Evolutionary analysis in Geraniaceae reveals correlation of rates between plastid- and nuclear-encoded genes, providing a plausible mechanism for plastome-genome incompatibility in this family.
Abstract
Although gene coevolution has been widely observed within individuals and between different organisms, rarely has this phenomenon been investigated within a phylogenetic framework. The Geraniaceae is an attractive system in which to study plastid-nuclear genome coevolution due to the highly elevated evolutionary rates in plastid genomes. In plants, the plastid-encoded RNA polymerase (PEP) is a protein complex composed of subunits encoded by both plastid (rpoA, rpoB, rpoC1, and rpoC2) and nuclear genes (sig1-6). We used transcriptome and genomic data for 27 species of Geraniales in a systematic evaluation of coevolution between genes encoding subunits of the PEP holoenzyme. We detected strong correlations of dN (nonsynonymous substitutions) but not dS (synonymous substitutions) within rpoB/sig1 and rpoC2/sig2, but not for other plastid/nuclear gene pairs, and identified the correlation of dN/dS ratio between rpoB/C1/C2 and sig1/5/6, rpoC1/C2 and sig2, and rpoB/C2 and sig3 genes. Correlated rates between interacting plastid and nuclear sequences across the Geraniales could result from plastid-nuclear genome coevolution. Analyses of coevolved amino acid positions suggest that structurally mediated coevolution is not the major driver of plastid-nuclear coevolution. The detection of strong correlation of evolutionary rates between SIG and RNAP genes suggests a plausible explanation for plastome-genome incompatibility in Geraniaceae.
INTRODUCTION
Although coevolution of gene sequences is a widely recognized phenomenon in biological systems, it has rarely been studied between the plastid and nuclear genomes of plants within a well-established phylogenetic framework. Coevolution may be detected within a single organism, such as gene pairs with known physical interactions in Escherichia coli (Pazos and Valencia, 2001), or between organisms, such as the correlated change of sequences between viral and host genes (Lobo et al., 2009). The coevolution of genes from organellar and nuclear genomes may be considered an intermediate case, in which the genes of interest are within the same organism but are encoded in different cellular compartments. Given that there is an order of magnitude higher mutation rate in nuclear genomes compared with plastid genomes in plants (Wolfe et al., 1987; Drouin et al., 2008), the detection of correlation in evolutionary rates, and how that correlation is maintained, presents an interesting area of study.
As gene function is expressed in amino acid sequences, coevolution between two genes is usually reflected in the encoded polypeptides. If mutual selective pressure exists between two genes, changes to the amino acid sequences encoded in one gene would be expected to cause corresponding changes in the other gene to maintain normal biological activity (Pazos and Valencia, 2008). Similarly, coevolution between two genes can be evaluated based on the rate of nonsynonymous substitutions (dN), which are nucleotide mutations that cause a change in the amino acid sequences. However, dN is also affected by local rate heterogeneity or local background mutation rates, represented by the rate of synonymous substitutions (dS), which do not result in amino acid changes. The correlation of dS between two genes is more likely due to a shared mutation rate than an indicator of coevolution.
Several factors can contribute to correlation of evolutionary rates (Lovell and Robertson, 2010), such as obligate physical interaction of gene products (Mintseris and Weng, 2005), shared functional constraint (Zhang and Broughton, 2013), or gene expression levels (Subramanian and Kumar, 2004). Because the evolutionary rate of the mammalian mitochondrial genome is much higher than that of the nuclear genome, studies of correlated evolution between organellar and nuclear genomes have focused on proteins of enzyme complexes with subunits encoded in each of these compartments. Using this approach, studies have shown that some nuclear genes that encode products that participate in mitochondrial-localized complexes have a corresponding higher evolutionary rate relative to cytosol targeted nuclear gene products (Willett and Burton, 2004; Osada and Akashi, 2012; Barreto and Burton, 2013; Zhang and Broughton, 2013).
The correlation of evolutionary rates between plastid and nuclear genomes has rarely been studied because plastid genome sequences are generally more highly conserved than those of the nuclear genome (Wolfe et al., 1987; Drouin et al., 2008), making it difficult to select appropriate taxa and genes for analyses of correlated rate acceleration. Studies in Silene (Sloan et al., 2014) identified elevated protein sequence divergence in organelle-targeted, but not cytosolic, ribosomal proteins in pairwise comparisons of species with rapidly evolving mitochondrial and plastid DNA, suggesting that coevolution occurs between different compartments. Like the Silene study, many investigations have adopted pairwise species comparisons, an approach that does not account for the effects of shared phylogeny on predictions of coevolution (Barreto and Burton, 2013).
Methods that incorporate a phylogenetic framework have proven more accurate in detecting coevolution among interacting proteins than pairwise comparisons (Clark and Aquadro, 2010). Various methods have been developed that incorporate the effects of phylogeny for detecting gene coevolution (Pazos and Valencia, 2008; de Juan et al., 2013; Rao et al., 2014). The mirror tree method (Pazos and Valencia, 2001) was originally introduced to predict protein-protein interactions, and it quantifies rate correlations by estimating the similarities of corresponding phylogenetic trees. For each gene tree, the evolutionary rates on each branch are extracted to form a rate vector, and Pearson correlation coefficients are calculated between the rate vectors of two genes. Despite its popularity, the original mirror tree method does not effectively account for underlying phylogenetic histories. Different modifications of this method were developed to remove the effects of shared phylogeny by introducing a correction factor (Pazos et al., 2005; Sato et al., 2005). A more recent likelihood based approach evaluates the coevolution between genes using normalized dN, or dN/dS (Clark and Aquadro, 2010). In this method, the likelihoods of three models (null, correlated, and free) are calculated and the correlation is quantified as the proportional improvement of the likelihood of the correlated model to the null model, with respect to the maximal possible improvement gained by the free model over the null model.
Studies of coevolution of amino acids adopt a different set of approaches, which assess coevolution between two sites by detecting similar amino acid frequencies or substitution patterns calculated from the multiple sequence alignment (Göbel et al., 1994; Neher, 1994; Taylor and Hatrick, 1994). Dutheil and Galtier (2007) developed an approach (CoMap) that examines the coevolution of given amino acid sites using the known phylogenetic history. In this method, the ancestral state of a given amino acid site is inferred from the phylogenetic history and the sequence of changes that occur across time (branches on a phylogenetic tree) form a substitution vector. Structurally mediated coevolution of any amino acid site is then evaluated using the substitution vector and a cluster-based approach (Dutheil and Galtier, 2007). Another approach studies the coevolved amino acids by incorporating a continuous-time Markov process model (Yeang and Haussler, 2007). Both of these approaches agree well with experimental results; however, the latter approach is computationally demanding and therefore more feasible for studies of small protein domains.
In plants, the plastid-encoded RNA polymerase (PEP) is a multisubunit enzyme complex (Shiina et al., 2005) containing subunits encoded by genes in both the plastid (RNAP: rpoA, rpoB, rpoC1, and rpoC2) and nuclear genomes (SIG: sigma factor 1-6). Studies in Geraniaceae have revealed highly elevated evolutionary rates in the plastid genome, especially in rpoB, rpoC1, and rpoC2 (Guisinger et al., 2008; Weng et al., 2012), and highly divergent rpoA sequences in the genus Pelargonium (Chumley et al., 2006). The interaction of SIG and RNAP gene products provides an attractive platform for the study of coevolution between the two genomes. Using transcriptomic and genomic data from 27 species with a well-established phylogenetic framework, the entire sigma factor gene family in Geraniaceae has been characterized and a systematic correlation analysis of evolutionary rates between plastid and nuclear genomes was conducted. Despite an order of magnitude difference in the mutation rate between these two genomes (Wolfe et al., 1987; Drouin et al., 2008), we detected a correlation of evolutionary rates among 27 species representing the entire family. Furthermore, analyses of interacting amino acid pairs suggest that structurally mediated coevolution plays a minimal role in maintaining the coordination of evolutionary rates. The identification of rate correlations between RNAP and SIG genes suggests a plausible explanation for the observed plastome-genome incompatibility within Pelargonium and possibly other genera of flowering plants.
RESULTS
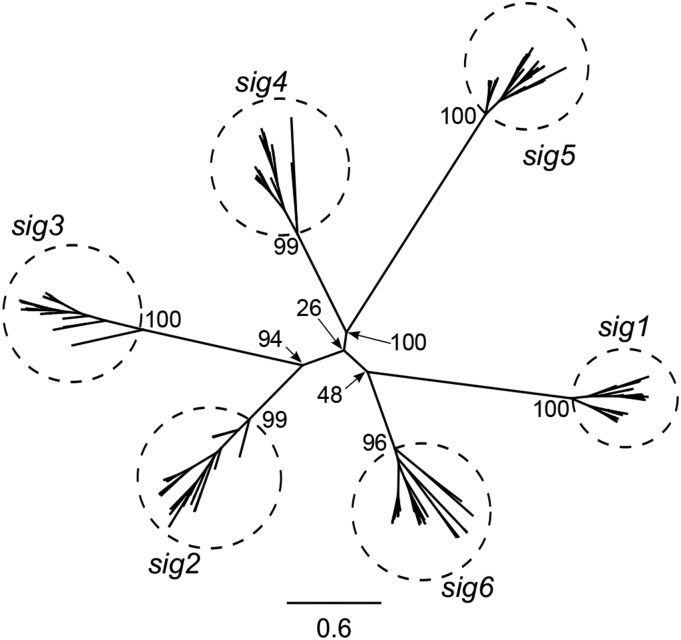
Transcriptome sequencing and assembly for 27 species was performed following Zhang et al. (2013). Sigma factor genes were extracted and accession numbers are provided in Supplemental Table 1. An amino acid maximum likelihood (ML) tree was generated to infer phylogenetic relationships among the 178 complete sigma factor (SIG) sequences identified from the 27 species in Geraniales and Arabidopsis thaliana (Figure 1; see Supplemental Data File 1 for alignments). The ML tree (-lnL = −65001.9) topology parsed the 178 sequences into six major clades. Two additional alignment algorithms (see Methods) were used and resulted in the same six major groups of sigma factor genes (Supplemental Figure 1; see Supplemental Data File 1 for alignments).
Figure 1.
Six Sigma Factor Families in Geraniales and Arabidopsis.
The unrooted amino acid-based ML tree was generated using 178 complete SIG sequences identified from 27 species of Geraniales and Arabidopsis. The ML tree (-lnL = −65001.9) topology parsed the 178 sequences into six subgroups (enclosed in labeled circles). Scale bar represents the number of amino acid substitutions per site. Numbers at nodes are bootstrap support values.
The copy number of individual SIG genes varied across different species (Supplemental Figure 2; see Supplemental Data File 1 for alignments). A single copy of sig1 and sig2 was found in all species except for Pelargonium transvaalense, Pelargonium tetragonum, and Geranium maderense, where two copies of sig2 were identified. A complete sig3 sequence was identified in all species except for P. tetragonum, Pelargonium myrrhifolium, and Pelargonium nanum. The sig4 sequence was detected in all Pelargonium and Geranium species, and, while a sig4 pseudogene missing the start codon was detected in Melianthus villosus, sig4 was not found in Francoa sonchifolia, Erodium chrysanthum, and Erodium gruinum. Two copies of sig5 and sig6 were identified in various species (Supplemental Figures 2E and 2F). Multiple copies of sig5 were identified in species of Geranium and Erodium and in California macrophylla, while two copies of sig6 were found in C. macrophylla and species of Erodium and Pelargonium. The sig6 gene of Hypseocharis bilobata contained multiple internal stop codons. RT-PCR confirmed 18 out of 21 bioinformatically identified gene duplication and pseudogenization events (Supplemental Table 1). Among the SIG gene families, 21 gene duplications and 10 losses were inferred with Notung (Durand et al., 2006) (Supplemental Figure 3 and Supplemental Table 2; see Supplemental Data File 1 for alignments) on the branches leading to the 27 species of Geraniaceae.
Evolutionary rates of each gene were estimated based on alignments from MAFFT (Katoh and Standley, 2013). To avoid biases of rates estimation specific to an alignment algorithm, rates based on alignments from two other tools, MUSCLE and ClustalW (Edgar, 2004; Larkin et al., 2007), were compared with MAFFT (see Supplemental Data File 1 for alignments). The agreement between rate estimates from the three alignment methods indicated that there was no or negligible bias due to the alignment method (Supplemental Table 3). Thus, MAFFT was used for all subsequent analyses.
Clade-specific rate acceleration was assessed for the four plastid RNA polymerase (RNAP) subunits, six nuclear-encoded SIG genes, and 20 control genes (Figure 2; Supplemental Figures 4 and 5). While 10 control genes from nuclear and plastid genomes were selected for all coevolution analyses, two additional sets of 10 nuclear control genes were randomly selected from the APVO database (see Methods) for the mirror tree analysis to reduce any bias of nuclear control gene sampling (see Supplemental Data File 2 for detailed control gene information).
Figure 2.
Shared Clade-Specific Nonsynonymous Rate (dN) Acceleration in Geraniaceae.
RNAP and SIG genes are highlighted in the key in blue and orange, respectively. Blue numerals on the constraint tree indicate shared dN acceleration in RNAP genes of the Pelargonium C clade. For a more detailed version of the constraint tree, see Supplemental Figure 5. Numbers at nodes indicate accelerated dN in corresponding gene from the key at left (0 to 14, plastid genes; 15 to 29, nuclear genes). Scale bar represents the number of nucleotide substitutions per codon. The long branch leading to E. chrysanthum and E. gruinum was interrupted for ease of visualization.
Although dN for the Pelargonium C clade was accelerated in all four RNAP genes and two SIG genes (rpoA, rpoB, rpoC1, rpoC2, sig1, and sig2) in an initial rank sum test, acceleration of rates of rpoA/B/C1 only remain significant after correction for multihypothesis testing (Figure 2), while two control genes have significant acceleration of rates in the Geranium and Erodium clades. Significant acceleration of dS in the Pelargonium C clade was observed for the rpoA gene alone (Supplemental Figure 4). Elevated dS in sig6 and five nuclear control genes was observed in Geranium, with no acceleration detected within other clades (Supplemental Figure 4; see Supplemental Data File 1 for alignments).
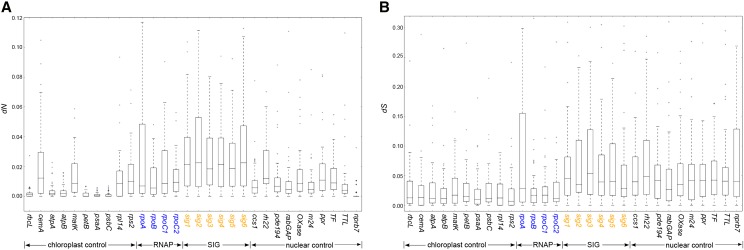
The values of dN and dS for each gene from all branches were used to analyze the rate correlation between gene pairs from RNAP, SIG, and control genes. The highest average values for dN were found in SIG genes followed by RNAP genes (Figure 3A; Supplemental Figure 5). Four plastid genes (cemA, matK, rpl14, and rps2) and one nuclear gene (rh22) had similar average dN values to the RNAP genes, and the other nuclear genes had slightly lower values. The lowest average dN values were found in the remaining plastid genes, which represent ATP synthase and photosynthetic genes. The dS values were similar among genes from the same cellular compartment (Figure 3B). The average dS values of nuclear genes were much higher than those of plastid genes except for rpoA, which had the highest dS value among plastid genes.
Figure 3.
Nonsynonymous (dN) and Synonymous (dS) Substitution Rates for Individual Genes.
Box plots represent the distribution of dN (A) or dS (B) value for each branch on the constraint tree (Supplemental Figure 5). While the dS values (B) were similar among genes from the same cellular compartment, the highest average values for dN (A) were found in SIG genes (orange) followed by RNAP genes (blue). The scale for dN and dS is different to facilitate visualization of the results.
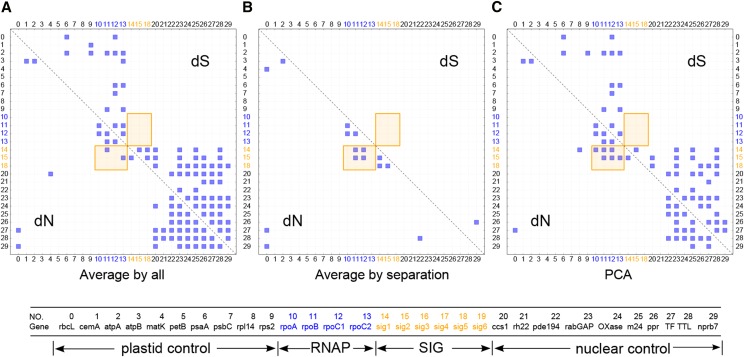
Correlation of dN and dS was evaluated for each gene pair by three variations of the mirror tree method, each of which adopts a different approach for removing the effect of shared phylogeny prior to tree similarity estimation (Pazos et al., 1997, 2005; Pazos and Valencia, 2001): average by all, average by separation, and principal component analysis (PCA; see Methods for detailed description). The sequences of sig1, sig2, and sig5 were grouped together for rate correlation analysis using complete sequences (Figure 4) or conserved domains (Supplemental Figure 6). Due to their absence in different species, the genes sig3, sig4, and sig6 were analyzed separately using complete sequences (Supplemental Figure 7) or conserved domains (Supplemental Figure 8). In addition to the initial 10 plastid and nuclear control genes, two additional sets of nuclear control genes were added to the sig1, sig2, and sig5 analyses (Supplemental Figures 9 and 10).
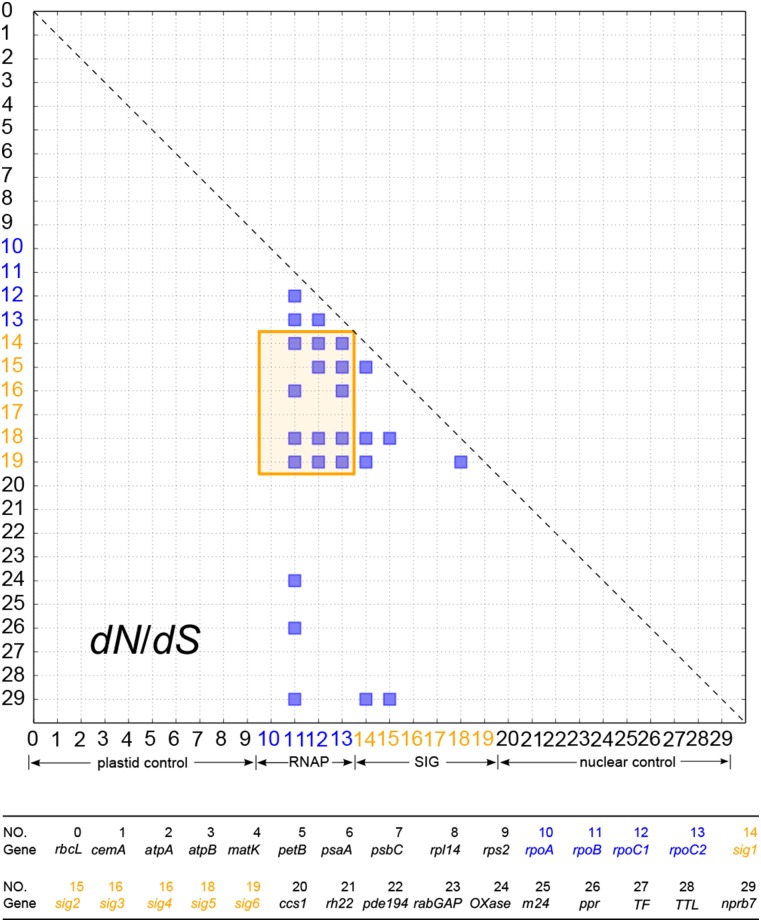
Figure 4.
Strong Correlation of Nonsynonymous (dN) but Not Synonymous (dS) Substitution Rates between sig1/2/5 and RNAP Genes Using Three Methods of Analysis.
The entire sequence of each gene was used in this analysis (see Methods). The correlation of dN and dS values were calculated by modifications of the mirror tree method average method (all) (A), average method (separation) (B), and PCA (C). All interaction pairs with a correlation coefficient of higher than 0.6 were considered significant and shown with a blue square. RNAP and SIG genes, highlighted in blue and orange fonts, respectively, show strong correlation of dN but not dS (highlighted in orange shaded box). Little to no correlation of dN between RNAP/SIG genes and the control genes (in black font) was detected. Gene names and cellular locations corresponding to each number are given below the diagram.
A cutoff of 0.6 for the Pearson correlation coefficient was used as an indicator of strong rate correlation (Sato et al., 2005). After removing the effects of shared phylogeny (see Methods), correlation of dN was detected between RNAP and sig1/2 genes (orange rectangle in Figures 4A and 4B), but not between RNAP/SIG and the control genes. The mirror tree methods did not detect a dN correlation between RNAP and sig3, sig4, or sig6 (Supplemental Figure 7). The correlation of dS was sensitive to the average method used in the analyses (Figures 4A and 4B). Correlation of dS was identified between certain plastid or nuclear gene pairs but not between the two groups when the average by all method was employed (Figure 4A), and only one pair of nuclear genes had correlated dS when the average by separation method was used (Figure 4B). Application of the PCA method produced correlations of dN and dS that were similar to those from the average by all method except that correlation of dN was detected between rpl14/rpoA and sig1 (Figure 4C). None of the three methods identified correlation of dS between RNAP and SIG genes (Figures 4A to 4C), suggesting that the correlation of dN was not due to the effects of background mutation rates. The number of gene pairs with positive rate correlations is shown in Table 1. Similar rate correlations were detected with the additional nuclear control genes (Supplemental Figures 9 and 10).
Table 1. The Number of Interaction Pairs with a Rate Coefficient of over 0.6 within Corresponding Genes Estimated by Three Mirror Tree Methods.
|
dN |
dS |
|||||
|---|---|---|---|---|---|---|
| Interactionsa | ρava | ρavs | ρpca | ρava | ρavs | ρpca |
| RNAP-RNAP (6) | 5 | 3 | 5 | 3 | 0 | 2 |
| RNAP-sig1 (4) | 1 | 2 | 3 | 0 | 0 | 0 |
| RNAP-sig2 (4) | 1 | 2 | 3 | 0 | 0 | 0 |
| RNAP-sig3 (4) | 0 | 0 | 0 | 0 | 1 | 0 |
| RNAP-sig4 (4) | 0 | 0 | 0 | 0 | 0 | 0 |
| RNAP-sig5 (4) | 0 | 0 | 0 | 0 | 0 | 0 |
| RNAP-sig6 (4) | 0 | 0 | 0 | 0 | 0 | 0 |
| RNAP-control (80) | 0 | 0 | 0 | 10 | 0 | 8 |
| sig1-control (20) | 0 | 0 | 1 | 4 | 0 | 3 |
| sig2-control (20) | 0 | 0 | 0 | 5 | 0 | 4 |
| sig3-control (20) | 8 | 5 | 1 | 3 | 5 | 3 |
| sig4-control (20) | 11 | 0 | 0 | 5 | 1 | 2 |
| sig5-control (20) | 0 | 0 | 0 | 8 | 0 | 5 |
| sig6-control (20) | 1 | 0 | 0 | 10 | 0 | 9 |
RNAP contains rpoA, rpoB, rpoC1, and rpoC2. Results of the average by all method (ρava), average by separation method (ρavs), and PCA method (ρpca) are shown.
The number in parentheses is the total number of interaction pairs within corresponding genes.
The correlation of dN and dS between RNAP and SIG genes using conserved domains was similar to that seen using the entire sequences; however, more gene pairs of RNAP and SIG were identified as correlated for dN (Supplemental Figures 6 and 8). The number of rate correlations of all gene pairs is shown in Supplemental Table 4.
The rate correlation coefficient between each individual gene and the RNAP genes was compared (Table 2). The dN correlation coefficients of RNAP and RNAP/sig1/sig2 genes were ranked significantly higher (P < 0.05 after correction for multihypothesis testing) than all other pairs by average by separation and PCA methods. Using the average by separation method, correlation coefficients of RNAP genes and sig6 were also ranked significantly higher than other pairs. No significantly higher rank was detected between RNAP and the plastid or nuclear control genes by any method. Synonymous substitution rate correlation coefficient ranking produced no significant result for any of the gene groups. The same tests were performed with rates calculated from the conserved domains of selected genes as described (Supplemental Table 5). Similar to the results generated using the entire sequences, the dN correlation coefficients of RNAP and RNAP/sig1/sig2/sig6 were ranked significantly higher than any other pairs by average by separation and PCA methods. The correlation coefficients of dN for RNAP and sig5 were ranked significantly higher using PCA method. The rank of dS correlation coefficients was the same as that using the entire sequences with no significant highly ranked gene groups detected.
Table 2. Rank Sum Test of Rate Correlation Coefficient.
|
dN |
dS |
|||||
|---|---|---|---|---|---|---|
| Interactions | ρava | ρavs | ρpca | ρava | ρavs | ρpca |
| RNAP-RNAP | + | + | + | − | − | − |
| RNAP-sig1 | − | + | + | − | − | − |
| RNAP-sig2 | − | + | + | − | − | − |
| RNAP-sig3 | − | − | − | − | − | − |
| RNAP-sig4 | − | − | − | − | − | − |
| RNAP-sig5 | − | − | − | − | − | − |
| RNAP-sig6 | − | + | − | − | − | − |
| RNAP-pt | − | − | − | − | − | − |
| RNAP-nu | − | − | − | − | − | − |
The entire sequence of each gene was used in the analysis. Correlation coefficients ranked significantly higher among all interaction pairs are indicated with “+” sign. Coefficients that are not ranked significantly higher are indicated with “−” sign. “pt” is the group of control genes from the plastid genome, and “nu” is the group of control genes from the nuclear genome. Results of the average by all method (ρava), average by separation method (ρavs), and PCA method (ρpca) are shown. Results from average method were estimated in two ways (all/separate; see Methods).
Correlation of normalized dN (dN/dS ratio) was evaluated with the proportional improvement method (Clark and Aquadro, 2010). Since the proportional improvement dN/dS test is more appropriate when dS is unsaturated (Clark and Aquadro, 2010), saturation was tested for each of the genes examined. To examine saturation of synonymous sites, values of dN and dS were plotted and linear/quadratic models were used to fit the data. If dS is saturated, the quadratic model with a concave curve should fit the data better. The two models were compared with the improvement of sum of squares explained by these models (Fares and Wolfe, 2003; Weng et al., 2014). Of the 30 genes tested, only rbcL showed significant improvement (P < 0.05) of sum of squares (Supplemental Table 6); however, rbcL is known to be a conservative gene with low dS values (dS < 0.15 for all branches) (Figure 3B).
Strong correlation (proportional improvement > 0.6) of dN/dS was identified between rpoB/C1/C2 and sig1/5/6 genes, between rpoC1/C2 and sig2 genes, and between rpoB/C2 and sig3 genes (Figure 5). Correlation of dN/dS was also identified among RNAP (between rpoB and rpoC1/C2; rpoC1 and rpoC2) and among SIG (between sig1 and sig2/5/6; sig2 and sig5) genes (Figure 5). A correlation of RNAP/SIG and control genes was lacking between most interaction pairs except for rpoB and three nuclear control genes (OXase, ppr, and nprb7) and sig1/2 and nprb7 genes (Figure 5). Compared with the mirror tree methods, more interaction pairs (proportional improvement, 13; average by all, 2; average by separation, 4; PCA, 6) were identified with strong rate correlation between RNAP and SIG genes, while fewer or comparable interaction pairs (proportional improvement, 5; average by all, 20; average by separation, 5; PCA, 2) were identified between RNAP/SIG and control genes.
Figure 5.
Strong Correlation of dN/dS between RNAP and SIG Genes.
The correlation of dN/dS was quantified using proportional improvement (Clark and Aquadro, 2010). Interaction pairs with a proportional improvement of higher than 0.6 were considered significant and shown with a blue square in the figure. RNAP and SIG genes, highlighted in blue and orange fonts, respectively, show strong correlation of dN/dS (highlighted in orange shaded box). Little to no correlation of dN/dS between RNAP/SIG genes and the control genes (in black font) was detected. Gene names corresponding to each number are given below the diagram.
To investigate the role of structurally mediated coevolution in the correlation of evolutionary rates between RNAP and SIG genes, CoMap (Dutheil and Galtier, 2007) was used to predict coevolved amino acid pairs by comparing the substitution vectors, weighted by the different amino acid properties (volume, charge, and polarity) at given positions (see Methods for more details). Since the β′ subunit of the cyanobacterial ancestor was split in the lineage leading to plants (Shiina et al., 2005), the relevant residues from the β′ and β′′ subunits in plants were combined for comparison with the β′ subunit in E. coli. The interaction sites between SIG and RNAP subunits were predicted by contact map analysis (Sobolev et al., 2005) and by estimation of the physical distance between two interacting residues. More interaction sites were predicted by contact map analysis than by distance estimation (Supplemental Table 7); however, few (0 to 20%) of the predicted coevolving amino acid sites overlapped with interaction sites (Supplemental Table 7). The analysis of distance distributions across the coevolved amino acid pairs suggested that among the 4223 residue pairs predicted to be involved in structurally mediated coevolution by CoMap, only one pair had a distance of <5 Å. The analyses of structurally mediated coevolution within amino acid pairs showed a minimal overlap between coevolved and interacting amino acid sites (Supplemental Table 7), with few of the coevolved residues in close physical proximity (<5 Å; Supplemental Figure 11).
DISCUSSION
Duplication and Loss of Sigma Factor Genes
Twenty-one duplicated SIG genes were identified across the Geraniaceae (Supplemental Figures 2 and 3 and Supplemental Table 2). Duplications of SIG genes have been documented in several angiosperms, including maize (Zea mays), rice (Oryza sativa), and poplar (Populus trichocarpa) (Lerbs-Mache, 2011). These duplicate copies may be functionally diversified to regulate gene expression at different developmental stages or under changing environmental conditions as seen for the duplicated sig1 of maize that is differentially expressed in etiolated leaves (Tan and Troxler, 1999; Lerbs-Mache, 2011). The pattern of SIG gene duplication in Geraniaceae could have arisen in several ways, including whole-genome duplication followed by elimination of some copies. Polyploidy is widespread across Geraniaceae (Widler-Kiefer and Yeo, 1987; Yu and Horn, 1988; Touloumenidou et al., 2007) and has likely contributed to duplication of SIG genes. However, an alternative explanation, that multiple, small-scale gene duplications have occurred (Davis and Petrov, 2004; Li et al., 2006) was supported by the pattern of SIG gene duplications observed in Geraniaceae (Supplemental Figures 2 and 3).
Multiple gene losses were detected in the SIG gene family in Geraniales. While it is possible that these genes are so lowly expressed as to fall below the level of detection, the high depth of coverage in transcriptome sequencing (Zhang et al., 2013) and the fact that the same genes were identified in transcriptomes of closely related species make this unlikely. The Sig4 protein specifically recognizes the promoter of ndhF, which encodes a subunit of NADH dehydrogenase in the plastid (Endo et al., 1999; Favory et al., 2005). The lack of sig4 transcripts in E. chrysanthum and E. gruinum is plausible given the loss of ndh genes from the plastid genomes of these species (Chris Blazier et al., 2011). The identification of a sig4 pseudogene in M. villosus also correlates with a recent loss of ndh genes in that species (Weng et al., 2014). The loss of both sig4 and ndh genes provides an explicit example of coevolution between the plastid and nuclear genomes.
Coevolution of Plastid and Nuclear Genomes
Genome coevolution is expected to produce correlated evolutionary rate changes between different genes. Studies of coevolution usually focus on protein sequences from multisubunit enzyme complexes. Correlated change of evolutionary rates has been widely observed in various organisms (Campo et al., 2008; Pazos and Valencia, 2008; Lovell and Robertson, 2010). A previous study showed that nuclear genes encoding subunits of enzyme complexes that assemble in the mitochondria with subunits encoded in the organelle have significantly higher evolutionary rates than genes whose products are targeted to the cytosol (Barreto and Burton, 2013). Likewise, analyses of coevolution between organellar and nuclear genomes have mainly focused on genes from mitochondrial and nuclear genomes (Zhang and Broughton, 2013; Barreto and Burton, 2013). A recent study of Silene (Sloan et al., 2014) suggested that coevolution occurred between plastid and nuclear genomes based on the observation of elevated protein sequence divergence in organelle targeted, but not cytosolic, ribosomal proteins for species with fast evolving mitochondrial and plastid genomes. The unusually high substitution rates of genes in the plastid genomes of Geraniaceae (Chumley et al., 2006; Guisinger et al., 2008; Weng et al., 2012) provide an attractive system for the study of coevolution. Using both transcriptome and genome data and a well-characterized phylogenetic framework, this systematic analysis revealed the existence of correlation of evolutionary rates, evidence of coevolution between plastid and nuclear genomes at different levels (dN and dN/dS).
Correlation of dN but not dS was detected in gene pairs between RNAP and SIG genes, suggesting that the correlation of dN was not due to shared background mutation rates. The absence of correlation of dN between RNAP/SIG and control genes indicates that the rate correlation between RNAP and SIG genes is likely due to coevolution between plastid and nuclear genome or a local functional constraint acting on RNAP and SIG genes, rather than a global constraint on dN of all genes. The case of dN correlation between rpoA and rpl14, detected exclusively by the PCA method, may be a result of factors other than direct physical interaction, such as a common functional role in gene expression in plastids (transcription for sig1 and translation for rpl14) (Agrafioti et al., 2005; Chen and Dokholyan, 2006).
Because correlation coefficients of gene pairs with known interactions are significantly higher than unrelated sequences (Clark et al., 2012), correlation coefficients between each SIG gene and the four RNAP genes should be higher than those between any control genes and the four RNAP genes. The significantly highly ranked correlation coefficients of dN between RNAP and sig1/2 by any method supports the conclusion that there is a strong rate correlation between RNAP and sig1/2 genes (Table 2). The significantly highly ranked rate correlation detected between RNAP and sig5/6 genes using conserved domain sequences in both average and PCA methods suggests that there might be correlation between RNAP and sig5/6 genes but that it is weaker than those between RNAP and sig1/2 (Supplemental Table 4).
A correlation analysis of normalized dN (dN/dS ratio) was performed (Clark and Aquadro, 2010), and this approach identified additional strong rate correlations between RNAP (rpoB/C1/C2) and sig3/5/6. The low number (5/400) of strong rate correlation pairs (Figure 5) between RNAP/SIG and control genes is the result of either weaker correlation or false discoveries.
Across all methods, strong rate correlations (dN and/or dN/dS) are present for RNAP and all SIG except sig4. Rate correlations among interacting genes are affected by several factors (Lovell and Robertson, 2010), such as physical interaction (Mintseris and Weng, 2005), functional constraint (Zhang and Broughton, 2013), or gene expression levels (Fraser et al., 2004). The sig4 gene is involved in the transcription of ndhF (Favory et al., 2005). Knockout studies of sig4 in Arabidopsis revealed no observable phenotypes (Lerbs-Mache, 2011), and the loss of it and the corresponding plastid encoded ndh genes (Chris Blazier et al., 2011) in multiple species in Geraniaceae suggests that sig4 is dispensable. Relaxed functional constraint may contribute to the absence of coevolution between RNAP and sig4 genes.
Possible phenomena that could underlie the rate correlations between RNAP and SIG genes include: (1) a cause-and-effect relationship between rate variation of RNAP and SIG genes or (2) a common factor affecting rates of both RNAP and SIG genes, such as relaxed functional constraint acting on both gene sets (Subramanian and Kumar, 2004; Mintseris and Weng, 2005; Lovell and Robertson, 2010; Zhang and Broughton, 2013). If the first explanation were correct, rate correlations between RNAP and SIG genes could be due to structurally mediated compensatory evolution. However, results suggest that structurally mediated coevolution plays a minor role in maintaining rate correlations between SIG and RNAP subunits and other factors contributing to compensatory evolution could not be excluded. Shared functional constraint is another agent that may be maintaining the observed correlations. Additional study is required to further elucidate the contribution of functional constraints, gene expression, or other factors in the correlation of evolutionary rates of PEP subunits in Geraniaceae.
Plastome-genome incompatibility (PGI), which was first documented in Pelargonium species (Smith, 1915), is observed across flowering plants (Schmitz-Linneweber et al., 2005; Greiner et al., 2008; Weihe et al., 2009; Greiner et al., 2011). Various mechanisms have been proposed for PGI, including impaired interactions between cis-elements and their cognate nuclear factors involved in transcription and/or transcript stability. In Oenothera, perturbations of photosystem II activity, presumed to be caused by changes in transcription of the psbB gene, contributes to PGI (Greiner et al., 2008). Likewise, steady state RNA levels of three PEP-controlled genes were severely reduced in leaf sections taken from variegated, interspecific hybrids of Zantedeschia (Yao and Cohen, 2000). The two nuclear genes included in the Zantedeschia study also evidenced low levels of mRNA; expression of cab and rbsS is known to be regulated by retrograde plastid to nuclear signals and would therefore be susceptible to the PGI phenotype (Ruckle et al., 2007). Correlation of accelerated nucleotide substitution rates between SIG and RNAP genes provides a plausible explanation for PGI in Geraniaceae (Weihe et al., 2009; Greiner et al., 2011). Specifically, the high evolutionary rates and rate correlation between SIG and RNAP genes within species could lead to interspecific incompatibilities, and such incompatibilities would reduce efficiency or even cause dysfunction of the PEP holoenzyme, impairing the transcription of essential plastid genes. The role of sigma factors in transcription initiation through cis-element binding and polymerase recruitment suggests similarity between the Geraniaceae, Zantedeschia, and Oenothera PGI systems.
METHODS
RNA Isolation, Transcriptome Sequencing, and Assembly
Total RNA was isolated from newly emerged leaves of 26 species in Geraniales (Supplemental Figure 5), and four tissues (emergent and expanded leaves, roots, and flowers) of Pelargonium × hortorum following the protocols described by Zhang et al. (2013). Transcriptome sequencing was performed on the HiSequation 2000 platform, and the sequence data were preprocessed and assembled as described by Zhang et al. (2013).
Identification of Sigma Factors
SIG sequences were extracted from transcriptome assemblies with reciprocal BLAST as described by Zhang et al. (2013). The orthologous genes of each class of SIG were determined by reciprocal BLAST and single-gene phylogeny. RT-PCR was performed to verify the problematic SIG gene sequences with internal stop codons or missing 5′ or 3′ ends. For any species in which multiple gene sequences were identified as the same class of sigma factor, RT-PCR was performed to verify the existence of all the gene sequences (for primers, see Supplemental Table 8). All RT-PCR products were subjected to Sanger sequencing to confirm the result.
Phylogenetic Analysis
Multiple sequence alignments were done using MAFFT (Katoh and Standley, 2013), MUSCLE (Edgar, 2004), and ClustalW (Larkin et al., 2007) with default parameters in Geneious 6.0 (Biomatters, http://www.geneious.com/) (see Supplemental Data File 1 for alignments). Amino acid-based ML trees for all SIG genes were constructed by RAxML (Stamatakis, 2006) with parameters “raxmlHPC-PTHREADS-SSE3 -f a -x 12345 -p 12345 -T 12 -m PROTGAMMAJTT -N 100.” A Perl script (http://sco.h-its.org/exelixis/web/software/raxml/hands_on.html) was used to examine all protein models and the model with the best likelihood score (JTT) was selected. ML trees of each class of SIG genes were constructed by RAxML (Stamatakis, 2006) with parameters “raxmlHPC-PTHREADS-SSE3 -f a -x 12345 -p 12345 -T 12 -m GTRGAMMAI -N 100.” Bootstrap values were generated using RAxML with 100 replicates and the above settings.
Evolutionary Rate Estimation
PAML’s codeml (Yang, 2007) was used to estimate dN, dS, and dN/dS using the codon frequencies model F3X4. Gapped regions were excluded with parameter “cleandata = 1.” The constraint tree was generated by RAxML using 12 plastid genes (atpA, atpB, atpI, ccsA, cemA, matK, petA, rbcL, rpoB, rpoC1, rpoC2, and rps2) with a total length of 21,500 bp. Bootstrap values were generated using RAxML with 100 replicates and the above settings. Ten plastid genes (rbcL, cemA, atpA, atpB, matK, petB, psaA, psbC, rpl14, and rps2) from different functional groups and 30 nuclear genes with three different subcellular targeting sites (plastid, mitochondria, and other), which are orthologous to genes in the APVO database (Duarte et al., 2010), were used as negative control groups. The APVO database was separated into three groups based on their subcellular locations (plastid, mitochondria, and other), and an approximately equal number of genes were selected randomly from each group. The plastid genes were extracted from the annotated plastid genome assemblies as described by Weng et al. (2014). Thirty Arabidopsis thaliana nuclear genes were downloaded from TAIR (Lamesch et al., 2012), and the corresponding accession numbers are in Supplemental Data File 2.
Analysis of Correlation of Evolutionary Rate
Rates along branches leading to and within Pelargonium A, B, and C clades, Monsonia, Geranium, and Erodium I and II clades were grouped separately for clade specific rate acceleration analysis. The rank sum test was performed to test clade-specific rate accelerations, and a P value of <0.05 was considered significant. Correction for multihypothesis testing was performed by adjusting the original P value with the false discovery rate correction method using the built-in p.adjust function (method=“fdr”) in the R software package (http://www.r-project.org). After correction, the false discovery rate among the significantly accelerated clades within each gene is <5%.
Correlation coefficients of evolutionary rates dN and dS between each gene pair were estimated using modified mirror tree methods (Pazos et al., 1997, 2005; Pazos and Valencia, 2001). Specifically, the evolutionary rates, dN or dS, on each branch of a given gene tree were collected to form a rate vector. The rate correlation was quantified using the Pearson correlation coefficient between the rate vectors of different genes. The rate vector of each gene pair was adjusted via vector projection by a correction vector representing the shared phylogenetic effects prior to the comparison. The correction vector was generated with different modifications of the mirror tree method, the average method, and PCA (Sato et al., 2005). In the average method, the correction vector was determined in two ways, the average by all and the average by separation, in which either the correction vector was defined as the average of rate vectors of all genes or two different correction vectors for nuclear and plastid genes were defined separately, as the average of rate vectors of corresponding gene groups. In PCA, the correction vector was calculated as the first principle component of the rate matrix formed by rate vectors of all genes.
The correlation of dN/dS ratio of most (447/450) interaction pairs, quantified as “proportional improvement” as described (Clark and Aquadro, 2010), was analyzed using HYPHY (Pond et al., 2005) with batch scripts downloaded from http://mbg.cornell.edu/cals/mbg/research/aquadro-lab/software.cfm (Clark and Aquadro, 2010). Specifically, the evolutionary rates and the likelihood of three (correlated, null, and free) models for the estimated rates of each gene pair were evaluated with HYPHY, and the proportional improvement method estimates correlation of dN/dS ratio by calculating the proportional improvement of likelihood of the correlated model over the null model, with respect to the maximal possible improvement gained by the free model over the null model (Clark and Aquadro, 2010). The remaining interaction pairs (3/450, psbC and psaA, psbC/psaA and nprb7) were analyzed using the same batch script template with modifications so that the likelihoods of the correlation model with different start points (−0.8, 1, and 1.3) were optimized separately rather than sequentially. The test for dS saturation was performed as described by Fares and Wolfe (2003) and Weng et al. (2014).
The conserved domains of RNAP and SIG genes were predicted by NCBI CDD (Marchler-Bauer et al., 2013). The predicted conserved domains were used for rate analysis, except for rpoC1 and rpoC2, because conserved domains were predicted to comprise the entire sequence for both of these genes. Rate corrections were done by custom python scripts and are available as Supplemental Data File 3. The Pearson correlation coefficients were calculated using the built in function in the python scipy module. A correlation coefficient value of 0.6 or above was used to indicate a positive rate correlation (Sato et al., 2005). The rate correlation of each gene with itself was removed from all analyses. The one-side Wilcoxon Rank Sum test and correction for multihypothesis testing was performed using R software package as described above.
Structurally mediated coevolution within groups of amino acids was evaluated for 28 plant species (27 Geraniales from this study plus Arabidopsis) and Escherichia coli using CoMap (Dutheil and Galtier, 2007). To evaluate structurally mediated coevolution between amino acids from different genes, three different features (volume, charge, and polarity) of amino acids were considered and amino acid substitutions at specific sites on each branch of the gene tree were quantified based on these features. For each gene, the changes of amino acids on all branches at each site were extracted to form a site-specific substitution vector. The site-specific substitution vectors from two different genes were compared with find structurally mediated coevolved changes (i.e., volume increase at site X of gene A and volume decrease at site Y of gene B) of sites from different genes. The interacting amino acid pairs between SIG and RNAP subunits were predicted with CMA (Sobolev et al., 2005) and by mapping the amino acid pairs with distance between them of <5 Å (Hu and Yan, 2009) in RNA polymerase of E. coli to those of Geraniales using custom python scripts (Supplemental Data File 3). The distribution of distances of the coevolved amino acid pairs identified in the structurally mediated evolutionary analysis and the overlap between coevolved and the interacting amino acid pairs were executed with custom python scripts (Supplemental Data File 3).
Accession Numbers
All sequences have been submitted to GenBank/EMBL data libraries under accession numbers KJ916247 through KJ917105, and KM461126 through KM461665.
Supplemental Data
Supplemental Figure 1. Phylogeny of the Sigma Factor Families in Geraniales and Arabidopsis.
Supplemental Figure 2. Copy Number of the Six SIG Genes Varies across Geraniales.
Supplemental Figure 3. Multiple Gene Duplication and Loss Events in Geraniales.
Supplemental Figure 4. Shared clade-specific synonymous rate (dS) acceleration in Geraniaceae.
Supplemental Figure 5. Maximum likelihood tree of 27 species from Geraniales and Arabidopsis.
Supplemental Figure 6. Strong Correlation of Nonsynonymous (dN) but Not Synonymous (dS) Substitution Rates between sig1/2/5 and RNAP Genes by Three Methods Using Conserved Domains.
Supplemental Figure 7. Little to No Correlation of Nonsynonymous (dN) or Synonymous (dS) Substitution Rate between sig3/4/6 and RNAP Genes by Three Methods.
Supplemental Figure 8. Little to No Correlation of Nonsynonymous (dN) or Synonymous (dS) Substitution Rate between sig3/4/6 and RNAP Genes by Three Methods Using Conserved Domains.
Supplemental Figure 9. Strong Correlation of Nonsynonymous (dN) but Not Synonymous (dS) Substitution Rates between sig1/2/5 and RNAP Genes by Three Methods Using the Entire Sequences and a Second Set of 10 Nuclear Control Genes.
Supplemental Figure 10. Strong Correlation of Nonsynonymous (dN) but Not Synonymous (dS) Substitution Rates between sig1/2/5 and RNAP Genes by Three Methods Using the Entire Sequences and a Second Set of 10 Nuclear Control Genes.
Supplemental Figure 11. The Distribution of Distance between Amino Acid Pairs Predicted to Be Involved in Structurally Mediated Coevolution.
Supplemental Table 1. Summary of Accession Numbers, RT-PCR Results, and Voucher Information for All Species Examined.
Supplemental Table 2. Summary of Gene Duplication and Loss Events.
Supplemental Table 3. Pairwise Comparison of Evolutionary Rates from Different Alignment Methods.
Supplemental Table 4. The Number of Interaction Pairs with a Rate Coefficient over 0.6 within Corresponding Genes.
Supplemental Table 5. Rank Sum Test of Rate Correlation Coefficient Using Conserved Domains.
Supplemental Table 6. Test of dS Saturation of 30 Genes.
Supplemental Table 7. Analysis of Interaction Sites and Overlap between the Coevolving and Interaction Sites.
Supplemental Table 8. Primer Pairs Used for Amplification of Sigma Factor Genes.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.m02v7.
Supplemental Data File 1. Alignments of Different Genes by Different Tools.
Supplemental Data File 2. List of Plastid and Nuclear Control Genes.
Supplemental Data File 3. Custom Scripts Used in This Study.
Supplementary Material
Acknowledgments
We thank Mao-Lun Weng for helpful discussions on rate analyses and the graphic icon photograph, Scott Hunicke-Smith and Heather Deiderick of the University of Texas GSAF for assistance with Illumina sequencing, Claus Wilke for suggestions on correction of multihypothesis testing, Mario Fares for suggestions on dS saturation test, Chen-Hsiang Yeang for suggestions on analysis of coevolution of amino acids, Nancy Moran, Ahmed Bahieldin, Greg Clark, and three anonymous reviewers for comments on an earlier version of the article, and Texas Advanced Computing Center for supercomputer access. Support was provided by the National Science Foundation (IOS-1027259 to R.J.K. and T.A.R.) and from Vice President for Educational Affairs Abdulrahman O. Alyoubi at King Abdulaziz University, Jeddah, Saudi Arabia, to J.Z. and J.S.
AUTHOR CONTRIBUTIONS
J.Z., T.A.R., and R.K.J. designed the research. J.Z. performed research and analyzed the data. T.A.R. isolated RNA and assisted with experiments. J.C.B. contributed plastid genome data. J.Z. wrote the article. T.A.R., J.S., J.C.B., and R.K.J. critically revised the article. All authors read and approved the article.
Glossary
- PEP
plastid-encoded RNA polymerase
- ML
maximum likelihood
- PCA
principal component analysis
- PGI
plastome-genome incompatibility
References
- Agrafioti I., Swire J., Abbott J., Huntley D., Butcher S., Stumpf M.P.H. (2005). Comparative analysis of the Saccharomyces cerevisiae and Caenorhabditis elegans protein interaction networks. BMC Evol. Biol. 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto F.S., Burton R.S. (2013). Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol. Biol. Evol. 30: 310–314. [DOI] [PubMed] [Google Scholar]
- Campo D.S., Dimitrova Z., Mitchell R.J., Lara J., Khudyakov Y. (2008). Coordinated evolution of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 105: 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Dokholyan N.V. (2006). The coordinated evolution of yeast proteins is constrained by functional modularity. Trends Genet. 22: 416–419. [DOI] [PubMed] [Google Scholar]
- Chris Blazier J., Guisinger M.M., Jansen R.K. (2011). Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol. Biol. 76: 263–272. [DOI] [PubMed] [Google Scholar]
- Chumley T.W., Palmer J.D., Mower J.P., Fourcade H.M., Calie P.J., Boore J.L., Jansen R.K. (2006). The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23: 2175–2190. [DOI] [PubMed] [Google Scholar]
- Clark N.L., Alani E., Aquadro C.F. (2012). Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 22: 714–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark N.L., Aquadro C.F. (2010). A novel method to detect proteins evolving at correlated rates: identifying new functional relationships between coevolving proteins. Mol. Biol. Evol. 27: 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.C., Petrov D.A. (2004). Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2: E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D., Pazos F., Valencia A. (2013). Emerging methods in protein co-evolution. Nat. Rev. Genet. 14: 249–261. [DOI] [PubMed] [Google Scholar]
- Drouin G., Daoud H., Xia J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49: 827–831. [DOI] [PubMed] [Google Scholar]
- Duarte J.M., Wall P.K., Edger P.P., Landherr L.L., Ma H., Pires J.C., Leebens-Mack J., dePamphilis C.W. (2010). Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 10: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D., Halldórsson B.V., Vernot B. (2006). A hybrid micro-macroevolutionary approach to gene tree reconstruction. J. Comput. Biol. 13: 320–335. [DOI] [PubMed] [Google Scholar]
- Dutheil J., Galtier N. (2007). Detecting groups of coevolving positions in a molecule: a clustering approach. BMC Evol. Biol. 7: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Shikanai T., Takabayashi A., Asada K., Sato F. (1999). The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 457: 5–8. [DOI] [PubMed] [Google Scholar]
- Fares M.A., Wolfe K.H. (2003). Positive selection and subfunctionalization of duplicated CCT chaperonin subunits. Mol. Biol. Evol. 20: 1588–1597. [DOI] [PubMed] [Google Scholar]
- Favory J.-J., Kobayshi M., Tanaka K., Peltier G., Kreis M., Valay J.-G., Lerbs-Mache S. (2005). Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Res. 33: 5991–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.B., Hirsh A.E., Wall D.P., Eisen M.B. (2004). Coevolution of gene expression among interacting proteins. Proc. Natl. Acad. Sci. USA 101: 9033–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel U., Sander C., Schneider R., Valencia A. (1994). Correlated mutations and residue contacts in proteins. Proteins 18: 309–317. [DOI] [PubMed] [Google Scholar]
- Greiner S., Rauwolf U., Meurer J., Herrmann R.G. (2011). The role of plastids in plant speciation. Mol. Ecol. 20: 671–691. [DOI] [PubMed] [Google Scholar]
- Greiner S., Wang X., Herrmann R.G., Rauwolf U., Mayer K., Haberer G., Meurer J. (2008). The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol. Biol. Evol. 25: 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger M.M., Kuehl J.V., Boore J.L., Jansen R.K. (2008). Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. USA 105: 18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Yan C. (2009). A tool for calculating binding-site residues on proteins from PDB structures. BMC Struct. Biol. 9: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011). Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol. Biol. 76: 235–249. [DOI] [PubMed] [Google Scholar]
- Li L., Huang Y., Xia X., Sun Z. (2006). Preferential duplication in the sparse part of yeast protein interaction network. Mol. Biol. Evol. 23: 2467–2473. [DOI] [PubMed] [Google Scholar]
- Lobo F.P., Mota B.E.F., Pena S.D.J., Azevedo V., Macedo A.M., Tauch A., Machado C.R., Franco G.R. (2009). Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS ONE 4: e6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S.C., Robertson D.L. (2010). An integrated view of molecular coevolution in protein-protein interactions. Mol. Biol. Evol. 27: 2567–2575. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41: D348–D352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintseris J., Weng Z. (2005). Structure, function, and evolution of transient and obligate protein-protein interactions. Proc. Natl. Acad. Sci. USA 102: 10930–10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. (1994). How frequent are correlated changes in families of protein sequences? Proc. Natl. Acad. Sci. USA 91: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N., Akashi H. (2012). Mitochondrial-nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome C oxidase complex. Mol. Biol. Evol. 29: 337–346. [DOI] [PubMed] [Google Scholar]
- Pazos F., Helmer-Citterich M., Ausiello G., Valencia A. (1997). Correlated mutations contain information about protein-protein interaction. J. Mol. Biol. 271: 511–523. [DOI] [PubMed] [Google Scholar]
- Pazos F., Ranea J.A.G., Juan D., Sternberg M.J.E. (2005). Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J. Mol. Biol. 352: 1002–1015. [DOI] [PubMed] [Google Scholar]
- Pazos F., Valencia A. (2008). Protein co-evolution, co-adaptation and interactions. EMBO J. 27: 2648–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F., Valencia A. (2001). Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 14: 609–614. [DOI] [PubMed] [Google Scholar]
- Pond S.L.K., Frost S.D.W., Muse S.V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679. [DOI] [PubMed] [Google Scholar]
- Rao V.S., Srinivas K., Sujini G.N., Kumar G.N.S. (2014). Protein-protein interaction detection: methods and analysis. Int. J. Proteomics 2014: 147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle M.E., DeMarco S.M., Larkin R.M. (2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Yamanishi Y., Kanehisa M., Toh H. (2005). The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics 21: 3482–3489. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Kushnir S., Babiychuk E., Poltnigg P., Herrmann R.G., Maier R.M. (2005). Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase α-subunit mRNA. Plant Cell 17: 1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T., Tsunoyama Y., Nakahira Y., Khan M.S. (2005). Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 244: 1–68. [DOI] [PubMed] [Google Scholar]
- Sloan D.B., Triant D.A., Wu M., Taylor D.R. (2014). Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol. Biol. Evol. 31: 673–682. [DOI] [PubMed] [Google Scholar]
- Smith L. (1915). Variegation in Pelargonium. Proc. R. Hortic. Soc. 41: 36. [Google Scholar]
- Sobolev V., Eyal E., Gerzon S., Potapov V., Babor M., Prilusky J., Edelman M. (2005). SPACE: a suite of tools for protein structure prediction and analysis based on complementarity and environment. Nucleic Acids Res. 33: W39–W43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Kumar S. (2004). Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Troxler R.F. (1999). Characterization of two chloroplast RNA polymerase sigma factors from Zea mays: photoregulation and differential expression. Proc. Natl. Acad. Sci. USA 96: 5316–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W.R., Hatrick K. (1994). Compensating changes in protein multiple sequence alignments. Protein Eng. 7: 341–348. [DOI] [PubMed] [Google Scholar]
- Touloumenidou T., Bakker F.T., Albers F. (2007). The phylogeny of Monsonia L. (Geraniaceae). Plant Syst. Evol. 264: 1–14. [Google Scholar]
- Weihe A., Apitz J., Pohlheim F., Salinas-Hartwig A., Börner T. (2009). Biparental inheritance of plastidial and mitochondrial DNA and hybrid variegation in Pelargonium. Mol. Genet. Genomics 282: 587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M.-L., Blazier J.C., Govindu M., Jansen R.K. (2014). Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31: 645–659. [DOI] [PubMed] [Google Scholar]
- Weng M.-L., Ruhlman T.A., Gibby M., Jansen R.K. (2012). Phylogeny, rate variation, and genome size evolution of Pelargonium (Geraniaceae). Mol. Phylogenet. Evol. 64: 654–670. [DOI] [PubMed] [Google Scholar]
- Widler-Kiefer H., Yeo P.F. (1987). Fertility relationships of Geranium (Geraniaceae): sect. Ruberta, Anemonifolia, Lucida and Unguiculata. Plant Syst. Evol. 155: 283–306. [Google Scholar]
- Willett C.S., Burton R.S. (2004). Evolution of interacting proteins in the mitochondrial electron transport system in a marine copepod. Mol. Biol. Evol. 21: 443–453. [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Li W.H., Sharp P.M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yao J.-L., Cohen D. (2000). Multiple gene control of plastome-genome incompatibility and plastid DNA inheritance in interspecific hybrids of Zantedeschia. Theor. Appl. Genet. 101: 400–406. [Google Scholar]
- Yeang C.-H., Haussler D. (2007). Detecting coevolution in and among protein domains. PLOS Comput. Biol. 3: e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.-N., Horn W.H. (1988). Additional chromosome numbers in Pelargonium (Geraniaceae). Plant Syst. Evol. 159: 165–171. [Google Scholar]
- Zhang F., Broughton R.E. (2013). Mitochondrial-nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol. Evol. 5: 1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ruhlman T.A., Mower J.P., Jansen R.K. (2013). Comparative analyses of two Geraniaceae transcriptomes using next-generation sequencing. BMC Plant Biol. 13: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.