Metabolite QTL for secondary metabolism in a Solanum pennelli introgression line population show different modes of inheritance and network properties linking the traits.
Abstract
A large-scale metabolic quantitative trait loci (mQTL) analysis was performed on the well-characterized Solanum pennellii introgression lines to investigate the genomic regions associated with secondary metabolism in tomato fruit pericarp. In total, 679 mQTLs were detected across the 76 introgression lines. Heritability analyses revealed that mQTLs of secondary metabolism were less affected by environment than mQTLs of primary metabolism. Network analysis allowed us to assess the interconnectivity of primary and secondary metabolism as well as to compare and contrast their respective associations with morphological traits. Additionally, we applied a recently established real-time quantitative PCR platform to gain insight into transcriptional control mechanisms of a subset of the mQTLs, including those for hydroxycinnamates, acyl-sugar, naringenin chalcone, and a range of glycoalkaloids. Intriguingly, many of these compounds displayed a dominant-negative mode of inheritance, which is contrary to the conventional wisdom that secondary metabolite contents decreased on domestication. We additionally performed an exemplary evaluation of two candidate genes for glycolalkaloid mQTLs via the use of virus-induced gene silencing. The combined data of this study were compared with previous results on primary metabolism obtained from the same material and to other studies of natural variance of secondary metabolism.
INTRODUCTION
Over the last 20 or so years, the adoption of quantitative trait locus (QTL) analysis of natural variation in segregating populations has become an increasingly popular approach (Jansen, 1993; Frary et al., 2000; Koornneef et al., 2004; Ashikari et al., 2005; Xue et al., 2008; Bagheri et al., 2012). While the majority of early studies focused on easy-to-measure morphological traits or simple chemical compositional analyses (reviewed in Koornneef et al., 2004; Fernie et al., 2006), more recently the arsenal of postgenomic tools has been brought to bear on such segregating populations. For instance, genome-wide evaluation in levels of gene expression and metabolite levels have been recently performed (Keurentjes et al., 2006; Fu et al., 2009; Lisec et al., 2009). While the majority of these studies have been performed in segregating populations of Arabidopsis thaliana, a number of studies at the metabolite level have also been performed in the crop species: tomato (Solanum lycopersicum), potato (Solanum tuberosum), maize (Zea mays), and rice (Oryza sativa) (Schauer et al., 2008; Carreno-Quintero et al., 2012; Matsuda et al., 2012; Hu et al., 2014; Wen et al., 2014).
In tomato, the majority of both natural variance and metabolite quantitative trait loci (mQTL) studies have focused on primary metabolism (Schauer et al., 2005, 2006, 2008; Stevens et al., 2007; Do et al., 2010; Maloney et al., 2010; Quadrana et al., 2013, 2014). They have revealed the critical importance of cell wall invertase and fruit yield (Fridman et al., 2004; Ruan et al., 2012) and have identified the genomic regions underlying vitamin content in fruit (Stevens et al., 2007; Fitzpatrick et al., 2013; Quadrana et al., 2013, 2014). In addition, these studies highlighted a strong negative correlation between fruit amino acid content and the harvest index (i.e., the ratio of total fruit weight to total plant weight; Schauer et al., 2006; Do et al., 2010). Targeted QTL analyses have also been performed on volatile organic compounds, pigments, cell wall components, sesquiterpenes, and acyl-sugars in the Solanum pennellii introgression line population (Liu et al., 2003; Tieman et al., 2006; Fraser et al., 2007; Schilmiller et al., 2010; de Godoy et al., 2013). Screens of natural variance have additionally focused on a similar range of compounds (Sallaud et al., 2009; Gonzales-Vigil et al., 2012; Schilmiller et al., 2012; Matsuba et al., 2013; Tikunov et al., 2013), but also of cuticle composition (Yeats et al., 2012). In some cases, the observed considerable variation in the contents of these chemical constituents has been related to the growth habit to which the wild species of tomato have adapted (Schauer et al., 2005; Yeats et al., 2012; Ichihashi and Sinha, 2014). Furthermore, these studies were able to elucidate the biosynthetic pathways of the volatiles phenylethanol and phenylacetaldehyde (Tieman et al., 2006), as well as specific glycoalkaloids (Itkin et al., 2011; Iijima et al., 2013; Itkin et al., 2013). Such research thus contributes considerably to the enhancement of our understanding of fruit specialized metabolism (Tohge et al., 2014).
In this study, a broader scale analysis of secondary fruit metabolite levels was performed in two independent harvests, including an analysis of lines heterozygous for the introgression of chromosomal segments from the S. pennellii genome in the second harvest. In doing so, it was possible to evaluate the heritability of the mQTLs of secondary metabolism. Furthermore, we were able to determine their mode of inheritance, a highly important characteristic to study from a breeding perspective, but one that has been overlooked in all but a handful of metabolic studies (Dhaubhadel et al., 2003; O’Reilly-Wapstra et al., 2005; Schauer et al., 2008). Given that these results indicated that only a few of introgression lines (ILs) harbored secondary metabolite QTL hot spots, we further evaluated two additional features. We first checked whether known gene clusters involved in the biosynthesis (or degradation) of the metabolites in question, for instance, the recently described glycoalkaloid cluster (Field and Osbourn, 2008; Itkin et al., 2013), colocalized to these hot spots. Second, we evaluated the expression level of transcription factors in a subset of the lines in which the most QTLs were found, since secondary metabolism is well documented as being under strict transcriptional control and the influence of a number of important transcription factors has already been well characterized (Butelli et al., 2008; Luo et al., 2008; Adato et al., 2009; Dubos et al., 2010; Chan et al., 2011; Shi et al., 2013). Finally, we compared the combined data, with those earlier obtained for morphological and primary metabolite traits, in an attempt to better understand network interactions between the various parts of fruit metabolism and fruit and plant growth. Performance of various cellular tasks ensuring viability of a given genotype is usually achieved by the capacity to change the phenotype with respect to a given trait while maintaining the phenotype of other traits under different environments, referred to as plasticity and robustness, respectively. These phenomena have already been studied to a certain degree for different molecular traits on various levels of cellular organization, from gene expression to protein abundances and metabolite levels. Robustness of phenotypic traits coincides with the concept of canalization: the ability to maintain a standard phenotype despite environmental and genetic perturbations, coined by Waddington (1942), and is an established theme in evolutionary genetics. However, the robustness and plasticity of the relationships between molecular traits, as manifested in the correlation patterns, remain less well understood. To this end, we distinguish the following: environmental robustness to changing conditions and genetic robustness to heritable mutations (Flatt, 2005). Analogously, one can define the notions of environmental and genetic plasticity. Phenotyping of interspecific introgression lines grown in different conditions provides a valuable resource to dissect the concept of environmental robustness and plasticity not only of individual traits, but also their relationships. In addition, the availability of homozygous and heterozygous lines allows them to be used to obtain insights in genetic robustness and plasticity underlying a phenotypic trait. Results are discussed both in the context of the regulation of secondary metabolism in tomato fruits and with respect to attempts at nutritional fortification and antinutrient reduction in crop breeding.
RESULTS
Identification of Conserved QTL for Secondary Metabolites in the S. pennellii IL Population
We previously reported 889 single-trait QTLs for metabolite accumulation following a gas chromatography-mass spectrometry (GC-MS)-based survey of a tomato IL population in which marker-defined regions of the wild species S. pennellii were replaced with homologous intervals of the cultivated variety S. lycopersicum M82 (Eshed and Zamir, 1995). This study was based on the evaluation of fruit pericarp material harvested from two independent field trials (2001 and 2004). As an initial approach to assess the variability in secondary metabolite, we inventoried which metabolites were present in mature pericarp extracts from S. pennellii or S. lycopersicum M82 by ultraperformance liquid chromatography-Fourier transform mass spectrometry (UPLC-FTMS) using the same protocol as described by Tohge et al. (2011). With minor optimizations for the tissue type, we were able to annotate the chemical structure of 43 metabolites, including nine flavanols, 23 hydroxycinnamate derivatives, six glycoalkaloids, two acyl-sugars, one amino acid, one polyamine, and one unclassified nitrogen containing compound. In addition, eight flavonols, 21 hydroxycinnamate derivatives, 16 glycoalkaloids, and three acyl-sugars were putatively identified on the basis of their mass spectral properties and literature information (for example, see Schwahn et al., 2014; Supplemental Data Set 1).
Having optimized the protocol for tomato pericarp, we next used it to evaluate extracts from the introgression lines from the exact same samples that we profiled previously for primary metabolite abundances (Schauer et al., 2006, 2008). Figure 1 provides an overlay heat map from samples harvested in the 2001 and 2004 seasons superimposed on one another in an additive manner such that consistently large increases, with respect to the S. lycopersicum content, are displayed in a deep red color, while those that display consistently large decreases in a deep blue color and those that increase in one year and decrease in the other are displayed in a purple color (a fully annotated version of the figure is available as Supplemental Figures 1 to 3 and Supplemental Data Set 2). In instances where the changes were smaller, a paler coloration was used. Interestingly, the range in content was far larger than that seen between the parental lines (Koenig et al., 2013; T. Tohge and A.R. Fernie, unpublished data) and additionally is beyond that recorded for primary metabolites in this population (with the secondary metabolites ranging from 0 [absent] to 95-fold that of the M82, while the primary metabolites only ranged between 0.18- and 17.7-fold of the M82 value; Schauer et al., 2006). This is additionally reflected in the distribution of log-fold changes, as seen in Figure 2, which shows that the median of log-fold changes of secondary metabolites is consistently lower than that of primary metabolites both in the 2001 and 2004 field trials (Figures 2A and 2B) and when the data are combined (Figure 2C).
Figure 1.
Overlay Heat Map of the Metabolite Profiles of Two Independent Studies of the Pericarp Metabolite Content of the ILs Compared with the Parental Control (M82).
Data represent measurements of material harvested in field trials performed in 2001 and 2004 and are presented as a heat map. Large sections of the map are white or pale in color, reflecting that many of the chromosomal segment substitutions do not have an effect on the amount of every metabolite. Regions of red or blue indicate that the metabolite content increased or decreased, respectively, after introgression of S. pennellii segments. Very dark coloring indicates that a large change in metabolite content was conserved across both harvests, whereas purple indicates an inconsistent change in that IL relative to M82. For each harvest, UPLC-FTMS was used to quantify 145 metabolites, including flavanols, hydroxycinnamate derivatives, glycoalkaloids, and acyl-sugars. Due to space constraints, this heat map is not annotated; however, fully annotated heat maps for the individual data sets are provided in Supplemental Figures 1 to 3. The introgression lines are presented in chromosomal order from top of chromosome 1 to base of chromosome 12 from left to right.
Figure 2.
Distributions of Log-Fold Changes.
Box plots of the log-fold changes of all primary and secondary metabolites, measured with GC-MS and LC-MS technologies, respectively, from the considered ILs with respect to M82 are shown. Note that the median of the log-fold changes of secondary metabolites is smaller than that of the primary metabolites.
(A) Data from 2001 field trail.
(B) Data from 2004 field trail.
(C) Data from both field trails combined.
As can be seen in the plot, a large number of the observed changes in abundance were conserved, with 15% being dark blue and 27% dark red, although some 11% were purple (suggesting that there was no strong genetic control underlying their changes in abundance). Indeed, carrying out a correlation analysis for all metabolite pairs across the whole spectrum of combinations revealed that 23% of these showed significant correlations (P ≤ 0.05) between the two harvests, further demonstrating the strong genetic influence on metabolite abundance. These features are discussed further below in the section on heritability of secondary metabolites.
QTLs were next determined using ANOVA tests. These were performed at two levels of significance hereafter referred to as permissive (P ≤ 0.05) and stringent (P ≤ 0.01) (the results are available as Supplemental Data Sets 3 and 4). Although the vast majority of QTLs reported here were previously unknown, it is important to note that QTLs for total phenolic content as well as total antioxidant content have been reported previously for tomato (Rousseaux et al., 2005; Di Matteo et al., 2013) as well as QTLs for specific compounds of the hydroxycinnamates, flavonol, and glycoalkaloid families (Itkin et al., 2013; Kim et al., 2014; Perez-Fons et al., 2014). We found a total of 679 QTL at the permissive and 340 QTL at the stringent threshold corresponding to between 0 permissive (0 stringent) and 26 permissive (19 stringent) per (putative) compound. When broken down into compound class, this corresponded to 147 permissive (67 stringent) hydroxycinnamates, 75 permissive (30 stringent) flavonols, 151 permissive (84 stringent) glycoalkaloids, 13 permissive (5 stringent) acyl-sugars, 111 permissive (67 stringent) N-containing compounds, 80 permissive (38 stringent) phenolics, and 102 permissive (49 stringent) unclassified compounds.
Analysis of the stable mQTLs of secondary metabolism from the perspective of their genomic location revealed that while they were generally well spread across the genome with all chromosomes, there were a few hot spots particularly notable being the loci on chromosomes 6 (IL6-2 and IL6-3), 8 (IL8-2 and IL8-2-1), and 10 (IL10-2 and IL10-3). These can be best seen in Figure 3 where the number of QTLs per IL is presented in a compound class dependent manner with the size of the circle next to the genome segment being proportional to the number of QTLs for each compound class and the circles being presented from left to right in the order: flavonoids, hydroxycinnamates, glycoalkaloids, N-containing, acyl-sugars, and others. In Figure 4, we zoomed in on a few select QTLs for nutritionally important metabolites, showing their levels in overlapping ILs against the M82 control.
Figure 3.
Metabolic Hot Spots.
Chromosome mapping of (P ≤ 0.01) QTLs location based on a genetic map of S. pennellii introgression lines (http://www.sgn.cornell.edu); circles next to the genome segment indicate the positions and are proportional to the number of QTLs for each compound class. The circle color is presented in the order of flavonoids, hydroxycinnamates, glycoalkaloids, N-containing, other phenolic, and acyl-sugars from left to right.
Figure 4.
Schematic Representation of S. pennellii ILs for Four Chromosomes, Showing the Effect of Genomic Regions on the Levels of Secondary Metabolites.
(A) Region on chromosome 1 (IL1-1) for dehydroesculeoside A or B.
(B) Region on chromosome 6 (IL6-2 and IL6-2-2 for lycoperoside F [B-1] and IL6-3 and IL6-4 for pregnane derivative [B-2]).
(C) Region on chromosome 7 (IL7-4 and IL7-4-1) for glycoalkaloids derivatives (1363.528 m/z).
(D) Region on chromosome 8 (IL8-2 and IL8-2-1) for acylated hexoses.
(E) Region on chromosome 10 (IL10-2 and IL10-3) for lycoperoside G and F or esculeoside A (IL10-3) for coumaric acid hexose.
(F) Region on chromosome 12 (IL12-4 and IL12-4-1) for chlorogenic acid isomers.
Data are shown as a fold changes compared with recurrent parent M82. All the QTLs are conserved in both harvests; all significance at P ≤ 0.01.
Heritability of Secondary Metabolites
When the combined metabolite data sets were evaluated, we noted that there was a trend toward negative QTLs, although this was somewhat dependent on the compound class with glycoalkaloids exhibiting twice as many positive as negative QTLs. This is in sharp contrast to what we previously observed for primary metabolites in the pericarp for the population, which were dominated by positive QTLs (Schauer et al., 2006, 2008). We next assessed the heritability of the various metabolic traits by statistical analysis. These analyses allowed us to calculate the broad sense heritability (H2); values for selected identified metabolites, as well as their E and G × E values, are presented in Figure 5, while those for all spectral features are given in Supplemental Data Set 5. In both instances, we classified the heritability as high, intermediate, or low using thresholds of >0.4, between 0.2 and 0.4, and below 0.2, respectively. Reassuringly, the only metabolite that we determined by GC-MS in our previous study (Schauer et al., 2008), namely, tryptophan, was determined to have a low heritability and thus to be highly reproducible in both data sets. Of the other metabolites, the two acyl-sugars display a high heritability, the 10 known flavonoids tended to display high heritability; however, two of them displayed intermediate and one, low heritability. Among the 23 hydroxycinnamates, 11 displayed high heritability, four intermediate, and eight low, while the N-containing panthothenic acid-hexose and the polyamine feroyltyramine both displayed intermediate heritability.
Figure 5.
Heritability of Secondary Metabolite Traits in the S. pennellii Introgression Population.
Environment (E) + E × genotype (G) effect of selected secondary metabolites using a mixed-effect model to combine the data from the two years (2004 and 2001).
Next, we evaluated these trends from the perspective of the metabolic network (Figure 6). When doing so, several trends emerge, the nontoxic glycoalkaloids (esculeosides) tended to have high heritability (e.g., dihydro-esculeoside A, esculeoside A, and lycoperoside G/F (Figure 6A), and naringenin, chlorogenic acid I, coumaric acid-hexose I, and caffeic acid-hexose 2 displayed high heritability (Figure 6B), whereas dihydroxybenzoic acid displays a low heritability similar in range to those of earlier precursors in the phenylalanine and tryptophan pathways. However, the patterns of heritability are by no means as closely related to metabolic pathway position as they were for the mQTLs of primary metabolism (Schauer et al., 2008). Given the nutritional properties of several of these compounds, such as naringenin, caffeoylquinic acids, and caffeoyl derivatives, and the antinutritional properties of some of the glycoalkaloids, the evaluation of their heritability is also of interest. In general, the heritability values of the known compounds we documented here are considerably higher than those of the primary metabolites determined previously, suggesting that metabolic engineering of the levels of these metabolites via breeding will be feasible. When evaluating the heritability of all determined features, a similar pattern emerged, with three of the putative flavonoids displaying low heritability, one high, and six very low heritability; similarly, eight putative glycoalkaloids showed high heritability, and seven low heritability, while three hydroxycinnamates showed high to medium heritability and five low heritability (Supplemental Data Set 5). We initially hoped that these analyses might aid in the better identification of the chemical nature of these metabolites; however, given that the heritability was not closely associated to position of the metabolites within the metabolic pathways of their synthesis, this was not the case. Given these findings, we also determined the extent to which heritability is reflected in the metabolic correlation networks extracted from the data (see below).
Figure 6.
Metabolites That Display High, Moderate, and Low Hereditability as Assessed from 2 Years of Growth Trials.
Data are taken from Figure 5 and displayed in a pathway-based manner: glycoalkaloids (A) and flavonoids (B). Metabolites marked in red were determined to be highly hereditable, those in yellow to display low hereditability, and those in orange to be intermediate. Heritability for metabolites marked in pale gray was not calculated. Values for shikinate and phenylalanine were taken from Schauer et al. (2008). Heritability is classified as high, intermediate, or low using thresholds of >0.4, between 0.2 and 0.4, and below 0.2, respectively.
Analysis of Secondary Metabolite Contents in a Population Heterozygous for the S. pennellii Introgression
The above-described experiments indicated the important genetic influence underlying many of the mQTLs of secondary metabolism. To extend our understanding of this, we next analyzed the metabolite content of the fruit pericarp in hybrids between ILs and M82, which were grown alongside the ILs in the 2004 field trial as described previously (Semel et al., 2006; Schauer et al., 2008). A heat map of the metabolic profiling results of the heterozygous introgression lines (ILHs) is presented in Figure 7 (with the full data sets available in Supplemental Data Set 6 and Supplemental Figure 4). It is immediately apparent that some of the changes in metabolites are conserved in the ILs and ILHs, while others are not. Furthermore, there are clear quantitative differences in those traits that are conserved. Some metabolites, such as coumaric acid hexose I (F203), homovanillic acid hexose II (F204) both in IL10-3 and coumaric acid hexose II (F304) in IL11-1, are present at lower levels. Others, such as ferulic acid hexose III (F612) in IL3-3, homovanillic acid hexose II (F619) in IL8-1, naringenin chalcone hexose I (F411) in IL10-1, and 1-caffeoylquinic acid I (F026) in IL7-4, are present at approximately the same level in the ILH as its parent IL. Some, such as naringenin chalcone hexose I (F411) in IL3-2, sinapic acid hexose (F214) in IL6-3, and naringenin hexose II (F061) in IL1-3 are present at even higher levels.
Figure 7.
Heat Map of the Metabolite Profiles of M82 Lines Heterozygous (ILH) for the Chromosomal Segmental Substitution from S. pennellii.
Results presented are pericarp metabolite content data obtained from the ILHs of the 2004 harvest. Regions of dark red or dark blue indicate that the metabolite content is increased or decreased, respectively, after introgression of S. pennellii segments. UPLC-FTMS was used to quantify 145 metabolites, including flavanols, hydroxycinnamate derivatives, glycoalkaloids, and acyl-sugars. Due to space constraints, this heat map is not annotated; however, a fully annotated heat map including the metabolite profiles of the ILHs from the 2004 harvest is provided in Supplemental Figure 2. The introgression lines are presented in chromosomal order from top of chromosome 1 to base of chromosome 12 from left to right
Assessment of the Mode of Inheritance of Secondary Metabolite QTL
To assess whether these changes are associated with a particular mode of inheritance, we subjected the combined data set to a QTL analysis in which the ILs and ILHs were compared with a common control and only lines in which the significance was below the 1% threshold were considered to harbor a QTL. In addition to allowing point-by-point analysis, the inclusion of ILHs in the analysis enables us to classify each putative wild species QTLs into the following mode-of-inheritance categories: recessive, additive, dominant, or overdominant (for detailed explanation, see Semel et al., 2006). Evaluation of the results of this classification, presented in Figure 8 (full data set available in Supplemental Data Set 7), reveals that several of the putative wild species QTLs have an increasing effect on metabolite content. Although, in sharp contrast to mQTLs of primary metabolism (Schauer et al., 2006), the majority have a decreasing effect on metabolite content. What was similar between the mQTLs of secondary metabolism presented here and those presented for primary metabolism previously was that in both instances the vast majority of the QTLs exhibit either dominant or additive modes of inheritance with only a minority displaying recessive modes of inheritance and no incidence of overdominance.
Figure 8.
Distribution of the QTL Mode of Inheritance for Metabolite Accumulation.
Each vertical bar represents the number of QTLs for a specific trait, colored according to mode-of-inheritance categories: additive, dominant, and recessive. The bars above the 0 line represent the number of increasing QTLs, whereas the negative bars represent the number of decreasing QTLs relative to M82. A fully annotated figure with the exact compound ID is provided in Supplemental Figure 5 and Supplemental Data Set 7.
When the distribution of mode of inheritance is compared across the different compound classes, some clear differences can be observed. χ2 tests also revealed significant differences across compound types in the level of both positive and negative dominant modes of inheritance (Table 1). We observed a relatively high proportion of additive negative QTLs for all compound classes. The proportion between all other mode-of-inheritance types was significantly different between the compound class types. For instance, glycoalkaloids displayed a particularly high proportion of positive additive and dominant QTLs, acyl-sugars and hydroxycinnamates showed a very high proportion of dominant negatives, and all other classifications exhibited a fairly even split between dominant and additive negative QTLs. Taken together, the total proportion of QTLs were dominant irrespective of classification with most other QTLs being additive and there was very little recessive behavior evident.
Table 1. Qualitative Distribution of Mode of Inheritance Showing the Numbers of QTLs That Were Classified in Each Category for Each Chemical Compound Class.
| Acyl-Sugars | Flavonoids | Glycoalkaloids | Hydroxycinnamates | N-Containing Compounds | Others (Phenolics) | Unspecified | ||
|---|---|---|---|---|---|---|---|---|
| (5 Traits) | (19 Traits) | (21 Traits) | (34 Traits) | (25 Traits) | (14 Traits) | (27 Traits) | P (χ2) | |
| Additive − | 34 (31.7) | 78 (29.4) | 41 (15.1) | 157 (31.1) | 84 (26.7) | 93 (40.6) | 117 (34.4) | 0.292336757440 |
| Additive + | 4 (3.7) | 33 (12.4) | 97 (35.7) | 32 (6.3) | 38 (12.1) | 22 (9.6) | 49 (14.4) | 0.002225045604 |
| Dominant − | 57 (53.2) | 95 (35.8) | 37 (13.6) | 238 (47.2) | 101 (32.1) | 83 (36.2) | 105 (30.8) | 0.000013253208 |
| Dominant + | 6 (5.6) | 38 (14.3) | 60 (22.1) | 27 (5.3) | 38 (12.1) | 14 (6.1) | 42 (12.3) | 0.000000000001 |
| Recessive − | 6 (5.6) | 11 (4.1) | 15 (5.5) | 21 (4.1) | 23 (7.3) | 8 (3.4) | 12 (3.5) | 0.000000000275 |
| Recessive + | 0 (0) | 10 (3.7) | 21 (7.7) | 29 (5.7) | 30 (9.5) | 9 (3.9) | 15 (4.4) | 0.000000000001 |
| Total | 107 (100) | 265 (100) | 271 (100) | 504 (100) | 314 (100) | 229 (100) | 340 (100) |
The numbers in parentheses represent the percentage of this category among all QTLs in that group. The signs that follow the mode of inheritance indicate whether it is an increasing (+) or decreasing (−) QTL relative to M82. A statistical comparison between the different metabolite groups was conducted in each mode of inheritance using a χ2 test (with 1 df) by classifying the QTL into those that belong to this mode of inheritance and those that do not belong for each group.
Comparisons of Secondary Metabolite-Secondary Metabolite, Secondary Metabolite-Primary Metabolite, and Secondary Metabolite-Yield-Associated Trait Correlation Networks
In previous studies, we investigated the network behavior of primary metabolite traits in this population in correspondence to yield-associated traits (Schauer et al., 2006, 2008). Here, we decided to expand such analyses to include the secondary metabolite traits evaluated within this study. We investigated two aspects of the metabolic correlation networks extracted from metabolic traits between seasons and genotypes: (1) the extent to which they differ and (2) how heritability of the traits can be analyzed within the network context.
Networks were generated from the profiles comprising the measured metabolite levels together with phenotypic data from the 74 tomato ILs and the recurrent parental control M82. Each node in the network represents a metabolite or a composite phenotypic trait; an edge is established between two nodes if the corresponding traits are correlated, based on the Pearson correlation coefficient, above a threshold which guarantees false discovery rate (FDR) of 0.05 (see Methods). Altogether, we used the profiles of 145 compounds from secondary metabolism, from the investigated homozygous introgression lines in two seasons, 2001 and 2004, as well as from the heterozygous introgression lines from season 2004. In addition, 47 compounds participating in pathways from primary metabolism, profiled by GC-MS technology, were available for homozygous lines in two different seasons, 2001 and 2004 (Schauer et al., 2008). Integration of data from 38 phenotypic traits from homozygous and heterozygous lines from 2004 was also performed.
Altogether, seven networks were extracted by combining different data types, resulting in the following three groups of networks: (i to iii) three networks based on the liquid chromatography-mass spectrometry (LC-MS) data from the homozygous lines in 2001, homozygous lines in 2004, and heterozygous lines in 2004, respectively (Figures 9A to 9C); (iv and v) two networks based on the combined LC-MS and phenotypic data from the homozygous and heterozygous lines in 2004 (Figures 10A and 10B); and (vi and vii) two networks based on the combined LC-MS and GC-MS data from the homozygous lines in 2001 and 2004, respectively (Supplemental Figures 6A and 6B). Seminal properties of these networks are presented in Table 2.
Figure 9.
Correlation Networks for the LC-MS Data Assessed in This Study.
The network comprises nodes representing primary metabolites, secondary metabolites, and phenotypic traits, denoted by the following colors: N-containing compounds (brown), hydroxycinnamate derivatives (blue), acyl-sugars (red), glycoalkaloids (orange), flavonoids (green), polyamines (coral), amino acids (yellow), organic acids (pink), sugars (white), and all other metabolites without compound class (gray). Homozygous lines ([A] and [B]) from 2001 and 2004, respectively, and heterozygous introgression lines from 2004 (C), denoted by HO1, HO4, and HE4 networks. The networks are sparsified by removing 30% of the edges at random for easy visualization, while maintaining the relative ordering of nodes based on their degrees in the original network. (D) shows a scatterplot and linear fit between absolute value of the difference in node degrees of HO1, HO4, and the heritability of the respective metabolites.
Figure 10.
Combined Correlation Networks for the LC-MS Data and Phenotypic Traits.
The network comprises nodes representing primary metabolites, secondary metabolites, and phenotypic traits, denoted by the following colors: N-containing compounds (brown), hydroxycinnamate derivatives (blue), acyl-sugars (red), glycoalkaloids (orange), flavonoids (green), polyamines (coral), amino acids (yellow), organic acids (pink), sugars (white), all other metabolites without compound class (gray), and composite traits (black) from homozygous (A) and heterozygous (B) introgression lines from 2004, denoted by HO and HE networks. The networks are sparsified by removing 30% of the edges at random for easy visualization, while maintaining the relative ordering of nodes based on their degrees in the original network.
Table 2. Network Properties.
| Threshold − | Threshold + | No. of Nodes | No. of Edges | Positive Weight | Negative Weight | Isolated Nodes | Connected Components | Largest Component | Average Degree | Density | Communities | Modularity | Rand Index Chemical Classes | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HO1 | −0.39 | 0.34 | 145 | 1567 | 1561 | 6 | 11 | 12 | 134 | 21.61 | 0.15 | 15 | 0.36 | 0.02 |
| HO4 | −0.36 | 0.33 | 145 | 1088 | 1032 | 56 | 2 | 3 | 143 | 15.01 | 0.10 | 7 | 0.51 | 0.08 |
| HE4 | −0.33 | 0.29 | 145 | 2401 | 2236 | 165 | 1 | 2 | 144 | 33.12 | 0.23 | 4 | 0.28 | 0.02 |
| HO | −0.38 | 0.34 | 183 | 1356 | 1267 | 89 | 1 | 2 | 182 | 14.82 | 0.08 | 7 | 0.50 | 0.17 |
| HE | −0.35 | 0.29 | 183 | 2719 | 2479 | 240 | 1 | 2 | 182 | 29.72 | 0.16 | 4 | 0.29 | 0.007 |
| HOc1 | −0.44 | 0.36 | 192 | 1569 | 1565 | 4 | 19 | 20 | 173 | 16.34 | 0.08 | 25 | 0.44 | 0.09 |
| HOc4 | −0.38 | 0.36 | 192 | 959 | 924 | 35 | 24 | 25 | 168 | 9.99 | 0.05 | 31 | 0.53 | 0.16 |
Summary of the seminal network properties for the three groups of networks. The columns correspond to the following properties: threshold value for negative correlations, threshold value for positive correlations, number of nodes, number of edges, number of edges with positive weight, number of edges with negative weight, number of isolated nodes, number of connected components, the number of nodes in the largest component, the average degree, density, number of communities (determined by the greed community finding algorithm based on modularity), the modularity of the corresponding partition into communities, and the Rand index for the correspondence between the determined communities and the chemical classes for the compounds.
For the first group of networks (i to iii), with the LC-MS data from homozygous lines, a FDR of 0.05 was ensured by threshold values of 0.34 and 0.33 in seasons 2001 and 2004, respectively. These two networks will be referred to as HO1 and HO4 (Figures 9A and 9B). For the heterozygous data from 2004, the threshold value was smaller, at a value of 0.28; this network is referred to as HE4 (Figure 9C). Despite the similarity in the thresholds, HO1 contained 1567 edges, while HO4 included only 1088 edges; in addition, HE4 included almost double the number of edges despite the small difference in the thresholds used for establishing the edges. Only 0.04% and 5.1% of the significant correlations were negative, respectively. Moreover, all except for 11 and two isolated nodes were mutually connected in both HO1 and HO4. By comparison, in HE4, 6.9% of the significant correlations were negative and only one node was isolated, while the rest were connected in a single component. Based on their connectivity, the largest component in HO1 and HO4 could be partitioned into four and five communities, respectively, based on greedy optimization of modularity (value of 0.36 and 0.51); however, these communities showed little correspondence with the partitions of metabolites into chemical classes, which is in line with the lack of concordance between changes of metabolites and their heritability in the context of the metabolic pathways. In addition, the modularity of the partition based only on the chemical compounds was very close to zero (<−2.298*10−4), indicating that there might be a significant crosstalk between pathways of different compound classes.
However, despite the similarities in the global structural properties, these networks differed in their fine structure: Only 26% of all edges in HO1 and HO4 were shared, and 64.9% and 49.5% were specific to the HO1 and HO4 networks (see Table 3 for additional parameters characterizing the network difference). Any pair of nodes adjacent in at least one of these networks contributed on average 0.37 to the difference of the total weight of network associations, quantified by the absolute value of the Pearson correlation coefficients. The shared edges on average differed by 0.13 with respect to their weights, indicating that the shared edges may be canalized, i.e., are robust. Indeed, there was no significant difference between the average correlation values of the shared edges in HO1 and HO4 (Kruskal-Wallis test, P value = 0.492). From the 549 shared correlations, 391 appeared to be canalized (Fisher z-transformation, P value > 0.05) and included a total of 125 compounds including representatives of all chemical classes measured.
Table 3. Difference between Networks.
| Jaccard Index | Specific to First | Specific to Second | Average Difference in Correlation | Average Difference in Shared Correlations | Difference in Degree | Difference in Closeness | Difference in Betweenness | Difference in Eigenvector Centrality | |
|---|---|---|---|---|---|---|---|---|---|
| HO1-HO4 | 0.26 | 0.65 | 0.49 | 0.37 | 0.13 | 11.620 | 0.001 | 91.66 | 0.28 |
| HO4-HE4 | 0.32 | 0.22 | 0.64 | 0.29 | 0.09 | 18.410 | 0.001 | 81.88 | 0.22 |
| HOc1-HOc4 | 0.23 | 0.49 | 0.69 | 0.40 | 0.13 | 9.625 | 7.46e−5 | 169.56 | 0.22 |
| HO-HE | 0.32 | 0.26 | 0.63 | 0.30 | 0.09 | 15.770 | 0.0003 | 121.01 | 0.19 |
The difference between the considered networks is assessed based on the seminal network properties, including the ratio of shared edges to the union of edges of the two networks (Jaccard index), proportion of edges specific to the first network, proportion of edges specific to the second network, the average absolute value of the difference of shared correlations, the absolute value of the difference between the degrees, closeness, and betweenness, and eigenvector centralities of the nodes.
In total, 32.4% of the edges were shared between HO4 and HE4, while 21.5% and 64.4% of the edges were specific to HO4 and HE4, respectively. In comparison to the difference between HO1 and HO4, the smaller contribution of shared edges between HO4 and HE4 suggested that the associations between metabolic profiles were more affected by seasonal differences (Table 3). Indeed, there was no significant difference between the average correlation values of the shared edges in HO1 and HO4 (Kruskal-Wallis test, P value = 0.464). Of 854 shared correlations, 733 appeared to be canalized (i.e., did not differ in value, Fisher z-transformation, P value > 0.05), and these correlations comprised 137 compounds including representatives of all chemical classes measured. Interestingly, 301 of the canalized correlations between HO4 and HE4 were also canalized between HO1 and HO4 and included 116 compounds from all different classes.
For the second group of networks (iv and v), the two networks are based on the combined metabolic and phenotypic data from the homozygous and heterozygous lines in 2004, referred to as HO and HE, respectively. They included 38 phenotypic variables in addition to the 145 metabolites measured via LC-MS (Figures 10A and 10B). Since the relationships between the metabolites were investigated in the previous section, here, we focused on identifying the phenotypic variables that can be best predicted by individual metabolites. From the 104 significant correlations between the phenotypic variables and compounds in HO, 76 were positive and the remaining 28 were negative. In addition, only 18 of the 38 phenotypic variables were correlated with the secondary metabolites, with on average approximately six regressors per phenotypic variable. The phenotypic variables with the largest number of associations were: EA, yield associated traits (19); BX, brix (18); HI, harvest index (16); PW, plant weight (9); SN-fruit unit, seed number/fruit unit (7); and SN-fruit; seed number/fruit (6), and only two of the correlations were >0.5. Additional properties and differences between HO and HE can be found in Tables 2 and 3, respectively.
The third group of networks (vi and vii) can be used to investigate the extent of concordance between primary metabolite profiles determined previously (Schauer et al., 2008), with the secondary metabolite profiles described here by constructing metabolic correlation networks of homozygous lines in 2001 and 2004, referred to as HOc1 and HOc4 (Supplemental Figures 6A and 6B). In HOc1, there are altogether 94 correlations of which all are positive on 70 nodes. The most connected compound is maltose, associated to 13 secondary metabolites followed by proline (12), succinate and trehalose (10), glycerol (nine), isoleucine (eight), and leucine and citramalate (five). More specifically, maltose is connected to five glycoalkaloids, two hydroxycinnamate derivatives, two nitrogen containing metabolites, and one acyl-sugar, while proline is connected to eight glycoalkaloids, two flavonoids, and one nitrogen-containing compound, while trehalose is connected to four glycoalkaloids, two flavonoids, and two nitrogen containing metabolites.
In HOc4, there are a total of 14 correlations, of which all are positive on 22 nodes. The most connected compounds are gluconate, isoleucine, and inositol; each of which is associated with two secondary metabolites followed by maltose, phenylalanine, succinate, threonate, tryptophan, lysine, galactunorate, and dehydroascorbate, which all are associated with a single secondary metabolite. More specifically, gluconate is connected to two flavonoids, inositol to two hydroxycinnamate derivatives, and isoleucine to one nitrogen-containing compound and one hydroxycinnamate derivative. Perhaps surprisingly in the case of the associations between primary and secondary metabolites, we did not find any shared correlations between the HO and HE networks, suggesting a greater environment dependence of the relationship and crosstalk between pathways of primary and secondary metabolism than that between yield-associated traits and secondary metabolism.
Next, in order to investigate whether changes and canalization of metabolite levels are manifested in the properties of the networks, we inspected the correlations between the broad-sense heritability and the differences in prominent types of node centralities (e.g., degrees, neighborhoods, betweenness, and closeness) in the networks from the two seasons, HO1 and HO4. This approach aimed at quantifying the extent to which correlation patterns between traits relate to heritability of the traits themselves. A significant correlation of −0.455 (P value = 4.593e−07) between the difference in node degrees and the broad-sense heritability of the corresponding metabolites thus demonstrated that the canalization of the metabolite levels is inversely proportional to the changes in the number of relationships in which metabolites participate; therefore, metabolites whose levels are maintained across seasons tend to alter their relationships (Figure 9D). This result provided the basis for relating heritability (i.e., degree of robustness of phenotype across environments) with the plasticity of the underlying networks, manifested in the changes of the correlation patterns. This was further confirmed by the significant correlation of −0.376 (p value = 4.391e−05) between the Jaccard distance (see Methods) in the first network neighborhoods of the corresponding nodes in HO1 and HO4. However, the Eigenvalue centrality as well as the centrality based on distribution of paths (betweenness and closeness, neglecting the weights due to the presence of negative correlations; Toubiana et al., 2013) do not capture this property, as manifested in the smaller and less significant correlations of −0.241, −0.102, and −0.016. The largest change in degree between the two networks is observed for glycoalkaloids (maximum of 31 and median of 12) in comparison to flavonoids (maximum of 37 and median of 10), hydroxycinnamate derivatives (maximum of 30 and median of 9), and nitrogen-containing metabolites (maximum 33 and median of 8). Altogether, our findings on the relationship between heritability of metabolic traits and their placement in metabolic correlation networks indicate that traits maintained across seasons can be detected from the respective changes of correlation patterns.
Profiling of Expression Levels of Transcription Factors and Secondary Metabolism-Related Genes
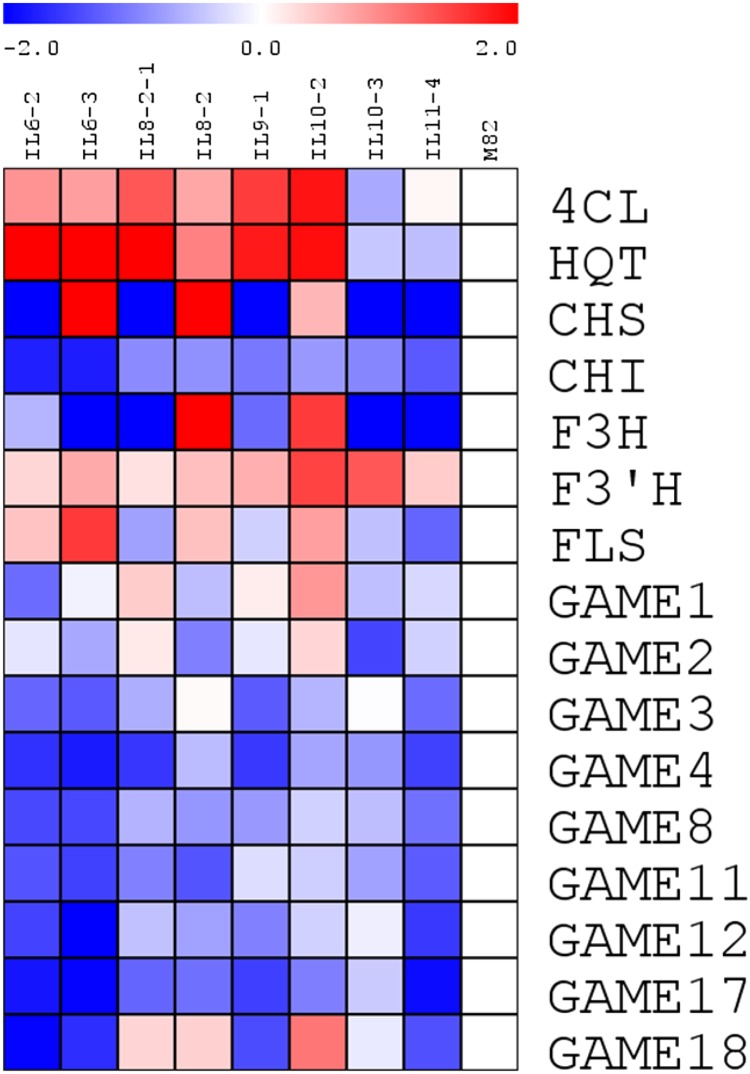
Having performed the above global network analyses, we next wanted to focus on beginning to delineate the genetic basis of some of the mQTLs of secondary metabolism that we determined here. From prior experience (Fridman et al., 2004), we know that the ultimate identification of the genetic causes underlying mQTLs remains an arduous task. For this reason, in order to get a first idea as to molecular event underlying the QTLs on the basis of the mQTL analyses described above, eight introgression lines (IL6-2, IL6-3, IL8-2, IL8-2-1, IL9-1, IL10-2, IL10-3, and IL11-4-1) and the recurrent parent M82 were selected for gene expression profiling of transcription factors (TFs) and secondary metabolism-related genes. This was achieved using the S. lycopersicum quantitative RT-PCR (qRT-PCR) transcription factor profiling platform that was recently established in our laboratory (Rohrmann et al., 2011). A total of 974 TFs were measured in this study. In addition, tomato genes encoding key enzymes involved in secondary metabolism (of the phenylpropanoid, flavonoids, and glycoalkaloid pathways) were obtained by BLAST searches using the sequence of previously characterized tomato genes. Using this approach, two phenylpropanoid biosynthetic genes (4-coumarate-CoA ligase [4CL]; Niggeweg et al., 2004), one hydroxycinnamate biosynthetic gene (hydroxycinnamoyl CoA quinate transferase [HQT]; Luo et al., 2008), five flavonoid biosynthetic genes (chalcone synthase [CHS1]; O’Neill et al., 1990; chalcone isomerase [CHI], flavanone-3-hydroxylase [F3H], flavonoid-3′-hydroxylase [F3′H], and flavonol synthase [FLS]; Groenenboom et al., 2013), and nine glycoalkaloid related genes (GAME1, 2, and 3; Itkin et al., 2011; GAME4, 8, 11, 12, 17, and 18; Itkin et al., 2013) were obtained for this analysis. Primers of secondary metabolism-related genes were designed by QUANTPRIME software (Arvidsson et al., 2008). The same pooled plant material as used for secondary metabolite analysis was used for gene expression profiling. The gene expression of TFs and secondary metabolism-related genes was visualized in the heat maps of Figures 11 and 12, respectively.
Figure 11.
TF Profiles of Tomato Fruits of Selected ILs.
Heat map showing the fold changes of 974 TFs relative to recurrent parent (M82) in 2001 (three biological replicates were measured). TFs grouped based on the TF families and sorted according to average of fold change within the same group (full data set available in Supplemental Data Set 8).
Figure 12.
Heat Map Showing the Relative Gene Expression of Phenylpropanoid, Flavonoid, and Glycoalkaloid Genes.
Expression levels were measured by qRT-PCR. 4CL, 4-coumarate-CoA ligase; HQT, hydroxycinnamoyl CoA quinate transferase; CHS1, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone-3-hydroxylase; F3′H, flavonoid-3′-hydroxylase; FLS, flavonol synthase; GAME, glycoalkaloid metabolism genes (GAME1, 2, and 3 [Itkin et al., 2011] and GAME4, 8, 11, 12, 17, and 18 [Itkin et al., 2013]). Values are average of three biological replicates and represent as log2 fold changes compared with M82.
As for the metabolite QTLs, expression QTLs were determined at both permissive (P ≤ 0.05) and stringent (P ≤ 0.01) thresholds using ANOVA tests. We found a total of 1347 QTL at the permissive and 533 at the stringent threshold corresponding to between 0 permissive (0 stringent) and 6 permissive (5 stringent) QTLs per TF. When broken down into TF families, 17 out of 59 families displayed a high number of QTLs. This corresponded to 113 permissive (47 stringent) QTLs for C3H TFs, 108 permissive (52 stringent) QTLs for AP2/ERF TFs, 87 permissive (37 stringent) QTLs for MYB-related TFs, 78 permissive (26 stringent) QTLs for C2H2 zinc finger TFs, 63 permissive (27 stringent) QTLs for HB/homeobox TFs, 71 permissive (35 stringent) QTLs for basic helix-loop-helix TFs, 61 permissive (23 stringent) QTLs for MADS box, 46 permissive (19 stringent) QTLs for MYB TFs, 55 permissive (31 stringent) QTLs for basic region/leucine zipper motif TFs, 36 permissive (8 stringent) QTLs for WRKY TFs, 39 permissive (10 stringent) QTLs for CCAAT TFs, 33 permissive (15 stringent) QTLs for heat shock factor TFs, 45 permissive (11 stringent) QTLs for NAC TFs, 39 permissive (19 stringent) QTLs for C2C2-DOF TFs, 34 permissive (16 stringent) QTLs for AUX/IAA TFs, 25 permissive (10 stringent) QTLs for GRAS TFs, and 69 permissive (23 stringent) QTL for orphan TFs. The full data set including QTLs of all TF families is available in Supplemental Data Sets 8 and 9.
The transcript of tomato MYB TF Solyc10g055410 (THM27) (Lin et al., 1996) showed higher expression in IL8-2-1 and IL10-3 compared with M82; by contrast, its expression was lower in IL6-2, IL11-4-1, and IL9-1 compared with M82. The C2H2 TF J0317, which has been reported as putative regulator of phenylpropanoid biosynthesis (Rohrmann et al., 2011), is upregulated in seven lines compared with M82. The tomato ortholog J0088 of the well characterized ORCA3 AP2/ERF TF known to regulate both primary and terpenoid alkaloid metabolites in Catharanthus roseus (van der Fits and Memelink, 2000), showed 4- to 38-fold higher expression in all introgression lines compared with M82. In addition, a MADS box TF (J0651) showed higher expression in all the lines with respect to M82. Expression of J0659 (Solyc06g059970, MADS box family), which is orthologous to PISTILLATA in Arabidopsis (Mara et al., 2010), was 15 times higher in IL11-4-1 compared with M82. Naringenin derivative (F061) showed significantly higher accumulation in this lines compared with other lines and M82. On the other hand, the MYB-related gene J0778 showed no expression in IL11-4-1. The MYB TF J0715 (tomato ortholog of MYB44 from Arabidopsis), previously reported to be correlated with 13 secondary metabolites (Rohrmann et al., 2011), showed no significant difference between ILs and M82. Furthermore, expression of the C2C2 TF J0310 known as a regulator of fruit size (Cong et al., 2008) and early response to ethylene during repining (Rohrmann et al., 2011) was significantly higher in IL10-2 than in M82, and no expression was observed in IL11-4-1. The TF SI_IAA9 (J0141), which plays an important role in fruit development (Wang et al., 2005), showed significantly lower expression in IL6-2 and IL10-2 compared with M82, while higher expression was found in IL8-2 and IL8-2-1 compared with other introgression lines and M82. The CNR_Colorless nonripening TF showed slightly higher expression in all lines except IL8-2-1. Significantly higher expression was found in IL6-2 and IL6-3 compared with M82 for NOR_non-ripening (J0824). In addition, the MADS box TF RIN (J0650) showed significantly higher expression in IL6-3 compared with M82.
Furthermore, IL6-2 and 6-3, which indicate most significant mQTL hot spots of several secondary metabolites, showed higher expression of the genes involved in the early step of hydroxycinnamate biosynthesis, such as 4CL and HQT (Figure 8), despite the fact that the expression level of glycoalkaloid biosynthetic genes clearly decreased. The introgression line IL8-2 and its subline IL8-2-1 showed slightly higher expression of phenylpropanoid biosynthesis genes, but flavonoid biosynthetic genes are highly expressed only in IL8-2, but other genes such as hydroxycinnamate and flavonoid biosynthetic genes are highly expressed only in IL10-2. In the IL9-1 region, the expression level of phenylpropanoid related genes such as 4CL and HQT were higher than M82. However, flavonoid biosynthetic genes (CHS, CHI, F3H, and FLS) were clearly decreased in IL9-1. On the other hand, IL11-4 showed lower expression of all genes involved in secondary metabolism
In order to provide additional support for the observed phenotypes, we conducted a pairwise comparison between M82 and S. pennellii to detect possible sequence polymorphisms at the level of TF-encoding genes. As a first approach, we defined the total number of genes in these eight ILs (this ranged from 103 and 1095), the number of predicted transcription factors (which ranged from 8 to 81), and the number of transcription factors for which we detected expression using our qRT-PCR platform (varied from 2 to 29; see Table 4 for details). Although it must be kept in mind that the qRT-PCR platform does not cover all TFs in tomato, this approach dramatically reduced the number of putative regulators in the ILs. Following this, we performed sequence analyses focused on the identification of SNPs and InDels at coding and promoter regions of all the TF genes contained within the exact physical location of each introgression (Chitwood et al., 2013). The list of TF-encoding genes along with a summary of detected polymorphisms is reported in Supplemental Data Set 11. Single intolerant amino acid changes, as predicted by SIFT BLink (P < 0.05), were found for 15 TF genes. On the basis of the expression data presented here, two of these were of high potential interest, namely, polymorphisms in highly conserved residues of the DNA binding domains (such as the Tyr-to-His change in the AP-2 ethylene-responsive TF gene, Solyc06g068570, from IL6-2) and specific residues maintaining the stable fold needed for phosphorylation or other posttranslational modifications (such as the Ile-to-Phe change in the response regulator MYB-like TF gene, Solyc11g072330, from IL11-4-1).
Table 4. Number of the Genes and Predicted Transcription Factors in Eight ILs.
| IL | Bins | Total No. of Genes in the Introgression | No. of TF Genes in the Introgression Predicted by PlantTFDB v3.0 | No. of TF Genes in the Introgression for Which We Have Expression Data |
|---|---|---|---|---|
| 6-2 | d6B, d6C, d6D, d6E | 1095 | 81 | 29 |
| 6-3 | d6E, d6F, d6G | 696 | 57 | 17 |
| 8-2 | d8D, d8E, d8F | 890 | 52 | 13 |
| 8-2-1 | d8D | 679 | 35 | 7 |
| 9-1 | d9A, d9B, d9C, d9D, d9E | 524 | 37 | 7 |
| 10-2 | d10C, d10D.1, d10E, d10D.2, d10F | 693 | 53 | 13 |
| 10-3 | d10F, d10G | 387 | 24 | 7 |
| 11-4-1 | d11H | 103 | 8 | 2 |
| Total | 5067 | 347 | 95 |
Wide(r) structural variation, involving, for example, several intolerant amino acid polymorphisms, or large insertion/deletions (InDels) in the coding sequences, was found in a few instances, such as in the GRAS TF genes from IL10-3. This is of interest given that members of the GRAS TF family have a role in gibberellin signaling, root/shoot development, and fruit set (Pysh et al., 1999; Carrera et al., 2012). In the case of Solyc10g086370, a large nucleotide insertion (87 bp) in the S. pennellii allele interrupted the GRAS domain, and several interspersed SNPs along the conserved portions led to 12 amino acid changes, three of which were predicted as intolerant substitutions for protein function. In the case of Solyc10g086380, also a member of the GRAS TF family, the occurrence of several InDels in the coding sequence of S. pennellii led to a truncated product: the predicted gene model lacked 165 amino acids from the C terminus and a number of additional interspersed SNPs resulted in five intolerant amino acid changes.
Pairwise analysis of the alignments of TF genes from all eight ILs was also extended to include noncoding sequences upstream their start codon ATG (up to around −1000 bp): Sequence variation in this region, in fact, may affect various conserved binding motifs and have a large impact on initiation of translation, stability of the downstream mRNAs, global gene expression level, and tissue specificity (Mignone et al., 2002). Following this analysis, large alignment gaps (at least one InDel with length > 30 bp) emerged for 16 (out of 107) TF genes (Supplemental Data Set 11): These large InDels frequently included core promoter elements (e.g., GATA/TATA box) and/or putative binding motifs for various TFs. The aligned upstream sequences of a G2-like TF gene from IL6-3 (Solyc06g076350), for example, showed several contiguous insertions of varying length in S. pennellii (from 8 to 101 bp), with the presence of at least 20 putative cis-acting elements. In the case of Solyc10g078700, a SBP (Squamosa binding protein) TF gene from IL10-2, the upstream sequences of S. lycopersicum and S. pennellii differed for the presence of three InDels (located from −900 to −640 bp with respect to the ATG and ranging in length from 22 to 39 bp), containing at least 12 different putative TF binding sites. In many cases (although not all), the extent of structural promoter variation was correlated with the magnitude of differential expression between a specific IL and S. lycopersicum M82 (Supplemental Data Set 11). However, the most important observation here is that a combination of expression and sequence analyses can be used to predict transcriptional regulators, although this is perhaps not as facile as would be expected. As a consequence, further (reverse) genetic experimentation will be required to validate all or even any of these candidate regulators of secondary metabolism.
Combined Analysis for Identification of Regulatory Circuits
In order to better understand the relationship between metabolic and gene expression changes, we next performed correlation network analysis (Supplemental Figure 7). The MADS box TF J0645 showed a positive correlation (>0.97) with an uncharacterized gene (Solyc06g062290, putative glycosyltransferase) involved in glycoalkaloid biosynthesis (Itkin et al., 2013). Significantly lower expression (P ≤ 0.01) of Solyc06g062290 was found in IL6-2, while a number of glycoalkaloid derivatives (lycoperoside G, F, or esculeoside A with m/z = 1268.592, 14.2 times higher; F407 with m/z = 1340.994, 3.5 times higher) were significantly higher in this line with respect to M82. F036, a glycoalkaloid derivative with m/z = 1224.565, was below the level of detection in this line. In addition, a strong positive correlation was found between the expression of Solyc06g062290 and the two TFs, TA38003_4081 and TA38817_4081. Moreover, a significant negative correlation of this gene was found with MYB TF J0713, while MYB TF J0707, homologous to MYB73 in Arabidopsis (responsive to salt stress, abscisic acid, and auxin stimuli; Kim et al., 2013; Zhao et al., 2014), is positively correlated (> 0.90) with an uncharacterized UDP-glycosyltransferase gene (Solyc10g085230) previously documented to be coexpressed with other genes involved in glycoalkaloid biosynthesis (Itkin et al., 2013). This gene is located in the overlapping region of IL10-2 and IL10-3 where it displayed significantly lower expression. A number of glycoalkaloid derivatives were found to be correlated to the expression of this gene and furthermore were lowly abundant or absent in these lines (e.g., lycoperoside G, F, or A, F063, and F229). Other glycoalkaloid derivatives showed high accumulation in these lines (e.g., F227, m/z = 1226.580). When taken together, these data identify TF J0707 as a putative regulator of the gene expression of Solyc10g085230 and consequently of the glycoalkaloid metabolism.
Evaluation of Genomic Polymorphism at Key mQTLs
In order to investigate key genes potentially involved in the mQTLs found in this study, we next attempted the prediction of candidate genes. First, we focused on the case of “transcriptional differences,” which were mainly caused by the differences within the promoter sequences of these genes. For this purpose, we focused on regions showing both global metabolic changes and a high density of secondary metabolite associated gene families, such as UGT, P450, and OMTs. Among the eight mQTL regions we discussed in the section on TF networks above, we focused on two QTL regions on chromosomes 6 and 10, which showed significant global metabolic changes [sinapic acid hexose (F214), homovanillic acid hexose II (F619), and 3-(2-hydroxy-4-methoxyphenyl] propanoic acid hexose [F027] metabolites were all altered) and a high density of secondary metabolism-related genes (such as P450s, laccase and oxidase genes; UGT, hydrolase and hydroxycinnamoyl transferase genes; OMT, chalcone isomerase, 4-coumarate-CoA ligase genes, and 2-OGDs). In the mQTL regions 695 (IL6-3) and 737 (in overlapping region of IL10-3 and IL10-2), genes were found by searching the parental genome sequences (Bolger et al., 2014). MapMan bin analysis revealed that 10 and 12 secondary metabolism-related genes are contained in these regions, respectively. Next, we performed genomic sequence analysis of the promoter regions (defined as 1000 bp upstream of start codon) of candidate genes and compared the promoter sequences of candidate genes by aligning the sequences from S. lycopersicum cv M82 and S. pennellii genome sequences (Bolger et al., 2014). Following this approach, three P450 genes, namely, Solyc06g076160, Solyc10g084590 (orthologs of AT3G26300/AtCYP71B34), and Solyc10g080870 (ortholog of AT4G39490), as well as OMT1 (Solyc10g084590, ortholog of AT5G54160/AtOMT1) were selected for detailed analysis. Solyc06g076160 was selected as candidate gene for QTLs located in IL6-2. For this gene there is a 44-bp deletion in the M82 sequence (at position −326 bp) and a further deletion of 11 bp at position −200 bp, in addition to other small gaps. Similarly, Solyc10g079540 was located within the overlapping region of IL10-2 and IL10-3. In addition, promoter sequence compression revealed a 60-bp deletion in M82 at position −362, in addition to an 8 bp at position −671 bp. However, both Solyc06g076160 and Solyc10g079540 were highly similar in sequence in their coding regions. Solyc10g080870 displayed high divergence in upstream sequences, with four deletions in M82 ranging between a few to up to 50 bp, in addition to other smaller gaps. The coding region of Solyc10g080870 is quite similar between M82 and S. pennellii (97%); however, it does contain a few polymorphisms, such as deletions in M82 at the first exon (length of deletion: 5 bp) and second exon (6 bp), while S. pennellii contains two 3-bp deletions in the first exon. Solyc10g084590 contained two clear deletions in the promoter region at −170 bp upstream (68 bp) and at −506 bp upstream (11 bp) of this gene. However, the coding region was very similar between M82 and S. pennellii with the exception of a 3-bp deletion in M82 at position 163.
Based on the genomic analysis of S. pennellii and M82 in the hot spot region, qRT-PCR was used to measure the expression of several genes related to secondary metabolism. As would be anticipated from the genomic sequence analysis of the promoter regions, the P450 genes Solyc10g084590 and Solyc10g080870 showed altered expression in IL10-2 and IL10-3, while little or no expression was found in the other lines and in M82 (Supplemental Figure 8). In addition, OMT1 (Solyc10g084590) was also highly expressed specifically in both IL10-2 and IL10-3. Cytochrome P450 gene Solyc06g076160 showed higher expression (5 times) in IL6-3, while little or no expression was found in the other lines and in M82 (Supplemental Figure 8).
Validation of Candidate Genes: A Case Study for Genes Associated with Glycoalkaloid Biosynthesis
As a first test of the involvement of Solyc06g062290 (annotated as a UDP-glycosyltransferase) and Solyc10g085230 (annotated as a UDP-glycosyltransferase) in the pathway of glycoalkaloid biosynthesis, we tested their expression levels in ILs that harbored QTLs for these metabolites (Figures 1 and 4). For this purpose, we selected ILs 6-2, 6-3, 10-2, and 10-3 and evaluated the levels of Solyc06g062290 (Figure 13A) and Solyc10g085230 (Figure 13B) by qRT-PCR. The expression of Solyc06g062290 was significantly decreased only in IL6-2, whereas Solyc10g085230 was significantly decreased in IL10-2 and 10-3. We next specifically independently silenced these two genes in tomato fruit using virus-induced gene silencing (VIGS). Following agroinjection, ripe tomatoes were harvested at 10 d after the breaker stage. qRT-PCR results revealed reductions of ∼71% and 76% in mRNA levels of Solyc06g062290 and Solyc10g085230, respectively, in silenced fruits compared with the fruits set from plants infiltrated with a pTRV2-empty vector (Figures 13C and 13D). Despite some variability, the mRNA levels of other major genes of glycoalkaloid biosynthetic pathway (GAME1, 4, and 12) were unchanged in these lines (Figures 13F to 13H).
Figure 13.
Gene Expression and Metabolite Changes in IL6-2, IL6-2, IL10-2, IL10-3, Solyc06g062290, and Solyc10g085230 Transiently Silenced Tomato Fruits.
(A) Relative levels of mRNA in M82, IL6-2, and IL6-3.
(B) Relative levels of mRNA in M82, IL10-2, and IL10-3.
(C) and (D) Relative levels of mRNA in control (empty vector) and silenced fruit of Solyc06g062290 (C) and Solyc10g085230 (D) harvested from VIGS agroinfiltrated tomato plants.
(E) Heat map of secondary metabolite changes in the control (empty vector) and silenced fruit of Solyc06g062290 and Solyc10g085230 harvested from VIGS agroinfiltrated tomato plants.
(F) to (H) Relative levels of mRNA in GAME1 (F), GAME12 (G), and GAME 4 (H) (GAME1; Itkin et al., 2011; GAME4 and 12; Itkin et al., 2013).
(I) Changes in major glycoalkaloids in control (empty vector) and silenced fruit of Solyc06g062290 and Solyc10g085230 (i), α-tomatin (ii), lycoperoside G/F (iii), esculeoside A + hexose (iv), dehydrolycoperoside G/F or dehydroesculeoside A (v), and unknown glycoalkaloids (vi) (m/z = 1341.1 and 1122.6). Data are mean ± se; asterisks indicate a significant difference at P ≤ 0.05.
Secondary metabolite profiles of methanol-extracted fruit pericarp were obtained from the same plant material by LC-MS analysis. Several glycoalkaloid derivatives were present at significantly different levels between silenced and nonsilenced fruits; the levels of α-tomatine in silenced mature ripe fruit of Solyc06g062290 and Solyc10g085230 were significantly higher compared with fruit infiltrated with empty vectors (Figure 13E). It is worth noting, however, that the levels of α-tomatine were much higher in fruit of silenced Solyc10g085230 compared with fruit of silenced Solyc06g062290. Importantly, a delay in fruit ripening was observed following silencing of Solyc10g085230, which may be of importance since it is often reported that green fruit accumulate considerably higher levels of α-tomatine compared with ripe fruit (Itkin et al., 2011, 2013; Iijima et al., 2013). That said, silencing of either gene resulted in significant changes in the levels of several other glycoalkaloids within the fruit (Figure 13E). Silencing Solyc06g062290 resulted in the accumulation of putative glycoalkaloid annotated as lycoperoside G/F (Figure 13I, ii) and reduced levels of the unknown glycoalkaloid of m/z = 1122.6 (Figure 13I, vi) or esculeoside A + hexose (Figure 13I, iii), suggesting that this enzyme is indeed involved in the glycoalkaloid biosynthetic pathway. By contrast, silencing Solyc10g085230 resulted in reduced levels of glycoalkaloid peak annotated as lycoperoside G/F or esculeoside A (Figure 13I, iv) and unknown glycoalkaloid of m/z = 1341.1 (Figure 13I, v) compared with the empty vector control. Interestingly, with the exception of a single ripening-related flavonoid (naringenin chalcone), which changed in Solyc10g085230-silenced plants, no other secondary metabolite changes were apparent in these lines.
Following the same approach as described above, a pairwise comparison between S. lycopersicum cv M82 and S. pennellii was conducted to detect possible sequence polymorphisms at the level of encoding and promoter regions of the candidate genes. For the first candidate, Solyc06g062290, which was annotated as a UDP-glucosyltransferase in the tomato ITAG 2.3, the sequence analysis revealed that the gene prediction in S. pennellii is that of a truncated product that corresponds to the terminal part of the S. lycopersicum protein. In addition, sequence alignments showed two open reading frames, which give partial alignments to the original S. lycopersicum protein; consequently, the predicted product(s) is a nonfunctional glucosyltransferase since it is lacking most of the functional domain. Furthermore, the sequence variation of the noncoding region −1000 bp upstream of the start codon ATG revealed the presence of a long terminal repeat-retrotransposon whose terminal sequence spreads over the promoter of the S. pennellii gene. Regarding the second candidate Solyc10g085230, annotated as a glycosyltransferase, our BLAST results indicate that no ortholog is present in the S. pennellii genome, consistent with the lack of expression of this gene in IL10-2 and IL10-3.
DISCUSSION
This study identified numerous mQTLs for secondary metabolite accumulation in tomato fruit pericarp. While only a handful of studies have used broad genetic crosses to identify mQTLs for a broad range of secondary metabolites (Kliebenstein et al., 2001a, 2001b; Keurentjes et al., 2006; Morreel et al., 2006; Khan et al., 2012; Matsuda et al., 2012; Routaboul et al., 2012; Gong et al., 2013; Wahyuni et al., 2014) and previous studies have largely been focused on Arabidopsis, considerable research has been focused on defining QTLs for volatile organic compounds from tomato fruit (Tieman et al., 2006; Mathieu et al., 2009; Mageroy et al., 2012; Rambla et al., 2014) and acyl-sugars in tomato leaf trichomes (Schilmiller et al., 2012, 2010). In addition, the levels of primary metabolites have been evaluated in multiple different breeding populations (Schauer et al., 2006, 2008; Do et al., 2010) as well as in association mapping panels (Sauvage et al., 2014). These previous studies provide an interesting context in which to evaluate the results of this study. We observed on average fewer mQTLs per metabolite for those secondary metabolite families studied here, 4.68, than we did for the primary metabolites we reported previously 11.86 (Schauer et al., 2006, 2008). When comparing our results with those for other secondary metabolite pathways in this population, we obtained considerably more QTLs than observed for the volatile organic compounds, 0.75 mQTLs per compound (Mathieu et al., 2009), but a similar amount in comparison to the trichome acyl-sugars, 4.57 per compound (Schilmiller et al., 2010).
These analyses likely reflect the observation that primary metabolism is characterized by a greater degree of redundancy than secondary with there often being several routes to the same end (for example, see Timm et al., 2011) and therefore exhibits a more complex genetic architecture. By contrast, the absolute variation in metabolite abundance across the population was much greater for secondary metabolites, which showed increases of up to 95-fold and decreases down to 0 (not produced) of the level found in the recurrent parental line (while for primary metabolites the maximal increases and decreases were 17.7 and 0.18, respectively). Two factors that may explain this are (1) that secondary metabolite abundance is under considerably less intricate control, which is dominated by transcriptional regulation; and (2) the prominence of unbranched pathways in secondary metabolism provides a better concordance of transcript and metabolite levels in these pathways (Fernie and Stitt, 2012). Additionally, the secondary metabolite data displayed a different pattern of change being dominated by decreases as opposed to the tendency to increase observed in the primary metabolite data set (Schauer et al., 2008). It is conceivable that this is due to differences in the natural variance of the primary and secondary metabolites between S. lycopersicum and S. pennellii. Many primary metabolites occur in higher concentrations in S. pennellii fruit (Schauer et al., 2005) and our own unpublished data suggest that mature fruit of S. pennellii also contain much higher levels of hydroxycinnamates, α-tomatine, and acyl-sugars, but contains lower levels of flavonols (T. Tohge and A.R. Fernie, unpublished data). Similarly, evaluation of the relative expression of key genes of secondary metabolism reveals that these have lower expression levels in fruit of S. pennellii than in the elite cultivar (Koenig et al., 2013; Bolger et al., 2014). A second possibility is that the divergence of pathways is considerably greater in S. pennellii, and this may lead to a rerouting of carbon into alternate metabolites thereby reducing the pool sizes of the core pathway intermediates. Given that transcriptional control of secondary metabolism has been demonstrated to be quite complex, at least occasionally (for instance, see Li et al., 2014), a third possibility is that primary metabolism is simply under more constrained evolutionary pressures, while secondary metabolism might be under more diversifying selection. While we favor the former two hypotheses, we cannot currently formally exclude any of them. Future isotope tracing studies and more detailed gene expression studies may allow this question to be addressed more comprehensively.
We also classified the metabolite heritability as high, intermediate, or low using thresholds of >0.4, between 0.2 and 0.4, and below 0.2, respectively. However, it is important to note that this study was based on only 2 years of data, and our experience has shown that these values could be anticipated to drop on addition of data from further harvests (Schauer et al., 2006, 2008). When looking at heritability on the basis of the individual compounds, a couple of interesting features were apparent. First, as was also observed for primary metabolites (Schauer et al., 2008), metabolites that are biochemically similar to one another in some instances display similar heritabilities, although it must be stressed that this linkage was by no means as strong for the secondary as the primary metabolites. Second, those metabolites that are known to be important stress protectants (Mintz-Oron et al., 2008, Itkin et al., 2011; Nakabayashi et al., 2014) appeared to have lower heritability. Similar to our findings described here, Matsuda et al. (2012) observed high hereditability for rice grain some secondary metabolites, especially in the case of flavonoids; however, most other metabolites were highly sensitive to environmental factors. Moreover, heritabilities of secondary metabolites in Arabidopsis tended to be considerably higher than those reported for primary metabolites (Kliebenstein et al., 2001a; Keurentjes et al., 2006; Wentzell et al., 2007; Rowe et al., 2008; Joseph et al., 2013). Although from a global perspective there are large differences in the number, direction, and magnitude of the mQTLs of tomato primary and secondary metabolism, and to a lesser extent their heritability in general, their mode or inheritance was very similar with the vast majority of secondary metabolite traits displaying dominant or additive inheritance. Intriguingly, this reinforces the conclusion of our earlier study that tomato metabolites do not exhibit overdominance and thus provide no support for proposed biochemical mechanisms of hybrid vigor (heterosis; Milborrow, 1998).
In addition to evaluating the metabolite traits on a one-by-one or even a compound class basis, we additionally performed more global analyses of trait correlations across the population. This is by no means the first time that network analyses have been used to evaluate the relationships between traits in wide genetic populations with many previous examples being published in Arabidopsis, potato, and maize (Rowe et al., 2008; Lisec et al., 2011; Carreno-Quintero et al., 2013), as well as for a range of different traits and tissue types in this very tomato population (Schauer et al., 2006, 2008; Toubiana et al., 2012). However, this study revealed several novel insights concerning the interrelation of traits from primary and secondary metabolism with yield-associated traits. Interestingly, when analyzed on the basis of their secondary metabolite traits, the ILHs and ILs were broadly similar in terms of their network structure albeit different in their fine structure, which is somewhat distinct from our previous comparisons on the basis of primary metabolite traits (Schauer et al., 2008). In addition, analysis of the correlations between primary and secondary metabolite traits revealed few correlations (either positive or negative) between secondary metabolites and the primary metabolites that act as their direct precursors. Indeed, the tricarboxylic acid cycle intermediate succinate was highly correlated with a large number of secondary metabolites, most likely reflecting the importance of this pathway for sustaining biosynthetic reactions (Fernie et al., 2004), but levels of the universal phenylpropanoid precursor phenylalanine showed no such correlation. The later observation is in keeping with previous studies in this population, which revealed that levels of phenylalanine-derived volatile organic compounds were not correlated with the levels of phenylalanine (Tieman et al., 2006). However, it is in sharp contrast to the results of several other studies suggesting that upregulation of secondary metabolism requires a coordinate upregulation of primary metabolism (Adato et al., 2009). In keeping with a coordinated regulation of primary and secondary metabolism is the observation that proline, glycerol, and succinate were strongly correlated to the glycoalkaloids given that they can be readily converted to the precursor of the glycoalkaloids, acetyl-CoA. These observations suggest that metabolic engineering of glycoalkaloid but not phenylpropanoid content could likely be achieved in tomato by strategies enhancing precursor supply. In addition, we used network analysis to address whether changes in metabolite levels (or by corollary the canalization of metabolite levels) observed on a point-by-point basis were also observable within the networks of correlated data patterns. Put another way, the heritability analyses discussed above do not take relationships between traits into account but rather consider the environment and only the behavior of individual traits. However, since metabolic networks were established for each season independently, we were interested in assessing whether heritability could be deduced from the changes in the networks between the seasons. For this purpose, we assessed the node centralities. The relationships between the heritability of metabolic traits and their position within networks suggested that conserved canalized traits may be detected by evaluating correlation patterns of the broader network. This approach may be useful in future studies on canalization.
Many of the compounds we detected here have important roles as nutrients or antinutrients; hence, the information obtained could prove interesting for informing attempts to produce healthier fruits, using either classical genetic or transgenic approaches (Martin et al., 2011; Fitzpatrick et al., 2013). As mentioned above, the cloning of QTLs from this population remains an arduous task, and we have genetically confirmed only a handful of the candidate genes we identified on the basis of our GC-MS screening most notably those for branched-chain amino acids (Maloney et al., 2010) and tocopherol (Quadrana et al., 2013, 2014). As a first step, given that secondary metabolism is predominantly under transcriptional control (Martin et al., 2010; Patra et al., 2013; Tohge et al., 2014), we used a previously established quantitative real-time PCR platform to define differences in transcription factor expression in eight of the ILs, as well as defining the expression level of a range of genes encoding key enzymes of secondary metabolism. The rationale behind this was that expression QTL analysis has previously been demonstrated to provide important information concerning the molecular basis of trait variance (for example, see Hansen and Houle, 2008).
Here, we used the approach as a first step in identifying the genetic basis of the observed changes in several metabolites notably the important nutrient chlorogenate and antinutrients of the glycoalkaloid class as well as some hydroxycinnnamates and acyl-sugars from trichome cells. Reassuringly, the mQTLs for dehydroesculeoside A or B we found in IL1-1 (Figure 4A) overlap with the identification of an isomer of dehydrotomatine (1363.85 m/z), which accumulated in IL1-1 trichomes but was not found in of M82 trichomes (Schilmiller et al., 2010). A single mQTL was found for the 693 m/z ion, which we annotated as a putative pregnane derivative (Figure 4B, B-2). Our mQTLs mapped to a small overlapping region of IL6-3 and IL6-4 at the end of chromosome 6; in our data set, the metabolite change is 14 and 12 times increased compared with M82. No genes involved in pregnane biosynthesis have been characterized until now; however, several strong candidate genes, O-methyltransferase (Solyc06g083450), duplicated genes of 3-ketoacyl-acyl carrier protein reductases annotated as tropan reductase (Solyc06g083470, Solyc06g083480, and Solyc06g083490), and ent-copalyl diphosphate synthase (Solyc06g084240), are present in this region. Another mQTL (that for a nontoxic form of glycoalkaloid derivatives) was mapped to chromosome 7 in the overlapping region of IL7-4 and IL7-4-1, in which the metabolite was absent. These QTLs colocalize to the cluster of glycoalkaloid metabolism genes (GAME) mapped in these regions (Itkin et al., 2013). However, direct proof concerning which gene controls the content of glycoalkaloids remains to be clarified in future studies. An acyl-sugar mQTL (345.1185 m/z) was found only in the overlapping region of IL8-2 and IL8-2-1 (Figure 4D). Two acyltransferases related to secondary metabolism Solyc08g075210 (ortholog of At3g30280 in Arabidopsis) and Solyc08g075180 (ortholog of At3g26040) localized to this region appear to be strong candidate genes for this QTL; however, they remain to be functionally validated. Furthermore, five UDP-glycosyltransferase 1 family (UGT1) genes (Solyc10g085730, Solyc10g085860, Solyc10g085870, Solyc10g085880, and Solyc10g086240) and one phenylalanine ammonia lyase (PAL; Solyc10g086180) were found in the mQTL region that showed higher accumulation of coumaric acid-hexoside in IL10-3 (Figure 4E, E-2). Finally, the mQTL region of one of the cholorogenic acid isomers (353 m/z) was found in IL12-4 and IL12-4-1 (Figure 4F) and does not contain strong candidates, except MYB genes (Solyc12g099130, Solyc12g099120, Solyc12g099140, Solyc12g099620, and Solyc12g099850). This last result suggests the presence of novel regulatory, or even structural, genes beyond those described to date in plants (Martin, 2013). Thus, the strategy used here may facilitate the identity of novel target genes for the metabolic engineering of this important nutraceutical. Furthermore, our VIGS data confirmed Solyc06g062290 and Solyc10g085230 as candidates in the control of glycoalkaloid accumulation, given that lycoperoside G/F increases and esculeoside A + hexose decreased in lines silenced in the former, and esculeoside A increased and the unknown glycoalkaloid of m/z = 1341.1 decreased in lines silenced in the latter. In addition, analysis of the metabolite profiles suggest that the gene product encoded by Solyc06g062290 likely acts as UDP-glucose and catalyzes the conversion of lycoperoside G/F to esculeoside A + glucose, while Solyc10g085230 catalyzes the conversion of esculeoside A to esculeoside A + hexose. These data essentially provide a proof-of-concept that at least for structural genes this approach allows in-depth characterization of the genetic architecture of secondary metabolic traits. However, our data concerning transcription factor networks suggest that these currently remain difficult to disentangle via the approaches used here and will require further (reverse) genetic studies for clarification.
In conclusion, the data presented here complement and extend that documented for traits of primary metabolism by Schauer et al. (2008). In this study, we were able to identify several hundred QTLs, some of which were of far greater magnitude than those described for mQTLs of primary traits, but the majority of which were negatively affected by the substitution of a portion of the S. pennellii genome into the background of the elite cultivar. Our results allow several important conclusions to be drawn regarding the metabolic networks of tomato fruits, including the relative importance of precursor supply for phenylpropanoid and glycoalkaloid metabolism, the lack of difference in network structure when lines heterozygous for the introgressions are compared with those homozygous for them, and the value of the use of network structure as a proxy for heritability analysis. In terms of directed metabolic engineering strategies, several positive QTLs were identified that could be used for biofortification of chlorogenic acid, whereas several negative QTLs for antinutrients such as those of the glycoalkaloids were also identified. This information could thus be directly useful for the design of breeding strategies to improve the nutritional quality of the fruit. One unexpected insight that this study afforded was the fact that many of the QTLs for secondary metabolite contents exhibited dominant negative mode of inheritance. This is counter to the received wisdom that secondary metabolite contents were reduced on domestication (Meyer et al., 2012) and as such is a highly interesting observation worthy of further study, as it emphasizes the potential importance of maintaining or improved stacking of domestic alleles in breeding for these compounds in addition to bringing in wild alleles.
All of the QTLs described, irrespective as to whether they are positive or negative, could ultimately (once the underlying gene has been cloned) provide further candidate genes for transgenic approaches toward fruit compositional improvement. Evaluation of global gene expression levels in the introgression lines would likely both deepen our understanding of the molecular basis of the QTLs described here as well as improve our chances of improving the nutritional quality of this crop.
METHODS
Growth Conditions
The metabolite data set presented is based on field-grown introgression lines (and in 2004 their respective heterozygous counterparts; Semel et al., 2006) over two harvests, 2001 and 2004. The field trials were conducted in the Western Galilee Experimental Station in Akko, Israel. Plants were grown in a completely randomized design with one plant per m2. Seedlings were grown in greenhouses for 35 to 40 d and then transferred to the field. Twelve seedlings of each homozygous IL and heterozygous ILH (IL*M82) were transplanted as well as 70 seedlings of M82. Eight ILs were not included in the analysis of 2004 because of poor germination (ILH2-4, IL3-1, ILH3-4, ILH6-2, ILH6-2-2, ILH6-4, ILH7-2, and ILH9-3-2). Fruit was harvested when 80 to 100% of the tomatoes were red (Eshed and Zamir, 1995). The field was irrigated with 320 m3 of water per 1000 m2 of field area throughout the season. Morphological and reproductive traits have previously been described for the 2001 harvest (Schauer et al., 2006) and for the 2004 harvest (Semel et al., 2006), and variance in primary metabolite traits has also been recorded for both harvests (Schauer et al., 2006, 2008).
Secondary Metabolite Profiling
Secondary metabolites were profiled by the Waters Acquity UPLC system coupled to the Exactive Orbitrap mass detector according to the previously published protocol (Giavalisco et al., 2009). The UPLC system was equipped with a HSS T3 C18 reversed phase column (100 × 2.1 mm i.d., 1.8-µm particle size; Waters) that was operated at a temperature of 40°C. The mobile phases consisted of 0.1% formic acid in water (Solvent A) and 0.1% formic acid in acetonitrile (Solvent B). The flow rate of the mobile phase was 400 µL/min, and 2 μL of sample was loaded per injection. The UPLC was connected to an Exactive Orbitrap (Thermo Fisher Scientific) via a heated electrospray source (Thermo Fisher Scientific). The spectra were recorded using full scan mode of negative ion detection, covering a mass range from m/z 100 to 1500. The resolution was set to 25,000, and the maximum scan time was set to 250 ms. The sheath gas was set to a value of 60, while the auxiliary gas was set to 35. The transfer capillary temperature was set to 150°C, while the heater temperature was adjusted to 300°C. The spray voltage was fixed at 3 kV, with a capillary voltage and a skimmer voltage of 25 and 15 V, respectively. MS spectra were recorded from minute 0 to 19 of the UPLC gradient. Molecular masses, retention time, and associated peak intensities were extracted from the raw files using RefinerMS (version 5.3; GeneData), Metalign (Lommen, 2012), and Xcalibur software (Thermo Fisher Scientific). Metabolite identification and annotation were performed using standard compounds, literature, and tomato metabolomics databases (Moco et al., 2006; Iijima et al., 2008; Tohge and Fernie, 2009, 2010; Rohrmann et al., 2011). Data are reported in a manner compliant with the standards suggested by Fernie et al. (2011).
Heat Maps
Heat maps were calculated using the “heat map” function of the statistical software environment R, version 1.9. False color imaging was performed on the log10-transformed metabolite data. We scaled data internally on a column basis to have a mean of 0 and a sd of 1. Metabolite data were only taken in instances in which metabolite content was determined in all six replicates of an introgression line (in any given harvest).
IL Mapping
This was performed exactly as described by Schauer et al. (2008). The broad-sense heritability (H2) was estimated by mixed effect models, with random effects for genotype (75), environment (years 2001 and 2004), and genotype-environment interaction. We used the lmer function from the lme4 package in the R environment. For QTL mapping in the ILHs, each IL and ILH was compared by t test with M82 as well as with each other. If either of them was significantly different to the M82 reference genotype, the introgression was considered as harboring a QTL. Correlation analysis was also performed across the entire population by means of the Pearson correlation coefficient in order to determine possible technical artifacts.
Mode-of-Inheritance Classification and Heritability
The phenotypic effect of a QTL was considered to be the effect of the significant line (IL or ILH) and was presented as percentage of M82 (positive values for increasing QTL in which the introgression was higher than M82 and negative values for decreasing ones). If both the IL and ILH were significant but in opposite directions relative to M82, the introgression was considered as harboring two QTL: One is increasing, and the other is decreasing. The mode of inheritance of a QTL was determined according to a decision tree (Semel et al., 2006) with a custom R script. In cases in which the IL was significantly different from M82 and the ILH phenotype was in between the IL and M82, there were three possibilities: (1) If the ILH was significantly different from the IL but not from M82, it was considered recessive; (2) If the ILH differed from both parents or did not differ from either of them, it was considered additive; and (3) If the ILH differed from M82 but not from the IL, the QTL was assigned as dominant. The last possibility is where the ILH was significantly higher or lower than both its parents, in which case it was considered overdominant. Broad-sense heritability was determined based on a mixed effect model implemented by the lmer function from the lme4 R package (Bates et al., 2014).
Transcription Factor Profiling
On the basis of the mQTL analyses described above, eight introgression lines (IL6-2,IL6-3, IL8-2, IL8-2-1, IL9-1, IL10-2, IL10-3, and IL11-4-1) and the recurrent parent M82 were selected for transcription factor profiling using the Solanum lycopersicum qRT-PCR transcription factor profiling platform capable of sensitive and reproducible quantification of a total of 974 transcription factors in developing tomato fruit (Rohrmann et al., 2011). Using the same pooled plant material as used for secondary metabolite analysis, total RNA was extracted from fruit pericarp as described by Bugos et al. (1995). DNA digestion and cDNA synthesis were performed as described previously, and PCR reactions were performed using an ABI Prism 7900 HT sequence detection system exactly as described using the same reference genes (Rohrmann et al., 2011).
Mining Sequence Variation of TF Genes
Genes predicted to encode TFs were obtained by querying the full list of genes contained in each introgression against the Plant Transcription Factor Database v. 3.0 (Plant TFDB; Jin et al., 2014). The deduced amino acid sequences of TFs from S. lycopersicum cv M82 and Solanum pennellii (Bolger et al.. 2014) were then aligned using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and polymorphic sequences were submitted to SIFT (Sorting Intolerant From Tolerant, http://sift.jcvi.org/www/SIFT_BLink_submit.html) to predict the impact of amino acid substitutions on protein function (Kumar et al., 2009). Promoter regions (up to 1000 bp upstream the predicted ATG) were aligned using Blast2seq (http://blast.ncbi.nlm.nih.gov) or NEEDLE (http://www.ebi.ac.uk/Tools/psa/emboss_needle/nucleotide.html) using default settings. To detect the presence of transcription factor binding motifs, upstream sequences were analyzed with PLACE (https://sogo.dna.affrc.go.jp; Higo et al., 1999) and PlantPAN (http://plantpan.mbc.nctu.edu.tw/; Chang et al., 2008).
VIGS
Vector construction, infiltration, and fruit harvesting procedures were performed as previously described (Orzaez et al., 2006; 2009). Briefly, 280- and 310-bp fragments of Solyc06g062290 and Solyc10g085230, respectively, were amplified from tomato fruit cDNA using Gateway-compatible primers and recombined into the pDONR207 vector (Invitrogen) by the BP reaction to generate an entry clone. The Entry vector was then recombined with the pTRV2-GW destination vector using an LR reaction to produce the expression clones pTRV2-Solyc06g062290 and pTRV2-Solyc10g085230. The sequenced expression vectors were then transferred into Agrobacterium tumefaciens strain GV3101:pMP90 by electroporation. Agroinfiltration was modified from the methods described previously as follows. In order to infiltrate fruit for VIGS, MicroTom tomato was used and the pTRV1 culture and pTRV2-Solyc06g062290 or pTRV2- Solyc10g085230 were mixed in a 1:1 ratio. Fruits were labeled and injected with 0.2 to 0.5 mL of bacterial mixture through the peduncule. Agroinfiltrated fruit were marked at the breaker stage, and samples were collected at 2 weeks after breaker.
Network Generation and Visualization
To establish presence of edges, we first determined sign-dependent thresholds ensuring FDR of 0.05 based on a permutation test, whereby the profile of each metabolite is shuffled independently of the others (Storey, 2003). Therefore, the thresholds are not hard coded and depend on the data considered in the network extraction (Toubiana et al., 2013). Positive and negative thresholds were determined separately due to the asymmetric distribution of values for the correlation coefficient. To visualize the networks, we made use of the chemical compound classes (Supplemental Data Set 1), and the metabolites of each compound class were plotted based on a circle layout, which were then merged. For ease of visualization, only a portion of edges present in the network are drawn, while approximating the proportions between the degrees of the adjacent nodes. The canalization of correlation is defined as correlations that do not change over seasons, as established via Fisher z-transformation.
Network Properties and Comparisons
Networks were characterized based on their global (weighted) structural properties, including threshold for positive and negative correlations, number of nodes, number of sign-dependent edges, number of isolated nodes and connected components, as well as the size of the largest component, average degree, density, and number of network communities (based on modularity). The correspondence between the communities and the partitioning of the metabolites based on the compound classes were quantified based on the adjusted Rand index (Hubert and Arabie, 1985). The comparison of the networks was conducted based on the following properties: structural difference based on the Jaccard index of the edge sets, graph difference, relative weighted Jaccard index, and the relative weighted difference of the intersection of edge sets, as well as the average difference based on the local-global network properties, including degree, closeness, betweenness, and Eigenvalue centrality (Newman, 2003). We additionally calculated the Pearson correlation values between selected genes and TFs. All pairs of gene-TF with high (r > 0.95) and significant (P < 0.05, FDR corrected) correlations were selected in order to construct the coexpression network. Further on, the correlation values between all gene (nonregulatory genes and TFs), and metabolite profiles were estimated and all pairs of gene-metabolite with significant correlations (P < 0.05, FDR corrected) were selected for further investigations.
Accession Numbers
The NCBI and SOL numbers for genes used in this study can be found in Supplemental Data Set 10.
Supplemental Data
Supplemental Figure 1. Annotated Overlay Heat Map of the Metabolite Profiles of the ILs in Comparison with That of the Parental Control (M82) from the Individual Data Sets of 2001 and 2004.
Supplemental Figure 2. Annotated Heat Map of Secondary Metabolite Profiles of the Introgression Lines in Comparison to That of the Parental Control (Solanum lycopersicum cv M82) from the Individual Data Set of 2001.
Supplemental Figure 3. Annotated Heat Map of Secondary Metabolite Profiles of the Introgression Lines in Comparison to That of the Parental Control (Solanum lycopersicum cv M82) from the Individual Data Set of 2004.
Supplemental Figure 4. Annotated Heat Map of the Metabolite Profiles of S. lycopersicum Lines Homozygous (IL) or Heterozygous (ILH) for Chromosomal Segmental Substitution from S. pennellii.
Supplemental Figure 5. Full Annotated Figure of Distribution of the QTL Mode of Inheritance for Metabolite Accumulation.
Supplemental Figure 6. Combined Correlation Networks for the Primary and Secondary Metabolites and Phenotypic Traits.
Supplemental Figure 7. Coexpression Network Based on Person Correlation between Values of Selected Genes (Structural Genes), Transcription Factors, and Metabolite Data of Eight Introgression Lines and the Recurrent Parent M82.
Supplemental Figure 8. Relative Gene Expression of Four Candidates Genes Was Measured by qRT-PCR.
Supplemental Data Set 1. All Detected Peaks, Putative Metabolite Name Identified.
Supplemental Data Set 2. Fold Changes of Metabolite Content of the ILs Compared with the Parental Control (M82).
Supplemental Data Set 3. QTL Detected at a Stringency of 5% for Each Metabolite.
Supplemental Data Set 4. QTL Detected at a Stringency of 1% for Each Metabolite.
Supplemental Data Set 5. Heritability of All Secondary Metabolite Traits in the S. pennellii Introgression Population, and Environmental Effect (E) and E × Genotype (G)
Supplemental Data Set 6. Fold Changes of Metabolite Content of the Heterozygous (ILH) Compared with the Parental Control (M82).
Supplemental Data Set 7. Qualitative Distribution of Mode of Inheritance Showing the Numbers of Major QTL That Were Classified in Each Category for Each Chemical Compound Class.
Supplemental Data Set 8. Results of the qRT-PCR Experiment for Tomato Fruits of Eight Introgression Lines.
Supplemental Data Set 9. Total Number of eQTL per TF Family.
Supplemental Data Set 10. TF Used in qRT-PCR.
Supplemental Data Set 11. Structural Variations of Transcription Factors in Eight Introgression Lines.
Supplementary Material
Acknowledgments
We thank Lothar Willmitzer (same institute) for helpful discussion and Cathie Martin and Yang Zhang (John Innes Centre, Norwich, UK) for help with the VIGS experiment. We also thank Änne Eckardt for help in sample analysis. Work in the Zamir and Fernie laboratories is funded in the framework of Deutsche Israeli Project FE 552/12-1, which is administered by the Deutsche Forschungsgemeinschaft. This research was supported in part by a grant from the German-Israeli Cooperation Project (DIP). Research activity on secondary metabolism of T.T. is supported by the Alexander von Humboldt Foundation.
AUTHOR CONTRIBUTIONS
D.Z., Z.N., and A.R.F. designed the research. Z.N. and A.R.F. wrote the article. S.A., T.T., and R.W. performed most of the research and data analysis. F.S., N.O., J.L., S.K., P.G., T.P., and B.M.-R. performed some aspects of the research and data analysis.
Glossary
- QTL
quantitative trait locus
- mQTL
metabolite quantitative trait loci
- IL
introgression lines
- GC-MS
gas chromatography-mass spectrometry
- UPLC-FTMS
ultraperformance liquid chromatography-Fourier transform mass spectrometry
- ILH
heterozygous introgression line
- FDR
false discovery rate
- LC-MS
liquid chromatography-mass spectrometry
- TF
transcription factor
- qRT-PCR
quantitative RT-PCR
- InDel
insertion/deletion
- VIGS
virus-induced gene silencing
Footnotes
Articles can be viewed online without a subscription.
References
- Adato A., et al. (2009). Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet. 5: e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riano-Pachon D.M., Mueller-Roeber B. (2008). QuantPrime – a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005). Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Bagheri H., El-Soda M., van Oorschot I., Hanhart C., Bonnema G., Jansen-van den Bosch T., Mank R., Keurentjes J.J.B., Meng L., Wu J., Koornneef M., Aarts M.G.M. (2012). Genetic analysis of morphological traits in a new, versatile, rapid-cycling Brassica rapa recombinant inbred line population. Front. Plant Sci. 3: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D., Maechler, M., Bolker, B., Walker, S. (2014). lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-7, http://CRAN.R-project.org/package=lme4.
- Bolger A., et al. (2014). The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46: 1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugos R.C., Chiang V.L., Zhang X.H., Campbell E.R., Podila G.K., Campbell W.H. (1995). RNA isolation from plant tissues recalcitrant to extraction in guanidine. Biotechniques 19: 734–737. [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H.P., Matros A., Peterek S., Schijlen E.G.W.M., Hall R.D., Bovy A.G., Luo J., Martin C. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26: 1301–1308. [DOI] [PubMed] [Google Scholar]
- Carreno-Quintero N., Bouwmeester H.J., Keurentjes J.J. (2013). Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends Genet. 29: 41–50. [DOI] [PubMed] [Google Scholar]
- Carreno-Quintero N., Acharjee A., Maliepaard C., Bachem C.W., Mumm R., Bouwmeester H., Visser R.G., Keurentjes J.J. (2012). Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol. 158: 1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E., Ruiz-Rivero O., Peres L.E.P., Atares A., Garcia-Martinez J.L. (2012). Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 160: 1581–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.K.F., Rowe H.C., Corwin J.A., Joseph B., Kliebenstein D.J. (2011). Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.C., Lee T.Y., Huang H.D., Huang H.Y., Pan R.L. (2008). PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics 9: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Kumar R., Headland L.R., Ranjan A., Covington M.F., Ichihashi Y., Fulop D., Jiménez-Gómez J.M., Peng J., Maloof J.N., Sinha N.R. (2013). A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25: 2465–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong B., Barrero L.S., Tanksley S.D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40: 800–804. [DOI] [PubMed] [Google Scholar]
- de Godoy F., et al. (2013). Galacturonosyltransferase 4 silencing alters pectin composition and carbon partitioning in tomato. J. Exp. Bot. 64: 2449–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaubhadel S., McGarvey B.D., Williams R., Gijzen M. (2003). Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol. Biol. 53: 733–743. [DOI] [PubMed] [Google Scholar]
- Di Matteo A., Ruggieri V., Sacco A., Rigano M.M., Carriero F., Bolger A., Fernie A.R., Frusciante L., Barone A. (2013). Identification of candidate genes for phenolics accumulation in tomato fruit. Plant Sci. 205-206: 87–96. [DOI] [PubMed] [Google Scholar]
- Do P.T., Prudent M., Sulpice R., Causse M., Fernie A.R. (2010). The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol. 154: 1128–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Zamir D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Stitt M. (2012). On the discordance of metabolomics with proteomics and transcriptomics: coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol. 158: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Aharoni A., Willmitzer L., Stitt M., Tohge T., Kopka J., Carroll A.J., Saito K., Fraser P.D., DeLuca V. (2011). Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Tadmor Y., Zamir D. (2006). Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 9: 196–202. [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Trethewey R.N., Krotzky A.J., Willmitzer L. (2004). Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5: 763–769. [DOI] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science 320: 543–547. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick T.B., Basset G.J., Borel P., Carrari F., DellaPenna D., Fraser P.D., Hellmann H., Osorio S., Rothan C., Valpuesta V., Caris-Veyrat C., Fernie A.R. (2013). Vitamin deficiencies in humans: can plant sciences help? Plant Cell 24: 395–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T. (2005). The evolutionary genetics of canalization. Q. Rev. Biol. 80: 287–316. [DOI] [PubMed] [Google Scholar]
- Frary A., Nesbitt T.C., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Tanksley S.D. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Enfissi E.M., Goodfellow M., Eguchi T., Bramley P.M. (2007). Metabolite profiling of plant carotenoids using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Plant J. 49: 552–564. [DOI] [PubMed] [Google Scholar]
- Fridman E., Carrari F., Liu Y.S., Fernie A.R., Zamir D. (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Fu J., et al. (2009). System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat. Genet. 41: 166–167. [DOI] [PubMed] [Google Scholar]
- Giavalisco P., Köhl K., Hummel J., Seiwert B., Willmitzer L. (2009). 13C isotope-labeled metabolomes allowing for improved compound annotation and relative quantification in liquid chromatography-mass spectrometry-based metabolomic research. Anal. Chem. 81: 6546–6551. [DOI] [PubMed] [Google Scholar]
- Gong L., Chen W., Gao Y., Liu X., Zhang H., Xu C., Yu S., Zhang Q., Luo J. (2013). Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. USA 110: 20320–20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Vigil E., Hufnagel D.E., Kim J., Last R.L., Barry C.S. (2012). Evolution of TPS20-related terpene synthases influences chemical diversity in the glandular trichomes of the wild tomato relative Solanum habrochaites. Plant J. 71: 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenenboom M., Gomez-Roldan V., Stigter H., Astola L., van Daelen R., Beekwilder J., Bovy A., Hall R., Molenaar J. (2013). The flavonoid pathway in tomato seedlings: transcript abundance and the modeling of metabolite dynamics. PLoS ONE 8: e68960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.F., Houle D. (2008). Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 21: 1201–1219. [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., et al. (2014). Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci. Rep. 4: 5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert L., Arabie P. (1985). Comparing partitions. Journal of Classification 2: 193–218. [Google Scholar]
- Ichihashi Y., Sinha N.R. (2014). From genome to phenome and back in tomato. Curr. Opin. Plant Biol. 18: 9–15. [DOI] [PubMed] [Google Scholar]
- Iijima Y., et al. (2008). Metabolite annotations based on the integration of mass spectral information. Plant J. 54: 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y., Watanabe B., Sasaki R., Takenaka M., Ono H., Sakurai N., Umemoto N., Suzuki H., Shibata D., Aoki K. (2013). Steroidal glycoalkaloid profiling and structures of glycoalkaloids in wild tomato fruit. Phytochemistry 95: 145–157. [DOI] [PubMed] [Google Scholar]
- Itkin M., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179. [DOI] [PubMed] [Google Scholar]
- Itkin M., et al. (2011). GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23: 4507–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.C. (1993). Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G., Luo J. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42: D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Corwin J.A., Li B., Atwell S., Kliebenstein D.J. (2013). Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2: e00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J.J., Fu J., de Vos C.H., Lommen A., Hall R.D., Bino R.J., van der Plas L.H., Jansen R.C., Vreugdenhil D., Koornneef M. (2006). The genetics of plant metabolism. Nat. Genet. 38: 842–849. [DOI] [PubMed] [Google Scholar]
- Khan S.A., et al. (2012). Genetic analysis of metabolites in apple fruits indicates an mQTL hotspot for phenolic compounds on linkage group 16. J. Exp. Bot. 63: 2895–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Matsuba Y., Ning J., Schilmiller A., Hammar D., Jones D., Pichersky E., and Last R. (2014). Analysis of natural and induced variation in tomato glandular trichome flavonoids identifies a gene not present in the reference genome. Plant Cell 26: 3272–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Nguyen N.H., Jeong C.Y., Nguyen N.T., Hong S.W., Lee H. (2013). Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 170: 1461–1465. [DOI] [PubMed] [Google Scholar]
- Kliebenstein D., Lambrix V., Reichelt M., Gershenzon J., Mitchell-Olds T. (2001a). Gene duplication in the diversification of secondary metabolism: Tandem 2-oxoglutarate–dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Kroymann J., Brown P., Figuth A., Pedersen D., Gershenzon J., Mitchell-Olds T. (2001b). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126: 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D., et al. (2013). Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 110: E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Vreugdenhil D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55: 141–172. [DOI] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P.C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Li X., Huang L., Zhang Y., Ouyang Z., Hong Y., Zhang H., Li D., Song F. (2014). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 14: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Hamilton W.D.O., Merryweather A. (1996). Cloning and initial characterization of 14 myb-related cDNAs from tomato (Lycopersicon esculentum cv Ailsa Craig). Plant Mol. Biol. 30: 1009–1020. [DOI] [PubMed] [Google Scholar]
- Lippman, Z.B., Semel, Y., Zamir, D. (2007). An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr. Opin. Gen. Dev. 545–552. [DOI] [PubMed]
- Lisec J., Römisch-Margl L., Nikoloski Z., Piepho H.P., Giavalisco P., Selbig J., Gierl A., Willmitzer L. (2011). Corn hybrids display lower metabolite variability and complex metabolite inheritance patterns. Plant J. 68: 326–336. [DOI] [PubMed] [Google Scholar]
- Lisec J., Steinfath M., Meyer R.C., Selbig J., Melchinger A.E., Willmitzer L., Altmann T. (2009). Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. Plant J. 59: 777–788. [DOI] [PubMed] [Google Scholar]
- Liu Y.S., Gur A., Ronen G., Causse M., Damidaux R., Buret M., Hirschberg J., Zamir D. (2003). There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol. J. 1: 195–207. [DOI] [PubMed] [Google Scholar]
- Lommen A. (2012). Data (pre-)processing of nominal and accurate mass LC-MS or GC-MS data using MetAlign. Methods Mol. Biol. 860: 229–253. [DOI] [PubMed] [Google Scholar]
- Luo J., Butelli E., Hill L., Parr A., Niggeweg R., Bailey P., Weisshaar B., Martin C. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56: 316–326. [DOI] [PubMed] [Google Scholar]
- Mageroy M.H., Tieman D.M., Floystad A., Taylor M.G., Klee H.J. (2012). A Solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. Plant J. 69: 1043–1051. [DOI] [PubMed] [Google Scholar]
- Maloney G.S., Kochevenko A., Tieman D.M., Tohge T., Krieger U., Zamir D., Taylor M.G., Fernie A.R., Klee H.J. (2010). Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol. 153: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara C.D., Huang T.B., Irish V.F. (2010). The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 22: 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. (2013). The interface between plant metabolic engineering and human health. Curr. Opin. Biotechnol. 24: 344–353. [DOI] [PubMed] [Google Scholar]
- Martin C., Butelli E., Petroni K., Tonelli C. (2011). How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Ellis N., Rook F. (2010). Do transcription factors play special roles in adaptive variation? Plant Physiol. 154: 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu S., Cin V.D., Fei Z., Li H., Bliss P., Taylor M.G., Klee H.J., Tieman D.M. (2009). Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. J. Exp. Bot. 60: 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba Y., et al. (2013). Evolution of a complex locus for terpene biosynthesis in solanum. Plant Cell 25: 2022–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F., Okazaki Y., Oikawa A., Kusano M., Nakabayashi R., Kikuchi J., Yonemaru J., Ebana K., Yano M., Saito K. (2012). Dissection of genotype-phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J. 70: 624–636. [DOI] [PubMed] [Google Scholar]
- Mignone, F., Gissi, C., Liuni, S., Pesole, G. (2002). Untranslated regions of mRNAs. Genome Biol. 3: reviews0004. [DOI] [PMC free article] [PubMed]
- Milborrow B.W. (1998). A biochemical mechanism for hybrid vigour. J. Exp. Bot. 49: 1063–1071. [Google Scholar]
- Mintz-Oron S., Mandel T., Rogachev I., Feldberg L., Lotan O., Yativ M., Wang Z., Jetter R., Venger I., Adato A., Aharoni A. (2008). Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147: 823–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S., Bino R.J., Vorst O., Verhoeven H.A., de Groot J., van Beek T.A., Vervoort J., de Vos C.H.R. (2006). A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 141: 1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Goeminne G., Storme V., Sterck L., Ralph J., Coppieters W., Breyne P., Steenackers M., Georges M., Messens E., Boerjan W. (2006). Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J. 47: 224–237. [DOI] [PubMed] [Google Scholar]
- Meyer R.S., DuVal A.E., Jensen H.R. (2012). Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196: 29–48. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R., Yonekura Sakakibara K., Urano K., Suzuki M., Yamada Y. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.J. (2003). The structure and function of complex networks. SIAM Rev. 45: 167–256. [Google Scholar]
- Niggeweg R., Michael A.J., Martin C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22: 746–754. [DOI] [PubMed] [Google Scholar]
- O'Neill S.D., Tong Y., Spörlein B., Gorkmann G., Yoder J. (1990). Molecular genetic analysis of CHALCONE SYNTHASE in Lycopersicon esculentum and an anthocyanin-deficient mutant. Mol. Gen. Genet. 224: 279–288. [DOI] [PubMed] [Google Scholar]
- O’Reilly-Wapstra J.M., Potts B.M., McArthur C., Davies N.W., Tilyard P. (2005). Inheritance of resistance to mammalian herbivores and of plant defensive chemistry in an Eucalyptus species. J. Chem. Ecol. 31: 357–375. [DOI] [PubMed] [Google Scholar]
- Orzaez D., Medina A., Torre S., Fernández-Moreno J.P., Rambla J.L., Fernández-Del-Carmen A., Butelli E., Martin C., Granell A. (2009). A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 150: 1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzaez D., Mirabel S., Wieland W.H., Granell A. (2006). Agroinjection of tomato fruits. A tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol. 140: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B., Schluttenhofer C., Wu Y., Pattanaik S., Yuan L. (2013). Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta 1829: 1236–1247. [DOI] [PubMed] [Google Scholar]
- Perez-Fons L., Wells T., Corol D.I., Ward J.L., Gerrish C., Beale M.H., Seymour G.B., Bramley P.M., Fraser P.D. (2014). A genome-wide metabolomic resource for tomato fruit from Solanum pennellii. Sci. Rep. 4: 3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18: 111–119. [DOI] [PubMed] [Google Scholar]
- Quadrana L., et al. (2014). Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 5: 3027. [DOI] [PubMed] [Google Scholar]
- Quadrana L., Almeida J., Otaiza S.N., Duffy T., Corrêa da Silva J.V., de Godoy F., Asís R., Bermúdez L., Fernie A.R., Carrari F., Rossi M. (2013). Transcriptional regulation of tocopherol biosynthesis in tomato. Plant Mol. Biol. 81: 309–325. [DOI] [PubMed] [Google Scholar]
- Rambla J.L., Tikunov Y.M., Monforte A.J., Bovy A.G., Granell A. (2014). The expanded tomato fruit volatile landscape. J. Exp. Bot. 65: 4613–4623. [DOI] [PubMed] [Google Scholar]
- Rohrmann J., et al. (2011). Combined transcription factor profiling, microarray analysis and metabolite profiling reveals the transcriptional control of metabolic shifts occurring during tomato fruit development. Plant J. 68: 999–1013. [DOI] [PubMed] [Google Scholar]
- Routaboul J.M., Dubos C., Beck G., Marquis C., Bidzinski P., Loudet O., Lepiniec L. (2012). Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 63: 3749–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.C., Hansen B.G., Halkier B.A., Kliebenstein D.J. (2008). Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.L., Patrick J.W., Bouzayen M., Osorio S., Fernie A.R. (2012). Molecular regulation of seed and fruit set. Trends Plant Sci. 17: 656–665. [DOI] [PubMed] [Google Scholar]
- Sallaud C., Rontein D., Onillon S., Jabès F., Duffé P., Giacalone C., Thoraval S., Escoffier C., Herbette G., Leonhardt N., Causse M., Tissier A. (2009). A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21: 301–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage C., Segura V., Bauchet G., Stevens R., Do P.T., Nikoloski Z., Fernie A.R., Causse M. (2014). Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 165: 1120–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Balbo I., Steinfath M., Repsilber D., Selbig J., Pleban T., Zamir D., Fernie A.R. (2008). Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24: 447–454. [DOI] [PubMed] [Google Scholar]
- Schauer N., Zamir D., Fernie A.R. (2005). Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J. Exp. Bot. 56: 297–307. [DOI] [PubMed] [Google Scholar]
- Schilmiller A., Shi F., Kim J., Charbonneau A.L., Holmes D., Daniel Jones A., Last R.L. (2010). Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 62: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Charbonneau A.L., Last R.L. (2012). Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc. Natl. Acad. Sci. USA 109: 16377–16382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwahn K., de Souza L.P., Fernie A.R., Tohge T. (2014). Metabolomics-assisted refinement of the pathways of steroidal glycoalkaloid biosynthesis in the tomato clade. J. Integr. Plant Biol. 56: 864–875. [DOI] [PubMed] [Google Scholar]
- Semel Y., Nissenbaum J., Menda N., Zinder M., Krieger U., Issman N., Pleban T., Lippman Z., Gur A., Zamir D. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103: 12981–12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.X., et al. (2013). The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol. 197: 468–480. [DOI] [PubMed] [Google Scholar]
- Stevens R., Buret M., Duffé P., Garchery C., Baldet P., Rothan C., Causse M. (2007). Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol. 143: 1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D. (2003). The positive false discovery rate: a Bayesian interpretation and the q-valie. Ann. Stat. 31: 2013–2035. [Google Scholar]
- Tieman D., Taylor M., Schauer N., Fernie A.R., Hanson A.D., Klee H.J. (2006). Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 103: 8287–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y.M., et al. (2013). Non-smoky glycosyltransferase1 prevents the release of smoky aroma from tomato fruit. Plant Cell 25: 3067–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S., Florian A., Jahnke K., Nunes-Nesi A., Fernie A.R., Bauwe H. (2011). The hydroxypyruvate-reducing system in Arabidopsis: multiple enzymes for the same end. Plant Physiol. 155: 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2009). Web-based resources for mass-spectrometry-based metabolomics: a user’s guide. Phytochemistry 70: 450–456. [DOI] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2010). Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5: 1210–1227. [DOI] [PubMed] [Google Scholar]
- Tohge T., Alseekh S., Fernie A.R. (2014). On the regulation and function of secondary metabolism during fruit development and ripening. J. Exp. Bot. 65: 4599–4611. [DOI] [PubMed] [Google Scholar]
- Tohge T., Ramos M.S., Nunes-Nesi A., Mutwil M., Giavalisco P., Steinhauser D., Schellenberg M., Willmitzer L., Persson S., Martinoia E., Fernie A.R. (2011). Towards the storage metabolome: profiling the barley vacuole. Plant Physiol. 157: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana D., Fernie A.R., Nikoloski Z., Fait A. (2013). Network analysis: tackling complex data to study plant metabolism. Trends Biotechnol. 31: 29–36. [DOI] [PubMed] [Google Scholar]
- Toubiana D., Semel Y., Tohge T., Beleggia R., Cattivelli L., Rosental L., Nikoloski Z., Zamir D., Fernie A.R., Fait A. (2012). Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 8: e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L., Memelink J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297. [DOI] [PubMed] [Google Scholar]
- Waddington C.H. (1942). Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Wahyuni Y., Stahl-Hermes V., Ballester A.R., de Vos R.C.H., Voorrips R.E., Maharijaya A., Molthoff J., Zamora M.V., Sudarmonowati E., Arisi A.C., Bino R.J., Bovy A.G. (2014). Genetic mapping of semi-polar metabolites in pepper fruits (Capsicum sp.): towards unravelling the molecular regulation of flavonoid quantitative trait loci. Mol. Breed. 33: 503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z.G., Frasse P., Delalande C., Regad F., Chaabouni S., Latche A., Pech J.C., Bouzayen M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Li D., Li X., Gao Y., Li W., Li H., Liu J., Liu H., Chen W., Luo J., Yan J. (2014). Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 5: 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell A.M., Rowe H.C., Hansen B.G., Ticconi C., Halkier B.A., Kliebenstein D.J. (2007). Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 3: 1687–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yeats T.H., Buda G.J., Wang Z., Chehanovsky N., Moyle L.C., Jetter R., Schaffer A.A., Rose J.K. (2012). The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J. 69: 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xing L., Wang X., Hou Y.J., Gao J., Wang P., Duan C.G., Zhu X., Zhu J.K. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7: ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.