Feedback regulation of the key gene in the ascorbate biosynthesis pathway, GDP Galactose Phosphorylase, is controlled at the level of translation by an upstream open reading frame with a noncanonical initiation codon.
Abstract
Ascorbate (vitamin C) is an essential antioxidant and enzyme cofactor in both plants and animals. Ascorbate concentration is tightly regulated in plants, partly to respond to stress. Here, we demonstrate that ascorbate concentrations are determined via the posttranscriptional repression of GDP-l-galactose phosphorylase (GGP), a major control enzyme in the ascorbate biosynthesis pathway. This regulation requires a cis-acting upstream open reading frame (uORF) that represses the translation of the downstream GGP open reading frame under high ascorbate concentration. Disruption of this uORF stops the ascorbate feedback regulation of translation and results in increased ascorbate concentrations in leaves. The uORF is predicted to initiate at a noncanonical codon (ACG rather than AUG) and encode a 60- to 65-residue peptide. Analysis of ribosome protection data from Arabidopsis thaliana showed colocation of high levels of ribosomes with both the uORF and the main coding sequence of GGP. Together, our data indicate that the noncanonical uORF is translated and encodes a peptide that functions in the ascorbate inhibition of translation. This posttranslational regulation of ascorbate is likely an ancient mechanism of control as the uORF is conserved in GGP genes from mosses to angiosperms.
INTRODUCTION
Ascorbate (vitamin C) is an essential biochemical found in most living organisms; it has a central role in regulating the redox potential of the cell (Asensi-Fabado and Munné-Bosch, 2010; Foyer and Noctor, 2011) as well as serving as an enzyme cofactor (Mandl et al., 2009). Humans lack the ability to synthesize ascorbate and obtain most of their vitamin C from plant sources. As most food is relatively low in vitamin C, there is an opportunity to improve human health by developing plant varieties with higher levels of ascorbate. To better understand how this can be achieved, we have investigated how the ascorbate biosynthetic pathway is regulated.
Ascorbate concentrations are regulated according to demand; for example, leaf ascorbate concentrations increase under high light intensities (Gatzek et al., 2002; Bartoli et al., 2006; Dowdle et al., 2007; Yabuta et al., 2007; Bartoli et al., 2009; Gao et al., 2011) when the need for ascorbate is greatest. This is due to increased light-driven production of reactive oxygen species in photosynthesis, and this response to light is associated with changes in gene expression for several l-galactose pathway genes (Figure 1) (Tabata et al., 2002; Dowdle et al., 2007; Yabuta et al., 2007; Gao et al., 2011; Massot et al., 2012). In nonphotosynthetic tissues, such as fruit, ascorbate concentrations are relatively stable within a cultivar, although ascorbate concentration does vary between cultivars (Ferguson and MacRae, 1992; Davey et al., 2006; Bulley et al., 2009) and this is related to changes in gene expression (Bulley et al., 2009).
Figure 1.
The l-Galactose Pathway of Ascorbate Biosynthesis.
PMI, phosphomannose isomerase; PMM, phosphomanno mutase; GMP, GDP mannose pyrophosphorylase; GME, GDP mannose epimerase; GGP, GDP galactose phosphorylase; GPP, galactose-1-P phosphatase; GDH, galactose dehydrogenase; GLDH, galactono lactone dehydrogenase.
Little is known about how ascorbate concentrations are rapidly regulated in response to environmental stress. Previous studies have concentrated on changes in gene expression (Gatzek et al., 2002; Tabata et al., 2002; Dowdle et al., 2007; Yabuta et al., 2007; Gao et al., 2011; Massot et al., 2012) often over a day or longer. We have previously shown that the enzyme GDP-l-galactose phosphorylase (GGP) (encoded by the gene GGP, also known as VTC2) is at a central control point of vitamin C biosynthesis as the first committed step; overexpression of this gene in plants results in a significant increase in tissue ascorbate concentrations (Laing et al., 2007; Bulley et al., 2009, 2012), suggesting it may serve a regulatory role, possibly balancing fluxes between ascorbate and GDP-l-fucose (Bonin et al., 1997). No other single genes in the l-galactose pathway have a significant effect on ascorbate (Bulley et al., 2012; Zhou et al., 2012), although GDP mannose epimerase (GME) acts synergistically with GGP to increase leaf ascorbate still further (Bulley et al., 2009). In addition, during fruit development, transcript levels of GGP and GME parallel the increase in ascorbate (Bulley et al., 2009). Analysis of Arabidopsis thaliana microarray data (e.g., from the Arabidopsis eFP browser, http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) and other data (Dowdle et al., 2007) show that transcripts of GGP undergo strong circadian rhythms with maximal expression predawn, presumably so transcript is available when photosynthesis begins with increasing light. Furthermore, GGP protein itself has been observed in the nucleus (Müller-Moulé, 2008), suggesting it may serve to control gene transcription. Thus, we focused on GGP and its gene as a likely control point for ascorbate biosynthesis.
In this article, we show that ascorbate, or a metabolite closely connected to ascorbate, downregulates the translation of the key regulatory gene, GGP, in ascorbate biosynthesis, setting up a feedback loop to regulate ascorbate concentrations. We show that this regulation occurs through a conserved noncanonical upstream open reading frame (uORF) in the long 5′ untranslated region (UTR) of GGP and takes place at the posttranscriptional level. We also present evidence that the control mechanism involves ribosome stalling on the uORF.
RESULTS
High Ascorbate Downregulates GGP Expression
To investigate if GGP is regulated by ascorbate, we fused the kiwifruit (Actinidia eriantha) GGP promoter including its 5′UTR to the luciferase (LUC) reporter gene and expressed the construct transiently in Nicotiana benthamiana leaves. We attempted to manipulate leaf ascorbate by injecting ascorbate or its precursors l-galactose and l-galactono lactone into attached leaves or feeding detached leaves through the petiole. However, sustained changes in leaf ascorbate were not observed over the 3 to 7 d needed for the transient expression of luciferase from the assay constructs. Instead, we raised leaf ascorbate by coexpressing the coding sequence (CDS) of kiwifruit (Actinidia chinensis) GGP (without its 5′ or 3′UTR) under a strong constitutive promoter or lowered leaf ascorbate by expressing a N. benthamiana endogenous GGP hairpin to silence the N. benthamiana endogenous GGP gene.
Ascorbate concentrations in control leaves of 24 mg/100 g fresh weight (FW) (equivalent to 1.35 µmol/g FW) are typical of glasshouse-grown Nicotiana species including N. benthamiana (Badejo et al., 2009; Hakmaoui et al., 2012). A doubling of ascorbate concentration from ∼20 mg/100 g FW to ∼40 mg/100 g FW was sufficient to reduce the relative LUC activity by 50%, and when ascorbate was increased above 100 mg/100 g FW, >90% of LUC activity was abolished (Figure 2A). Additionally, when the ascorbate content was reduced to ∼7 mg/100 g FW using GGP hairpin RNA interference (RNAi) constructs, LUC activity increased approximately by 50%. By contrast, high ascorbate had no inhibitory effect on LUC activity driven by a control promoter for a gene unrelated to ascorbate metabolism (TT8, an Arabidopsis bHLH transcription factor controlling polyphenolic biosynthesis), and this reduction in ascorbate content by RNAi actually decreased the activity of the control TT8 promoter, possibly due to stress induced by low ascorbate (Figure 2B).
Figure 2.
High Ascorbate Reduces Expression of GGP Promoter-Driven LUC Activity.
(A) LUC/REN ratio as a function of leaf ascorbate concentration for LUC expression driven by the GGP promoter.
(B) LUC/REN ratio as a function of leaf ascorbate concentration for LUC expression driven by the TT8 promoter.
(C) Effect of high ascorbate concentration on the amount of LUC protein as measured by an anti-LUC antibody in transiently transformed tobacco leaves driven by either the GGP promoter or the control TT8 promoter. The ascorbate concentration in the lanes marked +, where 35S-driven GGP was cotransformed, was 55 to 63 mg/100 g FW, while in the lanes without added GGP, it was 21 to 30 mg/100 g FW. Other details can be found in Methods.
(D) Comparison of the response of the reporter gene to ascorbate for full promoter (promoter and 5′UTR) constructs from kiwifruit and Arabidopsis GGP.
Note, in (A) to (D), the GGP overexpression construct was added to increase ascorbate and results in a range of ascorbate concentrations due to variation in transformation efficiency in different leaves. Error bars are se of the mean. Other details can be found in Methods.
Additional controls demonstrated that this downregulation of LUC activity by ascorbate was independent of the level of expression of the transgenes (Supplemental Table 1A), did not occur in a range of other promoters (Supplemental Table 1B), and was unaffected by expression of other unrelated genes (Supplemental Table 1C). The same effect was also seen with the GGP promoter and 5′UTR from Arabidopsis (VTC2) (Figure 2D), showing the mechanism was conserved between species.
It is possible that the effect on LUC expression is mediated through either increased ascorbate or through the feedback regulation via the GGP protein itself, as it has been found in the nucleus (Müller-Moulé, 2008). To separate the effects of ascorbate from a possible regulatory role of the GGP protein itself, we expressed GGP and GME separately and together. When the two genes are expressed together, there is a strong synergistic stimulation of ascorbate concentration (Bulley et al., 2009). Thus, by varying the infiltrated ratios of these two genes, ascorbate concentrations can be manipulated independently of the amount of GGP protein (Figure 3). LUC activity was negatively related to concentrations of ascorbate and not to the amount of GGP gene infiltrated (Figure 3C), showing that either ascorbate or a related metabolite, and not the GGP protein, is the factor reducing LUC activity.
Figure 3.
Ascorbate Concentration Not the GGP Enzyme Determines the Downregulation of GGP Promoter-Driven Reporter Gene by Ascorbate.
(A) The effect of different combinations of GGP and GME on the LUC/REN ratio.
(B) The effect of different combinations of GGP and GME on the ascorbate concentration. In (A) and (B), columns with the same letters are not significantly different at P < 0.05.
(C) The relationship between the individual leaf LUC/REN ratios and leaf ascorbate. In the key to (C), GGP/3 refers to one-third relative amount of GGP injected into the leaf. Note that the axes are on log scales.
Ascorbate concentration was varied independently of the amount of GGP enzyme used to increase the leaf ascorbate using GME to further enhance ascorbate at variable concentrations of GGP. Other details can be found in Methods.
Ascorbate Regulation of GGP Involves Sequences within the 5′UTR and Is at the Translational Level
To test whether the effect of ascorbate was mediated by the promoter or the 5′UTR (Supplemental Figure 1A), we undertook three experiments. First, we swapped the 5′UTR regions between the GGP and TT8 promoters. We transiently expressed these in leaves and measured the relative LUC activity. Increased ascorbate reduced the LUC activity (up to 6-fold) only when the 5′UTR from GGP was present (Supplemental Table 2). Second, we replaced the GGP promoter with a 35S promoter, leaving the GGP 5′UTR in place. Expression from this construct also was strongly inhibited by increased ascorbate (Figure 4A). Third, deleting two conserved regions within the 5′UTR (from 387 to 432 bp and 514 to 597 bp as well as 387 to 597 bp; Supplemental Figures 2A and 2B) caused the loss of the ability of ascorbate to downregulate the reporter gene expression (Figure 4B). These experiments show that the 5′UTR is necessary and sufficient for downregulation by ascorbate.
Figure 4.
The Effect of Ascorbate Does Not Depend on the Promoter but Only on the 5′UTR.
(A) The ascorbate inhibition is independent of the promoter. In the WT columns, the full promoter and 5′UTR of GGP drove the expression of LUC. In the 35S: columns, the GGP promoter was replaced with the 35S promoter leaving the GGP 5′UTR in place. Ascorbate averaged 27.3 ± 2 mg/100 g FW in the treatments without added GGP and 49.3 ± 8 mg/100 g FW in the “+” treatment with added GGP. Data are normalized to the wild type = 1.0.
(B) Deletions in the 5′UTR region of GGP negate the ascorbate inhibition of translation. Deletions 2 and 3 are as labeled in Supplemental Figures 2A and 2B, while deletion 1 is the deletion between the start of deletion 2 and the end of deletion 3. Ascorbate averaged 19.8 ± 1.3 mg/100 g FW in the treatments without added GGP and 134 ± 21 mg/100 g FW in the “+” treatment.
Columns with different letters are significantly different at the 0.05 level. Error bars are se of the mean. “+” refers to cotransformation with the 35S-driven GGP CDS in order to drive up the ascorbate levels.
To investigate whether ascorbate control was at the transcriptional or posttranscriptional level, we measured transcript concentrations of the reporter gene construct. Our data showed no effect of ascorbate on the concentrations of LUC mRNA (Table 1), but a strong effect on LUC activity (Table 1) and on LUC protein (Figure 2C), indicating that ascorbate, directly or indirectly, acts through the 5′UTR to control the translation of GGP. The TT8 control showed no effect of ascorbate on mRNA or LUC activity or protein level.
Table 1. Effect of Ascorbate Concentration on the RNA Concentrations for LUC Driven either by the GGP Promoter or TT8, a Control Gene.
| Promoter/Treatment | Gene Expression | LUC/REN | Ascorbate mg/100 g FW |
|---|---|---|---|
| GGP/low ascorbate | 6.0E-02 ± 8.8E-03 | 1.08 ± 0.05 | 25.3 ± 0.93 |
| GGP/high ascorbate | 5.3E-02 ± 7.4E-03 | 0.15 ± 0.03 | 92.4 ± 7.1 |
| TT8/low ascorbate | 8.5E-01 ± 5.7E-02 | 19.9 ± 0.59 | 26.1 ± 6.3 |
| TT8/high ascorbate | 9.3E-01 ± 1.1E-01 | 16.3 ± 0.83 | 84.6 ± 4.7 |
Accumulation of LUC mRNA was measured relative to the accumulation of REN mRNA in the same RNA preparation. Values are the mean of three biological replicates, each involving three combined leaves. Standard errors are shown. Within each promoter pair, there was no significant difference in gene expression for both promoters or the LUC activity for the TT8 promoter. The change in LUC activity for the GGP promoter was significant (P < 0.001), as were the changes in ascorbate for both promoters (P < 0.003).
To verify that the 5′UTR acts directly on GGP translation to affect leaf ascorbate concentrations, we made constructs in which a 35S promoter was used to express the GGP CDS with either the intact GGP 5′UTR or with only the last 37 bp immediately upstream of the CDS. Both constructs still had the short 35S 5′UTR (see Methods for details) and enhanced leaf ascorbate in the transient system, but the construct without the 5′UTR had ∼30% more ascorbate than the construct with the 5′UTR (Supplemental Figure 3). Furthermore, coinfiltrating GME (with GGP) into the leaf to increase the ascorbate even more than with GGP alone (Bulley et al., 2009) resulted in over 2-fold higher ascorbate in the construct without the 5′UTR compared with the construct with the 5′UTR. Thus, in high ascorbate conditions, the GGP 5′UTR limits both GGP production and ascorbate biosynthesis. Removal of this regulation therefore could provide a way to generate plants with higher ascorbate concentrations.
The 5′UTR of GGP Is Unusually Long and Contains a Noncanonical uORF
Given that the effect of ascorbate was mediated through the 5′UTR region of GGP, we examined its properties. GGP is unusual in having a long 5′UTR, over 500 bp in many species, with strongly conserved elements (Supplemental Figure 1A). Aligned GGP 5′UTRs from different species, including an alga and two mosses, revealed the potential presence of a highly conserved uORF encoding a 60- to 65-amino acid peptide that likely initiates at a noncanonical ACG initiation codon in all plants listed except the algae (Figure 5; Supplemental Figure 1B). While only a few examples of noncanonical translation initiation have been described in plants, it has been shown that a good Kozak consensus sequence around the noncanonical initiation site is required for efficient translation (i.e., a purine at −3 and a G at +4) (Wamboldt et al., 2009; Simpson et al., 2010) (Supplemental Figure 1 and Supplemental Table 3). Only the ACG codon lies in such a consensus with G at +4 and an A at −3 present in all plants. This noncanonical uORF was by far the best conserved uORF found in the 5′UTR (see later) and analyzing the DNA sequence upstream of the proposed ACG start codon to the first uORF in-frame stop codon showed poor conservation of peptide sequence and variable length upstream of the proposed ACG start codon (Supplemental Table 3).
Figure 5.
The Predicted Peptide Sequences of the Noncanonical uORF in the 5′UTR of GGP Is Conserved over a Wide Range of Species.
(A) Alignment of the dicotyledonous plants.
(B) Alignment line up that extends from algae to angiosperms.
Alignment was done using ClustalX (Thompson et al., 1997) as executed by Vector NTI. The suffixes are (dicotyledonous plants) as follows: Ad is Actinidia deliciosa; Cs is Cucumis sativus; Fv is Fragaria vesca; Gm is Glycine max; Md is Malus x domestica; Mt is Medicago truncatula; Nb is Nicotiana benthamiana; Pt is Populus trichocarpa; Sl is Solanum lycospersicum; and Vv is Vitis vinifera. The two sequences starting with At are from Arabidopsis. Suffices for other taxa are as follows: Cr, Chlamydomonas reinhardtii; Ps, Picea sitchensis; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; and Zm, Zea mays. The alphanumerical prefix before each name is the GenBank accession number except for SGN-U513234, which is from the Sol Genomics Network (http://solgenomics.net/organism/Nicotiana_benthamiana/genome). The genomic sequence of strawberry came from published resources (Shulaev et al., 2011).
To test if the uORF with the noncanonical initiation codon is required for ascorbate-dependent regulation of the GGP gene, we mutated the putative ACG initiation codon to TCG. We also separately changed the codon for a nonconserved Gly at residue 5 (GGA) into a stop codon (TGA) to create an early-terminating version of the uORF. Both these constructs abolished ascorbate downregulation of LUC activity (Figure 6), while significantly increasing LUC activity. To further examine the requirements of the uORF, we mutated a highly conserved His (CAC codon) at residue 30 to Leu (CTC). Again, this abolished ascorbate-dependent regulation (Figure 6). Interestingly, changing the start ACG of the uORF to an ATG almost abolished all LUC activity. Lastly, to provide evidence that it is only the peptide encoded by the uORF that is required for ascorbate control of GGP translation, changing the codon usage in the uORF region of the 5′UTR (Supplemental Figure 4) without changing the amino acid sequence did not disrupt repression by ascorbate. Furthermore, analysis of sequenced Arabidopsis revealed that of the 50 single nucleotide polymorphisms within the GGP 5′UTR (506 bp) only seven were within the uORF (193 bp), and none of these altered the uORF peptide sequence. This supports the observation that the most conserved the part of the 5′UTR is in the uORF sequence. Together, the results of these experiments demonstrate that the peptide encoded by the uORF, and not the nucleic acid sequence, is essential for the ascorbate-mediated regulation of GGP translation.
Figure 6.
Downregulation of GGP Promoter-Driven LUC Expression by Ascorbate Depends on a Noncanonical uORF.
The diagram underneath the figure shows the changes. “+” refers to the addition of 35S-driven GGP to the leaf to increase ascorbate. “1” is the synthesized wild-type variant 5′UTR with the wild-type promoter of GGP; “2” had the initiating ACG of the uORF changed to a TCG to make it no longer a start codon; “3” had the first His in a highly conserved region of the uORF (Figure 2) changed to a Leu; “4” was a combination of 3 and 4; “5” had the uORF truncated after four residues by insertion of a stop codon; “6” had 38% of the codons changed in the uORF without changing the amino acid sequence; “7” had the noncanonical start codon in the uORF changed to ATG, encoding a Met. 35S-driven GGP was added to manipulate the ascorbate concentrations (indicated by a +) and an RNAi vector was used to lower ascorbate (ko) as described in Methods. Error bars are the se of the mean. Letters above columns indicate whether there were significant differences between values at P < 0.05. The treatments “1 +” and “6 +” had a significantly lower LUC activity than their corresponding controls without GGP (P = 0.001), but the other five constructs were not significantly affected by ascorbate. Mean ascorbate concentration (mg/100 g FW) of the control treatments was 27 ± 0.8 and of the high GGP (+) ascorbate treatment was 83 ± 2.8. Low ascorbate induced by ko was 10 ± 2.7.
We also measured the mRNA transcript levels of these variant constructs in case the changes affected mRNA stability. The data in Supplemental Figure 5 clearly show that LUC mRNA transcript levels relative to REN were unaffected by ascorbate. However, there was a significant correlation (P < 0.02) between mRNA levels and LUC expression.
A conserved short canonical ORF potentially encoding a 10-amino acid uORF is also present but out of frame within the longer uORF (Supplemental Figure 1). To determine if this had functional significance, its start codon (ATG) was mutated to TTG, resulting in a nonconserved His-to-Leu change at residue 12 of the uORF. However, this did not change the relative sensitivity of the promoter to ascorbate concentration (Supplemental Figure 6) but did result in significantly increased LUC expression.
Further experiments to test how the uORF regulates translation were performed. The dicotyledonous plants had a conserved AUC codon upstream to the proposed ACG start codon (Supplemental Table 3). These bases form part of the Kozak sequence around the proposed ACG start, and we changed this to a CTC to disrupt the Kozak sequence. As shown in Figure 7, this resulted in a significant increase in LUC expression and reduced inhibition by ascorbate compared with the wild type. This supports the notion that the Kozak sequence is important in determining translation of the uORF (Wamboldt et al., 2009; Simpson et al., 2010).
Figure 7.
The uORF Starts at a Noncanonical Start Codon and Controls Translation.
WT refers to the native GGP promoter and 5′UTR, “+” refers to the addition of 35S-driven GGP to the leaf to increase ascorbate. “ATC to CTC” refers to the change of the codon immediately upstream of the proposed uORF start ACG from ATC to CTC, thus reducing the strength of the Kozak sequence (Supplemental Table 3 and Supplemental Figure 1). ACGuORF del refers to deleting all sequence between the uORF (including the stop codon) and the start codon of LUC. ATGuORF del is the same construct except using the uORF with an ATG start codon. Error bars are se of the mean. Mean ascorbate concentration (mg/100 gFW) of the control treatments was 25.2 ± 0.6 and of the high GGP (+) ascorbate treatment was 60.2 ± 5.4
We also directly deleted all sequence between the uORF and the start of the LUC gene so that there was no stop codon after the uORF, i.e., the uORF ran in frame into the LUC. We did this for both the wild-type ACG uORF and the variant uORF starting with ATG. In the case of the ACG uORF (Figure 7), the LUC expression was unchanged from the control, but the inhibition by ascorbate was reduced compared with the wild type. However, in the case of the ATG uORF, very little LUC activity was detected and no inhibition by ascorbate was observed (compared to Figure 6).
How Does the Noncanonical uORF Regulate GGP Translation?
To examine how this peptide regulates translation, we first tested whether the predicted uORF worked in a cis or trans configuration. We used the mutant that did not respond to ascorbate because it was lacking the uORF start codon to test whether expressing the uORF from a separate construct could recover ascorbate repression of LUC activity. Figure 8 shows that the presence of the uORF did not complement the mutant form of the uORF and had no effect on any treatment. This is consistent with the uORF working in a cis conformation with the GGP CDS.
Figure 8.
The Noncanonical uORF Acts in a cis Manner.
WT is the synthesized wild-type variant 5′UTR with the wild-type promoter of GGP; “ACG to TTG” refers to the mutated ACG codon of the uORF (nonresponding); “uORF” refers to the addition of the uORF driven by a 35S promoter; and “+” refers to the addition of the GGP CDS also driven by 35S to raise ascorbate. Error bars are se of the mean. Ascorbate concentrations in were 111 ± 12 and 24 ± 0.5 mg/100 g FW for leaves with and without 35S-driven GGP CDS coinfused. ANOVA was done on log-transformed data.
We examined if the uORF peptide directly interacted with ascorbate using surface plasmon resonance and NMR. However, no measurable interaction was observed. We also tried to measure the expression of the uORF protein in transformed leaves using liquid chromatography-mass spectrometry but were unable to observe the peptide. At least in humans, short peptides are degraded rapidly (Baboo et al., 2014).
In another series of experiments, we created deletions in the 5′UTR around the uORF without affecting the uORF itself (Supplemental Figure 7). In one construct, an accidental insertion occurred after the uORF, which allowed us to test the spacing between the uORF and the LUC gene. These data (Figure 9) demonstrate that a large deletion before the uORF of 92 bp reduced the inhibition by ascorbate, and short deletions after the uORF (2 to 61 bp) had little effect on ascorbate inhibition, while a larger deletion of 104 bp after the uORF abolished ascorbate inhibition. A large insert of DNA between the uORF and LUC, associated with a small 13-bp deletion, had no effect on ascorbate inhibition. None of the new potential uORFs created by the deletions (Supplemental Figure 7) have start codons with a strong Kozak context; therefore, they should not influence ascorbate inhibition. Together, the deletion data show that there are critical regions before and after the uORF that determine whether the uORF is recognized and translated or that affect the translation of the main CDS. It is also possible that the deleted sequences affect reinitiation of the short canonical uORF, which could alter LUC expression.
Figure 9.
The Sequence Surrounding the uORF Affects the Ascorbate Inhibition of the GGP Promoter-Driven LUC Translation.
Different areas of the DNA sequence around the uORF in the 5′UTR were deleted and the effect on LUC expression measured. The schematic below the graph shows the extent of deletions or insertions. The actual sequences of the changed 5′UTR are shown in Supplemental Figure 7. Error bars are se of the mean. Ascorbate concentrations (mg/100 g FW) were 66 ± 3.1 and 26 ± 0.9 for leaves with (+) and without the 35S-driven GGP CDS coinfused, respectively. ANOVA was done on log-transformed data.
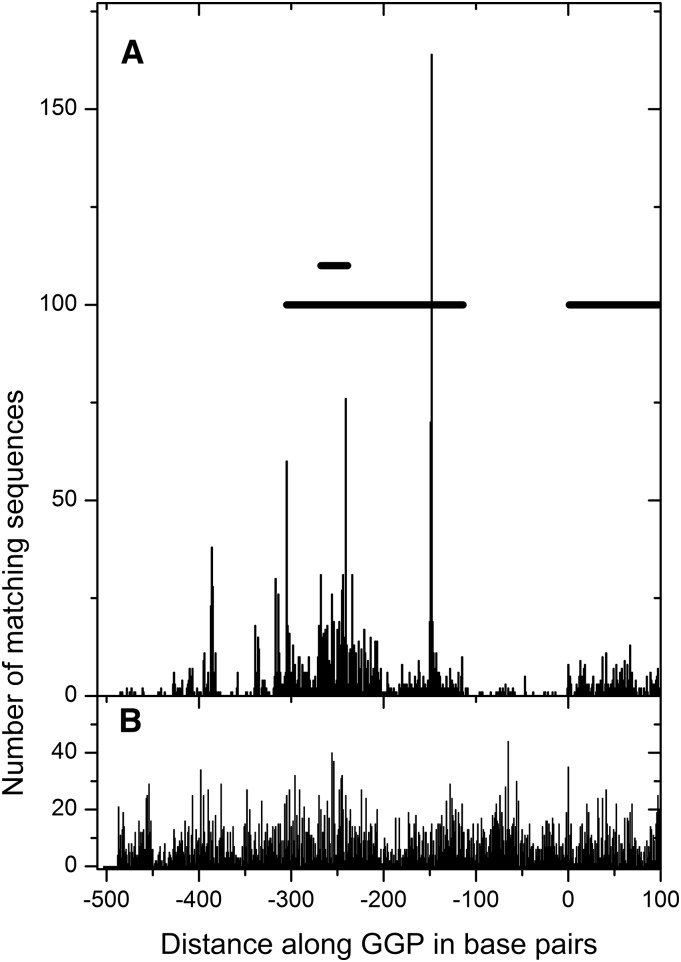
Some of our results suggested that the uORF is translated. To determine whether the uORF was translated by the ribosome, we analyzed published ribosome protection data (Liu et al., 2013; Juntawong et al., 2014) for the distribution of ribosomes along the GGP sequence. The data (Figure 10) clearly showed that the uORF has a high population of ribosomes associated with it, on average higher than the ribosome density on the main ORF (Supplemental Figure 8). At various positions along the gene (263 and 356 bp), the ribosome density was severalfold higher than the main ORF, and these positions were reproduced in three of the four replicates in a photomorphogenic study (Liu et al., 2013): The last replicate had too low sequence counts to allow meaningful comparisons. Another study (Juntawong et al., 2014) showed similar results although sequence counts were low compared with the photomorphogenic study and specific peaks were not detected. Notwithstanding, both these publications show that ribosomes associate with the noncanonical uORF and the main ORFs, and sequence counts are lower in noncoding regions. Together, these data indicate that ribosome stalling occurs on the translated uORF. A proposed mechanism is shown in Figure 11.
Figure 10.
Mapping of Ribosome Protection Sequencing Data to the Transcript of GGP Suggests the uORF Is Translated.
(A) The ribosome-protected sequences.
(B) The total mRNA data. The graph plots data from Arabidopsis exposed to 4 h light and shows only the first 100 bp of the main CDS of GGP. The average over the entire CDS was 3.5 counts (± 0.1) compared with 2.8 (± 0.07) over the first 100 bp. The thick horizontal lines represent the long ACG uORF, the short ATG uORF, and the first 100 bp of the main GGP CDS starting at 0. Data in Supplemental Figure 8 provide analysis on stretches of DNA covering the UTR, the uORF, and the CDS.
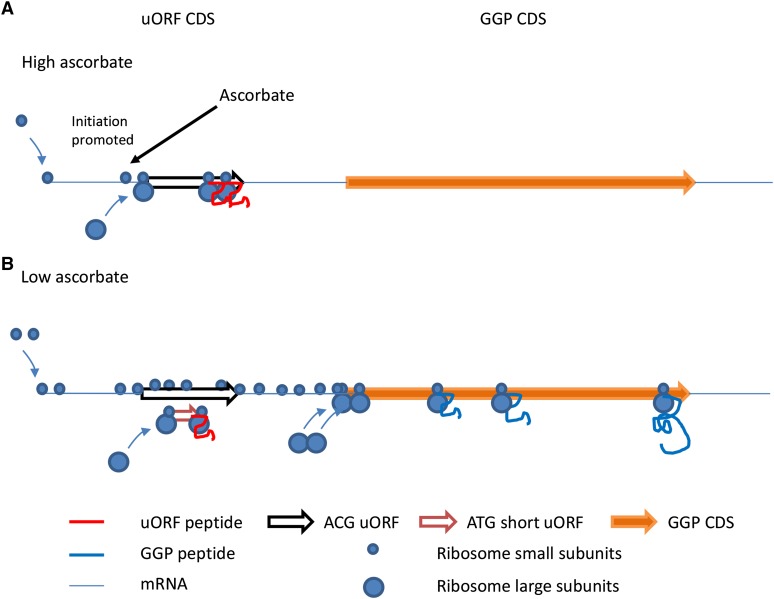
Figure 11.
Simplified Scheme for Ribosome Interaction with the GGP mRNA at the uORF and CDS.
The model assumes that high ascorbate (A) promotes recognition of the uORF ACG start codon, while under low ascorbate, the ribosomes skip the uORF (B) and start translation at the main GGP CDS. The likelihood that the short ATG uORF is translated is included in (B). This model is supported by the peaks of ribosomes seen on the uORF in Figure 10 and by the observation that inactivating the ATG uORF increases translation (Supplemental Figure 6).
This Noncanonical uORF Regulates Ascorbate Levels in Leaves
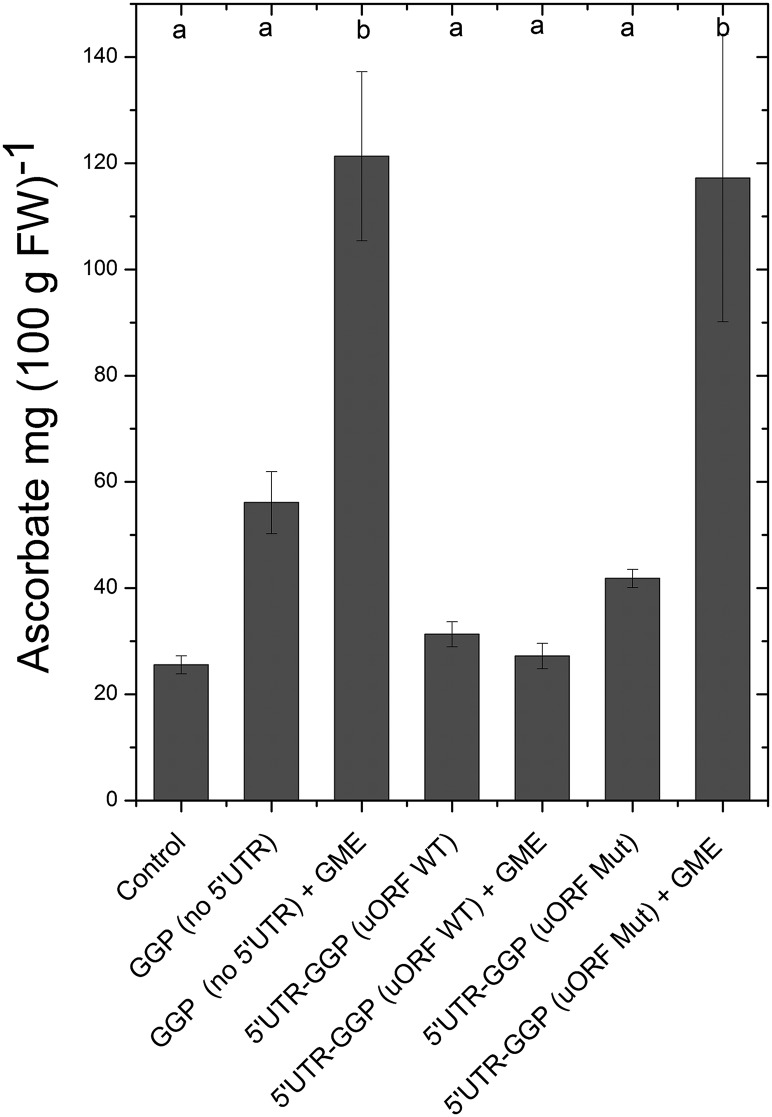
To test how the uORF might function in affecting ascorbate concentrations in vivo, we mutated the start ACG codon to TTG and tested the effect on ascorbate levels in transiently transformed tobacco leaves. We compared GGP driven by the 35S promoter without the uORF, with the uORF, and with the mutated start codon on the uORF (Figure 12). The data showed that GGP with a functional uORF in its 5′UTR does not increase ascorbate in leaves even with the addition of GME, while the enzyme with a mutated uORF start codon responded in a similar fashion to GGP without a 5′UTR.
Figure 12.
Effect of the Noncanonical uORF on Ascorbate Levels
N. benthamiana leaves were infiltrated with different constructs of the GGP coding sequence driven by different promoters and 5′UTR, and ascorbate was measured after 7 d as detailed in the Methods. The “control” was infiltrated with P19. “GGP (no 5′UTR)” had the 35S promoter and did not include the GGP 5′UTR. “GME” was coinfiltrated where indicated. “5′UTR-GGP (uORF WT)” had the 35S promoter in front of the native GGP 5′UTR. The columns with “5′UTR-GGP (uORF mut)” also used the 35S promoter in front of the 5′UTR of GGP, but the ACG start codon of the uORF was changed to a TCG.
Disruption of this uORF stopped the ascorbate feedback regulation of translation, and removal of the 5′UTR or inactivation of the uORF of GGP allowed increased ascorbate concentrations in leaves. Disrupting the mechanism that involves the uORF, including any interacting cofactors (e.g., a protein required to transmit the ascorbate concentration to the ribosome), is a potential method to increase ascorbate in plants and may explain why some fruits such as kiwifruit have very high ascorbate (Ferguson and MacRae, 1992).
DISCUSSION
Here, we reveal that the expression of GGP, the key gene in ascorbate biosynthesis, is regulated at the level of translation through the interaction of ascorbate and a conserved uORF with a noncanonical initiation codon. Reports of the regulation of gene expression in eukaryotes by products of a biosynthesis pathway are rare (von Arnim et al., 2014). Often, gene expression is regulated by signaling cascades that result in changes in mRNA concentrations or through posttranslational modification of the target proteins. While there are a number of reports of 5′UTR sequences that interact with small metabolites (riboswitches) to influence protein translation (Bocobza and Aharoni, 2008; Zhang et al., 2010; Liu et al., 2012; Dvir et al., 2013; Serganov and Nudler, 2013), regulation of eukaryotic translation by uORFs in response to a small molecule are uncommon, especially in plants (von Arnim et al., 2014). Furthermore, the only other eukaryotic example of metabolite regulation through a uORF initiated from a conserved noncanonical start codon is ornithine decarboxylase (Ivanov et al., 2008), and no plant examples have been reported.
Translation control provides a way to rapidly regulate GGP levels in response to environmental stress without the need to alter gene transcription. Transcript levels of GGP are regulated in plants, with circadian rhythms of GGP mRNA being observed and increased levels of GGP transcript being measured under increased light (Dowdle et al., 2007). The mechanism we describe here involving the uORF regulating GGP translation provides a more immediate and sensitive response to changing ascorbate levels.
We present the following evidence that the peptide encoded by the GGP uORF is critical in determining ascorbate repression of translation: The uORF encodes a peptide highly conserved over a wide range of plant taxonomies (Figure 5); ascorbate repression depends on the amino acid sequence and not the DNA (Figure 6); and ascorbate repression is abolished by a range of amino acid changes in the uORF including single amino acid mutations (Figure 6).
Interestingly, for the GGP uORF to be translated, initiation must occur at a noncanonical start codon. Translation initiation of the uORF likely occurs at an ACG codon, which is in a preferable context (i.e., purine at −3 and G at +4; Kozak) and is conserved in all plants from mosses to angiosperms. The only other codon that the uORF could potentially start from is next codon upstream, namely, an AUC. However, this codon is conserved in dicotyledonous species but not in monocots or other lower plants and is not in a beneficial context. The AUC sequence forms part of the ACG Kozak sequence and is likely conserved in dicotyledonous plants because it is necessary for efficient translation initiation. Changing this AUC to a CUC reduced the effect of ascorbate and increased expression of LUC (Figure 7), suggesting that uORF translation initiation is reduced. The initiation of translation at a noncanonical ACG codon may enable the GGP mRNA to be rapidly and selectively translated under stress to increase ascorbate concentrations due to high rates of ascorbate consumption.
While uORFs are estimated to occur in a high proportion of genes in eukaryotic organisms (Calvo et al., 2009; Takahashi et al., 2012), very few of these are known to encode conserved peptides (Hayden and Jorgensen, 2007; Tran et al., 2008). In almost all cases, the roles of these uORFs have not been characterized. Using a bioinformatics approach, 26 gene groups were found to possess a conserved canonical uORF in Arabidopsis and rice (Oryza sativa; Hayden and Jorgensen, 2007), and this has been subsequently expanded (http://arabidopsis.org/index.jsp). These analyses did not identify the uORF in the GGP gene, as their search protocol required an AUG initiation codon. This raises the possibility that there are other examples of uORFs that initiate using noncanonical codons that are yet to be identified in both plants and animals. A high proportion of eukaryote mRNA sequences contain canonical uORFs (Kim et al., 2007); however, few examples have been found of functional uORFs with a noncanonical start codon (Ivanov et al., 2011). Noncanonical ORFs in Arabidopsis are very rare (Simpson et al., 2010), and here we have established experimentally that uORFs can initiate from noncanonical codons in plants.
The discovery that uORFs can regulate translation by slowing or inhibiting the translation of the main CDS is relatively recent (Calvo et al., 2009; Medenbach et al., 2011; Ferreira et al., 2013; von Arnim et al., 2014). However, apart from the observation that they affect translation, little is known about their mechanism of action or their function (Andrews and Rothnagel, 2014). There are two proposed broad models (Figure 11) of how uORFs inhibit translation (Tran et al., 2008): the first where translation of the uORF influences the efficiency of translation including affecting reinitiation after the uORF (von Arnim et al., 2014) and the second where the nascent uORF peptide causes ribosome stalling during uORF translation (Fang et al., 2004). In a few cases, small molecules (metabolites) have been shown to interact with translation of the uORF to cause inhibition of downstream translation. Two examples in plants, polyamine and sucrose regulation (von Arnim et al., 2014), involve sequence-dependent small molecule-regulated uORFs.
In this work, we provide evidence that ascorbate, or a precursor of ascorbate, interacts either directly or indirectly through an intermediate, with a peptide produced by a noncanonical uORF (ACG uORF) in the 5′UTR of the key control gene of ascorbate biosynthesis, GGP. The uORF peptide inhibits translation of the GGP gene as truncation of the uORF or mutations within conserved residues (change of the proposed ACG start and change of a Leu to His) results in increased translation of LUC and the loss of ascorbate repression of translation (Figure 6), while synonymous base changes have little effect (Figure 6). Especially low ascorbate levels stimulated translation of the GGP gene, whereas the same conditions inhibited translation of a control gene (Figure 2). While it is possible that the redox state of the cell may be the critical factor (Supplemental Figure 9), we have interpreted our results in terms of ascorbate content.
A possible mechanism is that in the presence of ascorbate, the ACG uORF is translated and the uORF peptide causes the ribosome to stall, reducing translation of the downstream GGP sequence (Figure 11). It has been reported that positively charged peptides slow ribosomal velocity in an additive manner (Charneski and Hurst, 2013) and the uORF has up to six positively charged residues (Figure 5). Where the ascorbate acts is not clear: We suggest it is likely that either ascorbate promotes the recognition of the uORF ACG start codon (leaky scanning) or ascorbate causes the newly synthesized uORF peptide to stall. Ascorbate possibly slows the ribosomes on the long ACG uORF, and this increases the probability of translation initiation at the ACG so the ribosomes pile up. An alternative hypothesis is that rather than ascorbate influencing the rate of uORF translation, ascorbate inhibits the reinitiation of the ribosome after translation of the uORF. Reinitiation is affected by a range of factors (Schepetilnikov et al., 2013; Schleich et al., 2014). However, the observation that disabling the small 30-bp Met-uORF more than doubled the expression of LUC while ascorbate still repressed LUC expression (Supplemental Figure 6) suggests that ascorbate affects initiation of uORF translation in a leaky scanning model (the ribosome then stalls on the uORF) rather than the other alternatives.
That the uORF is translated is supported by the following: the conserved sequence of the uORF, the observation that changing a His to a Leu (Figure 6, construct 3) results in loss of ascorbate effect, the ribosome protection data (Figure 10; Supplemental Figure 8), the increase in LUC expression on disabling the uORF (Figure 6, constructs 2, 4, and 5), and the observation that reduction in the Kozak sequence strength increased LUC expression (Figure 7). It is also supported by the loss of ascorbate repression of LUC expression following the larger 5′UTR deletions before and after the uORF (Figure 9), suggesting the ribosome had less chance to “see” the uORF during its transit along the mRNA and less chance to reinitiate LUC translation after translating the uORF. The conserved His-to-Leu change (Figure 6) did not result in a significant increase in LUC expression or in ascorbate repression of LUC translation, suggesting the conserved His is a critical residue for ribosome stalling.
Studies of ribosome protection in plants show that ribosomes are associated with RNA sequences that are translated (Liu et al., 2013; Juntawong et al., 2014), including canonical uORFs. Analysis of their data shows that that ribosomes concentrate within the uORF at a much higher level than in the main coding region (Figure 10; Supplemental Figure 8), supporting our ribosome stalling model. The main peak of ribosome protection occurs close to residue 53 on the 62-residue uORF (Figure 10). This is at the end of the most conserved sequence of the uORF (Figure 5), and given that the ribosome tunnel can hold 30 to 40 residues (Ito and Chiba, 2013), this would leave at least 13 to 23 conserved N-terminal residues emerging from the tunnel. That the most conserved residues would be in the ribosome tunnel when the ribosome stalls suggests that they are critical for stalling.
We were unable to observe ascorbate directly interacting with a synthesized uORF peptide, as tested using surface plasmon resonance and NMR. It is therefore unclear whether ascorbate interacts directly with the uORF peptide to cause ribosome stalling or if ascorbate enhances noncanonical initiation and then the uORF peptide then stalls the ribosome. It is also possible that the uORF peptide may not directly sense ascorbate but that another factor interacts with ascorbate to cause either ribosome stalling or to promote uORF translation. Ascorbate concentration varies between fruit of different cultivars (Ferguson and MacRae, 1992; Davey et al., 2006; Bulley et al., 2009) and mutations affecting the functionality of the uORF or a factor that interacts with ascorbate might explain this variation. Identifying this unknown protein factor and/or discovering an allele GGP that has a mutated uORF are the obvious next step in this work.
In conclusion, we have shown that the concentration of ascorbate in a leaf can be regulated through ascorbate feedback through a taxonomically conserved noncanonical uORF in the long 5′UTR of the controlling gene of ascorbate biosynthesis, GGP. We provide evidence that this feedback acts posttranscriptionally by influencing the concentration of the GGP enzyme. We propose that this is a major mechanism by which ascorbate concentrations are regulated in the l-galactose pathway of ascorbate biosynthesis.
METHODS
Plant Materials and Assays
Nicotiana benthamiana was grown in a glasshouse with natural sunlight and supplemental lighting to extend the daylength to 16 h. Temperature was set to between 22 and 25°C. The N. benthamiana leaf transient reporter gene system using firefly (Photinus pyralis) luciferase (LUC) as the promoter-specific reporter and Renilla reniformis luciferase (REN) as the transformation reporter was as described previously (Hellens et al., 2005) using a P19 construct to prevent gene silencing. Once plants had at least five leaves, younger leaves (1 to 2 cm in diameter) were infiltrated. Two plants per treatment were infiltrated with Agrobacterium tumefaciens containing constructs of interest and experiments were repeated at least twice. Leaves were harvested for luminescence measurements 7 d after infiltration unless otherwise specified. Ascorbate concentration in the leaf was raised by infiltrating Agrobacterium tumefaciens containing a pGreen vector (Hellens et al., 2000) with the CDS for Actinidia chinensis GGP under the 35S promoter (Bulley et al., 2009) or lowered by infiltrating a RNAi vector constructed using as a template the GGP sequence from N. benthamiana assembled from seven ESTs in GenBank (Snowden et al., 2005). In addition, the version of the CDS of Actinidia eriantha GME described earlier (Bulley et al., 2009) was used to synergistically enhance ascorbate with GGP.
Ascorbate was measured in extracts of the same leaf using an HPLC-based assay as described (Rassam and Laing, 2005). Ascorbate was measured as total ascorbate by reducing extracts before HPLC (Rassam and Laing, 2005) or in unreduced extracts to measure reduced ascorbate (Supplemental Figure 9). The amount of LUC protein was measured using an antibody to LUC (Promega) in an immunoblot of 50 μg soluble cellular protein per lane (extracted in 40 mM phosphate buffer, pH 7.4, 150 mM NaCl). See Supplemental Methods for more details on tests of the system used herein.
Experiments were repeated at least twice with similar results. Statistical analysis was done by ANOVA using Minitab, and Tukey’s family error rate was used to calculate significant differences.
Gene Cloning and Plasmids
The A. eriantha GGP promoter was cloned from A. eriantha genomic DNA (gDNA) as follows. DNA (2 µg) was digested using seven blunt cutting restriction enzymes: DraI, EclII 136, EcoRV, HpaI, MScI, ScaI, SspI, and StuI. Digests were purified and eluted in 10 μL using PCR Clean and Concentrate spin columns (Zymogen). Double-stranded adapter sequences (Clontech) containing nested PCR primer sites were ligated onto cut fragments overnight at 16°C using T4 Rapid Ligase (Roche). Ligations were column-purified a second time and eluted in 30 µL. First-round PCR was performed using 1 μL each digest with primers 319998NRWLK1 (5′-CTTGCGTCGTACAGACCGGACATGT-3′), RPH-149 (5′-GTAATACGACTCACTATAGGGC-3′), and Ex Taq polymerase (Takara) using the following two-step cycling conditions. For the first high stringency step, one cycle of denaturation was performed at 94°C for 2 min, followed by seven cycles of 94°C for 25 s and elongation/annealing at 72°C for 3 min. The second step consisted of 32 cycles of 94°C for 25 s and 67°C for 3 min before a final 67°C extension for 3 min. The first-round products were run on a 1% agarose gel and 1 μL 1:50 dilution was used as a template for second round PCR with 319998NRWLK2 (5′-CCCTATGATGTTGCCTGAAAATAGTCATT-3′) and RPH-150 (5′-ACTATAGGGCACGCGTGGT-3′). Second-round PCR was also performed as a two-step PCR with an initial denaturation of 94°C for 2 min followed by five cycles of 94°C for 25 s and 72°C for 3 min. This was followed by 20 cycles of 94°C for 25 s and 67°C for 3 min before a final 67°C extension. Gel electrophoresis was used to identify PCR products between 0.5 and 2 kb in size for cloning into pGem T Easy vector (Promega) according to the manufacturer’s instructions. Clones were DNA sequenced and confirmed for overlap with the known 5′UTR. A second set of nested primers was designed to the end of the first promoter walk to extend the known A. eriantha promoter sequence to 2 kb. This 2-kb promoter sequence was then PCR amplified from A. eriantha gDNA using primers Eriantha gDNA PCR 5′ (5′-CTCCTCTTTTGAATTGACGTGACACATTTA-3′) and 319998NRWLK2 and sub-cloned into pGreen0800-5′_LUC (Hellens et al., 2000) using EcoRV and NcoI restriction enzymes. The final construct is called the GGP-promoter-pGreenII 0800-5 LUC vector.
The promoter for GGP from Arabidopsis thaliana (forward primer 5′-AAGCTTGATACTTGTCCTCTCGAAATTG-3′ and reverse primer 5′-CCATGGTCTCAAAAAAGACAAATTTCGAAGAG-3′) and control promoters from a range of sources were cloned by PCR based on published sequences. The control promoters were Arabidopsis TT8 (5′-AATTGTCGACTCCGAAAATGTCAAACTAATTCC-3′ and 5′-GCGGCCGC CGGAGATACGAAAACGTGGT-3′), EF1α (5′-TCTAGAATGGTACCTAATTACTTCAC-3′ and 5′-CTCTTTACCCATGGTTAGAGACTG-3′), Act2P (5′-CAGCCATTTTCCATGGGCTGCAAACAC-3′ and 5′-GCCATCAAAGCTTAAGAACTAATCC-3′), and Act7P (5′-TCACCATCGGCCATGGTTCACTAAAAAAAAAG-3′ and 5′-GATTATTGTTAAAGCTTTTAATCATTC-3′).
To test whether the effect of ascorbate was mediated by the promoter or the 5′UTR, we constructed two vectors where the 5′UTR regions were swapped between the GGP promoter and the TT8 promoter. The resulting constructs consisted of the TT8 core promoter (TT8P′) followed by the GGP 5′UTR (GGPUTR) and vice versa.
Generation of the inactivated start codons or other deletions and mutations in the uORFs of the 5′UTR was done by chemical synthesis (GenScript) of mutated and control sequences. In the inactivated versions, the ATG or ACG start codons of the uORFs were changed to a TTG. Other changes were made by site-specific mutagenesis. The version of the uORF where the DNA sequence was changed without a change in the amino acid sequence is shown in Supplemental Figure 4. There is a StuI site on the 5′ side of the UTR 28 bp into the 5′UTR that was used as the 5′ border of the synthesized fragment. We added an extra CC to the 3′ end of the synthesized genes to create a NcoI site (CCATGG) and removed the sequence equivalent to the synthesized fragment from the GGP-promoter-pGreenII 0800-5 LUC by digesting it with StuI and NcoI. Then, the synthesized fragments were separately cloned into the vector to create the two versions with (control) and without a uORF. All constructs were sequence verified before use.
To test the effect of the 5′UTR in front of the GGP CDS on ascorbate concentration, a different GGP gene with a full-length 5′UTR was used instead of the GGP used in other experiments. At the protein level, it was 96% identical to the standard GGP, and in the absence of the 5′UTR, raised ascorbate concentrations to similar levels seen for the usual GGP. The version with the 5′UTR had the full 5′UTR, while the version labeled without the UTR had all but 37 bp upstream of the starting ATG deleted using the XhoI restriction enzyme. In this region, at the 3′ end of the 5′UTR, there is little similarity between GGPs from different species (Supplemental Figure 1). Both versions were ligated into the pART277 vector (Gleave, 1992). The mutation to change the ACG start codon to a TTG was done by site-specific mutagenesis.
RNA Isolation and cDNA Synthesis
Total RNA was isolated from 100 mg leaf tissue using an RNeasy Plant Mini kit (Qiagen), and concentrations were measured using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). CDNA was then synthesized from 1 μg total RNA and random hexamers in a 10 μL total volume using a BluePrint reagent kit for real-time PCR (Takara Bio Company) following the manufacturer’s instructions. Following cDNA synthesis the preparation was diluted 75 times in preparation for quantitative real-time PCR.
Quantitative PCR
Quantitative PCR was performed in 5 μL total volume using a LightCycler 480 Real-Time PCR System (Roche Diagnostics) and the following primer pair: LUC1/2, 5′-TATCCGCTGGAAGATGGAAC-3′ and 5′-TCCACCTCGATATGTGCATC-3′. Primers were designed with annealing temperatures of 60°C using Primer3 (Rozen and Skaletsky, 2000). The luciferase primer pair amplifies regions from the 5′-end of the LUC open reading frame. Reaction components (using LightCycler 480 SYBR Green I Master Mix) were as follows: 2 μM each primer and 1.25 μL diluted cDNA preparation. The standard cycling protocol with a temperature of 60°C was used and relative quantification analysis normalized to REN transcripts was performed using LightCycler 480 software (Roche Diagnostics).
Transcript Analysis Using Galaxy
Data from Arabidopsis ribosome protection assay sequencing (GSE43703 file SRR974753.fastq uploaded onto the NCBI database; Liu et al., 2013) was analyzed by aligning sequence reads to the At4g26850 (GGP, VTC2) transcript using Galaxy (Goecks et al., 2010; https://usegalaxy.org/). Transcripts were trimmed of the 3′ 21 bp and then mapped to the target gene using BWA with default settings except with a maximum edit distance of 4 and number of first subsequences to take as seed of 26. Data were then filtered to remove unmapped reads and exported for graphing and further analysis. In addition, analysis was also done on a second ribosome protection data set (Juntawong et al., 2014).
Peptide Studies
A 60-amino acid peptide HPLC purified to over 95% purity with the sequence TAIIGVSRPLIHVRSVRRKGCVVESNPSPHGGRGALPSEGGSPSDLLFLAGGGHFAFSVY was purchased from Genscript. This was tested using a 600-MHz NMR machine and a T200 BiaCore in the presence and absence of ascorbate. For NMR, the ascorbate was adjusted to pH 6.0 in 200 mM KH2PO4 buffer immediately before adding it to the solution of the synthetic uORF. Ascorbate was titrated between 0 and 18 mM ascorbate with the uORF peptide at 0.75 mM in scans that lasted ∼1 h. In initial scans of 1D proton spectra as well as 2D TOCSY and NOESY spectra, no secondary structure was observed.
For BiaCore measurements, the peptide was dissolved in water and bound to a CM5 chip through the N-terminal NH2 group until a sufficient magnitude of resonance units were achieved. Then, after equilibration, ascorbate solutions in the range of up to 10 mM at pH 7.0 were passed through the chip using a blank chip as a control.
To measure the uORF peptide in leaves, ∼100 mg frozen powdered transiently transformed tobacco leaf was extracted using 50 mM BTP, pH 7.5, 2 mM DTT, and centrifuged at 16,000g twice to clarify the extract. Both tobacco transformed with the full GGP promoter driving LUC with high and low ascorbate (induced with 35S:GGP) as well as controls transformed with P19 and 35S:GGP were also processed. The sample was then concentrated using a 3000 molecular weight cutoff filter and the retentate washed with NH4HCO3. The sample was then digested with trypsin and liquid chromatography-mass spectrometry used to identify peptides based on the synthetic sequence performed as previously described (Laing et al., 2004). Both a start Thr and a start Met option were explored. As a separate approach, an extraction was made using 0.5 mL of 7 M urea and 2 M thiourea and the samples processed without filtering. Trypsin digestion was done after 5-fold dilution into NH4HCO3. In another experiment, the tissue samples were extracted in 0.5 mL NH4HCO3 and immediately boiled, centrifuged, and processed as above. Lastly, the tissue was extracted in 10 mL methanol/chloroform/water/formic acid (12/5/2/1, v/v/v/v) and centrifuged and the residue extracted in 10 mL 2% formic acid in 20% methanol and the extracts combined. Then, 2.5 mL chloroform and 3.5 mL water was added to the combined extract to split phases, the upper layer was removed, and the chloroform phase washed with 2% formic acid in 20% methanol. Then, the aqueous phase was dried down and digested. In all cases, samples spiked with peptide were run in parallel and the peptide was observed.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: A. eriantha GGP promoter, JX486682; Arabidopsis GGP promoter, At4g26850; A. eriantha GME, FG424114; A. chinensis GGP, FG528585; Arabidopsis TT8, AT4G09820; EF1α, AT1G07940; Act2P, AT3G18780; Act7P, AT5G09810. The kiwifruit (A. chinensis) GGP with a full-length 5′UTR (Figure 12) has accession number FG460629.
Supplemental Data
Supplemental Figure 1. Identification of Conserved Regions in the DNA Sequence of the 5′-UTR of GGP from Dicotyledonous Species.
Supplemental Figure 2. The Highly Conserved Region of the 5′UTR and the Three Deletions Used in Testing Their Functions.
Supplemental Figure 3. The Effect of the 5′UTR on Ascorbate in Tobacco Leaves.
Supplemental Figure 4. Alignment of the Codon Usage-Changed uORF (with an Unchanged Amino Acid Sequence Compared with the Wild Type) with the Wild-Type Synthesized Version of the uORF.
Supplemental Figure 5. Transcript Expression Levels for the Experiment Shown in Figure 5.
Supplemental Figure 6.The Small ATG uORF Is Not Involved in the Response to Ascorbate Concentration.
Supplemental Figure 7. Sequences of Constructs Made with Deletions in the 5′UTR but outside the uORF.
Supplemental Figure 8. Mapping of Ribosome Protection Sequencing Data to the Transcript of GGP.
Supplemental Figure 9. Reduction State of Ascorbate as a Function of Total Leaf Ascorbate.
Supplemental Table 1. Testing Alternative Explanations for the Effect of Ascorbate on the LUC Expression.
Supplemental Table 2. The Ascorbate Downregulation of the GGP Promoter Is Expressed through the 5′UTR Region of the Gene.
Supplemental Table 3. Potential Start Codons for the Proposed 5′UTR uORF.
Supplemental Methods. Control Tests of the System Used.
Supplementary Material
Acknowledgments
We thank Richard Espley for advice on quantitative PCR measurements, Andrew Gleave for developing the cloning strategy with and without the 5′UTR GGP expression vector, and Alistair Hall for advice on the binomial distribution. Ross Crowhurst undertook bioinformatic comparison of 5′UTRs between species. Cyril Brendolise, Niels Nieuwenhuizen, and Luke Luo also contributed to molecular cloning. Richard Newcomb advised on molecular phylogenics.
AUTHOR CONTRIBUTIONS
W.A.L., R.P.H., and S.M.B. designed the research. M.M.-S., M.A.W., A.P.D., M.R., D.B., D.W., S.M.B., R.S., and W.A.L. performed research. R.S. contributed new analytic/computational/etc. tools. W.A.L., M.M.-S., D.B., S.M.B., and R.C.M. analyzed data. W.A.L., R.C.M., R.P.H., and S.M.B. wrote the article.
Glossary
- uORF
upstream open reading frame
- UTR
untranslated region
- FW
fresh weight
- RNAi
RNA interference
- gDNA
genomic DNA
- CDS
coding sequence
References
- Andrews S.J., Rothnagel J.A. (2014). Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15: 193–204. [DOI] [PubMed] [Google Scholar]
- Asensi-Fabado M.A., Munné-Bosch S. (2010). Vitamins in plants: occurrence, biosynthesis and antioxidant function. Trends Plant Sci. 15: 582–592. [DOI] [PubMed] [Google Scholar]
- Baboo S., Bhushan B., Jiang H., Grovenor C.R.M., Pierre P., Davis B.G., Cook P.R. (2014). Most human proteins made in both nucleus and cytoplasm turn over within minutes. PLoS ONE 9: e99346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badejo A.A., Eltelib H.A., Fukunaga K., Fujikawa Y., Esaka M. (2009). Increase in ascorbate content of transgenic tobacco plants overexpressing the acerola (Malpighia glabra) phosphomannomutase gene. Plant Cell Physiol. 50: 423–428. [DOI] [PubMed] [Google Scholar]
- Bartoli C.G., Tambussi E.A., Diego F., Foyer C.H. (2009). Control of ascorbic acid synthesis and accumulation and glutathione by the incident light red/far red ratio in Phaseolus vulgaris leaves. FEBS Lett. 583: 118–122. [DOI] [PubMed] [Google Scholar]
- Bartoli C.G., Yu J., Gómez F., Fernández L., McIntosh L., Foyer C.H. (2006). Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J. Exp. Bot. 57: 1621–1631. [DOI] [PubMed] [Google Scholar]
- Bocobza S.E., Aharoni A. (2008). Switching the light on plant riboswitches. Trends Plant Sci. 13: 526–533. [DOI] [PubMed] [Google Scholar]
- Bonin C.P., Potter I., Vanzin G.F., Reiter W.-D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. USA 94: 2085–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulley S., Wright M., Rommens C., Yan H., Rassam M., Lin-Wang K., Andre C., Brewster D., Karunairetnam S., Allan A.C., Laing W.A. (2012). Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase. Plant Biotechnol. J. 10: 390–397. [DOI] [PubMed] [Google Scholar]
- Bulley S.M., Rassam M., Hoser D., Otto W., Schünemann N., Wright M., MacRae E., Gleave A., Laing W. (2009). Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J. Exp. Bot. 60: 765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Pagliarini D.J., Mootha V.K. (2009). Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA 106: 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski C.A., Hurst L.D. (2013). Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 11: e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.W., Kenis K., Keulemans J. (2006). Genetic control of fruit vitamin C contents. Plant Physiol. 142: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle J., Ishikawa T., Gatzek S., Rolinski S., Smirnoff N. (2007). Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 52: 673–689. [DOI] [PubMed] [Google Scholar]
- Dvir S., Velten L., Sharon E., Zeevi D., Carey L.B., Weinberger A., Segal E. (2013). Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc. Natl. Acad. Sci. USA 110: E2792–E2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Spevak C.C., Wu C., Sachs M.S. (2004). A nascent polypeptide domain that can regulate translation elongation. Proc. Natl. Acad. Sci. USA 101: 4059–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson A.R., MacRae E.A. (1992). Vitamin C in Actinidia. Acta Hortic. 297: 481–487. [Google Scholar]
- Ferreira J.P., Overton K.W., Wang C.L. (2013). Tuning gene expression with synthetic upstream open reading frames. Proc. Natl. Acad. Sci. USA 110: 11284–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2011). Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155: 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Badejo A.A., Shibata H., Sawa Y., Maruta T., Shigeoka S., Page M., Smirnoff N., Ishikawa T. (2011). Expression analysis of the VTC2 and VTC5 genes encoding GDP-L-galactose phosphorylase, an enzyme involved in ascorbate biosynthesis, in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 75: 1783–1788. [DOI] [PubMed] [Google Scholar]
- Gatzek S., Wheeler G.L., Smirnoff N. (2002). Antisense suppression of l-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated l-galactose synthesis. Plant J. 30: 541–553. [DOI] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207. [DOI] [PubMed] [Google Scholar]
- Goecks J., Nekrutenko A., Taylor J., Galaxy Team (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakmaoui A., Pérez-Bueno M.L., García-Fontana B., Camejo D., Jiménez A., Sevilla F., Barón M. (2012). Analysis of the antioxidant response of Nicotiana benthamiana to infection with two strains of Pepper mild mottle virus. J. Exp. Bot. 63: 5487–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden C.A., Jorgensen R.A. (2007). Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Ito K., Chiba S. (2013). Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 82: 171–202. [DOI] [PubMed] [Google Scholar]
- Ivanov I.P., Firth A.E., Michel A.M., Atkins J.F., Baranov P.V. (2011). Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 39: 4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Loughran G., Atkins J.F. (2008). uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. USA 105: 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-H., Cai X., Vaughn J.N., von Arnim A.G. (2007). On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 8: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing W.A., Bulley S., Wright M., Cooney J., Jensen D., Barraclough D., MacRae E. (2004). A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc. Natl. Acad. Sci. USA 101: 16976–16981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing W.A., Wright M.A., Cooney J., Bulley S.M. (2007). The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proc. Natl. Acad. Sci. USA 104: 9534–9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yin J., Xiao M., Gao C., Mason A.S., Zhao Z., Liu Y., Li J., Fu D. (2012). Characterization and evolution of 5′ and 3′ untranslated regions in eukaryotes. Gene 507: 106–111. [DOI] [PubMed] [Google Scholar]
- Liu M.-J., Wu S.-H., Wu J.-F., Lin W.-D., Wu Y.-C., Tsai T.-Y., Tsai H.-L., Wu S.-H. (2013). Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl J., Szarka A., Bánhegyi G. (2009). Vitamin C: update on physiology and pharmacology. Br. J. Pharmacol. 157: 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massot C., Stevens R., Génard M., Longuenesse J.-J., Gautier H. (2012). Light affects ascorbate content and ascorbate-related gene expression in tomato leaves more than in fruits. Planta 235: 153–163. [DOI] [PubMed] [Google Scholar]
- Medenbach J., Seiler M., Hentze M.W. (2011). Translational control via protein-regulated upstream open reading frames. Cell 145: 902–913. [DOI] [PubMed] [Google Scholar]
- Müller-Moulé P. (2008). An expression analysis of the ascorbate biosynthesis enzyme VTC2. Plant Mol. Biol. 68: 31–41. [DOI] [PubMed] [Google Scholar]
- Rassam M., Laing W. (2005). Variation in ascorbic acid and oxalate levels in the fruit of Actinidia chinensis tissues and genotypes. J. Agric. Food Chem. 53: 2322–2326. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M., Dimitrova M., Mancera-Martínez E., Geldreich A., Keller M., Ryabova L.A. (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32: 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S., Strassburger K., Janiesch P.C., Koledachkina T., Miller K.K., Haneke K., Cheng Y.-S., Küchler K., Stoecklin G., Duncan K.E., Teleman A.A. (2014). DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A., Nudler E. (2013). A decade of riboswitches. Cell 152: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G., Laurie R.E., Dijkwel P.P., Quesada V., Stockwell P.A., Dean C., Macknight R.C. (2010). Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell 22: 3764–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden K.C., Simkin A.J., Janssen B.J., Templeton K.R., Loucas H.M., Simons J.L., Karunairetnam S., Gleave A.P., Clark D.G., Klee H.J. (2005). The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K., Takaoka T., Esaka M. (2002). Gene expression of ascorbic acid-related enzymes in tobacco. Phytochemistry 61: 631–635. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Takahashi A., Naito S., Onouchi H. (2012). BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinformatics 28: 2231–2241. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran M.K., Schultz C.J., Baumann U. (2008). Conserved upstream open reading frames in higher plants. BMC Genomics 9: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A.G., Jia Q., Vaughn J.N. (2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214: 1–12. [DOI] [PubMed] [Google Scholar]
- Wamboldt Y., Mohammed S., Elowsky C., Wittgren C., de Paula W.B.M., Mackenzie S.A. (2009). Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell 21: 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y., Mieda T., Rapolu M., Nakamura A., Motoki T., Maruta T., Yoshimura K., Ishikawa T., Shigeoka S. (2007). Light regulation of ascorbate biosynthesis is dependent on the photosynthetic electron transport chain but independent of sugars in Arabidopsis. J. Exp. Bot. 58: 2661–2671. [DOI] [PubMed] [Google Scholar]
- Zhang J., Lau M.W., Ferré-D’Amaré A.R. (2010). Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry 49: 9123–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Tao Q.C., Wang Z.N., Fan R., Li Y., Sun X.F., Tang K.X. (2012). Engineering ascorbic acid biosynthetic pathway in Arabidopsis leaves by single and double gene transformation. Biol. Plant. 56: 451–457. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.