An alternative, tissue-specific pathway yielding a common homoterpene volatile in roots is implicated in defense against soil borne pathogens, revealing plasticity in volatile homoterpene biosynthesis and function.
Abstract
Plant-derived volatile compounds such as terpenes exhibit substantial structural variation and serve multiple ecological functions. Despite their structural diversity, volatile terpenes are generally produced from a small number of core 5- to 20-carbon intermediates. Here, we present unexpected plasticity in volatile terpene biosynthesis by showing that irregular homo/norterpenes can arise from different biosynthetic routes in a tissue specific manner. While Arabidopsis thaliana and other angiosperms are known to produce the homoterpene (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) or its C16-analog (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene by the breakdown of sesquiterpene and diterpene tertiary alcohols in aboveground tissues, we demonstrate that Arabidopsis roots biosynthesize DMNT by the degradation of the C30 triterpene diol, arabidiol. The reaction is catalyzed by the Brassicaceae-specific cytochrome P450 monooxygenase CYP705A1 and is transiently induced in a jasmonate-dependent manner by infection with the root-rot pathogen Pythium irregulare. CYP705A1 clusters with the arabidiol synthase gene ABDS, and both genes are coexpressed constitutively in the root stele and meristematic tissue. We further provide in vitro and in vivo evidence for the role of the DMNT biosynthetic pathway in resistance against P. irregulare. Our results show biosynthetic plasticity in DMNT biosynthesis in land plants via the assembly of triterpene gene clusters and present biochemical and genetic evidence for volatile compound formation via triterpene degradation in plants.
INTRODUCTION
Plants employ volatile compounds of diverse structure to interact with their environment. Volatile terpenes have been implicated with multiple functions in the attraction of insects, defense against herbivores or pathogens, and plant-plant interactions (Dudareva et al., 2006; Pichersky et al., 2006; Unsicker et al., 2009; Clavijo McCormick et al., 2012), and these activities often correlate with the tissue- and cell type-specific biosynthesis of terpenes. Independent of their tissue-specific origin, the specific classes of terpene compounds are supposed to be derived from the same central intermediates. For example, regular terpenes such as C10 monoterpenes, C15 sesquiterpenes, and C20 diterpenes are assembled from the 5-carbon units isopentenyl diphosphate and dimethylallyl diphosphate, which are condensed to the core C10, C15, and C20 trans- or cis-prenyl diphosphate substrates of terpene synthases (Chen et al., 2011; Tholl and Lee, 2011).
The C15 and C20 tertiary alcohols, (E)-nerolidol and (E,E)-geranyl linalool, have been shown to function as precursors of the irregular C11-homo/norterpene volatile (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) and its C16-analog (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene (TMTT), respectively (Tholl et al., 2011). DMNT and TMTT are common constituents of floral and herbivore- or pathogen-induced volatile blends of angiosperms, and they contribute to the deterrence or attraction of insect pests and their parasites or predators (Mumm and Dicke, 2010; Tholl et al., 2011). For example, both homoterpenes occur as floral volatiles of night-scented, moth-pollinated orchids (Kaiser, 1991; Donath and Boland, 1995). Several studies have supported a role of DMNT and TMTT together with other volatile compounds in indirect defense against spider mite attack by promoting the attraction of predatory mites (Dicke et al., 1990; de Boer et al., 2004; Kappers et al., 2005). Moreover, homoterpenes have been shown to induce defense gene expression in plant-plant interactions (Arimura et al., 2000). The formation of DMNT and TMTT in leaf or flower tissues usually occurs by the oxidative breakdown of (E)-nerolidol or (E,E)-geranyl linalool (Boland et al., 1998). The C-C cleavage reaction is catalyzed by cytochrome P450 monooxygenases, of which only a single enzyme, CYP82G1, has so far been identified in Arabidopsis thaliana (Lee et al., 2010). CYP82G1, which is induced in Arabidopsis leaves upon insect feeding damage, produces DMNT and TMTT in vitro but functions as a TMTT synthase in planta because of the presence of (E,E)-geranyllinalool (Herde et al., 2008) but not (E)-nerolidol in Arabidopsis leaves.
Here, we show that Arabidopsis can produce DMNT via an unexpected alternative pathway in a tissue-specific manner by the oxidative degradation of arabidiol, a tricyclic triterpene diol, in Arabidopsis roots. We demonstrate that CYP705A1, a member of the Brassicaceae-specific CYP705 family, cleaves the prenyl side chain of arabidiol to produce DMNT and a nonvolatile C19-ketone derivative. The reaction is induced by the root rot oomycete pathogen Pythium irregulare and treatment with the plant defense hormone jasmonic acid (JA) and contributes to Arabidopsis root defense. The CYP705A1 gene clusters with a gene encoding a pentacyclic triterpene synthase 1 (Pen1) named arabidiol synthase (here referred to as ABDS) (Xiang et al., 2006) as part of a larger triterpene biosynthetic gene cluster on chromosome 4 (Field et al., 2011; Castillo et al., 2013). Our results provide evidence for independent evolution and biosynthetic plasticity in the formation of functionally active volatiles. We discuss how alternative pathways in the production of the same homoterpene compound may evolve depending on the tissue-specific expression of terpene biosynthetic gene clusters. Our findings also support the notion that triterpenes, similar to C40 carotenoids, undergo enzyme-mediated degradation to serve as precursors of plant volatiles.
RESULTS
P. irregulare-Induced Production of DMNT in Arabidopsis Roots
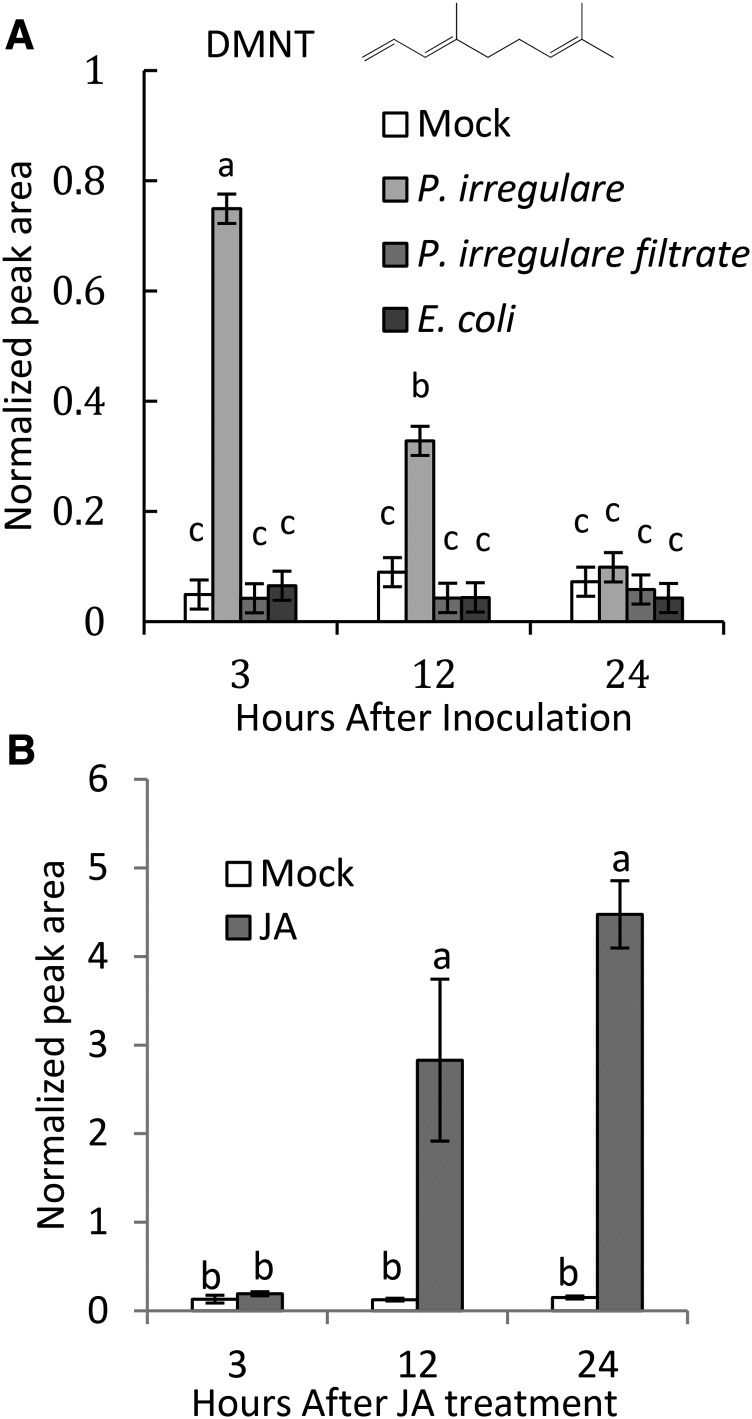
We investigated whether Arabidopsis roots release volatile compounds in response to infection by the soil-borne pathogen P. irregulare. P. irregulare is an oomycete pathogen causing seedling damping off, root rot, and vascular wilt disease in various plant species including Arabidopsis (Staswick et al., 1998). Arabidopsis plants were grown in liquid axenic culture and inoculated with a uniform suspension of mycelium and oospores of the oosporic P. irregulare isolate 110305. Root tissue was detached at different time points after inoculation, and root volatiles were analyzed by solid phase microextraction gas chromatography-mass spectrometry (SPME-GC/MS). We found that DMNT was transiently released in response to P. irregulare treatment (Figure 1A). Emission of DMNT was observed 3 h after inoculation with an ∼7-fold increase over constitutive background levels and then decreased to basal levels at 12 to 24 h (Figure 1A; Supplemental Figure 1). DMNT was detected at ∼10 ng/g root fresh weight at the peak of emission. Light microscopy analysis of P. irregulare-inoculated roots showed that the release of DMNT coincided with the germination of attached oospores to produce infection hyphae that penetrated the epidermal cells (Supplemental Figure 1).
Figure 1.
DMNT Is Emitted from Arabidopsis Roots upon Infection with P. irregulare or Treatment with Jasmonic Acid.
(A) Axenically grown Arabidopsis roots were infected with a suspension of P. irregulare mycelium and oospores, a P. irregulare filtrate, or with a suspension of E. coli. The structure of DMNT is shown.
(B) Treatment of axenically grown roots with 100 μM JA. Root volatiles were analyzed from detached roots at different time points by SPME-GC/MS. Normalized peak areas are shown. Values represent the mean ± se of three biological replicates. Different letters show significant differences based on two-way ANOVA and Tukey-Kramer HSD test; P < 0.001. The experiment was repeated at least two times with similar results.
Since P. irregulare produces elicitors such as lytic enzymes and phytotoxins (Latijnhouwers et al., 2003; Win et al., 2012), we tested the effect of a soluble P. irregulare filtrate on DMNT emission. Furthermore, we examined whether treatment with Escherichia coli, a non-plant pathogen, could cause induction of DMNT emission. Transient DMNT emission was observed only following inoculation with a suspension of P. irregulare containing mycelium and oospores and neither inoculation with the soluble filtrate nor treatment with an E. coli suspension could mimic this response (Figure 1A).
The importance of the defense hormone JA in plant defense against P. irregulare has been reported previously (Vijayan et al., 1998). To further understand the role of JA in DMNT formation in Arabidopsis roots, axenically grown Arabidopsis plants were treated with 100 μM JA. Roots were excised and headspace volatiles were measured by SPME-GC/MS. DMNT emission was induced at 12 h after the beginning of treatment and further increased at 24 h to ∼70 ng/g fresh weight (Figure 1B).
Identification of CYP705A1 as a Root-Specific Gene Involved in DMNT Biosynthesis
Previous studies on the formation of TMTT in Arabidopsis leaves demonstrated a two-step biosynthetic pathway consisting of the formation of the tertiary alcohol (E,E)-geranyllinalool (GL) catalyzed by the GL synthase TPS04 (Herde et al., 2008) and the subsequent breakdown of GL to (E)-TMTT catalyzed by CYP82G1 (Lee et al., 2010). Even though the CYP82G1 enzyme can convert (E)-nerolidol into DMNT in vitro (Lee et al., 2010) and possibly in planta (Kappers et al., 2005), CYP82G1 is not expressed in roots (Lee et al., 2010) and the null mutant cyp82g1-1 is not impaired in root-specific DMNT biosynthesis (Supplemental Figure 2), indicating that this P450 is not involved in the formation of DMNT in Arabidopsis roots. Since treatments of Arabidopsis hairy roots with the cytochrome P450-specific azole inhibitors miconazole and clotrimazole (St-Pierre and De Luca, 1995) severely reduced JA-induced emission of DMNT (Supplemental Figure 3), we hypothesized that DMNT is most likely produced by a different root-specific P450 enzyme.
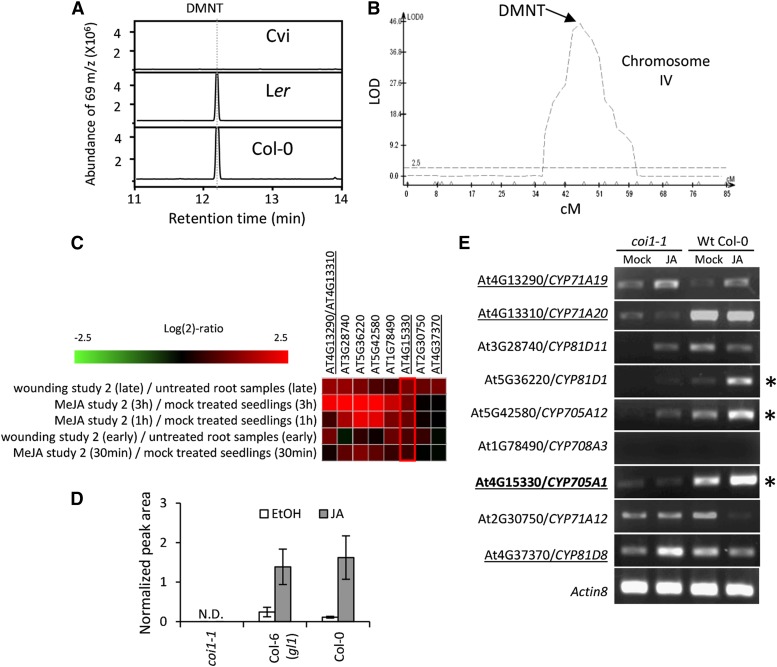
To identify P450 genes involved in DMNT formation, we performed quantitative trait locus (QTL) analysis of DMNT emission using recombinant inbred lines of the accession Cape Verdi Island (Cvi), a non-DMNT emitter, and the DMNT-emitting Arabidopsis accession Landsberg erecta (Ler) (Alonso-Blanco et al., 1998) (Figure 2A). We were able to map a QTL regulating this trait using a single measurement per line because the bimodal distribution indicates the underlying locus behaves in a qualitative Mendelian fashion with large effect (Supplemental Table 1 and Supplemental Figure 4). However, this also means that we may have missed smaller effect loci that would require replication to have been identified. Our analysis suggested that a single region on chromosome 4 contributes to the natural variation in DMNT biosynthesis (Figure 2B). To further refine this analysis, we searched for the expression of all Arabidopsis P450 genes in publically available microarray data sets under the treatment with methyl jasmonate or wounding assuming that the expression of the target P450 gene would be induced in roots under these conditions. This approach again excluded CYP82G1 and resulted in a list of nine candidate genes (Figure 2C). We then conducted a comparative RT-PCR analysis of transcripts of the selected candidate genes with and without JA treatment in roots of wild-type plants and the JA-insensitive mutant coronatine insensitive1 (coi1-1) (Xie et al., 1998). As a result, two P450 genes, CYP705A1 and CYP81D1, were found, whose transcripts accumulated upon JA treatment in wild-type but not coi1-1 roots (Figures 2D and 2E). Of these two genes, only CYP705A1 resides in the identified QTL region on chromosome 4.
Figure 2.
Selection of DMNT Synthase Candidate Genes.
(A) GC/MS chromatograms of induced emissions of DMNT from roots of the Arabidopsis accessions Ler, Cvi, and Col-0 in response to 24 h of JA treatment. No DMNT was detected from the Cvi accession, while the volatile was produced by roots of Col-0 and Ler.
(B) Identification of the QTL region for DMNT formation in the Ler × Cvi RILs on chromosome 4. The y axis is in log of the odds (LOD) units, and the x axis is in centimorgans (cM); the horizontal lines represent the 0.05 significance threshold determined by 1000 permutations.
(C) A screening was conducted of publically available microarray data sets using Genevestigator for all Arabidopsis P450 genes expressed upon treatment with methyl jasmonate (MeJA) or wounding assuming that the expression of the target P450 gene would be induced in roots under these conditions. Expression of selected P450 candidate genes upon wounding and methyl jasmonate treatments is shown (At4g13290 and At4g13310 share the same probe number).
(D) Volatile analysis of roots of coi1-1, its corresponding background genotype Col-6 (gl1), and wild-type Col-0. No DMNT was detected in hydroponically grown coi1-1 plants upon JA treatment. Values represent the mean ± se of three replicates.
(E) RT-PCR analysis of candidate gene expression in coi1-1 and wild-type Col-0 plants under JA and mock (ethanol) treatment. JA-inducible and coi1-1-regulated genes are marked with asterisks. Candidate genes on the selected QTL region on chromosome 4 are underlined. Only the CYP705A1 locus overlaps with the DMNT QTL and shows JA-inducible and coi1-1-dependent expression.
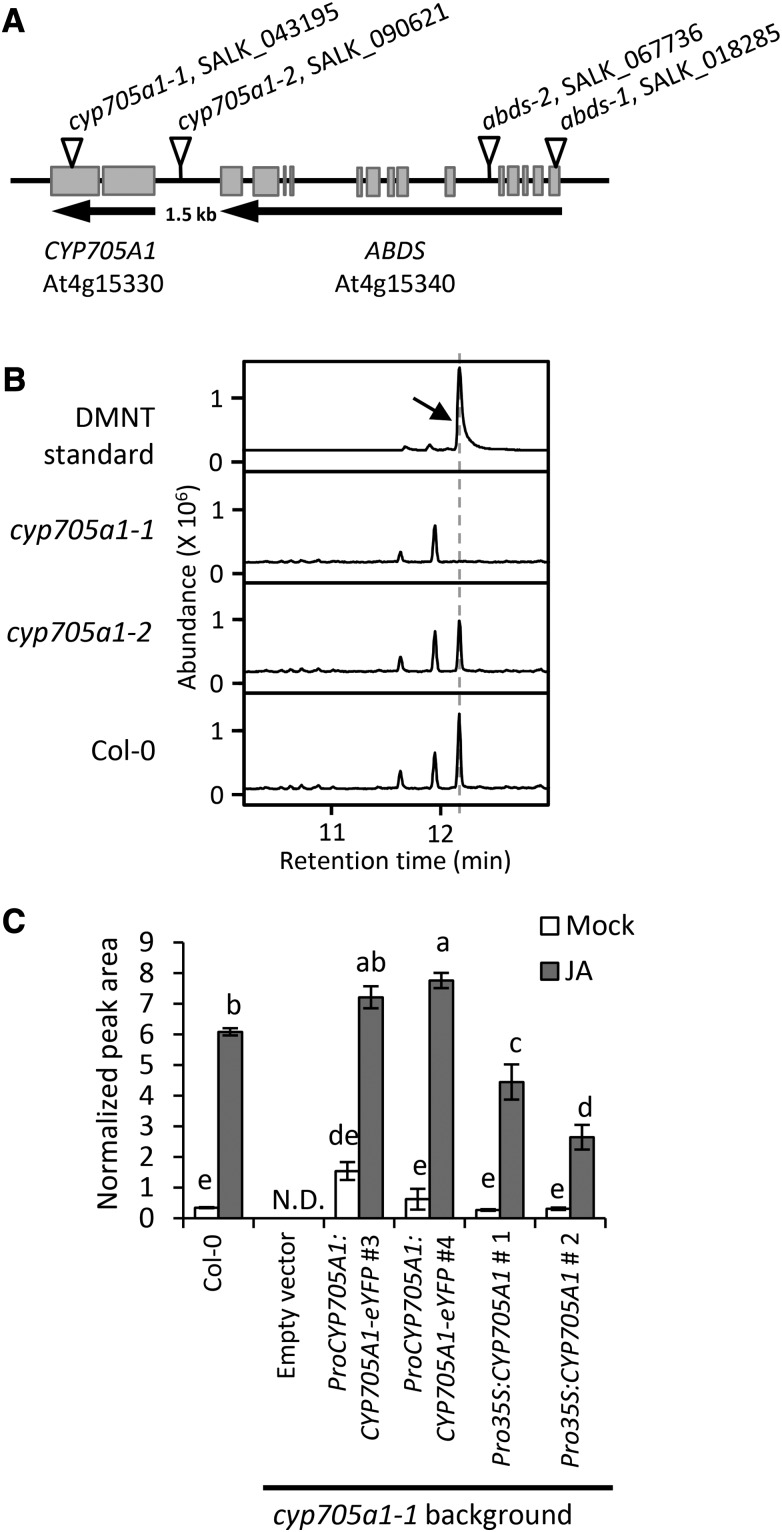
SPME-GC/MS analysis of volatiles emitted from roots of the gene knockout line cyp705a1-1 (SALK_043195) with a T-DNA insertion in the second exon (Figure 3A) lacked emission of DMNT upon JA treatment, while a second line, cyp705a1-2 (SALK_090621), carrying a T-DNA insertion in the CYP705A1 promoter region produced wild-type levels of DMNT (Figure 3B) and the CYP705A1 transcript (Supplemental Figure 5A). This finding suggested that CYP705A1 is involved in DMNT biosynthesis in Arabidopsis roots. Quantitative RT-PCR showed that treatment of wild-type roots with JA led to a 5-fold increase of CYP705A1 transcript levels 12 h after basal levels at the onset of treatment (Supplemental Figure 6). By comparison, inoculation with P. irregulare caused a 2-fold induction of CYP705A1 transcript abundance 1 h after inoculation prior to a decline to levels similar to those before inoculation, which is in agreement with the transient nature of DMNT production upon P. irregulare infection (Supplemental Figure 6).
Figure 3.
Identification of CYP705A1 as a DMNT Synthase.
(A) Schematic showing the genomic locus of Arabidopsis CYP705A1 (At4g15330) in tandem with the ABDS gene. Exons are represented by gray boxes. Introns and intergenic regions are represented by the black line. Insertion sites of the T-DNA mutants used in this study are marked with inverted triangles.
(B) DMNT emission in roots of wild-type and cyp705a1 mutants after 24 h of JA treatment. The retention time for the DMNT authentic standard (indicated by the arrow) is marked with a dashed line.
(C) DMNT emission in mock- and JA-treated plants in wild-type background compared with representative transgenic lines. Volatiles were collected from roots after 24 h of JA treatment and analyzed by SPME-GC/MS. Mock controls were treated with ethanol. Normalized peak areas are shown and the values represent the mean ± se of three biological replicates. Different letters show significant differences based on two-way ANOVA and Tukey-Kramer HSD test, P < 0.001. N.D. indicates that no volatile was detected.
To further study the role of the CYP705A1 gene in DMNT production, the cyp705a1-1 mutant was complemented with a full-length CYP705A1 cDNA in C-terminal fusion to enhanced yellow fluorescent protein (eYFP) under the control of a 1.5-kb fragment of the native CYP705A1 promoter. Additionally, the full-length CYP705A1 transcript was fused to the CaMV 35S promoter for ectopic expression in the cyp705a1-1 mutant background. In ProCYP705A1:CYP705A1-eYFP lines, accumulation of the full-length CYP705A1 transcript was detected upon JA treatment, while only basal transcript levels of the gene were observed in ProCaMV35S-CYP705A1 lines with no JA-dependent increase of transcript abundance (Supplemental Figure 5B). Consistent with the transcript accumulation, DMNT volatile formation was restored in both transgenic lines (Figure 3C), further supporting the role of CYP705A1 in the DMNT biosynthetic pathway. Despite the basal constitutive expression of CYP705A1 in the ProCaMV35S-CYP705A1 lines, an increase of DMNT emission was observed in these lines in response to treatment with JA, which suggests a JA-enhanced formation of the CYP705A1 enzymatic substrate.
Arabidiol Is the Precursor in DMNT Biosynthesis in Arabidopsis Roots
Despite our prediction that DMNT would be produced from (E)-nerolidol, we were unable to detect this precursor in the headspace or in organic extracts of JA-treated roots. In addition, an analysis of T-DNA insertion lines of several root-expressed terpene synthase genes (Vaughan et al., 2013) did not lead to the identification of a gene involved in DMNT formation. However, treatment of Arabidopsis hairy roots with lovastatin, an inhibitor of the isopentenyl diphosphate producing mevalonate pathway in the cytosol, severely reduced jasmonate-induced emission of DMNT, while this was not the case when fosmidomycin, an inhibitor of the plastidial methylerythritol phosphate pathway, was applied (Supplemental Figure 7). This result suggested a major contribution of the mevalonate pathway and a possibly farnesyl diphosphate-derived precursor in the DMNT biosynthetic pathway.
In line with these findings, a recent study by Castillo et al. (2013) on triterpene-modifying enzymes in Arabidopsis showed that the C30-triterpene diol, arabidiol, can be degraded by CYP705A1 to a C19 ketone product (14-apo-arabidiol). Arabidiol is produced by Pen1, also called ABDS (Xiang et al., 2006), which is highly coexpressed with CYP705A1 (Supplemental Table 2). Both genes are clustered in tandem on chromosome 4 (Figure 3A). Arabidiol has a 6,6,5-tricyclic ring system with two hydroxyl groups, whose tricyclic backbone is covalently linked to a C13-prenylalcohol side chain (Figure 4A). We predicted that the tertiary hydroxyl group of this side chain made arabidiol a suitable substrate for oxidative degradation by CYP705A1 to produce a compound resembling DMNT and the previously identified C19 degradation product (Figure 4A).
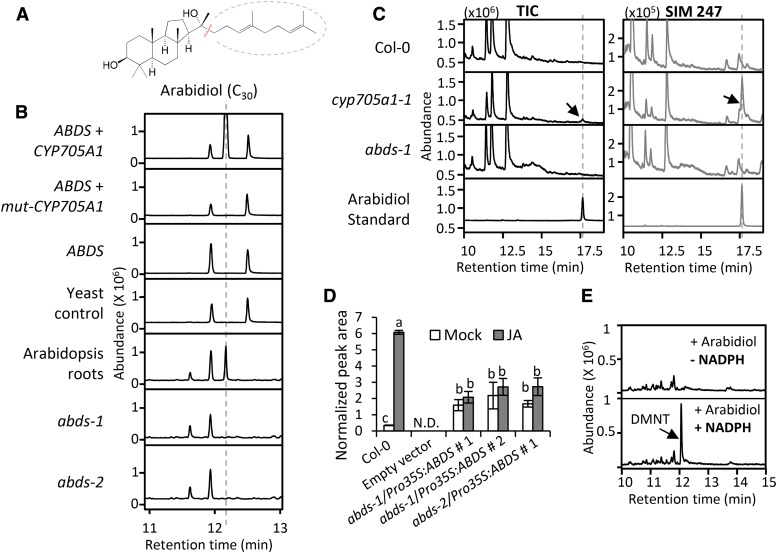
Figure 4.
Arabidiol Is the Substrate for DMNT Biosynthesis.
(A) Structure of arabidiol.
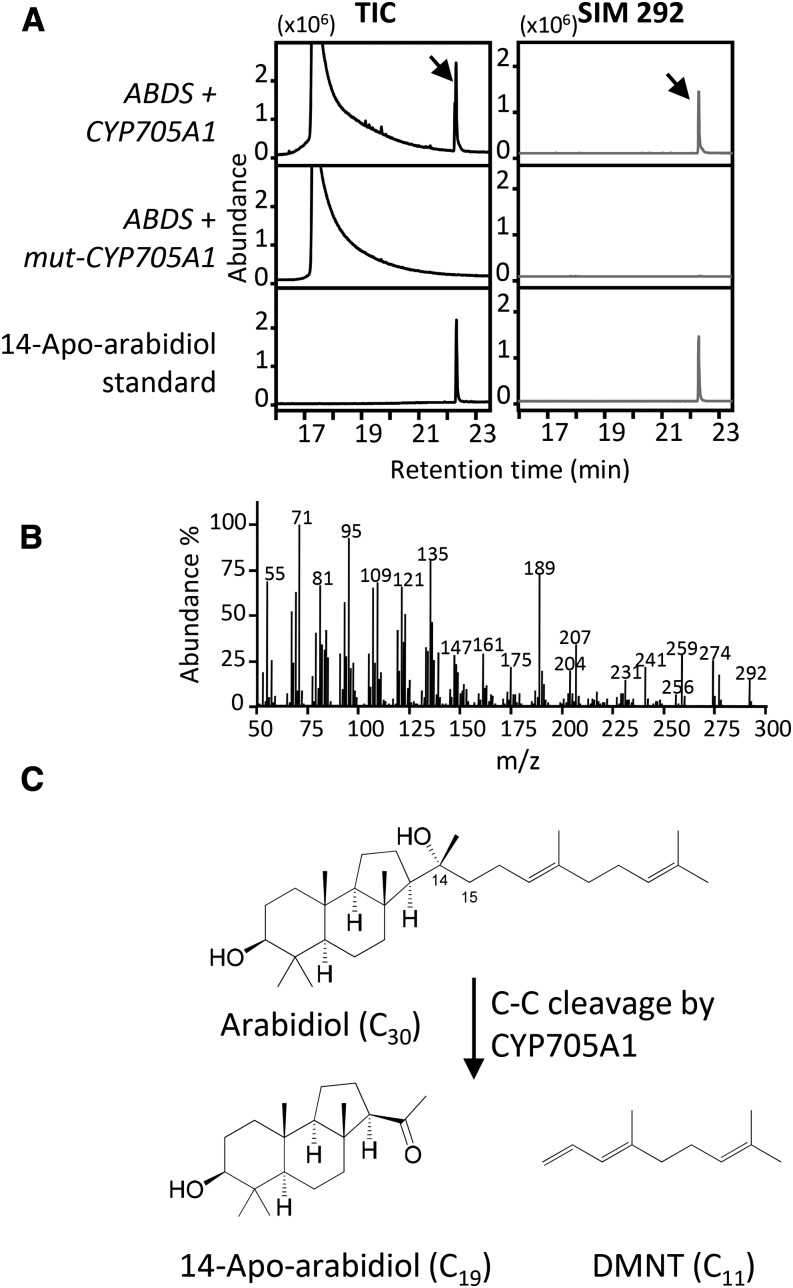
(B) DMNT emission in yeast and Arabidopsis plants. DMNT was detected from WAT11 yeast cells coexpressing ABDS with a wild-type CYP705A1 cDNA. In yeast cells expressing ABDS alone or coexpressing ABDS with a mutated version of CYP705A1 (mut-CYP705A1), no DMNT was observed. The yeast control line was transformed with only the empty vector used for expression of CYP705A1. Volatile analysis of Col-0 roots and abds mutants after 24 h of 100 μM JA treatment is shown. No DMNT was detected from abds mutants. Volatile products were analyzed in the yeast culture or plant tissue headspace by SPME-GC/MS. The retention time for DMNT is marked with a dashed line.
(C) Arabidiol detection from Arabidopsis roots. GC/MS chromatograms of liquid extracts from 1 g JA-treated roots of wild-type Col-0, abds-1, and cyp705a1-1 are depicted. Arabidiol (indicated by arrows) was only detected in the cyp705a1-1 mutant after JA treatment as shown in TIC (total ion chromatogram) and in single ion monitoring (SIM) mode for m/z 247.
(D) DMNT emission from roots of wild-type Col-0 and three Pro35S-ABDS overexpression lines in two different abds mutant backgrounds treated with JA for 24 h. Normalized peak areas are shown. Values represent the mean ± se of three biological replicates. Different letters show significant differences based on two-way ANOVA and Tukey-Kramer HSD test, P < 0.001. N.D. indicates that no volatile was detected.
(E) Microsomal preparations expressing CYP705A1 converted arabidiol to DMNT (indicated by the arrow) in the presence of 2.4 mM of the P450 cofactor NADPH.
Volatile headspace analysis of yeast (Saccharomyces cerevisiae) WAT11 cell cultures coexpressing the CYP705A1 and ABDS genes led to the detection of DMNT after induction with galactose (Figure 4B). No DMNT was found when ABDS was expressed alone or with a truncated CYP705A1 gene lacking the heme binding domain (mut-CYP705A1; Figure 4B). Involvement of arabidiol in DMNT biosynthesis was further confirmed by the absence of JA-induced DMNT emission in two independent T-DNA insertion mutants of the ABDS gene, SALK_018285 (abds-1) and SALK_067736 (abds-2) (Figures 3A and 4B), both of which lack a full-length ABDS transcript (Supplemental Figure 8A). To detect arabidiol from Arabidopsis plants, we performed organic solvent extraction from JA-treated roots of wild-type and DMNT biosynthetic mutants (Figure 4C). Arabidiol was not detected in the abds-1 mutant or in wild-type plants presumably due to an immediate conversion into DMNT. However, an accumulation of arabidiol was observed in cyp705a1-1 missing a functional arabidiol degradation enzyme. Additionally, complementation of the two abds mutants with the full-length ABDS cDNA driven by the CaMV 35S promoter led to a restoration of DMNT emission in both mutants (Figure 4D). Treatment with JA only slightly enhanced volatile emission from these lines most likely because of the absence of the JA-responsive ABDS promoter (see below; Supplemental Figure 8B). In addition, purified microsomal fractions from yeast expressing only CYP705A1 converted arabidiol into DMNT in the presence of the cofactor NADPH (Figure 4E). In conclusion, these findings suggested that both CYP705A1 and ABDS are necessary and sufficient for the biosynthesis of DMNT.
ABDS transcript abundance was very low in untreated roots of wild-type plants (mostly undetectable by RT-PCR; Supplemental Figure 8) but increased 10-fold at 12 h after treatment with JA (Supplemental Figure 7). No significant changes in transcript levels of ABDS were found at the early stage of infection with P. irregulare (Supplemental Figure 6), suggesting that DMNT biosynthesis is, at least in part, regulated by the expression of CYP705A1 but not ABDS.
Gas chromatography-mass spectrometry (GC/MS) analysis of ethyl acetate extracts of yeast coexpressing ABDS and CYP705A1 also showed the presence of the 14-apo-arabidiol cleavage product at tR = 22.65 min with the expected molecular ion of m/z 292.25 (Figures 5A and 5B). The compound was absent in lines expressing the nonfunctional CYP705A1 gene (Figure 5A). The pseudomolecular ion peak of the purified compound at m/z 275.2362, [M-OH]+ (calculated for C19H31O: 275.2369), observed in high-resolution electrospray ionization mass spectrometry analysis confirmed the proposed molecular formula with four degrees of unsaturation (Supplemental Figure 9A and Supplemental Methods). The 14-apo-arabidiol structure was then further confirmed using proton 1H-NMR (Supplemental Figure 9B and Supplemental Methods), HSQC, HMBC, NOESY, and COSY (Supplemental Figures 10 to 13 and Supplemental Methods). Proton and carbon chemical shifts are listed in Supplemental Table 3, and key correlations are shown in Supplemental Figure 14. Together, this analysis clearly demonstrated that CYP705A1 catalyzes a C-C cleavage of arabidiol at its C14-C15 bond. A double bond is introduced in the isoprenyl side chain to produce DMNT, and the tertiary hydroxyl group at C14 is converted into a ketone group resulting in 14-apo-arabidiol (Figure 5C).
Figure 5.
Detection of 14-Apo-Arabidiol in the Yeast Coexpression System.
(A) The C19 degradation product 14-apo-arabidiol was detected (indicated by arrows) in WAT11 yeast cells expressing ABDS and CYP705A1 but not mut-CYP705A1. The GC chromatogram of the purified degradation product is depicted. TIC, total ion chromatogram; SIM, single ion monitoring.
(B) MS spectrum of 14-apo-arabidiol with a m/z 292 molecular ion.
(C) The pathway for arabidiol degradation to DMNT and 14-apo-arabidiol. The molecular structure of 14-apo-arabidiol was determined by NMR analysis.
Spatial Pattern of DMNT Biosynthetic Gene Expression
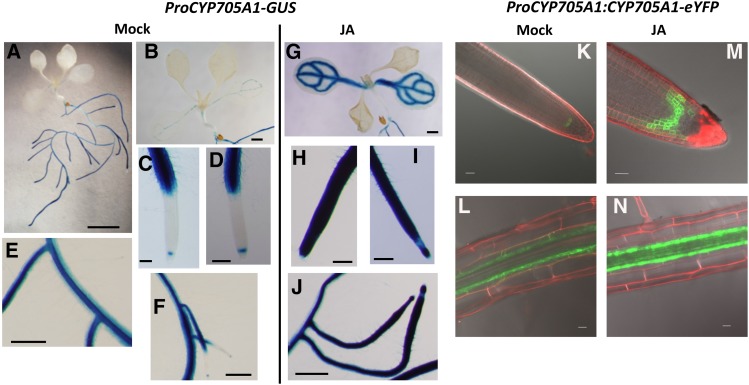
We further examined the tissue-specific expression of the DMNT biosynthetic genes by performing promoter activity assays in Arabidopsis transgenic lines expressing the β-glucuronidase gene (GUS) under the control of the native CYP705A1 and ABDS promoters. In 12-d-old seedlings grown on Murashige and Skoog (MS) medium, promoter activities of both genes were primarily observed in roots, and only weak ProCYP705A1-GUS and ProABDS-GUS activity was found in the vasculature of cotyledons and in true leaves, respectively (Figures 6A and 6B; Supplemental Figure 15A). In roots of ProCYP705A1-GUS lines, GUS staining was mainly detected in the stele and to some extent in the cortex and epidermis at the root differentiation zone and in the mature root (Figures 6C to 6F). No GUS activity was observed in the cell elongation area, but, curiously, cell type-specific expression was found in the quiescent center at the root meristematic zone (Figures 6C, 6D, and 6F). Similar expression patterns were detected in primary and lateral roots (Figures 6C to 6F). Despite the low transcript abundance of the ABDS gene in untreated roots, we found substantial GUS activity driven by the ABDS promoter, which largely colocalized to the same areas where ProCYP705A1-GUS activity was observed. However, ProABDS-GUS activity was confined to the stele in the differentiation and elongation zones and occurred in all cells of the root meristematic zone (Supplemental Figures 15B to 15D).
Figure 6.
Expression of CYP705A1 Is Localized to the Root Stele and Meristematic Zone and Responds to Treatment with JA.
(A) to (J) GUS activity in 12-d-old mock and JA-treated ProCYP705A1-GUS transgenic lines.
(A) Whole seedling. Bar = 5 mm.
(B) and (G) Cotyledon and true leaves. Bars = 1 mm.
(C) and (H) Main root tip. Bars = 200 μm.
(D) and (I) Lateral root tip. Bars = 200 μm.
(E), (F), and (J) Lateral, main root attachment site. Bars = 0.5 mm.
(K) to (N) Confocal microscopy analysis of roots of 12-d-old mock and JA-treated ProCYP705A1:CYP705A1-eYFP plants. Mock-treated roots show localization of the CYP705A1-eYFP protein in the quiescent center in the root meristematic zone (K) and in the pericycle in the root hair zone (L). A localized induction of protein after JA treatment is observed ([M] and [N]). Results are representative for at least three independent transgenic lines. Bars = 20 μm.
To further evaluate the tissue specificity of constitutive DMNT production at the protein level, we examined the cell type-specific localization of the CYP705A1 protein in ProCYP705A1:CYP705A1-eYFP lines. Confocal microscopy analysis of three independent transgenic lines suggested that expression of the CYP705A1 protein is confined primarily to the pericycle in the root differentiation zone (Figures 6K and 6L). Observation of a YFP signal in the quiescent center confirmed CYP705A1 expression in the root meristem (Figure 6K) under normal conditions.
Since we observed DMNT at high levels upon JA treatment, we also analyzed spatial patterns of GUS activity and YFP fusion protein expression in 12-d-old transgenic seedlings treated with 100 μM JA for 12 h. JA treatment induced strong CYP705A1 promoter activity in the root meristem and cell elongation zone in main and lateral roots (Figures 6H to 6J). Enhanced ProCYP705A1-GUS activity was also observed in the vasculature of cotyledons but not in true leaves, suggesting that the induced breakdown of arabidiol and emission of DMNT is largely confined to roots (Figure 6G). Patterns of JA-induced ABDS promoter-GUS activity were similar to those found for the CYP705A1 promoter except that no changes were detected in leaves in comparison to mock controls (Supplemental Figures 15E to 15H).
In contrast to the more widespread response of the ProCYP705A1-GUS activity, JA treatment appeared to enhance CYP705A1-YFP protein expression primarily in the area of the pericycle at the root differentiation zone without major systemic effects in this root zone (Figure 6N). Likewise, in the root meristematic zone, expression of the CYP705A1-YFP protein expanded only locally from the quiescent center to the endodermis and cortex but was excluded from the stele (Figure 6M). The observed differences in cell-type specificity of the ProCYP705A1-GUS activity and the CYP705A1-YFP protein could be attributed to a more widespread promoter activity and cell-specific posttranscriptional/translational restrictions, although possible effects of diffusion of the GUS product into nonexpressing neighboring cells or tissues cannot be entirely excluded.
DMNT Negatively Effects P. irregulare Oospore Germination and Growth
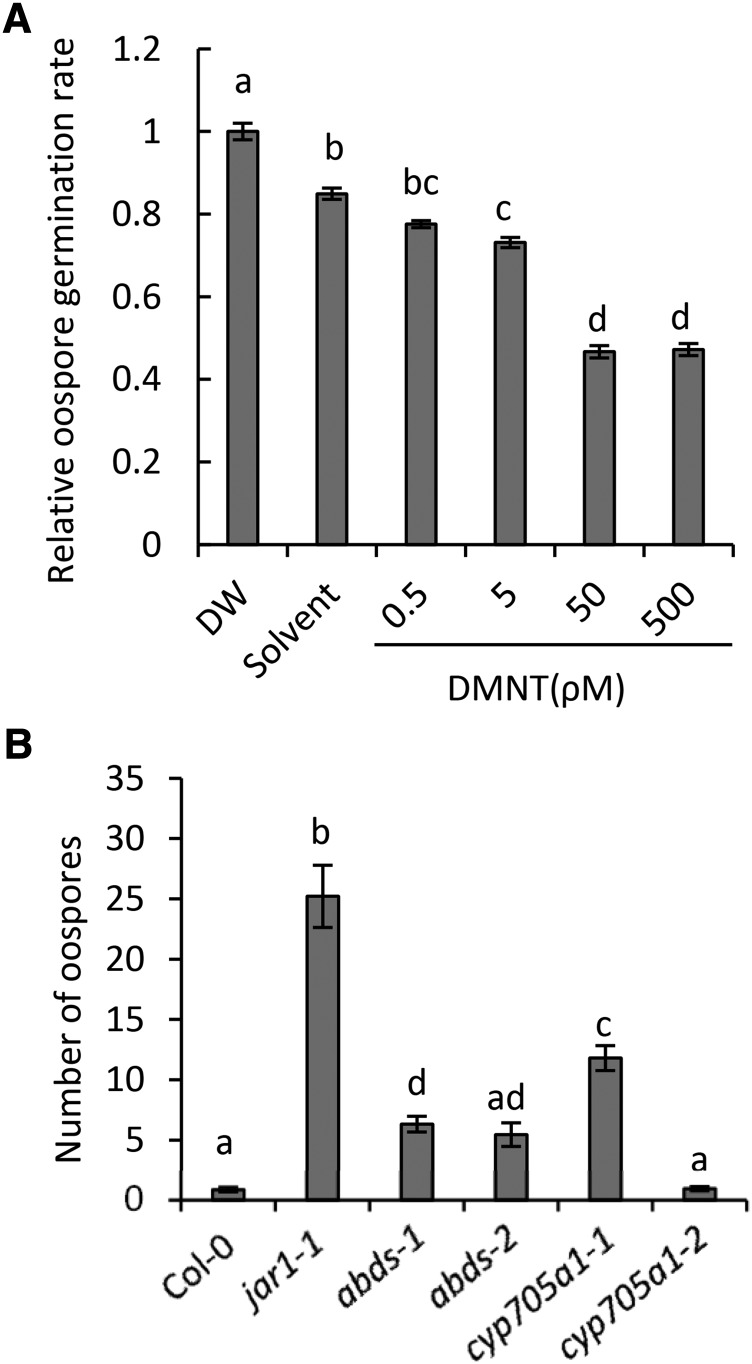
Since we had observed highest emission levels of DMNT within hours of oospore germination and germ tube penetration, we examined a possible effect of DMNT on P. irregulare oospore germination. Since an accurate quantitative assessment of these effects under in vivo conditions proved too difficult, we observed oospore germination rates in vitro on maize (Zea mays) meal agar plates supplemented with DMNT at different concentrations. Oospore germination was inhibited by ∼50% at concentrations as low as 50 pM DMNT (Figure 7A). Higher DMNT concentrations caused only minor additional inhibitory effects, which may suggest a dose-specific response. We also tested whether DMNT had any effect on the mycelium growth rate of the oomycete by comparing the growth area of the mycelium on potato dextrose agar at different DMNT concentrations (Supplemental Methods). Incubation with 10 nM DMNT resulted in ∼30% reduction of P. irregulare growth (Supplemental Figure 16), but again no strong dose-specific responses were observed at higher concentrations. By contrast, no growth reduction was observed when arabidiol or 14-apo-arabidiol were applied at similar concentrations (Supplemental Figure 16).
Figure 7.
DMNT Negatively Effects P. irregulare Oospore Germination and Formation.
(A) Effect of DMNT on oospore germination of P. irregulare. DMNT was applied at different concentrations in 10 mL of maize meal agar containing streptomycin. Oospore suspensions were added into each plate and incubated at 27°C in the dark. Germination rates were determined 24 h after inoculation. Thirty percent of the oospores germinated in the control treatment. The results were plotted relative to distilled water (DW), and oospore germination rate for distilled water was arbitrary set to 1.
(B) Root infection assay of P. irregulare in the wild type, DMNT biosynthetic mutants (abds-1, abds-2, and cyp705a1-1), and the control line cyp705a1-2. Representative root segments were taken 3 weeks after infection of soil-grown plants with the pathogen to measure oospore formation. Oospores were counted ∼10 mm behind the root tips. Statistical analysis was done using one-way ANOVA and Tukey-Kramer HSD test. The values represent the mean ± se of five replicates (P < 0.01) for (A) and at least six root segments from three different plants (P < 0.05) for (B). Different letters above the bars show significant differences. The experiments were repeated at least twice with similar results.
We further investigated whether the formation of arabidiol or its nonvolatile breakdown product had any long term effects on the root infection level of wild-type and abds and cyp705a1 mutant plants grown in potting substrate. Roots of 6-week-old plants were harvested 3 weeks postinoculation and stained with acid-fuchsin lactophenol to observe and count oospores inside root tissues in a 10-mm zone behind the root tip (Figure 7B). Whereas few oospores were found in roots of wild-type plants and the cyp705a1-2 line (DMNT wild-type phenotype), both the abds and the cyp705a1-1 mutants exhibited higher levels of oospores inside infected root tissues (significant for abds-1 and cyp705a1-1). However, the abundance of oospores in these mutants was significantly lower than that of the highly susceptible jar1-1 mutant, which is deficient in the formation of the JA-Ile conjugate (Staswick et al., 1998). Whether the somewhat enhanced susceptibility of the cyp705a1-1 mutant compared with the abds mutants is related to possible indirect effects caused by an accumulation of arabidiol requires further analysis. In summary, the results indicate that DMNT can inhibit oospore germination and to some extent retard P. irregulare growth at low concentrations under in vitro conditions. Moreover, arabidiol biosynthesis and breakdown contribute to Arabidopsis resistance against P. irregulare infection in vivo.
DISCUSSION
Arabidopsis Roots Produce DMNT via the Breakdown of a Triterpene Precursor
Our results provide evidence for an alternative pathway in the formation of DMNT by oxidative degradation of the triterpene alcohol, arabidiol, in Arabidopsis roots in contrast to the production of DMNT from (E)-nerolidol in aboveground tissues (Boland et al., 1998; Kappers et al., 2005; Lee et al., 2010). The formation of volatile compounds by degradation of nonvolatile terpene precursors is primarily known in the oxidative cleavage of carotenoids to produce norterpenes (irregular terpenes) called apocarotenoids such as ionones (Winterhalter and Rouseff, 2001; Walter et al., 2010). Interestingly, volatile homologs of ionones called irones are assumed to be produced by oxidative degradation of iridal triterpenes in rhizomes of Iris species (Jaenicke and Marner, 1990). Our results present homoterpenes as a group of volatile norterpenes derived from triterpenes and suggest that the role of catabolic reactions in the formation of volatile specialized metabolites may be underestimated.
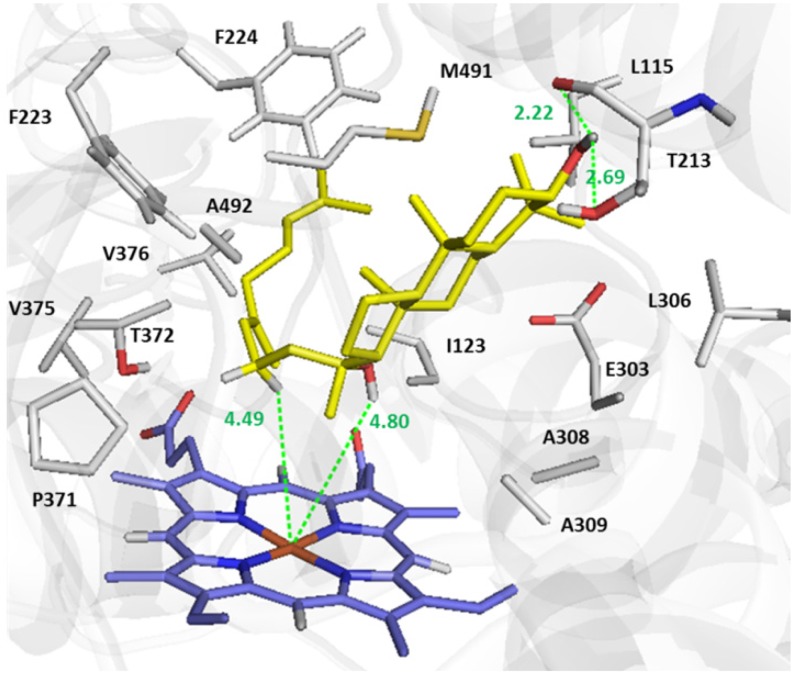
To explore the mechanism of arabidiol cleavage, docking of arabidiol to the active site of a homology-based protein model of CYP705A1 was performed (Figure 8; Supplemental Methods). The C3-OH group of arabidiol makes a H-bond to both the main and side chain of Thr-213. This H-bond and the orientation of the tricyclic moiety in the active site are comparable to the positioning of abiraterone (an sterol based inhibitor) in the active site of CYP17A1, the template used for homology modeling (PDB:3RUK) (DeVore and Scott, 2012). Two Phe residues (Phe-223 and Phe-224) mainly shape the active site and along with Pro-371, Thr-372, and Val-376 are in hydrophobic interaction to the alkyl chain of arabidiol (Figure 8). The predicted coordination of arabidiol in the active site supports a C-C bond cleavage reaction through the radical attack by [Fe(III)-O-O]⋅ and hydrogen abstraction from C16 of the arabidiol molecule followed by an internal atomic rearrangement that leads to the C-C bond breakage (Figure 9). The relative position of C16 indicates that the hydrogen abstraction is probably a syn-elimination reaction since the hydrogen atom on the same side of the hydroxyl group is closer to the Fe atom (Figure 9). Although CYP705A1 shares only 31% amino acid sequence identity with the Arabidopsis CYP82G1, its suggested reaction mechanism is equivalent to the oxidative C-C cleavage of (E)-nerolidol and (E,E)-geranyllinalool catalyzed by CYP82G1 (Lee et al., 2010) and similar reactions in the dealkylation of 22-hydroxycholesterol (Akhtar and Wright, 1991) and the formation of the furanocoumarin psoralen from its precursor (+)-marmesin (Larbat et al., 2007). While it is not yet well understood whether CYP82G1 produces DMNT or TMTT in a single cleavage step or in two sequential steps, our results clearly demonstrate that CYP705A1 synthesizes DMNT by a one-step cleavage reaction with 14-apo-arabidiol as the second product (Figure 5C).
Figure 8.
Docked Conformation of Arabidiol in the Active Site of Modeled CYP705A1.
Docking was performed using AutoDock-Vina. The structure of the arabidiol molecule to be docked into the active site of modeled CYP705A1 was taken form ZINC database (ZINC 59211647). The most energetically favorable orientation of arabidiol with −8.9 Kcal/mol binding affinity is shown. The side chains of residues in 5 Å of substrate are illustrated. The main chain of Thr-213 forms a H-bond to C3-OH of arabidiol. The figure was prepared with PyMol.
Figure 9.
The Proposed Mechanism for Oxidative Degradation of Arabidiol by CYP705A1.
The coordination of arabidiol in the active site based on docking experiments is shown. Radical attack by [Fe(III)-O-O]⋅, hydrogen abstraction, and subsequent internal atomic rearrangement are proposed for the C-C bond breakage of arabidiol to form 14-apo-arabidiol and DMNT.
DMNT Biosynthetic Genes Are Expressed in Specific Cell Types of the Arabidopsis Root and Respond to Jasmonate and Pathogen Treatment
According to their promoter GUS activities, both DMNT biosynthetic genes, ABDS and CYP705A1, are coexpressed primarily in the root vasculature under constitutive conditions. High-resolution root gene expression maps (Birnbaum et al., 2003; Brady et al., 2007) indicate a predominant expression of CYP705A1 in the endodermis and pericycle at the root differentiation/maturation zone, which was confirmed in our study by the tissue-specific localization of the CYP705A1-eYFP fusion protein (Figure 6). In addition, a highly cell type-specific expression of the P450 protein in the quiescent center of the meristematic zone (Figure 6) is in agreement with gene expression profiles by Birnbaum et al. (2003) (see BAR Arabidopsis eFP browser, http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi). In contrast to our results, transcriptome maps by Brady et al. (2007) (see BAR Arabidopsis eFP browser, http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi) also indicate some expression of the CYP705A1 gene in the endodermis/pericycle of the elongation zone, suggesting that the role of additional tissue-specific regulatory elements cannot be entirely excluded. In contrast to the CYP705A1 gene, ABDS appears to be expressed in the entire meristematic zone. It is possible that the specific activity of CYP705A1 in the root stem cells is required to remove the arabidiol precursor from this cell area because of putative inhibitory effects. This idea is supported by a study showing stunted growth and root hair deficiency in oat (Avena sativa) triterpene biosynthetic mutants that accumulate triterpene intermediates in the epidermis (Mylona et al., 2008). Effects of triterpenes on root development have also been demonstrated recently with the role of β-amyrin in determining patterns of epidermal root hair cells in oats (Kemen et al., 2014). Additionally, a pericycle-specific expression at the root differentiation zone could be important both under constitutive and induced conditions as a barrier to avoid vasculature invasion by root pathogens. A similar pericycle-specific expression profile was found for rhizathalene synthase in Arabidopsis roots (Vaughan et al., 2013). The expanded expression in the elongation zone, an area that is preferred by microbial pathogen infection, positively correlates with potential defensive functions of arabidiol-derived compounds. However, it was difficult to observe induced gene expression with promoter-GUS assays in vivo upon P. irregulare infection primarily due to the transient nature of infection and the overall weaker response compared with JA treatment.
Treatment with JA resulted in a 5- and 10-fold transcript induction for CYP705A1 and ABDS, respectively, whereas inoculation with P. irregulare caused only a 2-fold transient increase in expression of CYP705A1 but had no effect on ABDS expression. While we did not measure DMNT formation directly from soil-grown, P. irregulare-infested roots, we assume a similar induction of the DMNT biosynthetic pathway under these conditions because of the previously demonstrated JA-dependent nature of response of soil grown plants to P. irregulare (Vijayan et al., 1998). Although CYP705A1 and ABDS are required for the production of DMNT (Figure 5C), the induced formation of DMNT may not be regulated at gene transcript levels alone. Besides posttranslational modifications, an increase in metabolite flux toward squalene and 2,3-oxidosqualene (Fulton et al., 1994), the common precursor in triterpene biosynthesis, might contribute to the enhanced formation of arabidiol and its breakdown products.
We observed only a transient expression of CYP705A1 and a transient formation of DMNT at the time when oospores produce germ tubes and start to penetrate the epidermis. A similar transient expression was found for marker genes of JA- and salicylic acid-dependent pathways (e.g., PR1 and PDF1.2) within the first 5 h of inoculation of Arabidopsis roots with the oomycete, Phytophthora parasitica (Attard et al., 2010). In accordance with these observations, the transient nature of DMNT production might result from the activity of pathogen effectors that suppress host-specific defense responses within several hours after penetration of the pathogen. Studies on the infection of Arabidopsis seedlings with P. irregulare suggest an infection process similar to that of a hemibiotrophic pathogen such as Phytophthora (Schlink, 2010), with the formation of biotrophic appressoria and haustoria at the beginning of the infection followed by a nectrotrophic movement of hyphae through the vasculature and invasion of all tissues (Adie et al., 2007).
Arabidiol Breakdown Products Are Involved in the Defense against P. irregulare
Our results support a role of DMNT in chemical defense against P. irregulare, showing that DMNT reduces P. irregulare mycelium growth and oospore germination in vitro. Although it is somewhat difficult to compare the actual concentration of DMNT released at the root surface with those in the in vitro assays, the concentrations of DMNT with inhibitory effects were in the range of the amounts emitted per gram fresh weight of axenically grown roots. Due to the highly lipophilic nature of terpenes, it is assumed that terpenes exhibit antimicrobial activity by interfering with cell membrane integrity and function (Mann et al., 2000; Kalemba et al., 2002; Bakkali et al., 2008; Field and Osbourn, 2008). We did not find a clear dose–response effect on oospore germination and P. irregulare growth at higher DMNT concentrations, suggesting that the inhibitory effect could be dependent on a defined concentration range as has been observed with other terpenes (Inoue et al., 2005). Moreover, higher concentrations of DMNT could induce detoxification mechanisms that counteract its inhibitory effects.
Despite the fact that the breakdown of arabidiol seems to occur only within the first hours of infection, our comparative studies on long term disease assessment using oospore counting assays in wild-type and arabidiol and DMNT biosynthetic mutants demonstrate that metabolites of the arabidiol degradation pathway have a partial contribution from the onset of infection to slow down the infection process in roots. The volatile and nonvolatile breakdown products might exhibit different activities in this process. While DMNT appears to be primarily effective at the stage of oospore germination and penetration, it may also have signaling effects as demonstrated in plant-plant interactions (Arimura et al., 2000). Since 14-apo-arabidiol did not have inhibitory activity in vitro, we assume that its derivatives are functionally more important defense compounds. Strong antifungal activities have been described for root-produced triterpene saponins, such as avenacin, which is secreted from oat roots (Crombie and Crombie, 1986). We should note that the abds and cyp705a1 mutants did not show any abnormal growth or developmental phenotypes that could indirectly affect metabolite production and disease resistance.
Since the concentrations of arabidiol and other triterpenes in Arabidopsis are low compared with those in the roots of other plants such as oat, they represent only one component in the chemical defense machinery of Arabidopsis roots. Bednarek et al. (2005) reported that the infection of Arabidopsis roots with P. sylvaticum in axenic culture induced changes in the concentrations of secondary metabolites, including indole glucosinolates and phenylpropanoids. Together, these specialized metabolites combined with other chemical and physical responses (Adie et al., 2007; Oliver et al., 2009) may contribute to the comparatively mild pathogenicity of the P. irregulare strain investigated in this work on mature wild-type Arabidopsis plants.
Evolution of DMNT Biosynthesis in Arabidopsis via Triterpene Gene Cluster Assembly
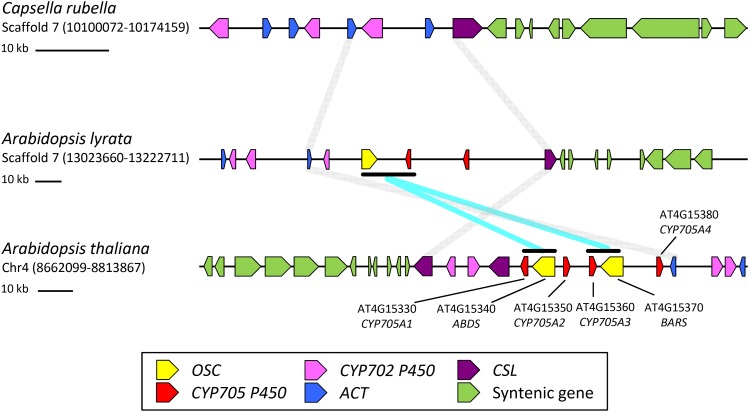
A closer analysis of the chromosomal region of CYP705A1 and ABDS shows that both genes are part of a larger triterpene biosynthetic gene cluster that contains another triterpene synthase gene encoding baruol synthase (BARS1) (Figure 10; Field et al., 2011). BARS1 shares 84% amino acid sequence identity with the ABDS protein. Several studies have demonstrated that genes involved in diterpene and triterpene biosynthesis are coordinated in the form of gene clusters (Qi et al., 2004; Shimura et al., 2007; Field and Osbourn, 2008; Swaminathan et al., 2009; Field et al., 2011; Krokida et al., 2013). In Arabidopsis, two other highly coordinated triterpene biosynthetic gene clusters have been described. The thalianol synthase (THAS1) gene cluster is responsible for the formation and modification of the triterpene thalianol in roots (Field and Osbourn, 2008), while enzymes encoded by the related marneral synthase (MRN1) gene cluster synthesize and modify marneral, a monocyclic triterpene aldehyde (Field et al., 2011). Thalianol has a tricyclic structure similar to that of arabidiol. However, because of the lack of the tertiary hydroxyl group in the prenyl side chain, thalianol is not cleaved by CYP705A1 and instead undergoes hydroxylation of the tricyclic moiety and desaturation at the prenyl side chain (Field and Osbourn, 2008). Chromosomal analysis of the THAS1 and MRN1 clusters revealed that their assembly occurred in dynamic chromosomal regions prior to a whole-genome duplication event in the Brassicales; this scenario also seems to be the case for the ABDS/BARS1 cluster (Field et al., 2011).
Figure 10.
Comparative Genome Analysis Maps of the ABDS Gene Cluster.
Synteny map for ABDS cluster regions between C. rubella, A. lyrata, and A. thaliana is shown. No triterpene (oxidosqualene) cyclase (OSC) gene ortholog was found on a highly syntenic region in C. rubella. One OSC and two CYP705 P450 members were present in A. lyrata. In A. thaliana, two OSC and four CYP705 members were found. A gene duplication event followed by inversion of the CYP705 gene is likely to be the source of DMNT biosynthetic gene evolution. Different gene families are color coded. Syntenic regions flanking the ABDS gene cluster are connected with gray lines. The syntenic regions for the DMNT biosynthetic genes are shown with cyan lines. ACT, acyltransferase; CSL, cellulose synthase-like; syntenic neighboring genes are colored green.
In line with a previous analysis of the THAS and MRN clusters (Field et al., 2011), we compared the chromosomal region of the ABDS/BARS cluster to those in the two close relatives of A. thaliana, Arabidopsis lyrata, and Capsella rubella, to obtain further insight to the evolution of the DMNT biosynthetic pathway (Figure 10). In A. thaliana, ABDS and BARS cluster with four members of the Brassicaceae-specific CYP705 family, CYP705A1, A2, A3, and A4 (Figure 10). Neither a triterpene synthase nor a CYP705 member was found on the ABDS syntenic region in C. rubella, but in A. lyrata, a single triterpene synthase gene with 92% amino acid sequence identity to BARS and two P450s (closely related to CYP705A2 and CYP705A3) are present in this region, indicating that cluster assembly occurred already in the common ancestor of A. thaliana and A. lyrata (Figure 10). With support by a phylogenetic analysis of the CYP705A genes on the syntenic regions (Supplemental Figure 17 and Supplemental Data Set 1), a scenario can be assumed, in which the ABDS/CYP705A1 and BARS/CYP705A3 clusters evolved via a duplication event followed by a relative inversion of the CYP705A genes and gene neofunctionalization. This event may have occurred in A. thaliana after the divergence of A. lyrata and A. thaliana or, alternatively, CYP705A1 and ABDS may have been lost from the syntenic region in the A. lyrata genome (Figure 10).
To study the underlying evolution of arabidiol cleavage activity in CYP705A1, we used a substrate docking and comparative protein homology modeling approach to examine the properties of the CYP705A1 active site and other Arabidopsis CYP705 paralogs (Supplemental Methods). Sequence alignment and homology modeling indicate that only four residues (Leu-115, Phe-223, Pro-371, and Met-491 based on sequence numbering of CYP705A1) out of 18 active-site residues are conserved among CYP705A1-A5, suggesting that enzyme-substrate specificity has diverged extensively after gene duplication events (Supplemental Figure 18). Homology modeling of A2-A5 using the structure of CYP705A1 as the template indicates that several alterations in the active site may interfere with the binding of arabidiol (Supplemental Figure 19). Residue changes in the A2-A5 proteins include the replacement of Thr-213, which forms a hydrogen bond to the C3-OH group of arabidiol, by Val/Ala and a change of Phe-223, which faces toward the active site and plays a major role in shaping the binding cavity in CYP705A1, with smaller side chain residues (Ile, Leu, and His). In addition, the replacement of two tandem alanine residues (Ala-308 and Ala-309) with Gly and Thr reduces the hydrophobicity of the microenvironment and may affect the orientation of C14-OH of arabidiol. Docking of arabidiol into the active site of modeled CYP705A2-A5 proteins did not result in a reactive coordination of arabidiol. Instead, arabidiol binds in a reverse orientation compared with its docked configuration in the active site of A1 (Supplemental Figure 19), thereby making the C14 and C15 atoms inaccessible to oxygen activated heme. In conclusion, the ability of CYP705A1 to cleave arabidiol has evolved presumably by substitution of several amino acids in the active site after gene duplication events in the ABDS gene cluster region.
Our analysis of JA-treated roots of wild-type plants suggested a rapid degradation of arabidiol and further modifications of the 14-apo-arabidiol breakdown product. We assume that other genes of the ABDS gene cluster are involved in these modifications, such as hydroxylation, acylation, and/or glycosylation, although these genes are not as tightly coexpressed with CYP705A1 and ABDS, which is also the case in the thalianol cluster. The forces driving the evolution of gene cluster assembly in terpene metabolism are not fully understood; however, it has been suggested that gene clustering facilitates the regulation of multiple genes at the level of chromatin and prevents the accumulation of possible cytotoxic products (Field and Osbourn, 2008; Wegel et al., 2009; Field et al., 2011). In summary, the formation of DMNT in Arabidopsis roots evolved as part of a triterpene biosynthesis gene cluster indicating plasticity in the biosynthesis of homoterpene volatiles. The reason for the evolution of distinct metabolic routes in TMTT and DMNT biosynthesis in shoots and roots, respectively, remains elusive, but it might indicate tissue-dependent “micro-environments” that facilitate metabolic pathway evolution under particular selective pressures.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana mutants (abds-1, abds-2, cyp705a1-1, and cyp705a1-2) used in this study were from the wild-type Columbia-0 (Col-0) genetic background and were acquired from the ABRC stock center (Alonso et al., 2003). The coi1-1 mutant was kindly provided by John G. Turner (Xie et al., 1998). All plants were grown in short-day (10-h-light/14-h-dark photoperiod) under standard growth conditions (150 μmol m–2 s–1 PAR, 22°C, 55% RH).
Unless stated otherwise, plants were maintained in axenic liquid culture as described by Hétu et al. (2005). Briefly, Arabidopsis seedlings were grown for 7 d on a mesh on MS solid medium containing 1% sucrose. Seedlings were then transferred to MS liquid medium containing 2% sucrose and cultured for 14 d before changing the sucrose concentration to 1% 2 d prior to treatment with Pythium irregulare or JA followed by root harvest for volatile collection. JA treatment was done by applying 100 μM JA (Sigma-Aldrich) directly to root cultures in the liquid medium.
Arabidopsis hairy root cultures were grown on Gamborg’s B5 liquid media with 2% sucrose. Cultures were kept at room temperature under dark conditions and with constant shaking at 75 rpm for two weeks before treatment and analysis. JA treatment was done as described for axenic culture. P450 inhibitor treatments were conducted at 5 and 50 μM concentrations along with JA treatment for 24 h.
Volatile Collection and Analysis
One gram of roots (fresh weight) was detached from plants and placed in screw-capped vials (20 mL) containing 1 mL of distilled water with 10 ng of 1-bromodecane as a standard. Root volatiles were adsorbed in the headspace with a 100 μm polydimethylsiloxane solid phase microextraction (SPME) fiber (Supelco) for 30 min at room temperature followed by incubation at 30°C for 30 min. Volatile compounds were desorbed from the fiber at 240°C (4 min) with a splitless injection and analyzed with a Shimadzu GC/MS-QP2010S. Separation was performed on an Rxi-XLB column (Restek) of 30 m × 0.25 mm i.d. × 0.25 μm film thickness. Helium was the carrier gas (1.4 mL min–1 flow rate), and a temperature gradient was applied at 4°C min–1 from 40°C (2-min hold) to 220°C followed by a gradient of 5°C min–1 from 40 to 220°C and 20°C min–1 from 220 to 240°C (2-min hold). Identification of volatile compounds was achieved by comparison of their retention times and mass spectra with those of authentic standards and with mass spectra of the National Institute of Standards and Technology and Wiley libraries (John Wiley and Sons). DMNT peak areas were normalized against those of the 1-bromodecance standard. Calibration of the SPME-based assay was performed with DMNT and 1-bromodecane in the absence and presence of root tissue. Linear calibration was obtained in both cases between 5 and 50 ng for 1-bromodecane (R2 = 0.99) and 1 to 100 ng for DMNT (R2= 0.99).
Genotyping of Plant Material and QTL Analysis
T-DNA insertion lines of the CYP705A1 gene with an insertion in the second exon (cyp705a1-1, SALK_043195) and the CYP705A1 promoter (cyp705a1-2, SALK_090621) were obtained from the ABRC. Also, two independent T-DNA insertion lines, abds-1 (SALK_018285) and abds-2 (SALK_067736), with insertions in exon 1 and intron 5, respectively, were obtained for the ABDS gene. Homozygous mutants were confirmed by PCR, and the absence of full-length transcript in these mutant lines was verified by RT-PCR.
For QTL analysis of DMNT emission, 162 lines of a Ler × Cvi recombinant inbred line (RIL) population (Alonso-Blanco et al., 1998) plus the Ler and Cvi parental lines were grown in axenic culture, and the presence or absence of DMNT emission from JA-treated root tissue was determined by volatile analysis as described above. To map QTLs for DMNT accumulation, a previously reported genetic map for the Ler × Cvi RIL population was used (Alonso-Blanco et al., 1998). For QTL detection, composite interval mapping was implemented within QTL Cartographer (Wang et al., 2006). To control for genome-wide false positive rates, declaration of statistically significant QTLs was based on permutation-derived empirical thresholds using 1000 permutations (Churchill and Doerge, 1994; Doerge and Churchill, 1996). Composite interval mapping to assign significance based on the underlying trait distribution is robust at handling bimodal trait distributions within metabolite accumulation as we have previously shown (Rebai, 1997; Kliebenstein et al., 2001a, 2001b).
Construction and Analysis of Transgenic Plants
The full-length cDNAs of the CYP705A1 and ABDS genes were prepared by RT-PCR from 1 μg of total RNA using the proofreading enzyme Pfx Turbo Cx hot start (Stratagene) and full-length cloning primers listed in Supplemental Table 4. RNA was extracted using the TRI reagent (Fisher) (Huang et al., 2010) from axenically grown Col-0 roots treated with JA for 24 h. The amplified fragments were cloned into the pENTR/D-TOPO vector (Invitrogen). For construction of CaMV 35S overexpression lines, the ABDS and CYP705A1 cDNAs were subcloned into the pB7WG2 vector (Karimi et al., 2002) using LR recombination (Invitrogen). Plant transformation was done with the Agrobacterium tumefaciens strain GV3101 using the vacuum infiltration method (Bechtold and Pelletier, 1998). Transgenic plants were identified by spraying soil grown plants with 0.01% BASTA solution. For construction of promoter-GUS fusion vectors, 1.5- and 2.6-kb fragments upstream of the start codon for CYP705A1 and ABDS, respectively, were amplified from genomic DNA, cloned into pENTR/D-TOPO, and recombined into pKGWFS7 (Karimi et al., 2002). Transformants were screened on half-strength MS plates with 1% (w/v) sucrose and 75 μg mL−1 kanamycin. Histochemical GUS assays were performed as previously described (Vitha et al., 1993) for at least three independent lines in the T2 generation. GUS staining was observed with an Olympus SZX16 microscope.
To construct the ProCYP705A1:CYP705A1-eYFP lines, the 1.5-kb promoter region upstream of the start codon was subcloned into the pDONR P4-P1R vector (Invitrogen) via a BP reaction using primers P5 and P6 (Invitrogen). Then, the LR reaction was done using pDONR P4-P1R and pENTR/D-TOPO vectors carrying promoter and gene fragments, respectively, with the pB7Y24WG binary destination vector (Tholl Lab) carrying the eYFP coding sequence. pB7Y24WG was constructed by replacing the attR1 and promoter element region in pB7YWG2 with the attR4 region from pK7m24GW using EcoRI and SacI digestion and ligation reactions. Upon introduction of the ProCYP705A1:CYP705A1-eYFP construct into Agrobacterium strain GV3101, cyp705a1-1 plant transformation was done by floral vacuum infiltration (Bechtold and Pelletier, 1998). Transgenic plants were identified by spraying soil grown plants with 0.01% BASTA solution. Root samples from three independent T2 transgenic lines were mounted on a microscope slide with distilled water and visualized using a Zeiss Axiovert 200 inverted fluorescence microscope with FITC (lex = 480 nm; lem = 535 nm), Texas Red (lex = 570 nm; lem = 625 nm) fluorescent filter sets, an attached MRc5 Axiocamcolor digital camera, and an LD Achroplan 40× objective. Propidium iodine staining was done in liquid growth medium by incubation with 10 μg/mL of propidium iodine up to 30 min before confocal microscopy analysis.
Yeast Expression, 14-Apo-Arabidiol Purification, and Enzyme Assays
For establishing yeast coexpression lines, the full-length cDNA of CYP705A1 was amplified using primers P7 and P8, directionally cloned into the multiple cloning site 1 region of the pESC-TRP vector (Stratagene), and expressed under the galactose-inducible promoter GAL10. The mut-CYP705A1 cDNA with a truncation in the heme binding domain was amplified using primers P7 and P9 and subcloned into pESC-TRP as described above. The ABDS full-length cDNA was recombined into the Gateway yeast expression vector YEp352-GW under control of the constitutive ADH1 promoter (Takahashi et al., 2007). Both vector constructs were simultaneously transformed into the yeast line WAT11 (Urban et al., 1997) by following the protocol described by the provider of the pESC-TRP vector (Stratagene). Transgenic yeast strains were grown in yeast selective media (SGI) and protein expression was done as described previously (Takahashi et al., 2007). For DMNT production in yeast expression lines, 4 mL of yeast culture induced with 2% galactose for 16 h was transferred to a screw cap SPME vial and allowed to grow for another 4 h at 28°C at 220 rpm followed by direct headspace volatile analysis using SPME-GC/MS as described above. Protein expression for microsomal purification was done according to Takahashi et al. (2007), and enzyme assays were performed as described by Lee et al. (2010) with 55 μM arabidiol substrate purified as described previously (Xiang et al., 2006).
To produce 14-apo-arabidiol in large quantities, we used transgenic yeast lines expressing ABDS and CYP705A1. Two liters of yeast culture was prepared in YPI medium and induced with 2% of galactose. DMNT levels were monitored every day to ensure continuous degradation of arabidiol and production of 14-apo-arabidiol. After 3 d of galactose induction, 0.5 liters of acetone was added to the yeast culture to burst open cells followed by three times extraction with 1 liter of ethyl acetate. The ethyl acetate extracts were combined and organic solvent was removed to dryness under low pressure using a rotary evaporator (Cole-Parmer). The extract was subjected to flash chromatography over silica gel (Merck grade 9385, pore size 60 Å, 230 to 400 mesh; Sigma-Aldrich). First, a slurry was prepared by adding 50 mL of ethyl acetate and 3 g of silica gel to the round bottom flask followed by rotary evaporation to obtain dried silica particles attached to the extract. Then, the silica gel with the extract was loaded onto a silica gel flash chromatography column preconditioned with 5:2 ethyl acetate:hexane and several fractions were collected. Upon verification of the presence of 14-apo-arabidiol by GC/MS analysis, aliquots of fractions containing 14-apo-arabidiol were individually loaded on preparative thin-layer chromatography plates (20 × 20-cm silica gel pore size 60 Å [250 μm] with 2.5 × 20-cm concentration zone) and developed with a 5:2 mixture of hexane:ethyl acetate. The silica gel was scraped from the plates at an Rf value similar to that of 14-apo-arabidiol and the purity of the extracted compound was evaluated by GC/MS analysis.
Transcript Analysis by RT-PCR and Quantitative RT-PCR
Total RNA was extracted from 100 mg of root tissue using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Two micrograms of total RNA was treated with DNase I (Promega) and subsequently converted into cDNA using SuperScript II (Invitrogen) according to the manufacturer’s instructions. The expression of P450 candidates in root tissues was monitored by RT-PCR according to Huang et al. (2010) using gene-specific primers designed by Prime-BLAST (Ye et al., 2012) listed in Supplemental Table 4. Homozygous mutants were identified using PCR-based genotyping and full-length ABDS and CYP705A1 transcript expression was analyzed by RT-PCR using corresponding gene specific full-length primer pairs (Supplemental Table 4).
For quantitative RT-PCR, 1 μg of total RNA was converted to cDNA by GoScript Reverse Transcriptase (Promega) and treated with DNase I (Promega). Quantitative RT-PCR was performed using cDNA equivalent to 20 ng of total RNA in 20 μL of Power SYBR green Master Mix (Applied Biosystems) in a 7300 Real-Time PCR System (Applied Biosystems) following the manufacturer’s protocol. Concentrations of cDNAs and primers were first optimized according to the instrument user’s manual. Relative gene expression was analyzed for three independent biological replicates each including three technical replicates. Threshold cycle (CT) values for ABDS and CYP705A1 genes in every sample were normalized to that of ubiquitin conjugating enzyme (UBC21) and fold change differences in gene expressions were calculated relative to the control using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Primer sequences are listed in Supplemental Table 4.
Growth and Bioassay Conditions of P. irregulare
P. irregulare 110305 was grown and maintained as described by Huffaker et al. (2006) and kindly provided by Clarence A. Ryan (Washington State University). One-week-old P. irregulare cultures were collected from the plates into sterile water and lightly ground with a mortar and pestle to yield a uniform suspension. Aliquots (300 μL) of the suspension (∼2.475 × 103 propagules), its filtrate, or water (used as a control) were added to the growth medium of axenically grown cultures containing ∼20 plants per flask. For inoculation of axenically grown plants with Escherichia coli, TOP10 cells (Invitrogen) were grown in Luria-Bertani medium at 37°C to an OD600 of 0.5. An aliquot of the bacterial suspension was then added to the plant culture medium to reach an OD600 of 0.01 (∼5 × 106 cells/mL).
Microscopy analysis of Arabidopsis root infection was done using 5-d-old Arabidopsis seedlings grown on six-well plates containing 3 mL of half-strength MS medium with 1% sucrose under short-day conditions infected with P. irregulare as described previously (Adie et al., 2007). To observe P. irregulare on root tissues, lactophenol-trypan blue staining (Koch and Slusarenko, 1990) was performed and followed by sample mounting in 50% glycerol and observed under a Olympus SV-16 stereomicroscope.
Disease assessment with P. irregulare on wild-type and DMNT biosynthetic mutants was performed by measuring oospore abundance 18 d after infection of plants in potting substrate. Plants were grown in jiffy pots (Φ 5 cm, height 6 cm) for 3 weeks under short-day conditions (10 h light/14 h dark). Jar1-1 (jasmonate signaling mutant) was used as a positive control for disease assessment due to its high susceptibility to P. irregulare (Staswick et al., 1998). Randomly selected individual plants were then transplanted along with the jiffy pot into single pots (6 × 6 × 8 cm3) containing P. irregulare-infested potting substrate (Sunshine mix 1). P. irregulare-infested substrate was prepared by slicing a plate of potato dextrose agar (PDA) containing 1-week-old P. irregulare mycelium, mixing it with the substrate (∼4.5 liters with 2 liters of deionized water), and incubating the mixture for 2 d for a uniform infestation. For mock treatments, sliced PDA pieces without P. irregulare were mixed with the substrate. Disease assessment was performed by counting the number of oospores ∼10 mm behind the root tips after staining roots with acid-fuchsin lactophenol (Vijayan et al., 1998).
Oospore Isolation and Germination of P. irregulare
Oospores were prepared following previous studies with minor changes (Yuan and Crawford, 1995; Manici et al., 2000). Briefly, an agar plug from 4-d-old cultures on PDA was inoculated on a V8 juice agar plate (Campbell Juice) and incubated at room temperature under dark conditions. After 10 d, V8 agar plugs containing mycelium were transferred into distilled water and further incubated for 10 d under dark conditions. The cultures containing abundantly produced oospores were comminuted (crushed) in distilled water by a Polytron tissue homogenizer. The homogenized mycelial and oospore mixture was filtered through two layers of cheesecloth, and the filtrate was subjected to centrifugation (4500g for 10 min). The pellet was suspended in distilled water, and the concentration of oospores was determined using a hemacytometer.
To determine the germination rate of oospores according to chemical treatment, oospore germination conditions were applied as described by Ruben and Stanghellini (1978). Oospores were induced to germinate directly on maize meal agar (Difco) containing different concentrations of DMNT with 15 μg mL−1 streptomycin. One hundred microliters of oospore suspension were applied to the surface of an agar plate (∼200 oospores per plate) and incubated for 24 h in an incubator at 27°C under dark conditions. The oospore germination rate was measured by counting oospores with emerging germ tubes using light microscopy.
Phylogenetic Analysis
Multiple sequence alignment was constructed using MUSCLE (Edgar, 2004), and the resulting alignment was used for further analysis using the MEGA version 5 (Tamura et al., 2011). For constructing the phylogenetic tree of selected members of CYP705A, the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992) was used. Initial trees were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 1.5170)). Bootstrap analysis was performed with 1000 replicates. A similar tree was obtained when neighbor-joining inference with 1000 bootstrap replicates was performed using MEGA5.
Statistical Analysis
Statistical data analysis for each experiment has been described along with individual experiments. One-way and two-way ANOVA and Tukey-Kramer HSD test were performed using JMP statistical analysis software (SAS Institute).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: CYP705A1, At4g15330; CYP705A2 (A. thaliana), At4g15350; CYP705A2 (A. lyrata), XM_002870201; CYP705A3 (A. thaliana), At4g15360; CYP705A3 (A. lyrata), XM_002870200; CYP705A4, At4g15380; CYP705A5 (A. thaliana), At5g47990; CYP705A5 (A. lyrata), XM_002863793.1; CYP82G1, At3g25180; ABDS, At4g15340; BARS1, At4g15370; MRN1, At5g42600, THAS1, At5g48010; and UBC21, At5g25760.
Supplemental Data
Supplemental Figure 1. Infection of Arabidopsis Roots with P. irregulare and DMNT Emission.
Supplemental Figure 2. The cyp82g1-1 T-DNA Insertion Mutant Is Not Impaired in JA-Induced Production of DMNT in Roots.
Supplemental Figure 3. Pharmacological Study of DMNT Formation in Arabidopsis Hairy Root Culture.
Supplemental Figure 4. Frequency Distribution of DMNT Emission Levels from JA-Treated Cvi × Ler Recombinant Inbred Lines.
Supplemental Figure 5. RT-PCR Analysis of CYP705A1 Transcript Levels in cyp705a1 and abds Mutants and in CYP705A1 and ABDS Transgenic Lines.
Supplemental Figure 6. qRT-PCR Analysis of CYP705A1 and ABDS Transcript Levels.
Supplemental Figure 7. DMNT Formation in Arabidopsis Roots Is Dependent on the Mevalonate Pathway for Precursor Biosynthesis.
Supplemental Figure 8. RT-PCR Analysis of ABDS Transcript Levels in cyp705a1 and abds Mutants and in CYP705A1 and ABDS Transgenic Lines.
Supplemental Figure 9. HRESIMS Analysis and 1H-NMR Spectrum of 14-Apo-Arabidiol.
Supplemental Figure 10. HSQC NMR Spectrum of 14-Apo-Arabidiol.
Supplemental Figure 11. Full HMBC NMR Spectrum of 14-Apo-Arabidiol.
Supplemental Figure 12. NOESY NMR Spectrum of 14-Apo-Arabidiol.
Supplemental Figure 13. COSY NMR Spectrum of 14-Apo-Arabidiol.
Supplemental Figure 14. Structure of 14-Apo-Arabidiol.
Supplemental Figure 15. ABDS Promoter-GUS Gene Expression Patterns.
Supplemental Figure 16. Effect of Arabidiol, 14-Apo-Arabidiol, and DMNT on the Growth of P. irregulare 110305.
Supplemental Figure 17. Molecular Phylogenetic Analysis of A. thaliana and A. lyrata CYP705 Members of the ABDS and THAS Gene Clusters.
Supplemental Figure 18. Sequence Alignment of Active Site Residues of CYP705A1 to CYP705A5.
Supplemental Figure 19. Docked Conformations of Arabidiol into the Active Site of CYP705A2-A5.
Supplemental Table 1. DMNT Emission Levels in Cvi × Ler Recombinant Inbred Line Population and Parental Lines.
Supplemental Table 2. Candidate Genes Coexpressed with CYP705A1 Evaluated Based on the ATTED-II Database.
Supplemental Table 3. Proton and Carbon Chemical Shifts for 14-Apo-Arabidiol and Key Correlations from NMR Spectra.
Supplemental Table 4. Sequences of Primers Used in Different Experiments.
Supplemental Data Set 1. Alignment of Protein Sequences Used for Construction of Phylogenetic Tree Shown in Supplemental Figure 17.
Supplementary Material
Acknowledgments
We thank Wilhelm Boland (Max Planck Institute for Chemical Ecology, Jena, Germany) for providing a DMNT standard, Joe Chappell (University of Kentucky, Lexington, KY) for sharing the modified YEp352 (ADH1) vector, and Daniele Werck-Reichhart (Centre National de la Recherche Scientifique, Strasbourg, France) for providing the WAT11 yeast strain. We thank John Jelesko (Department of Plant Pathology, Physiology, Weed Science, Virginia Tech) for providing the Arabidopsis hairy root culture. This work was supported by National Science Foundation Grant MCB-0950865 and by National Research Initiative Competitive Grant 2007-35318-18384 from the USDA National Institute of Food and Agriculture (to D.T.).
AUTHOR CONTRIBUTIONS
R.S. designed the research, performed research, analyzed data, and cowrote the article. J.-H.H. designed the research, performed research, analyzed data, and cowrote the article. S.B. designed the research, performed research, analyzed data, and cowrote the article. L.H.R. performed NMR analysis, analyzed data, and cowrote the article. D.K. performed research and analyzed data. P.S. supported research and data analysis by S.B. D.T. designed the research, analyzed data, and cowrote the article.
Glossary
- DMNT
(E)-4,8-dimethyl-1,3,7-nonatriene
- TMTT
(E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene
- JA
jasmonic acid
- SPME-GC/MS
solid phase microextraction gas chromatography-mass spectrometry
- GL
(E,E)-geranyllinalool
- QTL
quantitative trait locus
- Cvi
Cape Verdi Island
- Ler
Landsberg erecta
- GC/MS
Gas chromatography-mass spectrometry
- MS
Murashige and Skoog
- Col-0
Columbia-0
- SPME
solid phase microextraction
- RIL
recombinant inbred line
- PDA
potato dextrose agar
References
- Adie B.A., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M., Wright J.N. (1991). A unified mechanistic view of oxidative reactions catalysed by P-450 and related Fe-containing enzymes. Nat. Prod. Rep. 8: 527–551. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Peeters A.J., Koornneef M., Lister C., Dean C., van den Bosch N., Pot J., Kuiper M.T. (1998). Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271. [DOI] [PubMed] [Google Scholar]
- Arimura G., Ozawa R., Shimoda T., Nishioka T., Boland W., Takabayashi J. (2000). Herbivory-induced volatiles elicit defence genes in lima bean leaves. Nature 406: 512–515. [DOI] [PubMed] [Google Scholar]
- Attard A., Gourgues M., Callemeyn-Torre N., Keller H (2010). The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol. 187: 449–460. [DOI] [PubMed] [Google Scholar]
- Bakkali F., Averbeck S., Averbeck D., Idaomar M. (2008). Biological effects of essential oils—a review. Food Chem. Toxicol. 46: 446–475. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266. [DOI] [PubMed] [Google Scholar]
- Bednarek P., Schneider B., Svatos A., Oldham N.J., Hahlbrock K. (2005). Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 138: 1058–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- Boland W., Gabler A., Gilbert M., Feng Z.F. (1998). Biosynthesis of C-11 and C-16 homoterpenes in higher plants; stereochemistry of the C–C-bond cleavage reaction. Tetrahedron 54: 14725–14736. [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Castillo D.A., Kolesnikova M.D., Matsuda S.P. (2013). An effective strategy for exploring unknown metabolic pathways by genome mining. J. Am. Chem. Soc. 135: 5885–5894. [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., Bohlmann J., Pichersky E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66: 212–229. [DOI] [PubMed] [Google Scholar]
- Churchill G.A., Doerge R.W. (1994). Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo McCormick A., Unsicker S.B., Gershenzon J. (2012). The specificity of herbivore-induced plant volatiles in attracting herbivore enemies. Trends Plant Sci. 17: 303–310. [DOI] [PubMed] [Google Scholar]
- Crombie W.M.L., Crombie L (1986). Distribution of avenacins A-1, A-2, B-1 and B-2 in oat roots: Their fungicidal activity towards ‘take-all' fungus. Phytochemistry 25: 2069–2073. [Google Scholar]
- de Boer J.G., Posthumus M.A., Dicke M. (2004). Identification of volatiles that are used in discrimination between plants infested with prey or nonprey herbivores by a predatory mite. J. Chem. Ecol. 30: 2215–2230. [DOI] [PubMed] [Google Scholar]
- DeVore N.M., Scott E.E. (2012). Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature 482: 116–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicke M., Sabelis M.W., Takabayashi J., Bruin J., Posthumus M.A. (1990). Plant strategies of manipulating predator-prey interactions through allelochemicals: Prospects for application in pest control. J. Chem. Ecol. 16: 3091–3118. [DOI] [PubMed] [Google Scholar]
- Doerge R.W., Churchill G.A. (1996). Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donath J., Boland W. (1995). Biosynthesis of acyclic homoterpenes: enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39: 785–790. [Google Scholar]
- Dudareva N., Negre F., Nagegowda D.A., Orlova I. (2006). Plant volatiles: Recent advances and future perspectives. Crit. Rev. Plant Sci. 25: 417–440. [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science 320: 543–547. [DOI] [PubMed] [Google Scholar]
- Field B., Fiston-Lavier A.S., Kemen A., Geisler K., Quesneville H., Osbourn A.E. (2011). Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. USA 108: 16116–16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton D.C., Kroon P.A., Threlfall D.R. (1994). Enzymological aspects of the redirection of terpenoid biosynthesis in elicitor-treated cultures of Tabernaemontana divaricata. Phytochemistry 35: 1183–1186. [Google Scholar]
- Herde M., Gärtner K., Köllner T.G., Fode B., Boland W., Gershenzon J., Gatz C., Tholl D. (2008). Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C16-homoterpene TMTT. Plant Cell 20: 1152–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hétu M.F., Tremblay L.J., Lefebvre D.D. (2005). High root biomass production in anchored Arabidopsis plants grown in axenic sucrose supplemented liquid culture. Biotechniques 39: 345–349. [DOI] [PubMed] [Google Scholar]
- Huang M., Abel C., Sohrabi R., Petri J., Haupt I., Cosimano J., Gershenzon J., Tholl D. (2010). Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS03. Plant Physiol. 153: 1293–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Hada T., Shiraishi A., Hirose K., Hamashima H., Kobayashi S. (2005). Biphasic effects of geranylgeraniol, teprenone, and phytol on the growth of Staphylococcus aureus. Antimicrob. Agents Chemother. 49: 1770–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke L., Marner F.J (1990). The irones and their origin. Pure Appl. Chem. 62: 1365–1368. [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kaiser R. (1991). Perfumes: Art, Science and Technology. (London: Elsevier Applied Science; ). [Google Scholar]
- Kalemba D., Kusewicz D., Swiader K. (2002). Antimicrobial properties of the essential oil of Artemisia asiatica Nakai. Phytother. Res. 16: 288–291. [DOI] [PubMed] [Google Scholar]
- Kappers I.F., Aharoni A., van Herpen T.W., Luckerhoff L.L., Dicke M., Bouwmeester H.J. (2005). Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science 309: 2070–2072. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kemen A.C., Honkanen S., Melton R.E., Findlay K.C., Mugford S.T., Hayashi K., Haralampidis K., Rosser S.J., Osbourn A. (2014). Investigation of triterpene synthesis and regulation in oats reveals a role for β-amyrin in determining root epidermal cell patterning. Proc. Natl. Acad. Sci. USA 111: 8679–8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Gershenzon J., Mitchell-Olds T. (2001a). Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159: 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Lambrix V.M., Reichelt M., Gershenzon J., Mitchell-Olds T. (2001b). Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokida A., Delis C., Geisler K., Garagounis C., Tsikou D., Peña-Rodríguez L.M., Katsarou D., Field B., Osbourn A.E., Papadopoulou K.K. (2013). A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis. New Phytol. 200: 675–690. [DOI] [PubMed] [Google Scholar]
- Larbat R., Kellner S., Specker S., Hehn A., Gontier E., Hans J., Bourgaud F., Matern U. (2007). Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of furanocoumarin biosynthesis. J. Biol. Chem. 282: 542–554. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M., de Wit P.J., Govers F. (2003). Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 11: 462–469. [DOI] [PubMed] [Google Scholar]
- Lee S., Badieyan S., Bevan D.R., Herde M., Gatz C., Tholl D. (2010). Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 21205–21210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Manici L.M., Lazzeri L., Baruzzi G., Leoni O., Galletti S., Palmieri S. (2000). Suppressive activity of some glucosinolate enzyme degradation products on Pythium irregulare and Rhizoctonia solani in sterile soil. Pest Manag. Sci. 56: 921–926. [Google Scholar]
- Mann C.M., Cox S.D., Markham J.L. (2000). The outer membrane of Pseudomonas aeruginosa NCTC 6749 contributes to its tolerance to the essential oil of Melaleuca alternifolia (tea tree oil). Lett. Appl. Microbiol. 30: 294–297. [DOI] [PubMed] [Google Scholar]
- Mumm R., Dicke M. (2010). Variation in natural plant products and the attraction of bodyguards involved in indirect plant defense. Can. J. Zool. 88: 628–667. [Google Scholar]
- Mylona P., Owatworakit A., Papadopoulou K., Jenner H., Qin B., Findlay K., Hill L., Qi X., Bakht S., Melton R., Osbourn A. (2008). Sad3 and sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.P., Castro A., Gaggero C., Cascón T., Schmelz E.A., Castresana C., Ponce de León I. (2009). Pythium infection activates conserved plant defense responses in mosses. Planta 230: 569–579. [DOI] [PubMed] [Google Scholar]
- Pichersky E., Noel J.P., Dudareva N. (2006). Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004). A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101: 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebai A. (1997). Comparison of methods for regression interval mapping in QTL analysis with non-normal traits. Genet. Res. 69: 69–74. [Google Scholar]
- Ruben D.M., Stanghellini M.E. (1978). Ultrastructure of oospore germination in Pythium aphanidermatum. Am. J. Bot. 65: 491–501. [Google Scholar]
- Schlink K. (2010). Down-regulation of defense genes and resource allocation into infected roots as factors for compatibility between Fagus sylvatica and Phytophthora citricola. Funct. Integr. Genomics 10: 253–264. [DOI] [PubMed] [Google Scholar]
- Shimura K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282: 34013–34018. [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Yuen G.Y., Lehman C.C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15: 747–754. [DOI] [PubMed] [Google Scholar]
- St-Pierre B., De Luca V. (1995). A cytochrome P-450 monooxygenase catalyzes the first step in the conversion of tabersonine to vindoline in Catharanthus roseus. Plant Physiol. 109: 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S., Morrone D., Wang Q., Fulton D.B., Peters R.J. (2009). CYP76M7 is an ent-cassadiene C11alpha-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Yeo Y., Greenhagen B.T., McMullin T., Song L., Maurina-Brunker J., Rosson R., Noel J.P., Chappell J. (2007). Metabolic engineering of sesquiterpene metabolism in yeast. Biotechnol. Bioeng. 97: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D. Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D., Sohrabi R., Huh J.H., Lee S. (2011). The biochemistry of homoterpenes—common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry 72: 1635–1646. [DOI] [PubMed] [Google Scholar]
- Unsicker S.B., Kunert G., Gershenzon J. (2009). Protective perfumes: the role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 12: 479–485. [DOI] [PubMed] [Google Scholar]
- Urban P., Mignotte C., Kazmaier M., Delorme F., Pompon D. (1997). Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272: 19176–19186. [DOI] [PubMed] [Google Scholar]
- Vaughan M.M., Wang Q., Webster F.X., Kiemle D., Hong Y.J., Tantillo D.J., Coates R.M., Wray A.T., Askew W., O’Donnell C., Tokuhisa J.G., Tholl D. (2013). Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25: 1108–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P., Shockey J., Lévesque C.A., Cook R.J., Browse J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Benes K., Michalova M., Ondrej M. (1993). Quantitative beta-glucuronidase assay in transgenic plants. Biol. Plant. 35: 151–155. [Google Scholar]
- Walter M.H., Floss D.S., Strack D. (2010). Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232: 1–17. [DOI] [PubMed] [Google Scholar]
- Wang S., Basten C.J., Zeng Z.-B. (2006). Windows QTL Cartographer 2.5. (Raleigh, NC: Department of Statistics, North Carolina State University; ). [Google Scholar]
- Wegel E., Koumproglou R., Shaw P., Osbourn A. (2009). Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21: 3926–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win J., Chaparro-Garcia A., Belhaj K., Saunders D.G., Yoshida K., Dong S., Schornack S., Zipfel C., Robatzek S., Hogenhout S.A., Kamoun S. (2012). Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77: 235–247. [DOI] [PubMed] [Google Scholar]
- Winterhalter P. and Rouseff R. (2001). Carotenoid-derived aroma compounds: An introduction. In Carotenoid-Derived Aroma Compounds, P. Winterhalter and R.L. Rouseff, eds (Washington, DC:American Chemical Society; ), pp. 1–17. [Google Scholar]
- Xiang T., Shibuya M., Katsube Y., Tsutsumi T., Otsuka M., Zhang H., Masuda K., Ebizuka Y. (2006). A new triterpene synthase from Arabidopsis thaliana produces a tricyclic triterpene with two hydroxyl groups. Org. Lett. 8: 2835–2838. [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W.M., Crawford D.L. (1995). Characterization of Streptomyces lydicus WYEC108 as a potential biocontrol agent against fungal root and seed rots. Appl. Environ. Microbiol. 61: 3119–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.