at 10−5 M; however, the calcium responses to CO4 were not equivalent to those induced by Myc-LCOs (Supplemental Figure 1), as previously described. We consider that equivalent concentrations of these molecules for activation of the signaling pathway are 10−10 M Nod factor, 10−9 M S-LCO 10−6 M NS-LCO, and, taking into account the differences in the structure of the calcium response, 10−5 M CO4. At these concentrations, ∼50% of cells respond with calcium oscillations.
Figure 2.
Dose–Response Curves for Myc-LCOs and CO4 in Seedling Roots and Lateral Roots of M. truncatula.
Quantification of M. truncatula calcium responses in seedling roots (A) and lateral roots (B) to S. meliloti Nod factor (blue), S-LCO (red), NS-LCO (green), and CO4 (purple). The y axis denotes the percentage of cells measured that show calcium oscillations relative to the concentration of the molecule denoted on the x axis.
The Structure of Myc-LCO-Induced Calcium Oscillations
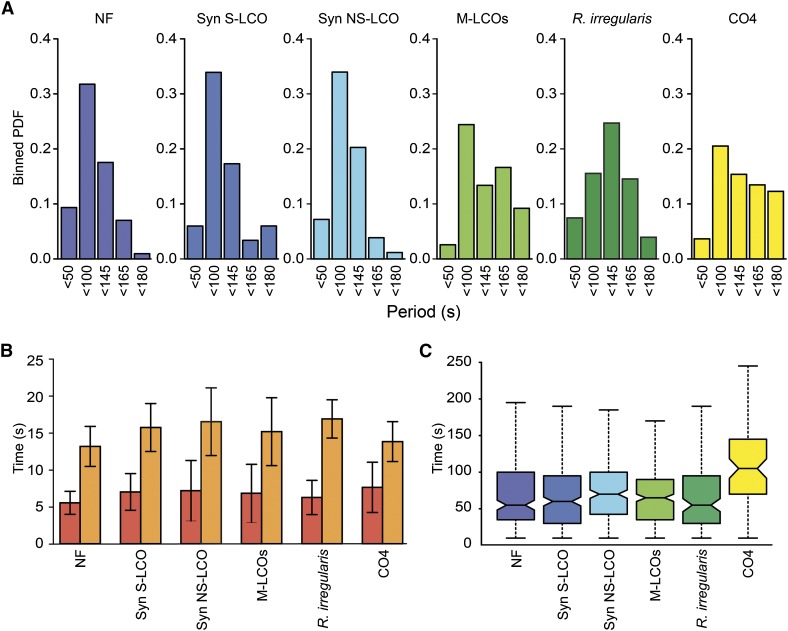
We previously reported differences in the structure of calcium oscillations induced by Nod factor and by AM fungi (Kosuta et al., 2008). While this difference was again apparent (Figure 3A), we observed no difference in the periodicity of the calcium oscillations induced by Nod factor, S-LCO and NS-LCO, with a predominant period between 50 and 100 s (Figures 3A and 3C). Calcium oscillations induced by M-LCOs had a less apparent preference for a specific period of oscillations, and this was similar to what we observed for R. irregularis-induced calcium oscillations (Figure 3A). CO4-induced calcium oscillations also showed longer and more variable periods compared with the Myc-LCO treatments (Figures 3A and 3C) that were more similar to the fungal induced calcium oscillations (Figure 3A). An assessment of the structure of each calcium transient revealed a high degree of similarity between responses induced by Nod factor, S-LCO, NS-LCO, M-LCOs, and CO4 (Figure 3B).
Figure 3.
The Structure of LCO-Induced Calcium Oscillations in M. truncatula.
(A) Bayesian spectrum analysis was used to generate binned probability distributions (PDF) of period length for 10 calcium traces from 10 M. truncatula plants, with each trace consisting of at least 2 h of imaging from each treatment: 10−10 M Nod factor (NF), 10−9 M S-LCO, 10−6 M NS-LCO, M-LCO (1:100 dilution), or 10−5 M CO4. Each sample size represents a group of periods, such that <50 = periods ranging from 1 to 50 s; <100 = periods ranging from 51 to 100 s, etc.
(B) The bar plots show the mean duration for upward (red) and downward (orange) phases of spikes, with their associated standard deviations. A minimum of three traces was used for each treatment, representing ∼80 spikes. No significant difference was observed in the structure of the calcium oscillations induced by the different signals.
(C) The box plots show the interspike intervals during the first 40 min of response. The whiskers mark the minimum and maximum values, and the box marks the lower and upper quartiles, so 50% of the values are inside the box. A thick black line is drawn at the median value. Each group consists of 10 traces that were detrended using the moving average method. The Mann-Whitney U-test detected a statistically significant difference (P < 0.05) in NS-LCO and CO4, compared with the other treatments; however, the NS-LCO responses were not significantly different when it was applied at 10−5 M NS-LCO.
Myc-LCOs and CO4 Activate the Common Symbiosis Signaling Pathway in M. truncatula
To demonstrate that the activation of calcium oscillations by the fungal LCOs was a function of the common symbiosis signaling pathway, we assessed mutants of M. truncatula in this pathway for their response to M-LCOs, S-LCO, and NS-LCO. For this analysis of mutants, we used concentrations of the signaling molecules that activated maximal responses in wild-type plants and thus were 10-fold greater than the concentrations we used previously (Figure 2). DMI1 (Ané et al., 2004) and DMI2 (Endre et al., 2002) were required for activation of calcium oscillations by M-LCOs, S-LCO, and NS-LCO, while Doesn't Make Infections 3 (DMI3), which functions downstream of calcium in symbiosis signaling (Lévy et al., 2004; Mitra et al., 2004), was not (Table 1). NFP was required for induction of calcium oscillations by M-LCOs, S-LCO, and NS-LCO (Table 1), and this result is consistent with what has been reported for the stimulation of root branching by Myc-LCOs that is at least partly NFP dependent (Maillet et al., 2011). The CO4 response was dependent on DMI1, but independent of NFP (Table 1; Supplemental Figure 2). The dependence on NFP is equivalent to previous reports (Genre et al., 2013) and consistent with the improved binding affinities of the closely related Lotus japonicus NFR5 receptor to LCOs versus COs (Broghammer et al., 2012). This work implies that the Myc-LCOs and CO4 activate the common symbiosis signaling pathway in M. truncatula and that this induction by Myc-LCOs involves recognition by a component of the Nod factor receptor, but perception of CO4 differs.
Table 1. Calcium Responses in Symbiosis Signaling Mutants.
| M-LCOs (1:100) | S-LCO (10−8 M) | NS-LCO (10−5 M) | CO4 (10−5 M) | |
|---|---|---|---|---|
| Wild type | 5/7 | 8/11 | 13/18 | 11/29 |
| nfp-1 | 0/12 | 0/9 | 0/17 | 3/10 |
| dmi1-1 | 0/14 | 0/15 | 0/10 | 0/12 |
| dmi2-1 | 0/8 | 0/9 | 0/7 | ND |
| dmi3-1 | 9/13 | 7/8 | 3/3 | ND |
Figures denote cells responding/total cells analyzed. ND, not determined.
Differential Gene Activation by Rhizobial and Myc-LCOs
Our work shows that the Myc-LCOs activate equivalent calcium oscillations in M. truncatula to those observed following Nod factor treatments, but higher concentrations of the Myc-LCOs were required. This coupled with the equivalent genetic requirements for Myc-LCO and Nod factor-induced calcium oscillations makes it difficult to ascertain whether the Myc-LCO treatments are simply activating Nod factor receptors, and this is particularly pertinent for S-LCO, which has close similarity to S. meliloti Nod factor. It has already been revealed that S. meliloti Nod factor, S-LCO, and NS-LCO activate overlapping, but have differential gene profiles in M. truncatula (Czaja et al., 2012), and this analysis provides a broad view of the gene sets induced by these signaling molecules. However, based on our analysis, the concentration of NS-LCO used by Czaja et al. (2012) should be insufficient to activate calcium oscillations, although the dependence on CCaMK by some of these induced genes suggests that a level of calcium signaling is activated. Considering the similarity within the structure of the calcium oscillations, we asked the question: Do the calcium oscillations induced by Nod factor, S-LCO, and NS-LCO activate equivalent sets of genes, or could the plant discriminate between these signaling molecules to induce different gene expression? In addition, we wanted to assess whether the gene induction by the different signaling molecules correlated with symbiont-specific gene induction. For this we used quantitative RT-PCR (qRT-PCR) on RNA isolated from seedling roots of M. truncatula treated with the different LCOs and CO4 and analyzed an array of genes that have been shown to be responsive to the Myc-LCOs or to AM fungi and/or rhizobia in both wild-type and mutant plants (Weidmann et al., 2004; Benedito et al., 2008; Gomez et al., 2009; Takeda et al., 2009; Talukdar et al., 2009; Czaja et al., 2012). We used the concentrations of the signaling molecules sufficient to induce calcium spiking in 50% of epidermal cells: 10−10 M Nod factor, 10−9 M S-LCO, 10−6 M NS-LCO, and 10−5 M CO4.
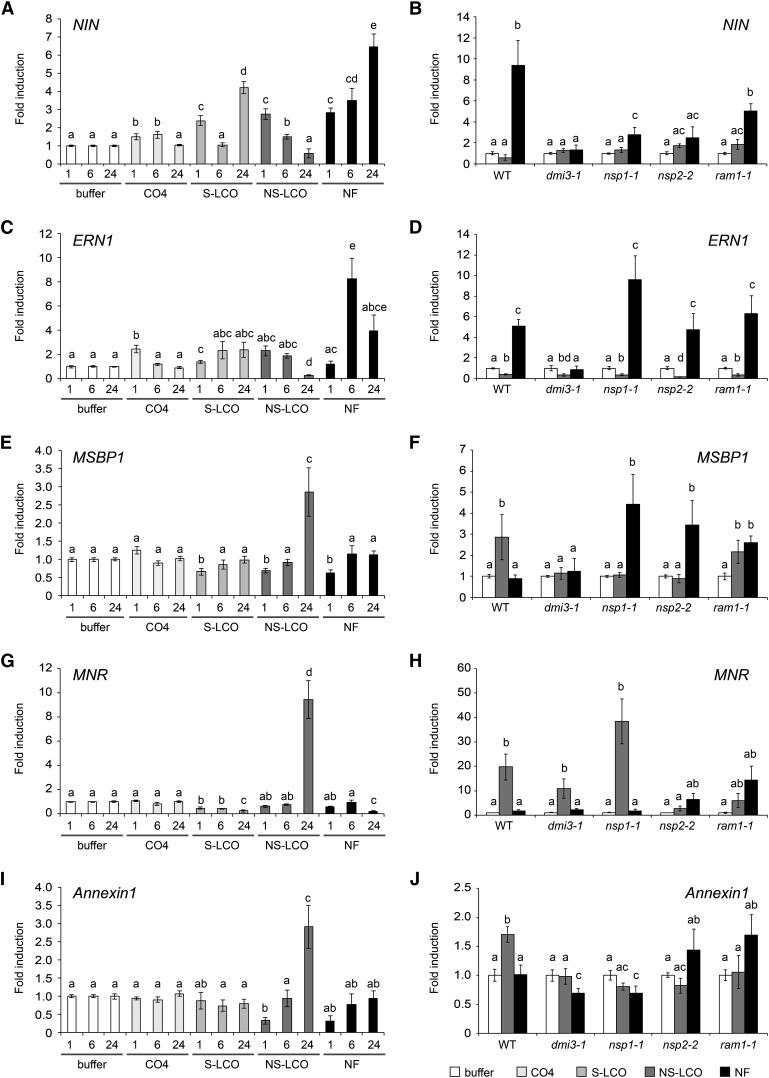
We looked at a variety of genes, the nodulation-associated transcription factors Nodule Inception (NIN) and ERF Required for Nodulation (ERN1) (Schauser et al., 1999; Marsh et al., 2007; Middleton et al., 2007); the rhizobial-induced Annexin1 (De Carvalho-Niebel et al., 2002); the mycorrhizal-induced nitrate reductase MNR (Weidmann et al., 2004); Vapyrin that is required during both rhizobial and mycorrhizal colonization (Reddy et al., 2007; Pumplin et al., 2010; Murray et al., 2011); the early mycorrhizal induced Membrane steroid binding protein (MSBP1) (Kuhn et al., 2010); and two genes reported to be activated by the Myc-LCOs (Czaja et al., 2012). Among the LCO treatments, ERN1 was specifically induced by Nod factor; NIN was induced by Nod factor and to a lesser extent by S-LCO; NS-LCO, MNR, MSBP1, and Annexin1 were only induced by NS-LCO (Figure 4); Vapyrin was induced by all three LCOs; DXS2 was induced by NS-LCO and Nod factor; and Mtr.41728.S1_at was induced by S-LCO and Nod factor (Supplemental Figure 3). CO4 showed only slight inductions of NIN and ERN1 at early time points and no induction of any of the other genes analyzed (Figure 4; Supplemental Figure 3). These differences in gene expression cannot be explained by the differences in the concentrations of the signaling molecules used, since the patterns of NIN and MSBP1 induction are retained even when we apply higher concentrations of S-LCO and Nod factor (Supplemental Figure 4). Of the genes analyzed, all showed a dependence on DMI3 (encoding the symbiotic calcium-activated kinase CCaMK), with the exception of MNR (Figure 4), which has been previously reported (Weidmann et al., 2004). While not exhaustive, this analysis shows that even though the calcium spiking profiles are very similar, the plant appears to be able to differentiate between the LCO and CO signals and activate different gene expression. However, there is not a precise correlation between the known expression patterns of the genes in a symbiotic context and their activation by the Nod factors and the Myc-LCOs.
Figure 4.
Specific Gene Induction in M. truncatula by the Different LCOs.
Induction of NIN ([A] and [B]), ERN1 ([C] and [D]), MSBP1 ([E] and [F]), MNR ([G] and [H]), and Annexin1 ([I] and [J]) following 1-, 6-, and 24-h treatments with 10−10 M Nod factor (NF), 10−9 M S-LCO, 10−6 M NS-LCO, and 10−5 M CO4 in seedling roots. For (B), (D), (F), (H), and (J), 6 h of Nod factor treatment (black bars) and 24 h of NS-LCO treatment (gray bars) were used to assess gene induction in the mutants. These time points were selected based on the intensity of gene induction observed for NIN and ERN with NF and MSBP1, MNR, and Annexin1 with NS-LCO. Values represent averages from three biological replicates and error bars are se. Samples are grouped according to their significance as denoted by letters, calculated with a two-tailed t test (P < 0.05).
Previous work has revealed a suite of GRAS protein transcription factors that function in symbiosis signaling (Kaló et al., 2005; Smit et al., 2005; Gobbato et al., 2012). NSP1 and NSP2 are essential for nodulation, and RAM1 is essential for mycorrhization (Catoira et al., 2000; Oldroyd and Long, 2003; Gobbato et al., 2012). While recent work has shown some role for NSP1 and NSP2 during mycorrhizal colonization (Maillet et al., 2011; Lauressergues et al., 2012; Delaux et al., 2013; Takeda et al., 2013), the mycorrhizal defects in these mutants are very minor compared with the nodulation phenotypes. To assess whether the gene induction observed was dependent on these GRAS proteins, we assessed induction in the mutants. For this we used a 6-h time point for Nod factor treatments, since at this time point we observed significant induction of both ERN1 and NIN, and a 24-h time point for NS-LCO treatments, since this was the only time point where we consistently observed significant gene induction. NIN induction by Nod factor was dependent on NSP1 and NSP2 (although there is still a significant but reduced induction of NIN in nsp1 mutants, which is consistent with previous work with this mutant; Catoira et al., 2000), but independent of RAM1, whereas ERN1 induction by Nod factor was independent of all three transcription factors (Figures 4B and 4D). NS-LCO induction of MNR was dependent on NSP2 and possibly RAM1, but independent of NSP1 (Figure 4H). Annexin1 induction by NS-LCO was dependent on all three transcription factors (Figure 4J). MSBP1 induction presents an interesting case, since the NS-LCO induction was dependent on NSP1 and NSP2, but independent of RAM1, and in all three transcription factor mutants, we observed a significant induction of MSBP1 following Nod factor treatments that was not observed in wild-type plants (Figure 4F). Overall, this presents a complex picture of gene induction by the different signaling molecules, with the different transcription factors. We conclude that the responses to the Myc-LCOs cannot be explained simply by an inappropriate activation of a Nod factor receptor, and while not exhaustive, our work highlights differential responsiveness to CO4 and the different LCO signals, which show differential dependence on the different transcription factors. However, we cannot correlate the specific symbiotic functions of the transcription factors, the symbiotic nature of the induced genes, and the origins of the signaling molecules.
Lateral Roots Are More Sensitive to LCOs and CO4 Than Seedling Roots
From the dose–response curves, it is apparent that M. truncatula seedlings require at least 10−5 M CO4 to activate calcium oscillations. This is consistent with previous work using M. truncatula seedlings (Oldroyd et al., 2001a), but contrasts with work in root organ cultures that revealed calcium responses at 10−8 M CO4 (Genre et al., 2013). It would appear that the major difference between these experiments is the material used: seedling roots versus root organ cultures. To address whether this discrepancy was a result of the different root types, we assessed the response to CO4 of root organ cultures of M. truncatula carrying nuclear-localized Yellow Cameleon 2.1 (Sieberer et al., 2009). As reported by Genre et al. (2013), we observed calcium oscillations in root organ cultures with treatments of 10−8 M CO4 (Supplemental Figure 5).
Considering this difference in sensitivity of seedling roots and root organ cultures, we assessed whether older roots also altered their sensitivity to CO4 and LCOs. Dose–response curves in M. truncatula lateral roots clearly showed that lateral roots were more sensitive to Nod factor, S-LCO, NS-LCO, and CO4 for induction of calcium oscillations as compared with seedling roots (Figure 2). We observed calcium responses in trichoblasts on lateral roots with treatments of 10−13 M Nod factor, a sensitivity only previously observed in the ethylene insensitive mutant skl (Oldroyd et al., 2001b). The greatest shift in sensitivity was for CO4, with a 1000-fold increase in the sensitivity of lateral roots compared with seedling roots (Figure 2B).
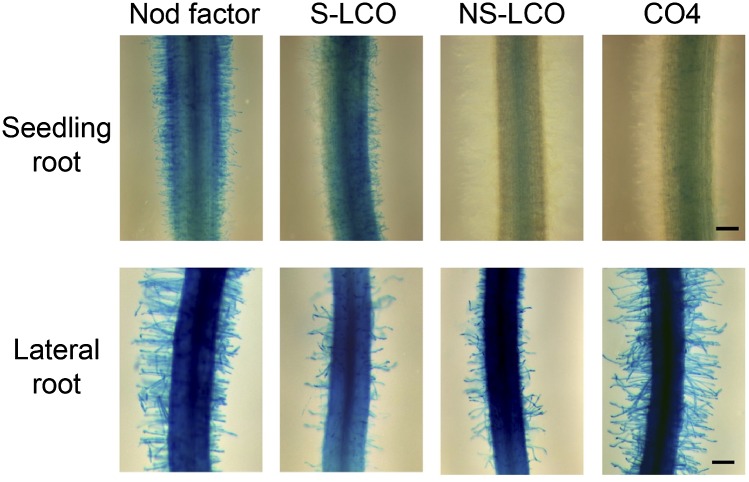
To validate this shift in sensitivity between seedling roots and lateral roots, we assessed induction of ENOD11-GUS, a reporter activated by the Myc-LCOs (Maillet et al., 2011) and by Nod factor (Journet et al., 2001). We treated seedling roots and lateral roots with concentrations of 10−7 M Nod factor, S-LCO, NS-LCO, and CO4; at this concentration, seedling roots show calcium spiking with Nod factors and S-LCO, but not NS-LCO or CO4 (Figure 2A), while lateral roots show calcium spiking in response to all four molecules (Figure 2B). Consistently, we observed ENOD11-GUS activity in seedling roots with 10−7 M Nod factor and S-LCO, but not NS-LCO or CO4, while lateral roots revealed a response to Nod factor, S-LCO, NS-LCO, and CO4 at 10−7 M (Figure 5). We conclude that the sensitivity to CO4 and LCOs is dependent on the age and developmental status of the root. Interestingly, lateral roots are preferentially colonized by AM fungi and in some cases by rhizobia (Heron and Pueppke, 1984; Gutjahr and Paszkowski, 2013), and this correlates well with the shifts in sensitivity we observed with the LCOs and CO4.
Figure 5.
Differential Induction of pENOD11-GUS in Seedling Roots versus Lateral Roots.
Induction of pENOD11-GUS in M. truncatula by 10−7 M Nod factor, S-LCO, NS-LCO, and CO4 in seedling roots (2-d-old plants) and lateral roots. These images are representative roots. The experiment was repeated three times with similar outcomes. Bars = 200 μm.
We further assessed the genetic dependencies on the activation of ENOD11-GUS in lateral roots (Supplemental Figure 6). We found that NSP1 and NSP2 were essential for ENOD11-GUS induction in lateral roots by all the signals. In contrast, RAM1 was not required for the activation of ENOD11-GUS by any of the signaling molecules. Surprisingly, we still observed some induction of ENOD11-GUS by CO4, S-LCO, NS-LCO, and Nod factor in the lateral roots of the dmi3-1 mutant; however, the majority of dmi3-1 roots did not respond and where we did observe some ENOD11-GUS in dmi3-1 lateral roots it was reduced compared with wild-type plants. We conclude that NSP1, NSP2, and DMI3 are required for ENOD11 induction by all the molecules tested.
Responses to Myc-LCOs and CO4 in L. japonicus
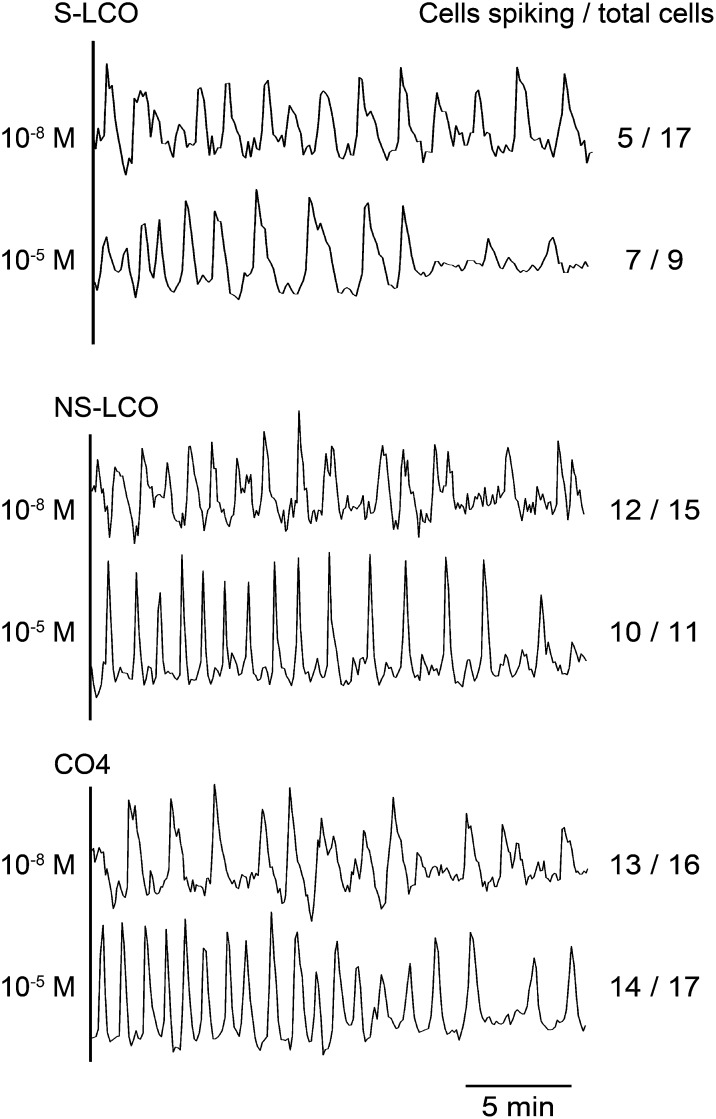
The Myc-LCOs were initially purified based on their ability to activate responses in M. truncatula that are also induced by Nod factor (Maillet et al., 2011). This complicates interpretations from the M. truncatula analyses, particularly when considering S-LCO, a molecule very similar to Nod factors produced by S. meliloti. In contrast, the rhizobial symbiont of L. japonicus, Mezorhizobium loti, does not produce sulfated Nod factors, rather producing Nod factors with carbamoyl, methyl, and acetyl-fucose groups that have varying importance for recognition by different Lotus species (Rodpothong et al., 2009). Considering these differences in Nod factor recognition between Medicago and Lotus, an analysis of L. japonicus responses to the Myc-LCOs should be informative with regard to the importance of the different modifications. We used lines of L. japonicus stably transformed with nuclear-localized Yellow Cameleon YC3.6 (Krebs et al., 2012) and initially treated plants with 10−5 M S-LCO, NS-LCO, and CO4. At this concentration, we observed robust calcium oscillations in atrichoblasts (epidermal cells that do not differentiate into root hair cells) treated with S-LCO, NS-LCO, and CO4 (Figure 6). Surprisingly, the CO4-induced calcium response was very periodic and much more similar to the LCO responses than we had previously observed in M. truncatula CO4 treatments (Supplemental Figure 1). To assess the sensitivity of L. japonicus to the Myc-LCOs and CO4, we tested a range of concentrations. We found that 10−8 M was the lowest concentration that gave a reliable response to S-LCO, NS-LCO, and CO4 in L. japonicus lateral roots, but at these concentrations, the quality of the calcium response was reduced for all three molecules (Figure 6). Unlike the response in M. truncatula, we saw no difference in the response of L. japonicus to S-LCO and NS-LCO, and this may reflect a preference in M. truncatula for sulfated LCOs. S-LCO has similarities to the Nod factors produced by S. meliloti; therefore, we tested the activation of calcium oscillations in L. japonicus by HPLC-purified S. meliloti Nod factor. We observed no calcium responses in the atrichoblasts or trichoblasts of L. japonicus treated with 10−8 M S. meliloti Nod factor (0 responsive cells out of 28 cells analyzed), but with 10−5 M S. meliloti Nod factor treatments, we observed calcium responses in both trichoblasts and atrichoblasts of L. japonicus (8 responsive cells out of 11 cells analyzed). This suggests a degree of specificity of recognition of these different LCO signals in L. japonicus atrichoblasts, with a high degree of sensitivity to Myc-LCOs that we did not observe with S. meliloti Nod factor. The fact that NS-LCO and S-LCO are equally active in L. japonicus supports the idea that both can act as Myc signals at least in some plant species.
Figure 6.
Calcium Responses in L. japonicus to the Myc-LCOs and CO4.
Representative calcium traces from L. japonicus atrichoblasts on lateral roots treated with either 10−5 M or 10−8 M S-LCO, NS-LCO, and CO4. The number of cells showing calcium responses, relative to the total number of cells analyzed, is indicated.
Responses to Myc-LCOs and CO4 in Rice
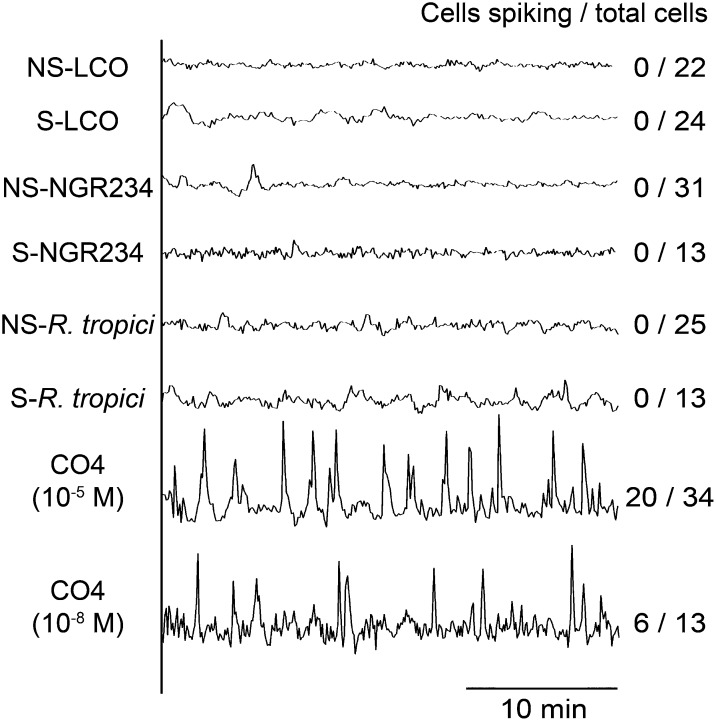
AM fungi associate with a wide range of plant species, and at least in rice this association is dependent on the common symbiosis signaling pathway (Chen et al., 2007; Banba et al., 2008; Chen et al., 2008; Gutjahr et al., 2008). Hence, we would propose that non-legumes should be able to recognize the Myc-LCOs and/or CO4, and for this we tested induction of calcium spiking in rice. To assess calcium responses in rice, we generated a stably transformed line of Nipponbare carrying YC3.6. AM fungi have been shown to predominantly colonize the major lateral roots (Gutjahr et al., 2009); therefore, we focused on this root type. No calcium oscillations were observed following treatment with 10−5 M S-LCO or NS-LCO, but strong calcium oscillations were observed in atrichoblasts following treatment with 10−5 M CO4 (Figure 7). To test an array of different LCOs, we analyzed Nod factor isolations from the broad host range rhizobial species, NGR234, as well as Rhizobium tropici, in addition to the Myc-LCOs. Considering that these LCO treatments were performed with 10−5 M, we are confident that rice does not respond to the LCOs tested. Treatments with 10−8 M CO4 still showed calcium oscillations in rice atrichoblasts, but the number of responsive cells was reduced (Figure 7). We observed this calcium spiking response in atrichoblasts on all root types of rice, with no apparent preference for major laterals.
Figure 7.
Calcium Responses in Rice to the Myc-LCOs, Nod Factors, and CO4.
Representative calcium traces from rice atrichoblasts on lateral roots treated with 10−5 M Myc-LCOs and LCO isolations from NGR234 and R. tropici, as well as 10−5 M and 10−8 M treatments of CO4. The number of cells showing calcium responses, relative to the total number of cells analyzed, is indicated.
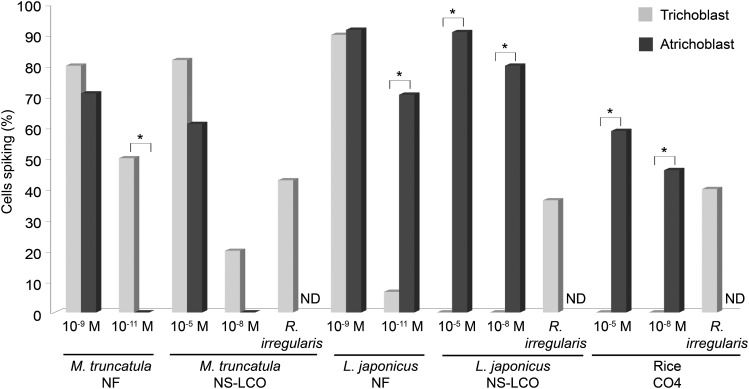
Atrichoblasts of L. japonicus and Rice Are Preferentially Responsive to Myc-LCOs and CO4
In our analysis of L. japonicus and rice, it was very clear that atrichoblasts, but not trichoblasts, were responsive to the Myc-LCOs and CO4. This was in contrast to what we had previously observed in M. truncatula where responses to Nod factor, NS-LCO, S-LCO, and CO4 occurred in both trichoblasts and atrichoblasts, but at lower concentrations only in trichoblasts (Figure 8). To validate this, we directly compared trichoblast and atrichoblast responses using high concentrations of NS-LCO in L. japonicus and CO4 in rice. Even with 10−5 M treatments, trichoblasts of L. japonicus showed no response to NS-LCO, while atrichoblasts responded well (Figure 8). L. japonicus trichoblasts showed no response to 10−8 M S-LCO treatment (0 responsive cells out of 12 cells analyzed), whereas atrichoblasts respond well at this concentration (Figure 6). Surprisingly, this preferential induction of calcium oscillations in atrichoblasts was also observed for M. loti Nod factor treatments of L. japonicus, with equivalent induction in both cell types at 10−9 M Nod factor, but preferential induction in atrichoblasts at 10−11 M Nod factor (Figure 8). The calcium oscillations that we observed in rice following treatments of CO4 were also restricted to atrichoblasts, with no responses in trichoblasts even with CO4 treatments of 10−5 M (Figure 8). This preferential nature of L. japonicus and rice atrichoblasts to respond to the AM signals is consistent with a preference for AM fungi to colonize the root via atrichoblasts (Chabaud et al., 2011).
Figure 8.
Calcium Responses in Trichoblasts and Atrichoblasts.
The percentage of calcium-responsive cells among trichoblasts (gray) and atrichoblasts (black) of M. truncatula, L. japonicus, and rice. For M. truncatula treatments of 10−11 M and 10−9 M S. meliloti Nod factor and 10−8 M and 10−5 M NS-LCO were analyzed, for L. japonicus 10−9 M and 10−11 M M. loti Nod factor, and 10−8 M and 10−5 M NS-LCO were analyzed, while for rice 10−8 M and 10−5 M CO4 was analyzed. For all plants, the response of trichoblasts in close proximity to R. irregularis hyphae was analyzed. Where indicated in the figure, the R. irregularis treatments were performed alone, without any additional signaling molecules added. Unfortunately, due to technical difficulties, it is not possible to assess atrichoblasts in close proximity to R. irregularis hyphae; therefore, responses in these cells were not determined (ND). The asterisks show significant differences (P < 0.05), measured using a χ2 test, between the data points indicated.
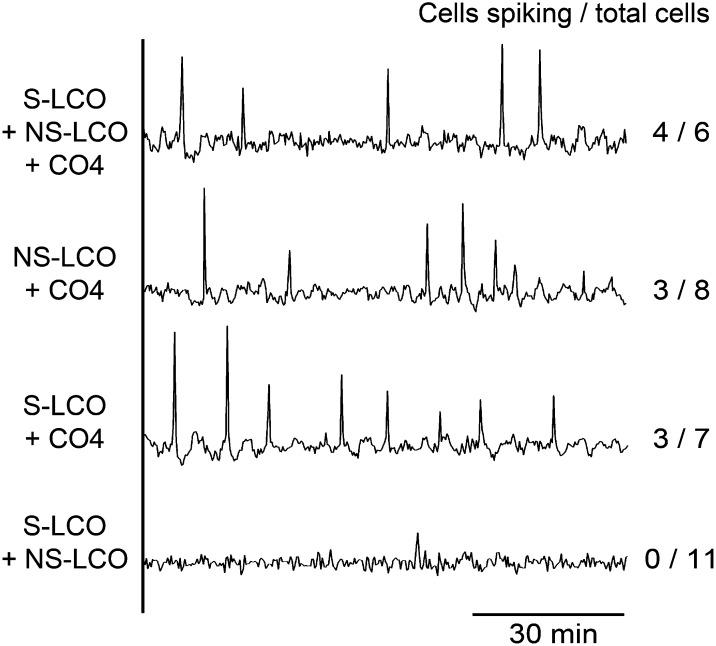
Rice Trichoblasts Respond to a Mix of CO4 and Myc-LCOs with Activation of Calcium Spiking
In previous analyses of M. truncatula, we observed calcium oscillations in trichoblasts in close proximity to R. irregularis hyphae. Our work with the Myc-LCOs suggests a difference in M. truncatula for cell type-specific responses. To assess whether trichoblasts were only responsive to mycorrhizal fungi in M. truncatula, we assessed calcium responses in trichoblasts of L. japonicus and rice when in close proximity to R. irregularis. We found that both L. japonicus and rice trichoblasts showed calcium oscillations when in close proximity to R. irregularis hyphae (Figure 8; Supplemental Figures 7 and 8 and Supplemental Table 1).
It is possible that either AM fungi produce signaling molecules other than S-LCO, NS-LCO, and CO4 that induce calcium oscillations in rice and L. japonicus trichoblasts or the mix of signaling molecules is important. To test this, we assessed induction of calcium oscillations by an equimolar mix of 10−5 M S-LCO, NS-LCO, and CO4. Strikingly, we observed calcium spiking in rice trichoblasts when treated with this mix of signals (Figure 9), yet these signals when applied individually had no effect in rice trichoblasts (Figure 7). To assess the relative importance of these different signals, we tested different combinations and found that when either NS-LCO or S-LCO was combined with CO4, we observed calcium oscillations in rice trichoblasts, but S-LCO and NS-LCO together had no affect (Figure 9). The induction of a calcium response in trichoblasts took considerably longer than the lag between the addition of CO4 and activation of a response in rice atrichoblasts (Table 2). Our work suggests that even though the Myc-LCOs alone are insufficient to activate the common symbiosis signaling pathway in rice, it appears that they are still recognized by rice trichoblasts when in combination with CO4 treatments. The long delay in activation of a response in rice trichoblasts could be associated with changes in the expression of receptors necessary for LCO recognition.
Figure 9.
Rice Trichoblasts Respond to Mixes of Myc-LCOs and CO4.
Representative calcium traces of rice trichoblasts treated with mixes of 10−5 M CO4, S-LCO, and NS-LCO. Note that mixes of Myc-LCOs with CO4 induced calcium oscillations, but the Myc-LCOs alone do not. The number of cells showing calcium responses, relative to the total number of cells analyzed, is indicated.
Table 2. The Lag to Calcium Oscillations in Rice Trichoblasts.
| Signaling Molecules | Average Time (min) |
|---|---|
| CO4* | 30 ± 7.7 |
| S-LCO + CO4 | 138 ± 4.5 |
| NS-LCO + CO4 | 134 ± 17.1 |
| S-LCO + NS-LCO + CO4 | 123 ± 3.6 |
n = 4 plants; sd is shown. *, Atrichoblast.
LCOs and CO4 Promote Root Development in Rice
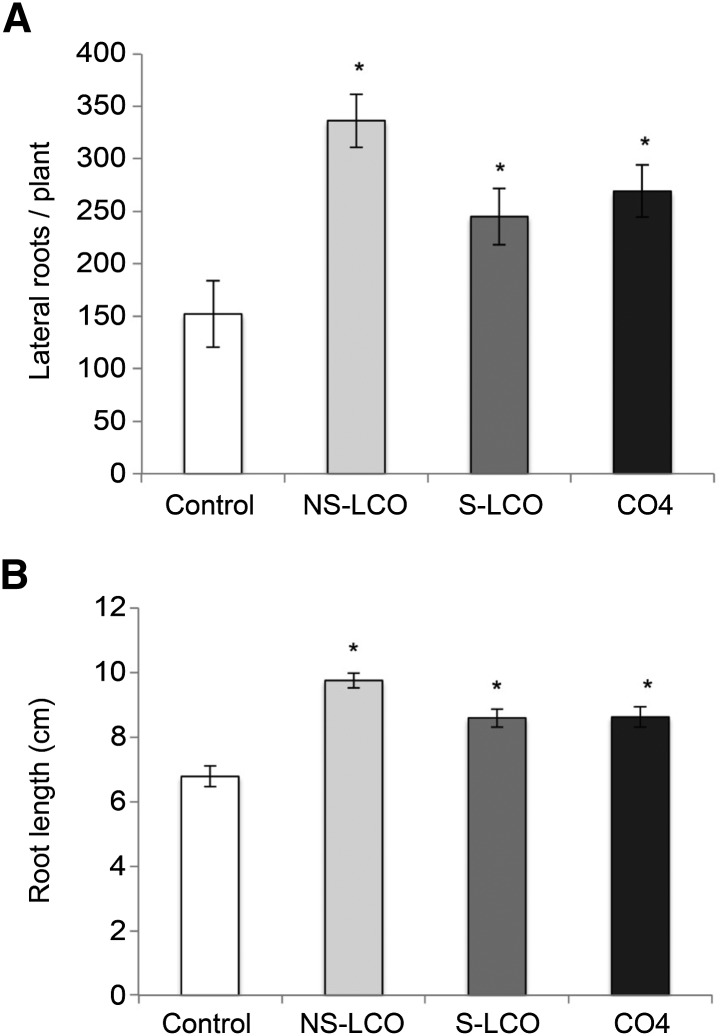
Rice has been shown to respond to AM fungi and to exudates from the spores of AM fungi, with changes to root structure, in particular the promotion of lateral root outgrowth (Gutjahr et al., 2009; Mukherjee and Ané, 2011). These responses are independent of the common symbiosis signaling pathway. We tested the promotion of lateral root outgrowth in rice by S-LCO, NS-LCO, and CO4 and found all three molecules could promote lateral root outgrowth and enhance overall root system growth (Figure 10). Surprisingly, these responses were dependent on the symbiosis signaling component CCaMK (Supplemental Figure 9), equivalent to DMI3 of M. truncatula. These results suggest that CO4 and Myc-LCOs may contribute to the active molecules present in germinated spore exudates that promote root architecture responses in rice (Mukherjee and Ané, 2011); however, additional signals must also be present that can promote this response independently of the symbiosis signaling pathway. In contrast to what we observed in rice, CO4 could not induce lateral root emergence in M. truncatula (Supplemental Figure 10), but the Myc-LCOs can activate this response (Maillet et al., 2011). Thus, it would appear that legumes and non-legumes differ in their response to the spectrum of Myc-LCOs and COs for activation of calcium oscillations and modifications to root architecture.
Figure 10.
Myc-LCOs and CO4 Promote Lateral Root Emergence in Rice.
The affect of 10−8 M CO4, S-LCO, and NS-LCO treatments on the mean number of lateral roots per rice plant (A) and the mean root system length (B). These results are based on two independent experiments. Values are means ± se. The P value was calculated using a t test, assuming a normal distribution of the data, or a Wilcoxon signed-rank test when a normal distribution was not observed. The asterisk indicates significant difference relative to the control (P < 0.05).
DISCUSSION
AM fungi signal to the host plant via diffusible signals (Kosuta et al., 2003, 2008; Chabaud et al., 2011; Mukherjee and Ané, 2011), primarily LCOs (Maillet et al., 2011) and COs (Genre et al., 2013). In this work, we show that the Myc-LCOs can activate calcium oscillations in M. truncatula and L. japonicus, while CO4 can induce calcium oscillations in M. truncatula, L. japonicus, and rice. S-LCO and NS-LCO were purified from exudates of R. irregularis based on their capability to activate symbiotic responses in M. truncatula that were dependent on the common symbiosis signaling pathway (Maillet et al., 2011). The fact that these LCOs are predominantly active for promotion of calcium oscillations on legumes and not rice may reflect this selectivity in their initial identification. However, it is clear that LCOs do play a symbiotic role in rice, since the mix of Myc-LCOs and CO4 is required to activate calcium oscillations in rice trichoblasts and since NS-LCO and S-LCO could promote lateral root emergence in rice. Therefore, the absence of calcium responses in rice to the LCO treatments does not indicate a lack of response to LCOs by rice. Our work reveals that LCOs and CO4 that are produced by AM fungi can activate a variety of responses in a variety of plant species.
The Structure of the Symbiotic Calcium Response
We and others have previously hypothesized that differences in the structure of the calcium oscillations may explain the discrimination between AM fungi and rhizobial bacteria (Kosuta et al., 2008; Genre et al., 2013). However, during the colonization of the root by AM fungi and by rhizobial bacteria, the structure of the calcium oscillations induced by these two symbionts is very similar (Sieberer et al., 2012). We also observed very similar structures in the calcium oscillations induced by Nod factor, S-LCO, and NS-LCO, even though we still observed differences in the calcium responses to AM fungi. Sieberer et al. (2012) report a switch in the frequency of calcium oscillations that is associated with infection by the symbiont: Prior to infection or in neighboring uninfected cells, poorly periodic calcium oscillations occur, but upon infection by the symbiont, rapid and periodic calcium oscillations ensue. Perhaps the relatively poor quality of calcium oscillations we observed with AM fungal inoculation reflects the fact that we are measuring the calcium responses in trichoblasts that will not be infected by AM fungi. An alternative hypothesis was put forward by Genre et al. (2013): The AM fungal response parallels the response observed by CO4 treatments, suggesting that the predominant signal from AM fungi is CO4 (Genre et al., 2013). However, in our analysis, we observed this “poor quality” calcium spiking in M. truncatula following treatment with both CO4 and with the M-LCOs (which do not contain COs). We believe that the most parsimonious explanation is that the nature of the calcium response is defined by the mix of signals provided and the relative concentrations of the individual signaling molecules. Such an explanation is consistent with a shift in the structure of the calcium oscillations as the symbionts approach the cell (Sieberer et al., 2012).
M. truncatula Can Differentiate between Nod and Myc Signals
Nod factor, S-LCO, and NS-LCO activated equivalent calcium oscillations, yet we observed different patterns of gene expression. This is supported by differences in the expression profiles of M. truncatula treated with these different LCOs (Czaja et al., 2012). It appears that the patterns of gene induction by the different LCOs do not correlate precisely with mycorrhizal versus rhizobial reporters. In spite of this, it is clear that the different LCO signaling molecules activate different gene expression patterns. Hence, despite the similarities in the calcium response, cells showed differential gene induction to the different LCO and CO signaling molecules, implying differential recognition of these signaling molecules, and this may involve a signal transduction mechanism parallel to the symbiosis signaling pathway. We proposed that the specificity of symbiont recognition may, at least in part, be controlled by the GRAS protein transcription factors NSP1, NSP2, and RAM1 (Gobbato et al., 2012; Oldroyd, 2013). In support of this, we observed a dependence on NSP1 and NSP2 for Nod factor induction of NIN and a dependence on NSP2 and RAM1 for NS-LCO induction of MNR. However, our own work points at a degree of complexity with regard to the relative dependence on these different GRAS protein transcription factors. Clearly, much more work is required to unpick the details of the gene expression profiles to these different signaling molecules and the roles of the GRAS protein transcription factors in their regulation.
The Influence of Cell Type and Developmental State for LCO and CO4 Responses
Our work has revealed a close correlation between the cell type and its developmental state, with its responsiveness to LCOs and CO4. We found that young seedling roots are less responsive to LCOs and CO4 than lateral roots on older plants. This shift in sensitivity was particularly pronounced for CO4, with a 1000-fold difference in the sensitivity of seedling roots versus lateral roots. Our results correlate well with preferential AM colonization of lateral roots (Gutjahr et al., 2009). We also observed that calcium responses to the AM signals were restricted to atrichoblasts in L. japonicus and rice. Clearly this restriction is not common to all plant species, since trichoblasts and atrichoblasts of M. truncatula responded to the Myc-LCOs. Again, this preferential response in atrichoblasts correlates well with a preferential colonization of atrichoblasts by AM fungi (Chabaud et al., 2011). However, despite the atrichoblast restriction of the response to the Myc-LCOs and CO4 in L. japonicus and rice, we still observed calcium oscillations in L. japonicus and rice trichoblasts in treatments with AM fungi. Our finding that in rice a mix of both Myc-LCOs and CO4 is required to elicit calcium oscillations in trichoblasts suggests that the rice trichoblast response could be explained by the mix of LCOs and CO4 secreted by the AM fungus. Interestingly, it was shown some years ago that a mix of Nod factor and COs was better at inducing symbiosis-associated gene expression in soybean (Glycine max) than Nod factor treatments alone (Minami et al., 1996). Perhaps these earlier observations reflect responses to AM fungi, rather than what was previously thought to be a rhizobial response. Alternatively, a mix of LCOs and COs may be relevant in rhizobial interactions as well as AM associations.
Complementarity of LCO and CO4 Signaling
Our work reveals that the Myc-LCOs and CO4 in different combinations can induce calcium oscillations in rice trichoblasts, as well as the promotion of lateral root emergence. While calcium oscillations are suggestive of the activation of the symbiosis signaling pathway in rice, it will be important to test mutants of this pathway for CO4 induction of calcium oscillations. Whereas in rice both LCOs and CO4 promote lateral root emergence, in the legume M. truncatula, only Myc-LCOs, and not CO4, are able to elicit this response.
It has already been shown that colonization of rice by AM fungi requires at least two separate signaling pathways (Gutjahr et al., 2008) and that the promotion of lateral root formation in rice by AM fungal spore exudates is independent of the common symbiosis signaling pathway (Mukherjee and Ané, 2011). It is very surprising that the promotion of lateral root emergence in rice by the Myc-LCOs and CO4 was dependent on the symbiosis signaling gene CCaMK, implying a different modality of signaling in the activation of this response by fungal spore exudates as compared with these single signaling molecules. We propose that the Myc-LCOs and CO4 present in AM fungal exudates are likely to be involved in the promotion of lateral root emergence, but additional, as yet uncharacterized, signals present in AM fungal exudates can activate this response in a manner independent of the symbiosis signaling pathway. In M. truncatula, the activation of calcium oscillations and lateral root promotion by LCOs are both dependent on the symbiosis signaling pathway (Wais et al., 2000; Oláh et al., 2005; Maillet et al., 2011), and our work draws parallels between the modes of signaling in M. truncatula and in rice.
AM fungi have the distinctive capability of colonizing an extremely broad group of plants. In this work, we reveal that part of this capability is reflected in the signaling molecules, CO4 and LCOs, which can activate symbiotic calcium oscillations in a broad range of plant species. This broad host range of AM fungi may be facilitated by the extent of symbiotic signaling molecules that they produce, with a mix of LCOs and COs and possibly other yet uncharacterized molecules. Clearly further work is necessary; however, we have shown that LCOs and CO4 form at least part of the spectrum of AM symbiotic signals that can be recognized by distinct plant lineages to activate symbiotic responses.
METHODS
Seed Preparation, Plant Growth Conditions, and Treatments with LCOs
Medicago truncatula cv Jemalong seeds and Lotus japonicus cv Gifu were scarified with sandpaper, surface-sterilized in 10% sodium hypochlorite solution for 2 to 3 min, rinsed in sterile water, and plated on 1% deionized water agar plates. After stratification at 4°C for 4 d, seeds were germinated overnight at room temperature. Seedlings were grown on Buffered Nodulation Media (BNM) plates for 24 h before treatment with LCOs. Rice (Oryza sativa cv Nipponbare) seeds were sterilized with 2% sodium hypochlorite solution for 20 min, rinsed in sterile water, and then imbibed overnight. Seeds were germinated on damp germination paper in the dark at 25°C for 7 d. Rice seedlings were grown on Fahraeus Medium plates for 5 d before being subjected to a 24-h LCO treatment for lateral root assays (Mukherjee and Ané, 2011). RNA preparations of 24 M. truncatula plants were used for each LCO treatment, and treatments were repeated in triplicate. LCO-treated roots were cut from the plants and frozen in liquid nitrogen for subsequent RNA extraction. For calcium imaging experiments, the seedling roots or lateral roots were fixed into a small chamber made on a cover glass using vacuum grease. The chamber was filled with 500 μL of BNM buffer, and the roots were treated with the different concentrations of LCOs and CO4.
Calcium Imaging
AM fungal-induced calcium responses were measured as described previously (Kosuta et al., 2008). For root culture experiments, hairy root cultures expressing nuclear-localized Yellow Cameleon (Sieberer et al., 2009) were produced by Agrobacterium rhizogenes Arqua1 (Boisson-Dernier et al., 2001). Cameleon was imaged on a Nikon Eclipse Ti inverted microscope equipped with an OptoLED Illuminator (model OptoLED; Cairn Research). Cameleon was excited at a wavelength of 455 nm using a royal blue LED and was captured with a CCD camera (model RETIGA-SRV; Qimaging). Emitted fluorescence was separated by an image splitter with a dichroic mirror (model Optosplit II; Cairn Research) and then passed through a Cameleon filter set. Images were collected every 5 s with 1-s exposure and analyzed using Metafluor (Molecular Devices). For the trichoblast and atrichoblast calcium imaging, the fluorescence was measured with a Zeiss LSM 510 Meta confocal scanning microscope equipped with a LD LCI Plan-Apochromat 0.80 25× oil-immersion objective (Zeiss). Cameleon was excited with an argon ion 458-nm laser and imaged using emission filters 476 to 486 nm for CFP and 529 to 540 nm for YFP. The images were acquired at 5-s intervals with a scanning resolution of 512 × 512 pixels. The images were processed with the time series analyzer V2.0 using ImageJ software (Abramoff et al., 2004).
Mathematical Analysis of Calcium Oscillations
For the Bayesian spectrum analysis, we computed the most probable periods in the time series following published procedures (Granqvist et al., 2011). To ensure that we were comparing equivalent regions of calcium oscillations, a single cell from a single plant was analyzed prior to and during the addition of the LCO or CO4. Calcium responses were measured for at least 2 h. Ten traces from trichoblasts from 10 different plants per treatment were analyzed. The joint distributions over the period were used to characterize each group. The plots show binned data to summarize the key periods. These 10 traces per treatment were also analyzed for interspike intervals. The point of maximum height for each spike was computed after detrending of the time series using a moving average algorithm. The distances between these maxima gave rise to an interspike distribution. We used the nonparametric Mann-Whitney U-test, also known as the Mann-Whitney-Wilcoxon test (Wilcoxon, 1945; Mann and Whitney, 1947) to test for significant differences between the distributions.
Three traces from three different plants, with altogether ∼80 spikes, per treatment were analyzed for calcium spike characteristics. The time series have an interval of 5 s between data points. The traces were detrended using a moving average algorithm (Brockwell and Davis, 2002). We then characterized the spikes by the time required for each upward and downward phase. This was computed by the number of data points it took from the maximum spike height to the baseline fluctuation of the trace. The plots show the mean value of the upward and downward phases for each treatment and the associated standard deviations are indicated by the error bars.
Gene Expression Analysis
M. truncatula seedlings were transferred to liquid BNM and treated with the LCOs at the concentrations stated. Seedlings were removed at set time points, and RNA was extracted from root tissues using the RNeasy Plant Mini Kit (Qiagen). Purified RNA was then treated with Turbo DNA-free DNase (Ambion) according to the manufacturer’s instructions. This RNA was subsequently tested for absence of contaminating genomic DNA by PCR with EF1α primers (Supplemental Table 2) and checked for quality by gel electrophoresis and the RNA quality control protocol for the QIAxcel system (Qiagen). RNA was quantified using a spectrophotometer (NanoDrop-1000; Thermo Scientific), and 1 µg total RNA was used for cDNA synthesis. cDNA synthesis was performed using the Superscript II first-strand synthesis system (Invitrogen) using oligo(dT)17, following the manufacturer’s instructions. One microliter of RNasin Ribonuclease Inhibitor (Promega) was added to each cDNA synthesis reaction.
qRT-PCR was performed using a DNA Engine Opticon 2 real-time cycler (Bio-Rad) with Opticon Monitor 3 software (Bio-Rad). Reactions were performed in 96-well plates using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich), 10 nM of each gene-specific primer (Supplemental Table 2), and 1:10 (v/v) cDNA:water. PCR cycling conditions were as follows: 95°C for 4 min followed by 40 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The specificity and efficiency of primer pairs was confirmed by analysis of dissociation curves (65 to 95°C) and serial dilution, respectively. Results were expressed as a threshold cycle (CT) value. At least three technical replicates and three biological replicates were performed for each sample, and their CT values were averaged. The standard curve method was used to analyze gene expression in each sample as described (El Yahyaoui et al., 2004). EF1α was used as a reference gene, and fold induction was calculated for treated samples relative to untreated samples.
For ENOD11-GUS analysis, M. truncatula line 416K was grown on BNM media and then transferred to liquid BNM containing 10−7 M Nod factor, S-LCO, NS-LCO, or CO4 for 24 h at room temperature. After incubation, the roots were washed several times in liquid BNM and stained overnight in 50 mM potassium phosphate buffer, pH 7.0, with 1 mM EDTA, 0.1% Triton X-100, and 1 mg/mL X-Gluc. The experiment was repeated three times with equivalent results. Representative roots are shown.
Measurements of Lateral Root Emergence
Total lateral roots of rice were enumerated manually 2 weeks after application of 10−8 M COs and LCOs. As COs and LCOs were dissolved in 50% ethanol, the control was sterile deionized water containing the appropriate amount of ethanol. A minimum of 10 plants of either wild-type rice or the ccamk mutant was used in each biological replicate. Lateral roots were defined as large and fine lateral roots emerging from crown roots, as well as fine lateral roots emerging from large lateral roots. Lateral root counts were assessed for normality using the Shapiro-Wilk test (P = 0.01), and statistical significance was determined using a paired t test assuming unequal variances (P = 0.05). All statistical analysis was conducted using the R software package. A minimum of 30 plants were used for treatment.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: M. truncatula CCaMK (DMI3), Q6RET7; M. truncatula NSP1, Q4VYC8; M. truncatula NSP2, Q5NE24; and M. truncatula RAM1, AFK81971.
Supplemental Data
Supplemental Figure 1. CO4-Induced Calcium Responses in M. truncatula Trichoblasts.
Supplemental Figure 2. Dose–Response Curves for CO4 in the Wild Type and nfp-1.
Supplemental Figure 3. Specific Gene Induction in M. truncatula by the Different LCOs.
Supplemental Figure 4. Gene Induction in M. truncatula by the Different LCOs Is Retained at Higher Concentrations.
Supplemental Figure 5. CO4-Induced Calcium Responses in M. truncatula Root Organ Cultures.
Supplemental Figure 6. Induction of pENOD11-GUS in Lateral Roots.
Supplemental Figure 7. Calcium Oscillations in L. japonicus Trichoblasts Induced by R. irregularis.
Supplemental Figure 8. Calcium Oscillations in Rice Trichoblasts Induced by R. irregularis.
Supplemental Figure 9. Promotion of Root System Development in Rice by NS-LCO, S-LCO, and CO4.
Supplemental Figure 10. Promotion of Lateral Root Emergence in M. truncatula.
Supplemental Table 1. Cell and Plant Numbers Assessed in Figure 8.
Supplemental Table 2. Primers Used for qRT-PCR in This Study.
Supplementary Material
Acknowledgments
We thank Barbara Howlett and the members of her group in the Botany Department at the University of Melbourne for hosting G.E.D.O. during much of the writing of this article. We thank Véréna Poinsot for quality control of LCOs by mass spectrometry and Martin Parniske for providing YC3.6 L. japonicus lines. This work was supported by the European Research Council as “SYMBIOSIS,” the Bill and Melinda Gates Foundation, the Biotechnology and Biological Sciences Research Council as BB/J004553/1, Labex Arcane, ICMG as FR2607, and the Gatsby Charitable Foundation as a Sainsbury PhD fellowship to J.B.M.
AUTHOR CONTRIBUTIONS
J.S., J.B.M., E.M., A.W.-K., E.S., S.F., R.J.M., J.-M.A., J.D., and G.E.D.O. designed the research. J.S., J.B.M., E.G., A.W.-K., E.G., F.M., and M.V. performed the research. G.E.D.O. wrote the article with input from E.G., J.-M.A., and J.D.
Glossary
- AM
arbuscular mycorrhizal
- LCO
lipochitooligosaccharide
- CO
chitooligosaccharide
- CO4
tetra-acetyl chitotetraose
- qRT-PCR
quantitative RT-PCR
- BNM
Buffered Nodulation Media
Footnotes
Articles can be viewed online without a subscription.
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- Amor B.B., Shaw S.L., Oldroyd G.E., Maillet F., Penmetsa R.V., Cook D., Long S.R., Dénarié J., Gough C. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34: 495–506. [DOI] [PubMed] [Google Scholar]
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367. [DOI] [PubMed] [Google Scholar]
- Arrighi J.F., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banba M., Gutjahr C., Miyao A., Hirochika H., Paszkowski U., Kouchi H., Imaizumi-Anraku H. (2008). Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol. 49: 1659–1671. [DOI] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700. [DOI] [PubMed] [Google Scholar]
- Brockwell P., Davis R.A. (2002). Introduction to Time Series and Forecasting, 2nd ed. (New York: Springer: ). [Google Scholar]
- Broghammer A., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 109: 13859–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R., Galera C., de Billy F., Penmetsa R.V., Journet E.P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M., Genre A., Sieberer B.J., Faccio A., Fournier J., Novero M., Barker D.G., Bonfante P. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189: 347–355. [DOI] [PubMed] [Google Scholar]
- Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ané J.M., Zhu H. (2008). OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol. 180: 311–315. [DOI] [PubMed] [Google Scholar]
- Chen C., Gao M., Liu J., Zhu H. (2007). Fungal symbiosis in rice requires an ortholog of a legume common symbiosis gene encoding a Ca2+/calmodulin-dependent protein kinase. Plant Physiol. 145: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja L.F., Hogekamp C., Lamm P., Maillet F., Martinez E.A., Samain E., Dénarié J., Küster H., Hohnjec N. (2012). Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiol. 159: 1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho-Niebel F., Timmers A.C., Chabaud M., Defaux-Petras A., Barker D.G. (2002). The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. Plant J. 32: 343–352. [DOI] [PubMed] [Google Scholar]
- Delaux P.M., Bécard G., Combier J.P. (2013). NSP1 is a component of the Myc signaling pathway. New Phytol. 199: 59–65. [DOI] [PubMed] [Google Scholar]
- Dénarié J., Debellé F., Promé J.C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65: 503–535. [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Wais R., Long S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681. [DOI] [PubMed] [Google Scholar]
- El Yahyaoui F., Küster H., Ben Amor B., Hohnjec N., Pühler A., Becker A., Gouzy J., Vernié T., Gough C., Niebel A., Godiard L., Gamas P. (2004). Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 136: 3159–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966. [DOI] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., Fournier J., Rochange S., Bécard G., Bonfante P., Barker D.G. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198: 190–202. [DOI] [PubMed] [Google Scholar]
- Gobbato E., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22: 2236–2241. [DOI] [PubMed] [Google Scholar]
- Gomez S.K., Javot H., Deewatthanawong P., Torres-Jerez I., Tang Y., Blancaflor E.B., Udvardi M.K., Harrison M.J. (2009). Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granqvist E., Oldroyd G.E., Morris R.J. (2011). Automated Bayesian model development for frequency detection in biological time series. BMC Syst. Biol. 5: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Paszkowski U. (2013). Multiple control levels of root system remodeling in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 4: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Casieri L., Paszkowski U. (2009). Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytol. 182: 829–837. [DOI] [PubMed] [Google Scholar]
- Gutjahr C., Banba M., Croset V., An K., Miyao A., An G., Hirochika H., Imaizumi-Anraku H., Paszkowski U. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.J. (2005). Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 59: 19–42. [DOI] [PubMed] [Google Scholar]
- Heron D.S., Pueppke S.G. (1984). Mode of infection, nodulation specificity, and indigenous plasmids of 11 fast-growing Rhizobium japonicum strains. J. Bacteriol. 160: 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E.-P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., Arnould C., Morandi D., Barker D.G., Gianinazzi-Pearson V. (2001). Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14: 737–748. [DOI] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kanamori N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103: 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Chabaud M., Lougnon G., Gough C., Dénarié J., Barker D.G., Bécard G. (2003). A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol. 131: 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M., Held K., Binder A., Hashimoto K., Den Herder G., Parniske M., Kudla J., Schumacher K. (2012). FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J. 69: 181–192. [DOI] [PubMed] [Google Scholar]
- Kuhn H., Küster H., Requena N. (2010). Membrane steroid-binding protein 1 induced by a diffusible fungal signal is critical for mycorrhization in Medicago truncatula. New Phytol. 185: 716–733. [DOI] [PubMed] [Google Scholar]
- Lauressergues D., Delaux P.M., Formey D., Lelandais-Brière C., Fort S., Cottaz S., Bécard G., Niebel A., Roux C., Combier J.P. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 72: 512–522. [DOI] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Maillet F., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63. [DOI] [PubMed] [Google Scholar]
- Mann H., Whitney D. (1947). On a test of whether one or two random variables is stochastically larger than the other. Ann. Math. Stat. 18: 50–60. [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E., Mun J.-H., Yeun L.H., Jayaraman D., Rougé P., Barre A., Lougnon G., Schornack S., Bono J.J., Cook D.R., Ané J.M. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant Microbe Interact. 20: 912–921. [DOI] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami E., Kouchi H., Carlson R.W., Cohn J.R., Kolli V.K., Day R.B., Ogawa T., Stacey G. (1996). Cooperative action of lipo-chitin nodulation signals on the induction of the early nodulin, ENOD2, in soybean roots. Mol. Plant Microbe Interact. 9: 574–583. [DOI] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E., Long S.R. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101: 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Ané J.M. (2011). Germinating spore exudates from arbuscular mycorrhizal fungi: molecular and developmental responses in plants and their regulation by ethylene. Mol. Plant Microbe Interact. 24: 260–270. [DOI] [PubMed] [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252. [DOI] [PubMed] [Google Scholar]
- Nars A., et al. (2013). Aphanomyces euteiches cell wall fractions containing novel glucan-chitosaccharides induce defense genes and nuclear calcium oscillations in the plant host Medicago truncatula. PLoS ONE 8: e75039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oláh B., Brière C., Bécard G., Dénarié J., Gough C. (2005). Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J. 44: 195–207. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Long S.R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131: 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E., Mitra R.M., Wais R.J., Long S.R. (2001a). Evidence for structurally specific negative feedback in the Nod factor signal transduction pathway. Plant J. 28: 191–199. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Engstrom E.M., Long S.R. (2001b). Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13: 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Mondo S.J., Topp S., Starker C.G., Gantt J.S., Harrison M.J. (2010). Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61: 482–494. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Reddy D.M.R.S., Schorderet M., Feller U., Reinhardt D. (2007). A petunia mutant affected in intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi. Plant J. 51: 739–750. [DOI] [PubMed] [Google Scholar]
- Rodpothong P., Sullivan J.T., Songsrirote K., Sumpton D., Cheung K.W.J.T., Thomas-Oates J., Radutoiu S., Stougaard J., Ronson C.W. (2009). Nodulation gene mutants of Mesorhizobium loti R7A-nodZ and nolL mutants have host-specific phenotypes on Lotus spp. Mol. Plant Microbe Interact. 22: 1546–1554. [DOI] [PubMed] [Google Scholar]
- Saito K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Chabaud M., Fournier J., Timmers A.C., Barker D.G. (2012). A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J. 69: 822–830. [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Chabaud M., Timmers A.C., Monin A., Fournier J., Barker D.G. (2009). A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Morgan D.R., Swensen S.M., Mullin B.C., Dowd J.M., Martin P.G. (1995). Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. USA 92: 2647–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962. [DOI] [PubMed] [Google Scholar]
- Takeda N., Sato S., Asamizu E., Tabata S., Parniske M. (2009). Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58: 766–777. [DOI] [PubMed] [Google Scholar]
- Takeda N., Tsuzuki S., Suzaki T., Parniske M., Kawaguchi M. (2013). CERBERUS and NSP1 of Lotus japonicus are common symbiosis genes that modulate arbuscular mycorrhiza development. Plant Cell Physiol. 54: 1711–1723. [DOI] [PubMed] [Google Scholar]
- Talukdar T., Gorecka K.M., de Carvalho-Niebel F., Downie J.A., Cullimore J., Pikula S. (2009). Annexins - calcium- and membrane-binding proteins in the plant kingdom: potential role in nodulation and mycorrhization in Medicago truncatula. Acta Biochim. Pol. 56: 199–210. [PubMed] [Google Scholar]
- Wais R.J., Galera C., Oldroyd G., Catoira R., Penmetsa R.V., Cook D., Gough C., Denarié J., Long S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.A., Viprey V., Downie J.A. (2000). Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc. Natl. Acad. Sci. USA 97: 13413–13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann S., Sanchez L., Descombin J., Chatagnier O., Gianinazzi S., Gianinazzi-Pearson V. (2004). Fungal elicitation of signal transduction-related plant genes precedes mycorrhiza establishment and requires the dmi3 gene in Medicago truncatula. Mol. Plant Microbe Interact. 17: 1385–1393. [DOI] [PubMed] [Google Scholar]
- Wilcoxon F. (1945). Individual comparisons by ranking methods. Biom. Bull. 1: 80–83. [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.