Using RNA-Seq and end-mapping, we showed that two Arabidopsis Mini-III proteins, which are putative double-stranded plastid endonucleases, process rRNA, introns, and a plastid noncoding RNA.
Abstract
RNase III proteins recognize double-stranded RNA structures and catalyze endoribonucleolytic cleavages that often regulate gene expression. Here, we characterize the functions of RNC3 and RNC4, two Arabidopsis thaliana chloroplast Mini-RNase III-like enzymes sharing 75% amino acid sequence identity. Whereas rnc3 and rnc4 null mutants have no visible phenotype, rnc3/rnc4 (rnc3/4) double mutants are slightly smaller and chlorotic compared with the wild type. In Bacillus subtilis, the RNase Mini-III is integral to 23S rRNA maturation. In Arabidopsis, we observed imprecise maturation of 23S rRNA in the rnc3/4 double mutant, suggesting that exoribonucleases generated staggered ends in the absence of specific Mini-III-catalyzed cleavages. A similar phenotype was found at the 3′ end of the 16S rRNA, and the primary 4.5S rRNA transcript contained 3′ extensions, suggesting that Mini-III catalyzes several processing events of the polycistronic rRNA precursor. The rnc3/4 mutant showed overaccumulation of a noncoding RNA complementary to the 4.5S-5S rRNA intergenic region, and its presence correlated with that of the extended 4.5S rRNA precursor. Finally, we found rnc3/4-specific intron degradation intermediates that are probable substrates for Mini-III and show that B. subtilis Mini-III is also involved in intron regulation. Overall, this study extends our knowledge of the key role of Mini-III in intron and noncoding RNA regulation and provides important insight into plastid rRNA maturation.
INTRODUCTION
Chloroplasts have their own genomes but are dependent on nucleus-encoded enzymes and accessory factors to generate functional transcripts. These enzymes and factors include a variety of RNases (Stoppel and Meurer, 2012) and large families of sequence-specific RNA binding proteins, such as those of the pentatricopeptide repeat (PPR) and RNA recognition motif families (Ruwe et al., 2011; Nakamura et al., 2012). Evidence from high-throughput RNA sequencing (RNA-Seq) and microarrays suggests that both strands of the ∼150-kb chloroplast genome are fully transcribed (Hotto et al., 2011; Zmieńko et al., 2011; Zhelyazkova et al., 2012), emphasizing the critical role of posttranscriptional processes in defining the steady state RNA population.
The critical function of multiple chloroplast RNases has been demonstrated through analysis of mutants defective for both endoribonucleases and exoribonucleases. For example, the absence of the endoribonuclease RNase E leads to severe chlorosis and the accumulation of long mRNA precursors (Walter et al., 2010), and deficiency of the endoribonuclease/exoribonuclease RNase J results in massive accumulation of antisense RNA (Sharwood et al., 2011a). Absence of either of the two prominent exoribonucleases, polynucleotide phosphorylase and RNase II, leads to defects in 3′ end maturation of mRNAs and rRNAs, and depletion of both causes embryo lethality in Arabidopsis thaliana (Germain et al., 2011, 2012). Clearly, accurate RNA metabolism and processing is essential for plastid biogenesis and function.
RNase III proteins are also important for RNA processing through their recognition of double-stranded RNA. The best-known plant RNase III enzymes are the Dicer-like proteins involved in RNA silencing that contain both a canonical RNase III domain and several double-stranded RNA binding domains (dsRBDs) (Liu et al., 2009). Bacterial RNase III, however, possesses one dsRBD (MacRae and Doudna, 2007), and more recently, a variant termed Mini-III was discovered that is limited to a single RNase III domain lacking the dsRBD (Redko et al., 2008). The Arabidopsis nuclear genome encodes four non-Dicer-like RNase III proteins with putative plastid-targeting sequences, designated here as RNC1 to RNC4 (reviewed in Olmedo and Guzmàn, 2008). RNC1 has a duplicated RNase III domain with multiple mutations in known catalytic residues. Maize (Zea mays) RNC1 has been shown to lack catalytic activity but is critical for chloroplast RNA splicing (Watkins et al., 2007). RNC2 (RTL2) was reported to be confined to the nucleus and cytoplasm and is involved in cleaving the 3′ external transcribed spacer of a pre-rRNA transcript (Comella et al., 2008). RNC3 and RNC4, the subjects of this study, resemble Mini-III and are candidates for a catalytic plastid RNase III. In addition to transit peptide predictions, RNC3 and RNC4 have been found in proteomic surveys of the chloroplast (Kim et al., 2009; Nishimura et al., 2013).
Potential functions for RNC3 and RNC4 include rRNA maturation. In Bacillus subtilis, Mini-III was shown to mature both the 5′ and 3′ ends of 23S rRNA (Redko et al., 2008), probably in a concerted fashion. RNC3 and/or RNC4 also might be required to resolve double-stranded RNAs found in mRNA-stabilizing inverted repeats or duplexes formed between sense and antisense RNAs. Here, we use null mutants generated by T-DNA insertions to show that chloroplast RNC3 and RNC4 are largely redundant but that a double mutant has multiple defects in maturation of the rRNA polycistron. Strand-specific RNA-Seq was used to validate the identified defects and also revealed the accumulation of a noncoding RNA complementary to a misprocessed rRNA operon intergenic region. Finally, several spliced introns show an abnormal degradation pattern in the double mutant, leading to the conclusion that chloroplast Mini-III acts on multiple classes of RNA, suggestive of a broad role in resolving RNA-RNA duplexes.
RESULTS
Evolutionary Analysis and Localization of Arabidopsis Mini-III Homologs
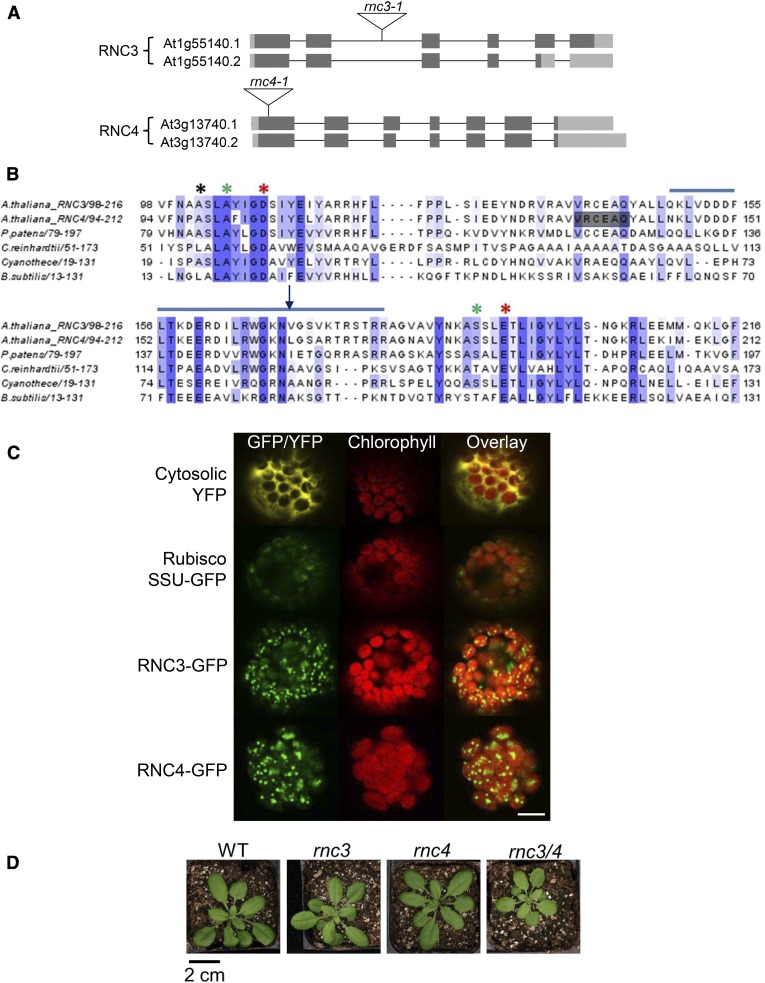
Sequence comparisons have previously revealed that Mini-III homologs are widespread in the green lineage. The Arabidopsis nuclear genome encodes two such proteins, RNC3 and RNC4 (Figure 1A), that have 75% sequence similarity (Olmedo and Guzmàn, 2008). Both gene models indicate that alternative splicing generates two variants. For RNC3, the splice variant At1g55140.2 lacks 67 C-terminal amino acids, including one catalytic residue (see below). In addition, six of the nine C-terminal amino acids differ compared with similar residues of At1g55140.1. The RNC4 splice variant At3g13740.2 lacks six residues of unknown function prior to the Mini-III specific domain. The full-length versions of RNC3 and RNC4 are predicted to be the dominant proteins by the TAIR genome browser; the biological significance of the minor variants is unknown.
Figure 1.
Domain Analysis and Localization of Arabidopsis Mini-III.
(A) Gene models for Arabidopsis RNC3 and RNC4, showing the locations of T-DNA insertions (triangles), and predicted protein splice variants, as discussed in the text. Dark gray boxes, exons; light gray boxes, 5′ or 3′ untranslated regions; thin lines, introns.
(B) Alignment of the RNase III domains from Mini-III proteins. Amino acid shading indicates sequence conservation, with dark blue designating fully conserved residues. Asterisks depict amino acids involved in catalysis according to the NCBI Conserved Domain Database (www.ncbi.nlm.nih.gov/cdd/) for cd00593 (ribonuclease III C-terminal domain). Red asterisks denote RNase III Asp-44 (D) and Glu-110 (E) involved in Mg2+ binding that are conserved in Mini-III. Green asterisks mark the locations of RNase III Glu-40 and Asp-107 that are not conserved in Mini-III. The blue horizontal line above the sequences highlights the Mini-III specific domain described by Redko et al. (2008). The black vertical arrow marks the C terminus of At1g55140.2, and gray shading highlights amino acids deleted in At3g13740.2.
(C) Confocal laser scanning microscopy of Arabidopsis protoplasts transiently expressing the first 100 amino acids of RNC3 or RNC4 fused to GFP. Chlorophyll autofluorescence of chloroplasts is shown in red. The images are overlaid to reveal the colocalization of both RNC3-GFP and RNC4-GFP to the chloroplast. Cytosolic YFP and GFP localized to chloroplasts by the Rubisco small subunit transit peptide are shown as negative and positive controls, respectively. Bar = 7.6 μm.
(D) Plants of the indicated genotypes after 27 d of growth on soil. WT, wild type. Bar = 2 cm.
Three of the four Mini-III variants are likely to be active RNase, as key catalytic residues are conserved between the well-characterized B. subtilis protein and those of Arabidopsis (Figure 1B). These include Asp-44 and Glu-110, which are canonical RNase III domain amino acids involved in coordinating an Mg2+ ion (Nicholson, 2014). RNC3 and RNC4, like B. subtilis Mini-III, lack Glu-40 and Asp-107, which are involved in binding a second Mg2+ ion, with Glu-40 replaced by Ala and Asp-107 replaced by Ser or Thr. They also possess a partly conserved Mini-III-specific domain and have strong sequence conservation surrounding the active sites, suggestive of functionality. A phylogenetic tree was assembled using sequences retrieved from the Phytozome version 9.1 (www.phytozome.net) database (Supplemental Figure 1) to determine whether the presence of two Mini-III proteins was a general feature of the plant kingdom. We found that most plant species analyzed encode a single Mini-III homolog, in contrast with the Brassicaceae family, although independent gene duplications are also present in Medicago truncatula and Linum usitatissimum. Given this, one objective was to determine whether the Arabidopsis proteins are redundant or subspecialized.
Both RNC3 and RNC4 have been found in the Arabidopsis chloroplast proteome (Kim et al., 2009; Nishimura et al., 2013), and we confirmed this through transient expression of the 100 N-terminal amino acids of RNC3 or RNC4 fused to green fluorescent protein (GFP) in Arabidopsis protoplasts. RNC3 and RNC4 harbor 47 and 55 amino acid transit peptides, respectively, as predicted by TargetP (Emanuelsson et al., 2000). Figure 1C shows a punctate GFP signal for both fusion proteins overlapping with chlorophyll autofluorescence, confirming chloroplast localization. The punctate pattern is consistent with a sublocalization to the plastid nucleoid, as was observed for known nucleoid-localized proteins (Terasawa and Sato, 2005; Kleinknecht et al., 2014). This interpretation is consistent with the presence of Mini-III in maize chloroplast nucleoid preparations (Majeran et al., 2012) and RNC4 in Arabidopsis nucleoids (Huang et al., 2013). Punctate staining, however, could also be an artifact of strong cauliflower mosaic virus 35S promoter-driven expression.
To characterize the in vivo function of RNC3 and RNC4, null mutants for both genes were obtained from the T-DNA insertion collection, termed rnc3-1 and rnc4-1, respectively (Figure 1A). The T-DNA insertions were confirmed via sequence analysis, and lack of transcript accumulation was verified by RT-PCR (Supplemental Figure 2). Homozygous mutants had no altered growth phenotype compared with a wild-type control (Figure 1D), although given the recent gene duplication and common chloroplast localization of the proteins, functional redundancy was possible. Therefore, homozygous rnc3-1/rnc4-1 (rnc3/4) mutants were created by genetic crosses, yielding slightly pale-green plants with reduced vigor. The pale-green phenotype correlates with a 10% reduction in chlorophyll a and b accumulation (P < 0.001, n = 20) in mature leaves, which was not observed in either single mutant. This suggests that chloroplast Mini-III is required for fully normal growth and development of Arabidopsis.
Mini-III Deficiency Causes Multiple Defects in rRNA Maturation
B. subtilis Mini-III simultaneously generates the 23S rRNA 5′ and 3′ ends by cleaving a double-stranded stem formed by the precursor transcript (Redko et al., 2008). In chloroplasts, an analogous structure has been proposed to form between the 5′ extension of 23S rRNA and the region immediately downstream of 4.5S rRNA (Massenet et al., 1987, and references therein), because the chloroplast 4.5S rRNA resembles the 3′ domain of bacterial 23S rRNA. If Mini-III targets were conserved between bacteria and chloroplasts, we would expect to observe defects in both 23S and 4.5S rRNA processing in the rnc3/4 mutant. In addition, the chloroplast 23S rRNA is internally processed at two hidden breaks, leading to the accumulation of seven distinct transcripts (Figure 2A). As the mechanism that generates the hidden breaks remains unknown, these sites were also investigated as potential chloroplast Mini-III targets.
Figure 2.
23S rRNA Abnormalities in Mini-III-Deficient Plants.
(A) Diagram of the 23S rRNA gene and flanking genes. Thin vertical lines within the 23S rRNA gene model represent processing sites. The major accumulating transcripts are lettered a to g, with lengths underneath in kb. Dotted red lines represent 5′ or 3′ extensions detected by RNA gel blot in rnc3/4. Locations of the probes (A to E) used in (B) are indicated to scale above the diagram. trnA, gene encoding tRNAAla.
(B) RNA gel blot analysis of 23S rRNA in the wild type, rnc3, rnc4, and rnc3/4 using the five dsDNA probes shown in (A). Transcripts are designated by letters referring to the 23S RNA fragment in (A). Transcript sizes are shown at left, and stained 28S rRNA is included at the bottom to reflect loading.
(C) 23S rRNA 0.5-kb fragment ends in the wild type (black bars and arrowheads) and rnc3/4 (red bars and arrowheads) deduced from cRT-PCR clones. Black arrows a to c indicate locations and directions of cRT-PCR primers. The 5′ ends mapped using primers a + b are displayed above the gene model with the number of clones indicated, along with a stained gel of the PCR products. The 3′ ends mapped from a + b are shown below the gene model, with each bar representing a single clone. The 5′ ends mapped using primers b + c are shown in the nucleotide sequence below the gene model, along with a stained gel of the PCR products. The black box denotes the location of a small RNA sequenced by Zheng et al. (2010).
23S rRNA 5′ Ends Are Heterogeneous in the Absence of Mini-III
23S rRNA maturation was first examined by RNA gel blot analysis (Figure 2B). Five probes were used: one to detect all segments of the mature rRNA (probe C), and the others designed to detect either the 5′ or 3′ end or extensions thereof (probes A/B and D/E, respectively). For each probe, the rnc3 and rnc4 single mutants resembled the wild type, consistent with functional redundancy, while the double mutant differed from the wild type in each case. In rnc3/4, wild-type-sized transcripts were present, but probes specific to the 5′ or 3′ end or their flanking regions revealed abnormal transcripts. Because the extended transcripts are of low abundance, they are readily visible with probes outside the coding region but are masked by wild-type-like transcripts when coding region probes are used. As described below, the misprocessed transcripts are extended versions of transcripts b, c, e, f, and g (Figure 2A), indicating that the absence of Mini-III affects 23S rRNA maturation efficiency, as in B. subtilis.
To determine precisely the nature of the 23S rRNA 5′ ends in rnc3/4, circular RT-PCR (cRT-PCR) was performed on the 0.5-kb fragment (transcript c) using two different primer pairs (Figure 2C). Simultaneously, the 3′ end of the first hidden break was assessed. With respect to the hidden break, the ends were staggered and similar between the wild type and rnc3/4, indicating that Mini-III does not contribute to processing at this site. The wild type and rnc3/4 did differ, however, near the 5′ end of the 0.5-kb fragment. Using primers within the mature rRNA (a + b), we found that the single wild-type 5′ end was replaced by shorter, staggered ends at positions +1 through +14 in rnc3/4 (Figure 2C, top nucleotide sequence). This is consistent with exonucleolytic trimming in rnc3/4 replacing a Mini-III-mediated endoribonucleolytic cleavage in the wild type (see Discussion). In addition, a single rnc3/4 clone mapped to position −53, contributing to the pool of 5′ extensions detected by RNA gel blot probes A and B. As PCR preferentially amplified wild-type-like fragments, a second amplification (b + c) was performed using a primer just upstream of the coding region to assess the longer 23S 5′ end precursors. This amplified a fragment that mapped 73 nucleotides upstream of the mature 5′ end and did not differ in position between the wild type and rnc3/4 (Figure 2C, bottom nucleotide sequence). This transcript, however, appears to be more abundant in rnc3/4, based on staining intensities of the PCR products combined with the RNA gel blot results, and maps directly upstream of a small RNA (sRNA) (Figure 2C, black box), indicative of an RNA binding protein footprint or secondary structure.
To analyze 5′ ends of the total 23S rRNA population, 5′ rapid amplification of cDNA ends (RACE) was used with a single primer internal to the mature 23S rRNA (Supplemental Figure 3). At the same time, to determine whether these RNAs are primary or processed transcripts, 5′ ends were analyzed with or without prior treatment with tobacco acid pyrophosphatase to remove the triphosphate from primary transcript ends. Tobacco acid pyrophosphatase treatment had no effect on 5′ RACE products, confirming their nature as processed RNAs. In the wild type, 5′ RACE amplified a single transcript corresponding to the mature 23S RNA 5′ end identical to that mapped by cRT-PCR, while in rnc3/4, three distinct products were observed. Clones from the shorter amplicons mapped to staggered positions near the mature 5′ end, and the longest product mapped to position −70, consistent with the cRT-PCR results. This suggests a role for Mini-III in processing the 23S rRNA 5′ end.
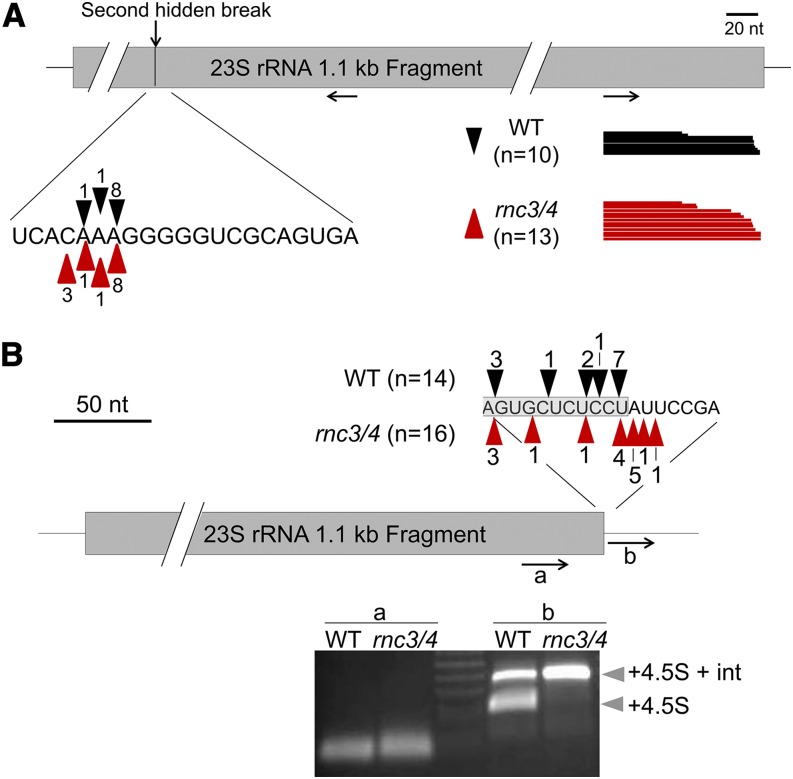
Mature 23S rRNA 3′ Ends Are Unaffected by Mini-III Deficiency
To analyze the 23S 3′ end extensions observed with probe E in Figure 2B, cRT-PCR was performed using primers within the terminal 1.1-kb fragment of the mature 23S rRNA (Figure 3A). Neither the 5′ nor 3′ end mapped from the resultant clones was substantially different between the wild type and rnc3/4, suggesting that processing at the 23S second hidden break is independent of Mini-III. For the 3′ end, however, it was possible that the RNA circularization step required for cRT-PCR was biased toward smaller, processed 1.1-kb fragments. As a complementary technique, 3′ RACE was employed using two different primers, one within the coding region (primer a; Figure 3B) and one just downstream of the coding region (primer b). The ends mapped using primer a were staggered around the annotated 3′ end in both the wild type and rnc3/4, with no discernible difference (Figure 3B, top nucleotide sequence), confirming the cRT-PCR results. Products of the PCR employing primer b differed, however, with two bands visible for the wild type and a single, longer product seen for rnc3/4. Sequence analysis revealed that the shorter wild-type band corresponded to a 23S 3′ extension including the 4.5S coding region, while the longer product in both samples includes the 4.5S coding region as well as a segment of the intergenic region between the 4.5S and the downstream 5S rRNA genes. None of the 3′ sequences shown in Figure 3 correspond to the presumably short 3′ extensions seen by RNA gel blot, implying that they make up a minor component of the 23S rRNA population compared with the mature 23S rRNA and the longer precursors that include the 4.5S rRNA.
Figure 3.
23S rRNA 3′ End Processing Is Similar in the Wild Type and rnc3/4.
(A) 23S rRNA 1.1-kb fragment ends in the wild type (black arrowheads and black bars) and rnc3/4 (red arrowheads and red bars) deduced from cRT-PCR clones. Numbers correspond to the clones mapped to a given position, and each bar represents a single clone. Black arrows indicate the positions and directions of the cRT-PCR primer pair.
(B) 23S rRNA 1.1-kb fragment 3′ ends deduced from 3′ RACE clones, annotated as in (A). The 3′ ends mapped with primer a are displayed above the gene model with the gray shaded region depicting the 23S rRNA gene. The stained PCR products using primers a and b are shown below the gene model. The bands produced from primer b (gray arrowheads) included the 23S-4.5S rRNA intergenic region plus the 4.5S rRNA coding region with (top band; +4.5S + int) or without (bottom band; +4.5S) the 4.5S-5S rRNA intergenic region.
Mature 4.5S rRNA Is Greatly Diminished in the rnc3/4 Mutant
Given that the chloroplast 4.5S rRNA resembles the bacterial 23S rRNA 3′ domain and that the rnc3/4 3′ RACE only identified 23S rRNA extensions containing the 4.5S rRNA plus downstream sequences, this transcript was suspected to be a Mini-III target. 4.5S rRNA processing was first assessed by RNA gel blot using a coding region probe (Figure 4A, probe A). This revealed reduced accumulation of the 102-nucleotide mature transcript in rnc3/4 compared with the wild type and both single mutants. In addition, a longer precursor form (p4.5S) predominated in rnc3/4 and was observed in both single mutants, but was absent in the wild type. Thus far, this is the only instance in which we have not observed full redundancy of RNC3 and RNC4. When the region downstream of the coding sequence was analyzed (probe B), only the longer species was detected, demonstrating that it was a 3′ extension, as anticipated based on the 23S rRNA 3′ RACE results (Figure 3B).
Figure 4.
A 4.5S rRNA Precursor Accumulates in rnc3/4.
(A) RNA gel blot detection of the mature and precursor (p4.5S) 4.5S rRNA transcripts using the dsDNA probes indicated by gray bars in (B). Stained 28S rRNA is shown to reflect loading.
(B) 4.5S rRNA 5′ and 3′ ends deduced from cRT-PCR clones, with annotation as in Figure 3. Nucleotide positions of 3′ ends extending to the middle of the intergenic region are not shown. Black arrows indicate the locations and directions of the cRT-PCR primer pair.
The precise 4.5S rRNA ends were determined by cRT-PCR, as shown in Figure 4B. In general, the 5′ ends did not differ between the wild type and rnc3/4, although two wild-type clones did contain part of the 23S rRNA sequence, which was not observed for rnc3/4. Also, the shorter wild-type 5′ ends mainly clustered around two positions (9 of 11 clones) compared with the staggered 5′ ends found in rnc3/4 around five positions (15 of 16 clones). This indicates a possible role for Mini-III in processing the 4.5S 5′ end. For the 4.5S rRNA 3′ end, two types of variation were seen. First, more transcripts with long 3′ extensions were recovered for rnc3/4 compared with the wild type (nine rnc3/4 clones versus two wild-type clones), which mapped ∼90 nucleotides upstream of the 5S rRNA 5′ end. Second, there were fewer mature-sized 4.5S rRNA 3′ ends in rnc3/4 (7 clones) compared with the wild type (11 clones), consistent with RNA gel blot analysis. Seven of the 11 mature-size clones in the wild type had the same 3′ end, while in the rnc3/4 mutant, the mature-sized 4.5S rRNA 3′ ends were more heterogeneous. This is consistent with endoribonuclease processing in the wild type and exoribonuclease trimming in rnc3/4. In contrast with the observations for 4.5S rRNA, no effects were found for the maturation or accumulation of 5S rRNA (data not shown).
rRNA Deficiencies Observed in rnc3/4 Can Be Complemented by an RNC Transgene
Transgenic complementation was used to establish conclusively that either RNC gene was capable of supporting rRNA processing functions. To do so, rnc3/4 was transformed with constructs designed to express either RNC3-HA (containing a hemagglutinin tag) or RNC4-FLAG (containing a FLAG tag) under the control of their native promoters. Following selection of transformants (see Methods), expression of the RNC transgene was verified by PCR and immunoblot analysis. Plants expressing either transgene recovered wild-type-like plant growth (Supplemental Figure 4A) and the rnc3 or rnc4 RNA phenotype. Specifically, 5′ extensions of the 23S rRNA and 3′ extensions of the 4.5S rRNA were diminished or fully recovered (Supplemental Figure 4B). This confirms that RNC3 and RNC4 are responsible for the traits examined but that overall expression of a single gene is insufficient to recover a fully wild-type phenotype.
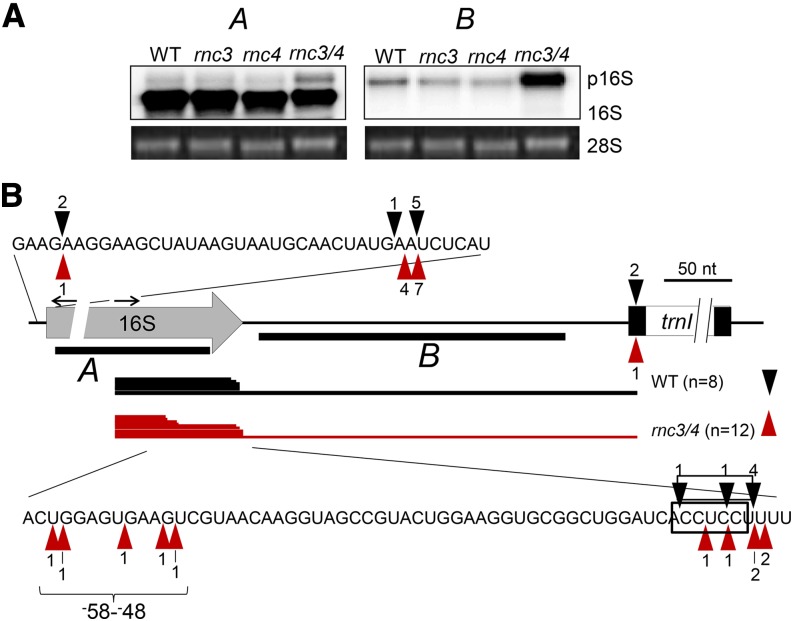
Aberrant Maturation of the 16S rRNA Does Not Affect Ribosome Loading
The 16S rRNA was analyzed, as it is also encoded by the rRNA operon and the precursor is an RNase III substrate in bacteria (Gegenheimer and Apirion, 1980). Figure 5A shows that the double mutant accumulates an increased amount of 3′-extended 16S rRNA, whose 3′ end abuts the RNase P site of tRNAIle (Figure 5B). This 16S rRNA precursor is often observed in chloroplast gene expression mutants, and its presence may be a pleiotropic consequence of slow ribosome assembly (reviewed in Germain et al., 2013). Unexpected heterogeneity was noted surrounding the mature 16S rRNA 3′ end; however, when transcript ends were analyzed by cRT-PCR (Figure 5B), no difference was observed for the 5′ end. In particular, 7 of 12 clones sequenced from the double mutant (but only 2 of 6 from the wild type) lacked all or part of the putative anti-Shine-Dalgarno (SD) motif, which stimulates translation initiation in bacteria by pairing with a complementary sequence found ∼10 nucleotides upstream of mRNA start codons. The spread of 3′ ends is most easily interpreted as nibbling by an exoribonuclease(s) in rnc3/4.
Figure 5.
Accumulation of 3′-Extended 16S rRNA in rnc3/4.
(A) RNA gel blot detection of mature and precursor (p16S) 16S rRNA using the two dsDNA probes (A and B) indicated by thick black bars in (B). Stained 28S rRNA is shown to reflect loading.
(B) 16S rRNA ends deduced from cRT-PCR clones, with annotation as in Figure 3. Nucleotide positions of 3′ ends extending to the 5′ end of trnI are not shown. Black arrows indicate the locations and directions of the cRT-PCR primer pair. The black box indicates the anti-SD sequence at the 16S rRNA 3′ end, and the bracket labeled −58 to −48 highlights the truncated 3′ ends found in rnc3/4.
The importance of chloroplast SD sequences, and by implication the anti-SD motif, varies among genes and species (Lim et al. 2014), but the necessity of an SD sequence for effective translation of at least some chloroplast transcripts is indisputable (Hirose et al., 1998; Kuroda et al., 2007; Drechsel and Bock, 2011). Therefore, it was possible that SD-dependent transcripts would bind to ribosomes less efficiently than their SD-independent counterparts in rnc3/4. To test this, the polysome association of mRNAs in both categories was examined by RNA gel blot following sucrose gradient sedimentation of stromal extracts. No difference in the polysome distribution of these transcripts was observed, however, suggesting that sufficient full-length 16S rRNA was present to support normal rates of ribosome loading (Supplemental Figure 5).
Improperly Processed rRNA Precursors Are Incorporated into Ribosomes in rnc3/4
Although the presence of a subpopulation of 16S rRNA lacking the anti-SD sequence did not appear to impact overall ribosome loading of the mRNAs tested, the rnc3/4 mutant accumulates additional 5′- and 3′-extended rRNAs that may affect efficient plastid translation and therefore plant growth. To assess whether the abnormal or unprocessed rRNAs were engaged in translation, RNA gel blot analysis was used following sucrose density gradient fractionation with and without puromycin treatment, as shown in Figure 6. Puromycin dissociates mRNA-ribosome complexes and was used to calibrate the migration of rRNAs not engaged in translation (<80S) versus polysomal fractions.
Figure 6.
Polysomal Association of rnc3/4 rRNA Precursors.
Polysomes were fractionated through a 15 to 55% sucrose step gradient as described in Methods. Total RNA from the 12 fractions collected was analyzed by RNA gel blot with dsDNA probes as indicated at right. Probe locations are found in Figure 2 (trnA-23S int., probe A; 23S rRNA, probe C; 23S-4.5S int., probe E), Figure 3 (4.5S, probe A), and Figure 4 (16S, probe A). Transcript sizes are shown at left, and precursors are marked by asterisks at right. Representative ethidium bromide (EtBr)-stained gels are shown to represent loading. The free ribosomal fractions (<80S; lanes 1 to 6) and polysomal fractions (lanes 7 to 12) were determined using a puromycin control (bottom) as described in the text.
Analysis of the 23S rRNA revealed that the wild type and rnc3/4 accumulate internally processed transcripts (i.e., 0.5, 1.1, 1.3, and 1.8 kb) in both free ribosomes and polysomal fractions, with longer transcripts (i.e., 2.4, 2.9, and 3.2 kb) favored in the free ribosome fractions. To determine whether the precursor transcripts found predominantly in rnc3/4 are incorporated into polysomes, gel blots were probed for the intergenic regions upstream (trnA-23S int.) or downstream (23S-4.5S int.) of the 23S coding sequence. In both cases, the rnc3/4 23S rRNA extensions were distributed in the polysome gradient. Similarly, the 3′-extended 4.5S rRNAs did not appear to vary in polysome distribution from the mature transcript. Therefore, complete 23S and 4.5S rRNA processing is not required for polysome assembly. A different result was obtained for the 16S rRNA. In this case, 3′-extended RNAs were limited to nonpolysomal fractions for both the wild type and rnc3/4. The exclusion of incompletely 3′-processed 16S rRNA from polysomal fractions has been reported previously in the mutant rbf1, which accumulates high levels of such molecules (Fristedt et al., 2014), and it is clear that mature 16S rRNA is the dominant polysome species in both the wild type and rnc3/4. Overall, this points toward unimpeded assembly of functional ribosomes in the absence of chloroplast Mini-III.
The fact that rRNA substrates of RNC3 or RNC4 can assemble into ribosomes raises the question of whether Mini-III processes the 23S and/or 4.5S transcripts prior to or subsequent to polysome assembly. For example, the final maturation steps for Escherichia coli 23S rRNA occur only after polysome formation (Srivastava and Schlessinger, 1988), and B. subtilis Mini-III activity is much higher in the presence of ribosomal protein L3 (Redko and Condon, 2009). If chloroplast Mini-III were active in fully assembled polysomes, it would migrate in those sucrose gradient fractions. To test this, total plant extracts from the wild type and rnc3/4:RNC4-FLAG were fractionated with and without puromycin treatment. Immunoblot analysis of the fractions revealed that RNC4 only migrated at the top of the gradient (Supplemental Figure 6), indicating that rRNA maturation by Mini-III likely occurs before polysome integration.
RNA-Seq Validates a Role for Mini-III in rRNA Maturation
To gain a global view of Mini-III function in chloroplast RNA metabolism, two wild-type and two rnc3/4 strand-specific RNA-Seq libraries were prepared from purified chloroplasts. More than 107 reads were obtained from each sample, of which ∼75% aligned to the chloroplast genome (Supplemental Table 1), an increase over the 30 to 50% of chloroplast-specific reads obtained from total leaf RNA (Hotto et al., 2011). Reads per kilobase of transcript per million mapped reads (RPKM) values for exons and introns showed that expression levels between replicates were highly reproducible (Supplemental Figure 7; r ≥ 0.93). Coverage of the entire plastid genome was extracted in sliding 100-nucleotide intervals on each strand to give a snapshot of the transcriptional landscape (Supplemental Figure 8). For areas of greater interest, single-base coverage was extracted according to Castandet et al. (2013).
Comparison of the exonic RPKM values between the wild type and rnc3/4 showed that only the 4.5S rRNA gene is differentially expressed between the two genotypes (Figure 7A), a result that is mirrored in the 100-nucleotide window coverage (Supplemental Figure 8). Further analysis of the rRNA operon using either the 100-nucleotide window or single-base resolution confirmed additional rRNA processing defects in rnc3/4. First, the 5′ extension of the 23S rRNA is overrepresented in rnc3/4 compared with the wild type beginning ∼97 nucleotides upstream of the 23S start site (Figure 7B), which is similar to the ends mapped by 5′ RACE and cRT-PCR (Figure 2C; Supplemental Figure 3). Additionally, coverage of the 23S-4.5S intergenic region is overrepresented in the rnc3/4 data set (Figure 7C). There was no sharp increase/decrease in reads, indicative of an endonuclease cleavage site apparent in the 23S-4.5S rRNA region, consistent with the results obtained by 3′ RACE. Finally, comparison of the 16S rRNA 3′ downstream region showed increased reads in rnc3/4 compared with the wild type, corresponding to the 3′ extensions observed by RNA gel blot and mapped by cRT-PCR (Figure 7D). Furthermore, a small rise in rnc3/4 RNA-Seq reads occurs ∼100 nucleotides upstream of the annotated 16S rRNA 3′ end compared with the wild type, consistent with the shorter 3′ ends, lacking the anti-SD sequence, identified by cRT-PCR. Together, these results corroborate the RNA-Seq profile with the previously identified rnc3/4 targets.
Figure 7.
RNA-Seq Analysis of rnc3/4 Exons.
(A) Differential expression of exons between the wild type and rnc3/4 by comparison of average RPKM values (n = 2 each). Exons present in the large inverted repeat were included only once. The arrow indicates the 4.5S rRNA data point.
(B) Normalized read coverage for an average of two wild-type (blue trace) or rnc3/4 (red trace) RNA-Seq replicates in the 23S rRNA 5′ end genomic region. A gene model is shown to scale, and the beginning of the region where coverage of the wild type and rnc3/4 diverges is indicated by the red arrowhead.
(C) Normalized read coverage for the 23S rRNA 3′ end annotated as in (B).
(D) Normalized read coverage for the 16S rRNA 3′ end annotated as in (B). The region where the wild type and rnc3/4 begin to diverge prior to the annotated 3′ end is indicated by the red arrowhead.
A Noncoding RNA Accumulates in rnc3/4
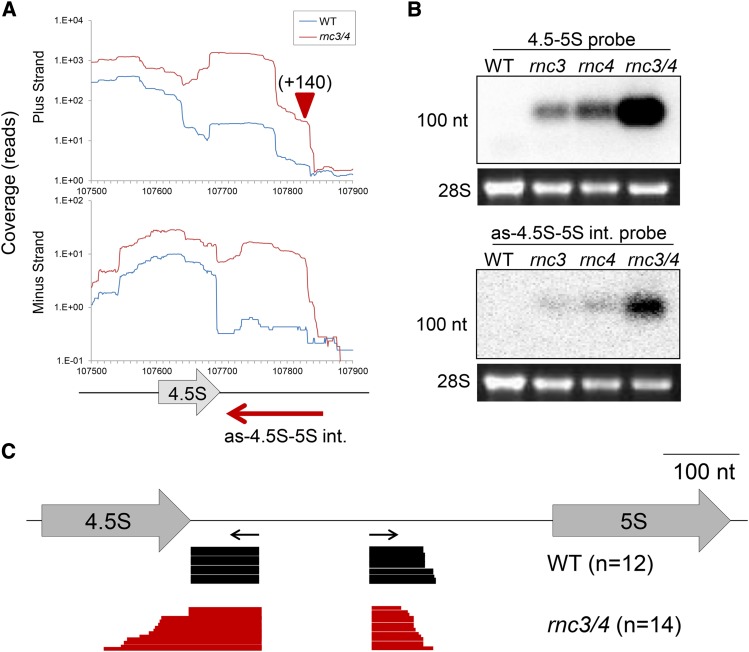
As described earlier, aberrant RNA processing occurs in the 4.5S rRNA region in rnc3/4. Analysis of the 4.5S-5S rRNA intergenic region RNA-Seq profile revealed differences between the wild type and rnc3/4 (Figure 8A). For the 4.5S precursor, an increased number of RNA-Seq reads extended ∼140 nucleotides downstream of the 4.5S coding region in rnc3/4, similar in distance to ends mapped by cRT-PCR (Figure 4B). In addition, there were more rnc3/4 reads on the minus strand, corresponding to the plastid noncoding RNA (pncRNA) denoted as-4.5S-5S int. There was a general divergence of the RNA-Seq profiles between the wild type and rnc3/4 extending from the 5′ end of as-4.5S-5S int. to its 3′ end, which is complementary to the middle of the 4.5S coding region in rnc3/4. RNA gel blot analysis using strand-specific probes confirmed the accumulation of as-4.5S-5S int. in rnc3/4 and showed minor accumulation of the transcript in both rnc3 and rnc4 single mutants (Figure 8B). The pncRNA profile correlated with the sense-strand profile for all four genotypes, suggesting a possible mechanistic link for their accumulation, although the total number of antisense reads was 1 to 5% of the sense reads. Strand-specific cRT-PCR was used to map as-4.5S-5S int. and showed that a 159-nucleotide transcript accumulates in the wild type (Figure 8C, black bars). A striking observation was that, in the wild type, all clones sequenced had the same 3′ end, whereas the 5′ end was only slightly staggered. For rnc3/4, this was not the case. The 5′ ends of as-4.5S-5S int. were staggered, and the 3′ ends were highly variable and generally longer than the wild-type 3′ end, extending into the region complementary to the 4.5S gene, as suggested by RNA-Seq. These results strongly point to a role for Mini-III in dictating the structure and abundance of as-4.5S-5S int.
Figure 8.
Accumulation of a Noncoding RNA Antisense to the 4.5S-5S rRNA Intergenic Region.
(A) Normalized read coverage in the 4.5S rRNA region for either the plus (top graph) or minus (bottom graph) strand. A gene model is shown below, with the red arrow depicting the extent of as-4.5S-5S int. in the wild type.
(B) RNA gel blot analysis using the indicated strand-specific probes. Stained 28S rRNA is shown to indicate loading, and the 100-nucleotide position is indicated at left.
(C) Results of sequencing from cRT-PCR clones, with horizontal black arrows showing primer positions. Each horizontal bar represents a single clone for the wild type (black) or rnc3/4 (red).
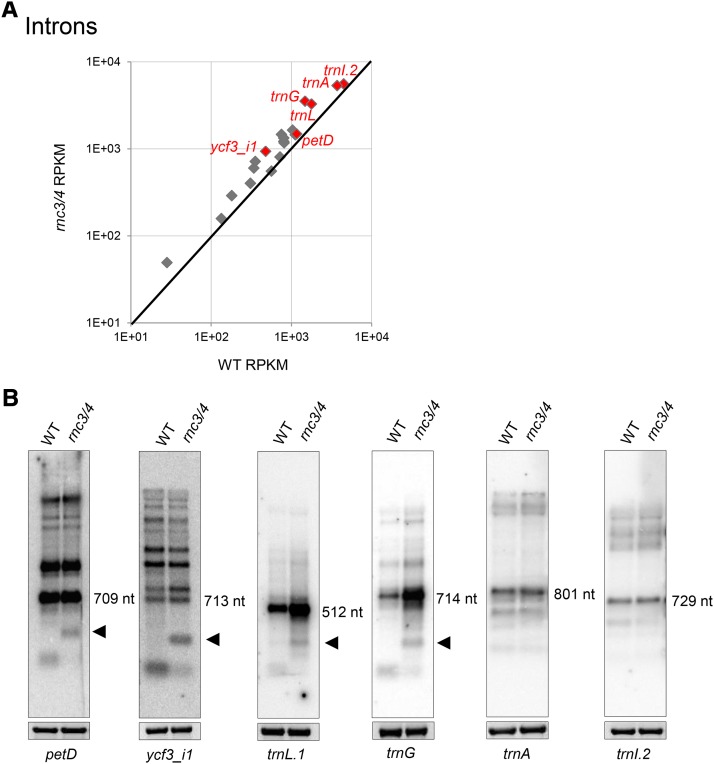
Introns Are Improperly Degraded in rnc3/4
Analysis of the intronic RPKM values for the wild type and rnc3/4 showed some variation in intron accumulation (Figure 9A), but when splicing efficiency was calculated from the RNA-Seq data, no differences were observed (Supplemental Figure 9A). Previously, however, it was proposed that RNase III was involved in chloroplast intron degradation, because sequenced intron fragments had termini consistent with cleavages in bulged double-stranded RNA domains (del Campo and Casano, 2008). Indeed, RNA gel blot analysis of intron accumulation patterns differed between the wild type and rnc3/4 (Figure 9B). For trnL.1, trnG, and ycf3i1, there was increased accumulation of the intact, excised intron in rnc3/4, consistent with the trend seen in RPKM values. There were also qualitative differences. For four introns (petD, ycf3i1, trnL.1, and trnG), a degradation product was observed that appeared to be a longer version of its wild-type equivalent (Figure 9B, black arrowheads). No differences were observed, however, for trnA and trnI.2. Taken together, these results indicate that Mini-III influences both the kinetics and the pathway of intron degradation.
Figure 9.
Intron Expression Differs between the Wild Type and rnc3/4.
(A) Differential expression of introns between the wild type and rnc3/4 by comparison of average RPKM values (n = 2 each). Introns present in the large inverted repeat were included only once. Introns represented by gel blots in (B) are colored red.
(B) RNA gel blot analysis of introns using the dsDNA probes indicated at the bottom. Bands marked with sizes in nucleotides (nt) are the intact, excised introns. Products specifically accumulating in rnc3/4 are marked by black arrowheads. Stained 28S rRNA is included to reflect loading.
Excised Introns Accumulate in B. subtilis Lacking Mini-III
While a role for Mini-III in B. subtilis rRNA maturation is well established, its influence on intron metabolism has not been examined previously. B. subtilis harbors a prophage called SPβ, which possesses two group I introns in adjacent genes encoding the α- and β-subunits of ribonucleotide reductase, yosP and nrdEB (Lazarevic et al., 1998). Expression of prophage genes and excision of the yosP and nrdEB introns can be induced with the drug mitomycin C (Zahler et al., 1977). B. subtilis strain W168 either possessing or deleted for the Mini-III gene (∆mrnC) was treated with mitomycin C for 60 min, and the two excised introns were analyzed by RNA gel blot (Figure 10A). The nrdEB and yosP introns could be detected in both strains following induction; however, the levels were ∼3-fold higher in ∆mrnC cells. To show that the effect of the ∆mrnC mutation was on intron stability rather than transcription, we measured the remaining amounts of each intron at different times after inhibition of transcription by rifampicin. Intron half-life was visibly increased in the absence of Mini-III in both cases, showing clearly that, as in chloroplasts, B. subtilis Mini-III is involved in intron turnover.
Figure 10.
Absence of Mini-III in B. subtilis Slows Intron Degradation.
(A) RNA gel blot showing the levels of the nrdEB and yosP introns in wild-type and ∆mrnC mutant cells. Mitomycin C was added to half of the culture at an OD600 of 0.3 (T0), and both treated and untreated cells were harvested for RNA preparation 60 min later (T60). The blots were reprobed for 5S rRNA for normalization.
(B) RNA gel blot showing the decay of nrdEB and yosP introns in wild-type and ∆mrnC mutant cells at times after the addition of rifampicin. The blots were reprobed for 5S rRNA for normalization.
DISCUSSION
RNase III is found in all kingdoms of life except Archaea (Court et al., 2013) and thus was present in the bacterial progenitor of the chloroplast. As shown in this study, Arabidopsis chloroplasts possess two largely redundant Mini-RNase III variants, similar to proteins found in cyanobacteria (Supplemental Figure 1) as well as B. subtilis. Many other plant species possess a single Mini-III gene with predicted plastid localization, for which functional information is lacking. Unlike B. subtilis, chloroplasts appear to lack a canonical RNase III, which contains a dsRBD in addition to the catalytic domain found in Mini-III. Therefore, chloroplast Mini-III might be expected to carry out a broader range of activities than the rRNA-specific functions reported previously for its bacterial counterpart. Indeed, our results document additional roles in rRNA maturation as well as the regulation of a pncRNA. We also found that Mini-III is involved in spliced intron accumulation and degradation in Arabidopsis, a finding that was confirmed for B. subtilis. RNA editing, on the other hand, was unaffected (Supplemental Figure 9B).
rRNA Maturation by Plastid Mini-III: Is It Similar or Different in B. subtilis?
The major substrate for B. subtilis Mini-III is the 23S rRNA precursor (Redko et al., 2008; Redko and Condon, 2009), which forms an ∼70-nucleotide double-stranded stem from precursor 5′ and 3′ extensions (reviewed in Deutscher, 2009). The stem is processed first by RNase III, and mature 5′ and 3′ ends are completed by Mini-III. In the absence of Mini-III, wild-type-like ends are formed via an alternative pathway through the combined 5′→3′ exoribonucleolytic activity of RNase J1 at the 5′ end and 3′→5′ exoribonuclease activity of RNases PH and YhaM at the 3′ end (Redko and Condon, 2010).
In chloroplasts, the precise secondary structure of the 23S-4.5S rRNA precursor remains ambiguous. Predictive structures suggest that the 5′ extension of 23S rRNA and the 3′ extension of 4.5S rRNA form a stem of ∼13 nucleotides (MacKay, 1981; Massenet et al., 1987, and references therein), as depicted in Figure 11B (blue and black lines). If this were the functional equivalent of the bacterial Mini-III substrate, chloroplast Mini-III mutants should be defective in processing these regions, and this is just what we observed in rnc3/4 plants. This short stem is characteristic of the B. subtilis Mini-III processing site, as Mini-III does not require the long stem and bulge that is essential for RNase III cleavage. In addition, the chloroplast 23S rRNA 5′ end and the majority of 4.5S 3′ ends are homogeneous in the wild type, consistent with a single endonuclease cleavage (Figures 2C and 4B). Finally, there is a two-nucleotide overhang between the mature 23S 5′ and 4.5S 3′ ends that is the expected result of Mini-III cleavage. In the absence of Mini-III, the accumulation of wild-type-like 23S rRNA 5′ and 4.5S rRNA 3′ ends must occur through a different pathway. For example, the 23S rRNA 5′ end may be matured by RNase J and the 4.5S rRNA 3′ end by PNPase-catalyzed exonuclease trimming, similar to the B. subtilis model (Redko and Condon, 2010).
Figure 11.
The Role of Mini-III in Chloroplast rRNA Maturation.
(A) rRNA polycistron processing following transcription from its single upstream promoter. Initial endonucleolytic cleavage by RNase P and RNase Z releases the tRNAs, and an unknown endonuclease acts to release and mature the 5S rRNA 5′ end. Subsequently, a combination of exonucleases and endonucleases creates mature 5′ and 3′ ends. For the 16S rRNA, a putative exonuclease and endonuclease (possibly RNase J) processes the 5′ end, while RNase II processes the 3′ end with a potential role for Mini-III in either processing or stabilization. The 23S-4.5S rRNA dicistron is processed at its 5′ and 3′ ends by Mini-III. An unknown endonuclease releases the 4.5S from the dicistron, potentially processing the 5′ end simultaneously. Following this, PNPase trims the 23S rRNA 3′ end, and unknown factors cleave the 23S rRNA at the two hidden breaks.
(B) Putative secondary structure of the 4.5S rRNA (black line), the 4.5S-5S intergenic region (gray line), and the 5S rRNA (red line) (adapted from Figure 5 of Leal-Klevezas et al., 2000a). Base pairing of the 23S rRNA 5′ end (dark blue line) and extension (light blue line) to the 4.5S rRNA 3′ end are shown. Suspected Mini-III cleavage sites are marked (a; black arrowheads), as are the relative locations of two sRNAs (orange and green lines). The 4.5S-5S rRNA intergenic region processing sites identified previously are marked b and c, with site b corresponding to the 4.5S rRNA 3′ extension observed in rnc3/4.
With respect to the 23S rRNA 5′ end, analysis of an Arabidopsis sRNA data set (Zheng et al., 2010) identified a 36-nucleotide sequence mapping precisely at the mature 23S rRNA 5′ end (Figure 11B, green line). This sequence is not predicted to form a stable secondary structure on its own, but it could arise through protection by an RNA binding protein, possibly a ribosomal protein such as L22 that binds nearby sequences in the bacterial ribosome (Klein et al., 2004). In the wild type and rnc3/4, it is possible that this putative protein could guide maturation by Mini-III or a secondary pathway.
Prior to Mini-III cleavage, several putative processing steps occur that are distinct from the B. subtilis model. Within the trnA-23S rRNA intergenic region, a 25-nucleotide sRNA (Figure 2C) was identified (Zheng et al., 2010), and pre-23S rRNA reads mapped exactly to the 5′ end of this sRNA in the wild type, rnc3/4, and rnr1-3, a 3′→5′ exoribonuclease mutant (Bollenbach et al., 2005). This indicates that, subsequent to the release of trnA by RNase Z, processing occurs to the 5′ end of the footprint, which may be a PPR or other helical repeat protein signature. This 5′ end processing could be catalyzed by RNase J, which has been shown to be arrested by PPR proteins during mRNA 5′ end maturation (Luro et al., 2013).
For the 4.5S rRNA 3′ end, 4.5S-5S rRNA intergenic processing likely precedes Mini-III cleavage (Figure 11A). Evidence for this model includes S1 nuclease protection mapping (Strittmatter and Kössel, 1984; Leal-Klevezas et al., 2000b) as well as transcripts mapped by cRT-PCR in this study. Given the structural predictions and experimental results, we propose that Mini-III catalyzes cleavage at site a, subsequent to processing at site b, ∼90 nucleotides upstream of the 5S rRNA coding region, and site c, the mature 5S rRNA 5′ end (Figure 11B). In the absence of Mini-III, processing to site b still occurs, resulting in accumulation of the 4.5S + intergenic region transcript (Figure 4A, p4.5S). A minor population of wild-type-like 4.5S transcripts accumulates in rnc3/4, possibly occurring via inefficient exonuclease processing from site b. An sRNA of ∼44 nucleotides maps to the 4.5S rRNA 3′ end (Figure 11B, orange line), consistent with the predicted stem structure that could serve as a processing signal, as described for spinach (Spinacia oleracea; Audren et al., 1987). Whether ribosomal protein L3, which binds 23S rRNA and stimulates Mini-III activity in B. subtilis (Redko and Condon, 2009), influences processing in the chloroplast remains to be determined. In both the wild type and rnc3/4, 5S rRNA processing at site c proceeds normally, indicating that this cleavage is independent of Mini-III.
Aberrant 16S rRNA transcripts accumulated in rnc3/4 with more heterogeneous 3′ ends (Figure 5). Structural evidence argues against 16S rRNA being a Mini-III substrate, as no duplex is predicted to form between the 16S rRNA 3′ end and either the 16S rRNA 5′ end or a pncRNA. Additionally, analogous 16S rRNA processing defects were not observed in B. subtilis lacking Mini-III (data not shown); therefore, the effects we observed may be indirect. In yeast, for example, the accumulation and correct processing of the small rRNA are dependent on the large subunit through an ambiguous quality control mechanism (Venema and Tollervey, 1999). Similarly, misprocessed plastid rRNAs frequently accumulate as a secondary defect when chloroplast protein import, photosynthetic activity, translation, or ribosome assembly is perturbed (Germain et al., 2013).
A pncRNA Is Processed by Mini-III
In bacteria, RNase III has a significant role in the processing and regulation of noncoding RNAs (Stead et al., 2011; Lasa et al., 2012), suggesting that chloroplast Mini-III also may have noncoding RNA targets. One such target was identified via RNA-Seq analysis, termed as-4.5S-5S int., which is antisense to the 4.5S-5S intergenic region. The wild-type as-4.5S-5S int. transcript has a precise 3′ end, indicative of endonuclease processing, which maps precisely to the mature 4.5S rRNA 3′ end on the opposite strand. In rnc3/4, these ends are longer and more variable, consistent with exonuclease trimming from a longer precursor. Consequently, we speculate that Mini-III normally processes the 3′ end of as-4.5S-5S int. In bacteria, the formation of sense-antisense duplexes creates RNase III recognition sites, either for processing or regulation (Durand et al., 2012; Lybecker et al., 2014). One possibility is that as-4.5S-5S int. anneals to the complementary strand, competing with the proposed 4.5S-5S secondary structure and binding to the 23S 5′ end (Figure 11B). The increased accumulation of as-4.5S-5S int. and its complement in rnc3/4 may suggest that duplex formation creates a Mini-III recognition site, although the two-nucleotide overhang characteristic of Mini-III cleavage was not observed. In this way, the pncRNA could regulate RNA processing, as postulated previously for the complementary chloroplast 5S and AS5 transcripts (Hotto et al., 2010; Sharwood et al., 2011b). We have not yet found evidence for increased accumulation of other pncRNAs in rnc3/4, however, raising the possibility that the 4.5S-5S region is one of a small number of Mini-III targets.
Mini-III Functions in Intron Recycling
Whereas chloroplast RNA splicing has been intensively studied genetically and biochemically (Germain et al., 2013), the fate of introns subsequent to splicing is not well understood. Intact, and presumably mainly lariat-form introns, are readily visualized using RNA gel blots, and partially degraded introns accumulate in Arabidopsis plants lacking chloroplast polynucleotide phosphorylase (Germain et al., 2011). Here, we have shown that, as proposed earlier for chloroplast introns (del Campo and Casano, 2008), RNase III (specifically, Mini-III) also participates in intron degradation, a function that was conserved for B. subtilis Mini-III.
For most introns analyzed, the rnc3/4 mutant accumulated a longer product compared with the wild-type counterpart. This degradation intermediate is assumed to be a linear form of the intron following cleavage by one or more endoribonucleases and perhaps PNPase trimming. Since the smallest intron-only product in rnc3/4 is longer than that in the wild type, we suspect that Mini-III is one of the endonucleases that can open the intron lariat, fitting with the proposed mechanism for intron degradation. Chloroplasts appear to lack the classical intron lariat debranching enzyme, because the only Arabidopsis gene product appears to be cytoplasmic (Wang et al., 2004), although not all chloroplast introns form lariats during splicing (Vogel and Börner, 2002). A role for Mini-III in lariat cleavage would be consistent with the ability of yeast RNase III to cleave lariats as an alternative pathway for intron degradation (Danin-Kreiselman et al., 2003).
METHODS
Phylogenetic Analyses
Thirty-five plant Mini-III protein sequences were retrieved from the Phytozome database version 9.1 (www.phytozome.net) using RNC3 (At1g55140) as a query sequence. Using the Bacillus subtilis Mini-III protein sequence as a query against cyanobacterial sequences, the best 20 BLASTP hits were retrieved. Finally, Chlamydomonas and Volvox Mini-III protein sequences were obtained from their respective databases using a BLASTP search with the B. subtilis protein sequence. The sequence alignment was made using the default parameters for MUSCLE (Edgar, 2004) (gap penalties, −2.9, 0, and 1.2 for gap open, gap extend, and hydrophobicity multiplier, respectively). All the sequences and the resulting alignment can be found in Supplemental Data Set 1. The phylogenetic tree was built using the neighbor-joining method (Saitou and Nei, 1987) with 100 replicates. All positions containing gaps and missing data were eliminated, giving a total of 75 positions in the final data set. Evolutionary distance is the number of amino acid substitutions per site. The tree was rooted using the B. subtilis Mini-III protein sequence as an outgroup. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013).
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia was used as the wild-type control for this study. Insertion lines for RNC3 (gene At1g55140; T-DNA line SALK_073005) and RNC4 (gene At3g13740; T-DNA line SALK_006516.55.75.x) were identified via the SIGnAL website and obtained from the ABRC (Alonso et al., 2003). Insertions were verified by PCR (Supplemental Figure 2) and genomic locations confirmed by sequencing. Plants were grown on soil under fluorescent lighting (150 μmol m−2 s−1) with a 16-h-light/8-h-dark photoperiod at 25°C. Tissue was analyzed from 25- to 27-d-old plants.
Chlorophyll Determination
A 0.1-cm2 mature rosette leaf disc was homogenized with a Wheaton homogenizer in 1 mL of methanol. Plant tissue was pelleted for 3 min in a microcentrifuge at 4°C. The supernatant absorbance was measured at A666 (peak for chlorophyll a), A653 (peak for chlorophyll b), and A750 (cell debris). Total chlorophyll was calculated as described (Porra et al., 1989).
Transient Expression of RNC3 and RNC4 in Arabidopsis Protoplasts
The cDNAs corresponding to the first 100 amino acids of RNC3 or RNC4 were cloned into pMDC83 (Curtis and Grossniklaus, 2003). Transient expression of GFP or yellow fluorescent protein (YFP) fusions was achieved in Arabidopsis protoplasts isolated from 5- to 7-d-old seedlings grown on Murashige and Skoog-agar under continuous light by digestion with 15 mL of enzyme solution (Murashige and Skoog medium containing 0.4 M mannitol, 1% cellulase, and 0.25% Macerase [EMD Millipore] at pH 5.8) overnight in the dark. Protoplasts were released by gentle pipetting using a wide-bore tip until the solution turned dark green, then filtered through a 37-μm funnel. The flow-through was spun at 35g for 5 min in a swinging-bucket rotor at 4°C. The supernatant was removed and the protoplast pellet resuspended in 4 mL of W5A solution (0.6 M mannitol, 1 mM MgCl2, 5 mM glucose, 154 mM NaCl, 1.2 mM CaCl2, 5 mM KCl, and 0.1% MES, pH 5.8). The protoplast solution was placed on a 21% sucrose cushion, then spun at 35g for 5 min in a swinging-bucket rotor at 4°C. Intact protoplasts were removed from the upper layer and washed with 10 mL of W5A solution. The protoplast pellet was resuspended in 5 mL of W5A solution, incubated on ice for 30 min, and spun at 35g for 5 min. The cells were resuspended in 1 mL of MMg solution (15 mM MgCl2, 0.1% MES, and 0.4 M mannitol, pH 5.6) and quantified using a hemacytometer.
Protoplast transformation was completed according to Yoo et al. (2007). Transformed protoplasts were incubated at 23°C overnight in the dark, after which GFP or YFP fluorescence was detected from 505 to 541 nm, and chlorophyll autofluorescence was detected from 656 to 707 nm using a Leica TCS SP5 laser scanning confocal microscope with an argon laser (excitation wavelength of 488 nm). Images were processed with either LAS AF Lite or ImageJ software (Leica).
Complementation of rnc3/4
RNC3 and RNC4 genes driven by their native promoters were inserted into a pGWB Gateway binary vector (Nakagawa et al., 2007) adding a C-terminal epitope tag. The native promoters were defined as extending from the region ∼2 kb upstream of the start codon until exon 2, including the first intron. The coding sequence beginning with exon 2 was amplified separately from cDNA. The 3′ end of the promoter and the 5′ end of the coding region fragments were annealed through restriction digest overhangs in exon 2, BglII for RNC3 or BsaI for RNC4. Confirmed pENTR-RNC3 or pENTR-RNC4 clones were used in a subsequent Gateway LR cloning reaction with pGWB510 or pGWB513 destination vector to add a C-terminal FLAG or HA tag, respectively. Agrobacterium tumefaciens GV3101 was used to transform rnc3/4 plants by floral dip (Desfeux et al., 2000). Transformants were selected on 30 μg/mL hygromycin, and positive transformants were confirmed by RNA gel blot analysis of 4.5S or trnA-23S rRNA gene regions. T4 generation plants were germinated on soil after homozygosity was confirmed.
RNA Extraction and Analysis
Total RNA was extracted using TRI reagent (Molecular Research Center) with modifications according to Hotto et al. (2011). To evaluate transcript processing, 1 to 5 μg of total RNA was analyzed by RNA gel blots probed with either a double-stranded DNA (dsDNA) or RNA probe as indicated in the figure legends. For polysome analysis, 200 mg of leaf tissue was extracted and fractionated through a 15 to 55% sucrose density step gradient (Barkan, 1998). Twelve fractions were collected, and the RNA was extracted and analyzed by gel blot.
The cRT-PCR method (Zimmer et al., 2012) was modified by the addition of a tobacco acid phosphatase (25 units) treatment prior to circularization. Circularized RNA (2 to 5 μg) was reverse transcribed using SuperScript III (Invitrogen) with either random hexamers or a gene-specific oligonucleotide positioned in the reverse orientation near the 5′ end of the gene of interest. Amplification was with 0.5 units of GoTaq polymerase (Promega). 5′ RACE was completed as described previously (Hotto et al., 2011). For 3′ RACE, an RNA adapter was ligated to the 3′ end, and cDNA was synthesized using SuperScript III and 2 μM of 3′ RACE primer. The cDNA was amplified using gene-specific primers and the 3′ RACE primer. All primers used in the study are listed in Supplemental Table 2.
Strand-Specific RNA Sequencing
Chloroplasts were isolated from rnc3/4 and wild-type leaf tissue as described previously (Gruissem et al., 1986) with minor modifications. Tissue collected in 10-g lots was immediately ground in 1× GR to maximize RNA stability. Intact chloroplasts collected at the interface between the 40 and 80% sucrose cushions were washed with 1× GR and resuspended in a minimal volume of 1× GR. Aliquots of 500 μL were pelleted at 5900g at 4°C, the supernatant was removed, and the pellet was stored at −80°C. RNA was extracted from two samples of wild-type and rnc3/4 chloroplast pellets using the Qiagen RNeasy plant RNA extraction kit (www.qiagen.com). Pelleted chloroplasts were resuspended in the RLT buffer provided, and the remainder of the protocol was followed as outlined by the manufacturer starting at step 3. RNA was treated with DNase (Ambion), following which rRNAs were depleted from 5 μg of chloroplast RNA by two treatments with the RiboMinus plant kit (Invitrogen). The RNA-Seq library was prepared from 50 ng of rRNA-depleted DNA-free RNA using the ScriptSeq version 2 RNA-Seq library preparation kit (Epicenter; http://www.epibio.com/). The protocol provided was followed, and the ScriptSeq Index PCR primers (Epicenter Set 1) were used for the final amplification step. Library integrity was visualized using a Bioanalyzer, after which samples were pooled and run on a single lane using Illumina HiSeq 2000 with 100-bp single-end reads.
Following quality control, strand-specific reads with a quality higher than 30 and length greater than 60 were aligned to the Arabidopsis chloroplast genome (TAIR10) using TopHat2 (Kim et al., 2013). Up to two locations were accepted for sequence reads to account for the large inverted repeat in the chloroplast genome. Following alignment, coverage on both strands was calculated using the BEDTools suite (Quinlan and Hall, 2010). This was performed for the complete genome using a 100-nucleotide window overlapping by 50 nucleotides, and single-base coverage was extracted when needed, similar to Castandet et al. (2013). Coverage for each sample was normalized to the total number of reads aligned to the chloroplast genome. Data presented here are averages of two replicates for the wild type and rnc3/4.
RPKM values for exons and introns were calculated according to the formula
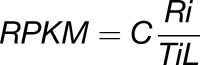 |
where C is a constant (C = 109), Ri is the number of reads that intersect the region of interest, Ti is the total number of reads of the sample, and L is the length of the region. The splicing efficiency (SE) was determined according to the formula
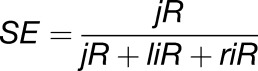 |
where jR is the number of reads spanning the exon/exon junction, liR is the number of reads spanning the 5′ exon/intron boundary, and riR is the number of reads spanning the 3′ intron/exon boundary. The number of reads used for the calculations was the average of two replicates for the wild type and rnc3/4.
Protein Isolation and Immunoblot Analysis
To analyze RNC4 association with polysomes, total protein was precipitated from pooled polysome fractions 1 to 3, 4 to 6, 7 to 9, and 10 to 12 (for polysome protocol, see above) from rnc3/4:RNC4-FLAG lines. The protein was precipitated with methanol and chloroform. The final protein was resuspended in a minimal volume of water and 4× SDS-PAGE loading buffer (200 mM Tris-Cl, pH 6.8, 400 mM DTT, 8% SDS, 0.4% bromophenol blue, and 40% glycerol). Total proteins were separated on a 13.5% SDS-PAGE gel and then transferred onto polyvinylidene difluoride membranes. Detection used the polyclonal anti-FLAG antibody (Sigma-Aldrich; http://www.sigmaaldrich.com/) and chemiluminescence with ECL2 (Peirce; http://www.fishersci.com/) according to the manufacturer’s instructions.
RNA Analysis in B. subtilis
B. subtilis strain W168 was grown in 2xYT medium (1.6% peptone, 1% yeast extract, and 1% NaCl) to an OD600 of 0.3 at 37°C. Mitomycin C was added to 10 μg/mL to induce prophage gene expression, and the culture was incubated for another 1 h before harvesting. For half-life measurements, rifampicin was added at 150 μg/mL. One milliliter of cells was centrifuged for 1 min at 15,700g, the supernatant was removed, and the pellet was frozen on dry ice. RNA was isolated by the RNAsnap method described previously (Stead et al., 2012). RNA gel blotting was performed as described previously (Durand et al., 2012) using the oligonucleotide probes indicated in Supplemental Table 2.
Accession Numbers
The Arabidopsis genes analyzed in this study are At1g55140 (RNC3) and At3g13740 (RNC4). T-DNA mutants used were SALK_073005 (rnc3-1) and SALK_006516.55.75.x (rnc4-1). RNA-Seq data from this article can be found at the NCBI with the BioProject number 268035 (http://www.ncbi.nlm.nih.gov/bioproject/268035).
Supplemental Data
Supplemental Figure 1. Phylogenetic Tree of Mini-III Proteins Using the Neighbor-Joining Method.
Supplemental Figure 2. Analysis of Arabidopsis T-DNA Lines for Homozygosity and Transcript Accumulation.
Supplemental Figure 3. Analysis of the 23S rRNA 0.5 kb Fragment by 5′ RACE.
Supplemental Figure 4. Complementation of rnc3/4 Restores the rnc3 or rnc4 Phenotype.
Supplemental Figure 5. Polysome Association of mRNA Transcripts Is Similar between WT and rnc3/4.
Supplemental Figure 6. RNC4 Is Not Associated with Polysomes.
Supplemental Figure 7. Reproducibility of Exon and Intron Expression Levels between WT and rnc3/4 Replicates.
Supplemental Figure 8. Normalized Sliding Window Average of RNA-Seq Coverage across the Chloroplast Genome in WT and rnc3/4.
Supplemental Figure 9. RNA-Seq-Based Determination of Splicing and Editing Efficiencies.
Supplemental Table 1. RNA-Seq Alignment Summary for Two WT and rnc3/4 Samples.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank the Boyce Thompson Institute Plant Cell Imaging Facility and Bioinformatics Help Desk for assistance with experimental methods and analysis. This work was supported by Grant DE-FG02-10ER20015 to D.B.S. from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, of the U.S. Department of Energy. C.C. was supported by the Agence Nationale de la Recherche (Grant subtilRNA2 and Labex Dynamo). Support for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation.
AUTHOR CONTRIBUTIONS
A.M.H., B.C., C.C., and D.B.S. contributed to the experimental design, data analysis, and writing. A.M.H., B.C., L.G., and A.H. performed the research.
Glossary
- PPR
pentatricopeptide repeat
- RNA-Seq
RNA sequencing
- dsRBD
double-stranded RNA binding domain
- cRT-PCR
circular RT-PCR
- sRNA
small RNA
- RACE
rapid amplification of cDNA ends
- SD
Shine-Dalgarno
- RPKM
reads per kilobase of transcript per million mapped reads
- pncRNA
plastid noncoding RNA
- dsDNA
double-stranded DNA
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Audren H., Bisanz-Seyer C., Briat J.F., Mache R. (1987). Structure and transcription of the 5S rRNA gene from spinach chloroplasts. Curr. Genet. 12: 263–269. [DOI] [PubMed] [Google Scholar]
- Barkan A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297: 38–57. [Google Scholar]
- Bollenbach T.J., Lange H., Gutierrez R., Erhardt M., Stern D.B., Gagliardi D. (2005). RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 33: 2751–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castandet B., Hotto A.M., Fei Z., Stern D.B. (2013). Strand-specific RNA sequencing uncovers chloroplast ribonuclease functions. FEBS Lett. 587: 3096–3101. [DOI] [PubMed] [Google Scholar]
- Comella P., Pontvianne F., Lahmy S., Vignols F., Barbezier N., Debures A., Jobet E., Brugidou E., Echeverria M., Sáez-Vásquez J. (2008). Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3′ETS in Arabidopsis. Nucleic Acids Res. 36: 1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D.L., Gan J., Liang Y.-H., Shaw G.X., Tropea J.E., Costantino N., Waugh D.S., Ji X. (2013). RNase III: Genetics and function; Structure and mechanism. Annu. Rev. Genet. 47: 405–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman M., Lee C.Y., Chanfreau G. (2003). RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11: 1279–1289. [DOI] [PubMed] [Google Scholar]
- del Campo E.M., Casano L.M. (2008). Degradation of plastid unspliced transcripts and lariat group II introns. Biochimie 90: 474–483. [DOI] [PubMed] [Google Scholar]
- Desfeux C., Clough S.J., Bent A.F. (2000). Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol. 123: 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M.P. (2009). Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol. Transl. Sci. 85: 369–391. [DOI] [PubMed] [Google Scholar]
- Drechsel O., Bock R. (2011). Selection of Shine-Dalgarno sequences in plastids. Nucleic Acids Res. 39: 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S., Gilet L., Condon C. (2012). The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 8: e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fristedt R., Scharff L.B., Clarke C.A., Wang Q., Lin C., Merchant S.S., Bock R. (2014). RBF1, a plant homolog of the bacterial ribosome-binding factor RbfA, acts in processing of the chloroplast 16S ribosomal RNA. Plant Physiol. 164: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. (1980). Precursors to 16S and 23S ribosomal RNA from a ribonuclear III-strain of Escherichia coli contain intact RNase III processing sites. Nucleic Acids Res. 8: 1873–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain A., Herlich S., Larom S., Kim S.H., Schuster G., Stern D.B. (2011). Mutational analysis of Arabidopsis chloroplast polynucleotide phosphorylase reveals roles for both RNase PH core domains in polyadenylation, RNA 3′-end maturation and intron degradation. Plant J. 67: 381–394. [DOI] [PubMed] [Google Scholar]
- Germain A., Hotto A.M., Barkan A., Stern D.B. (2013). RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 4: 295–316. [DOI] [PubMed] [Google Scholar]
- Germain A., Kim S.H., Gutierrez R., Stern D.B. (2012). Ribonuclease II preserves chloroplast RNA homeostasis by increasing mRNA decay rates, and cooperates with polynucleotide phosphorylase in 3′ end maturation. Plant J. 72: 960–971. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Greenberg B.M., Zurawski G., Hallick R.B. (1986). Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 118: 253–270. [DOI] [PubMed] [Google Scholar]
- Hirose T., Kusumegi T., Sugiura M. (1998). Translation of tobacco chloroplast rps14 mRNA depends on a Shine-Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett. 430: 257–260. [DOI] [PubMed] [Google Scholar]
- Hotto A.M., Huston Z.E., Stern D.B. (2010). Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol. 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto A.M., Schmitz R.J., Fei Z., Ecker J.R., Stern D.B. (2011). Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the Arabidopsis transcriptome. G3 (Bethesda) 1: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Friso G., Nishimura K., Qu X., Olinares P.D., Majeran W., Sun Q., van Wijk K.J. (2013). Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships: The concept of orthoproteomics. J. Proteome Res. 12: 491–504. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P.D., van Wijk K.J. (2009). Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.J., Moore P.B., Steitz T.A. (2004). The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340: 141–177. [DOI] [PubMed] [Google Scholar]
- Kleinknecht L., Wang F., Stübe R., Philippar K., Nickelsen J., Bohne A.V. (2014). RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26: 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H., Suzuki H., Kusumegi T., Hirose T., Yukawa Y., Sugiura M. (2007). Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 48: 1374–1378. [DOI] [PubMed] [Google Scholar]
- Lasa I., Toledo-Arana A., Gingeras T.R. (2012). An effort to make sense of antisense transcription in bacteria. RNA Biol. 9: 1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V., Soldo B., Düsterhöft A., Hilbert H., Mauël C., Karamata D. (1998). Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPbeta. Proc. Natl. Acad. Sci. USA 95: 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal-Klevezas D., Martinez-Soriano J., Nazar R. (2000a). Transcription and processing map of the 4.5S–5S rRNA intergenic region (ITS3) from rapeseed (Brassica napus) chloroplasts. Plant Cell Rep. 19: 667–673. [DOI] [PubMed] [Google Scholar]
- Leal-Klevezas D.S., Martínez-Soriano J.P., Nazar R.N. (2000b). Cotranscription of 5S rRNA-tRNAArg(ACG) from Brassica napus chloroplasts and processing of their intergenic spacer. Gene 253: 303–311. [DOI] [PubMed] [Google Scholar]
- Lim K., Kobayashi I., Nakai K. (2014). Alterations in rRNA-mRNA interaction during plastid evolution. Mol. Biol. Evol. 31: 1728–1740. [DOI] [PubMed] [Google Scholar]
- Liu Q., Feng Y., Zhu Z. (2009). Dicer-like (DCL) proteins in plants. Funct. Integr. Genomics 9: 277–286. [DOI] [PubMed] [Google Scholar]
- Luro S., Germain A., Sharwood R.E., Stern D.B. (2013). RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res. 41: 9141–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker M., Zimmermann B., Bilusic I., Tukhtubaeva N., Schroeder R. (2014). The double-stranded transcriptome of Escherichia coli. Proc. Natl. Acad. Sci. USA 111: 3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay R.M. (1981). The origin of plant chloroplast 4.5S ribosomal RNA. FEBS Lett. 123: 17–18. [Google Scholar]
- MacRae I.J., Doudna J.A. (2007). Ribonuclease revisited: Structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 17: 138–145. [DOI] [PubMed] [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 158: 156–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet O., Martinez P., Seyer P., Briat J.F. (1987). Sequence organization of the chloroplast ribosomal spacer of Spinacia oleracea including the 3′ end of the 16S rRNA and the 5′ end of the 23S rRNA. Plant Mol. Biol. 10: 53–63. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yagi Y., Kobayashi K. (2012). Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs in plants. Plant Cell Physiol. 53: 1171–1179. [DOI] [PubMed] [Google Scholar]
- Nicholson A.W. (2014). Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA 5: 31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Asakura Y., Friso G., Kim J., Oh S.H., Rutschow H., Ponnala L., van Wijk K.J. (2013). ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo G., Guzmàn P. (2008). Processing precursors with RNase III in plants. Plant Sci. 175: 741–746. [Google Scholar]
- Porra R., Thompson W., Kriedemann P. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redko Y., Condon C. (2009). Ribosomal protein L3 bound to 23S precursor rRNA stimulates its maturation by Mini-III ribonuclease. Mol. Microbiol. 71: 1145–1154. [DOI] [PubMed] [Google Scholar]
- Redko Y., Condon C. (2010). Maturation of 23S rRNA in Bacillus subtilis in the absence of Mini-III. J. Bacteriol. 192: 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redko Y., Bechhofer D.H., Condon C. (2008). Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 68: 1096–1106. [DOI] [PubMed] [Google Scholar]
- Ruwe H., Kupsch C., Teubner M., Schmitz-Linneweber C. (2011). The RNA-recognition motif in chloroplasts. J. Plant Physiol. 168: 1361–1371. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sharwood R.E., Halpert M., Luro S., Schuster G., Stern D.B. (2011a). Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA 17: 2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood R.E., Hotto A.M., Bollenbach T.J., Stern D.B. (2011b). Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 17: 230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.K., Schlessinger D. (1988). Coregulation of processing and translation: Mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. USA 85: 7144–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead M.B., Agrawal A., Bowden K.E., Nasir R., Mohanty B.K., Meagher R.B., Kushner S.R. (2012). RNAsnap™: A rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 40: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead M.B., Marshburn S., Mohanty B.K., Mitra J., Pena Castillo L., Ray D., van Bakel H., Hughes T.R., Kushner S.R. (2011). Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 39: 3188–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R., Meurer J. (2012). The cutting crew—Ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 63: 1663–1673. [DOI] [PubMed] [Google Scholar]
- Strittmatter G., Kössel H. (1984). Cotranscription and processing of 23S, 4.5S and 5S rRNA in chloroplasts from Zea mays. Nucleic Acids Res. 12: 7633–7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K., Sato N. (2005). Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 46: 649–660. [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. (1999). Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261–311. [DOI] [PubMed] [Google Scholar]
- Vogel J., Börner T. (2002). Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 21: 3794–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Piepenburg K., Schöttler M.A., Petersen K., Kahlau S., Tiller N., Drechsel O., Weingartner M., Kudla J., Bock R. (2010). Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 64: 851–863. [DOI] [PubMed] [Google Scholar]
- Wang H., Hill K., Perry S.E. (2004). An Arabidopsis RNA lariat debranching enzyme is essential for embryogenesis. J. Biol. Chem. 279: 1468–1473. [DOI] [PubMed] [Google Scholar]
- Watkins K.P., Kroeger T.S., Cooke A.M., Williams-Carrier R.E., Friso G., Belcher S.E., van Wijk K.J., Barkan A. (2007). A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zahler S.A., Korman R.Z., Rosenthal R., Hemphill H.E. (1977). Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J. Bacteriol. 129: 556–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P., Sharma C.M., Förstner K.U., Liere K., Vogel J., Börner T. (2012). The primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Ryvkin P., Li F., Dragomir I., Valladares O., Yang J., Cao K., Wang L.S., Gregory B.D. (2010). Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 6: e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer S.L., McEvoy S.M., Menon S., Read L.K. (2012). Additive and transcript-specific effects of KPAP1 and TbRND activities on 3′ non-encoded tail characteristics and mRNA stability in Trypanosoma brucei. PLoS ONE 7: e37639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmieńko A., Guzowska-Nowowiejska M., Urbaniak R., Pląder W., Formanowicz P., Figlerowicz M. (2011). A tiling microarray for global analysis of chloroplast genome expression in cucumber and other plants. Plant Methods 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.